Abstract
Breast cancer is a group of clinically, histopathologically and molecularly heterogeneous diseases, with different outcomes and responses to treatment. Triple-negative (TN) breast cancers are defined as tumors that lack the expression of estrogen receptor, progesterone receptor and epidermal growth factor receptor 2. This subgroup accounts for 15% of all types of breast cancer and its prevalence is higher among young African, African-American and Latino women. The hypermethylation of CpG islands (CpGI) is a common epigenetic alteration for suppressing gene expression in breast cancer and has been shown to be a key factor in breast carcinogenesis. In this study we analyzed the hypermethylation of 110 CpGI within 69 cancer-related genes in TN tumors. For the methylation analysis, we used the methyl-specific multiplex-ligation probe amplification assay. We found that the number of methylated CpGI is similar between TN and non-TN tumors, but the methylated genes between the groups are different. The methylation profile of TN tumors is defined by the methylation of five genes (that is, CDKN2B, CD44, MGMT, RB and p73) plus the non-methylation of 11 genes (that is, GSTP1, PMS2, MSH2, MLH1, MSH3, MSH6, DLC1, CACNA1A, CACNA1G, TWIST1 and ID4). We conclude that TN tumors have a specific methylation profile. Our findings give new information for better understanding tumor etiology and encourage future studies on potential drug targets for triple-negative breast tumors, which now lack a specific treatment.
Keywords: CpG island, triple negative breast tumors, methylation profile
Introduction
Breast cancer constitutes a group of diseases characterized by different morphologies, biological behaviors and clinical outcomes.1 During the previous years, classification of breast cancer has been based on the expression of different proteins and the therapeutic treatments have been enhanced by targeted strategies against specific receptors, that is, Tamoxifen for estrogen receptor (ER)-expressing tumors and Trastuzumab for epidermal growth factor receptor 2 (HER2)-expressing tumors.
Among the variety of breast tumors a subtype called triple negative (TN) is defined by what they are not: that is, by the absence of ER, progesterone receptor (PR) and HER2 in the tumor cells.1 TN cancer patients account for approximately 15% of total breast cancers and its prevalence is higher among young African, African-American and Latino women.2 TN tumors are more aggressive and present lower survival rates compared with other breast tumor types. One of the main reasons for this is that TN patients do not benefit from the anti-receptor therapies owing to the loss of known targets. Consequently, surgery and chemotherapy, separately or in combination, appear to be the only available treatments.3 TN tumors are typically high-grade ductal carcinomas with high mitotic index, and pushing borders of invasion, resulting in tumors of bad prognosis.4
During the last few years, the advances in proteomic studies have contributed to a more profound knowledge of the molecular etiology of TN tumors. TN tumors frequently express basal cytokeratins (particularly cytokeratins 5, 14 and 17), EGF receptor and myoepithelial markers (such as caveolins 1 and 2, c-kit and P-cadherin)4, 5, 6 and rarely express E-cadherin.7 Moreover, TN tumors present high expression of genes associated with proliferation, that is, Ki67 and TOP2A, high levels of cyclin E and low levels of cyclin D1. Regarding their DNA repair behavior, they are often p53 mutants and give evidence of genomic instability.8
Molecular assays have also shown that TN disease includes more than one molecular subtype and that the major components are the basal-like and the claudin-low-molecular subtype.4, 9 These findings reveal that TN cancer is not a single disease, making it a heterogeneous entity. Improved outcomes in TN disease require better understanding of the tumor etiology and the detection of potential drug targets.10
Cancer is the result of a multistep process characterized by the accumulation of genetic and epigenetic alterations leading to uncontrolled growth and eventually to the acquisition of metastatic potential. Among the epigenetic modifications, the most studied event in human cancers is the hyper-methylation of DNA. It is well established that widespread changes in DNA-methylation patterns occur during tumor progression.11 The epigenome of tumors is characterized by global DNA hypo-methylation and gene-specific hyper-methylation. Gene silencing by CpG island (CpGI) hyper-methylation in gene promoters can modulate pathways that control the basic function of the cell by acting directly on tumor suppressor genes and caretaker genes and indirectly on oncogenes through their regulators.11
Recent findings indicate epigenetic modifications as key factors in breast carcinogenesis. These modifications are quite attractive as targets for preventive care and therapeutics because of their potential for reversal.12 In this context, we considered that the epigenetic profile of TN tumors is of key importance for establishing potential therapeutic options for these patients. To further understand this area, we analyzed in TN tumors the methylation profile of 110 CpGI located within 69 cancer-involved genes, which are known to present epigenetic alterations in other human neoplasms. We hypothesized that TN tumors have a specific epigenetic signature, different from the epigenetic signature of other breast tumor subtypes. For this purpose we used the methylation-specific multiplex ligation-dependent probe amplification (MS-MLPA) methodology, which allows the detection of multiple methylated genes in a simultaneous manner.13, 14
By analyzing TN and non-TN tumors, our key findings revealed that the number of methylated CpGI is the same in both tumor groups, and that TN tumors present a specific methylation profile defined by a panel of 16 cancer-related genes.
Results
Methylation index and methylation profile of TN tumors
Given that the tumor cell content varies among the samples, we only considered a binary approach to determine the methylation status of each genetic region (0=unmethylated and 1=methylated). Based on our previous experiments analyzing cell lines, we established a cut-off threshold by considering a region to show methylation if the dosage ratio was >8%. Previous studies performed by our group have already confirmed that none of the 110 included CpGI presented a methylated status in blood. Studies performed on 49 of the included CpGI confirmed no methylation in normal breast tissue.15 In order to know whether the magnitude of the epigenetic deregulation differed among TN and non-TN tumors, we compared the number of methylated regions per single tumor (called Methylation Index), relativized to the total number of analyzed CpGI on the same sample. The minimum relative number of methylated regions in non-TN tumors was 4.08 and the maximum was 39.09 (mean=18.86, s.d.=7.99). The minimum relative number of methylated regions in TN tumors was 8.18 and the maximum was 33.64 (mean=17.85, s.d. =6.00). The Pearson's correlation test showed no significant difference between the relative MI of TN and non-TN tumors (P=0.467).
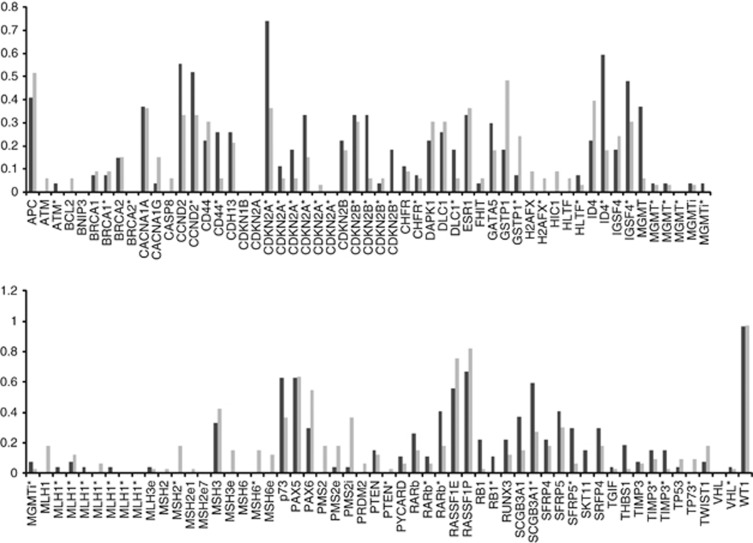
Although the MI results were similar, the methylation frequency of some of the 110 CpGIs was different in TN and non-TN tumors (Figure 1).
Figure 1.
Comparison of the methylation frequencies (methylation defined as dosage ratio >8%) for each of the 110 CpGI analyzed among TN and non-TN tumors. Black bars represent TN tumors and gray bars represent non-TN tumors. (Asterisk indicates that more than one CpGI of a gene were analyzed, i corresponds to introns and e to exons.)
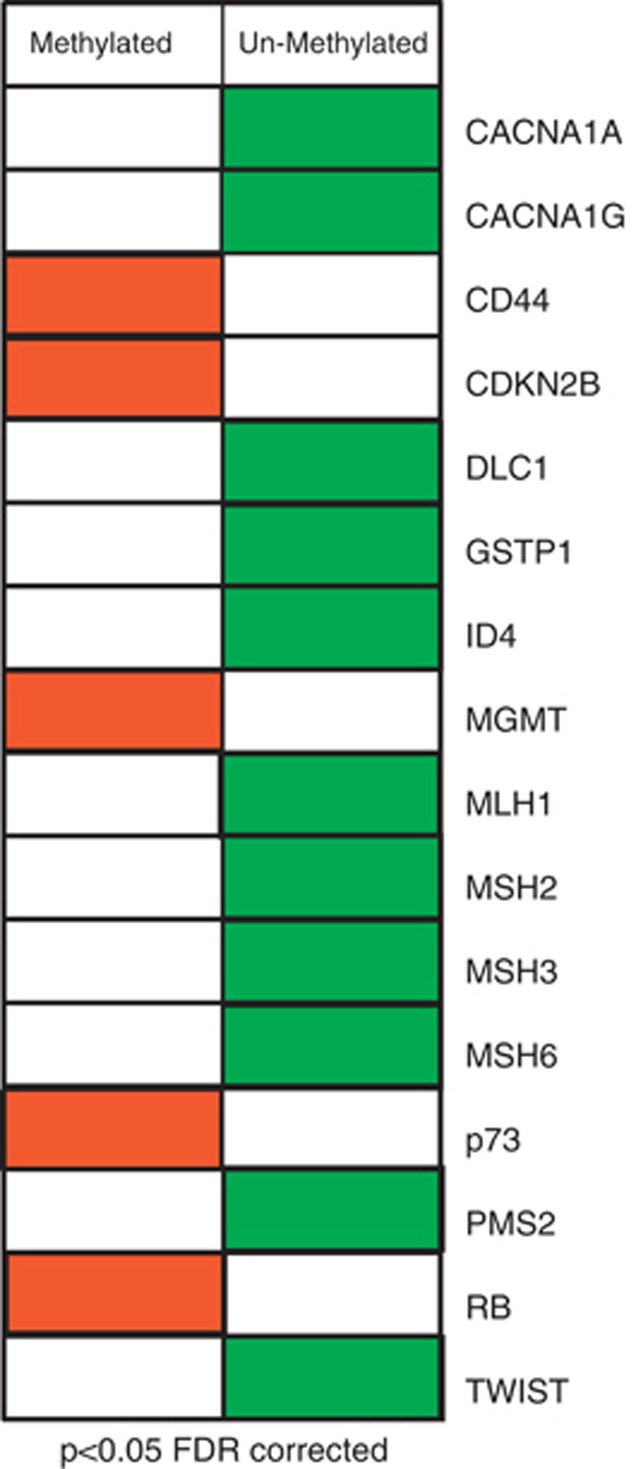
To analyze if the methylation status of each single CpGI was associated with TN tumors, we decreased the influence of unmethylation events (and therefore the consequent excess of zeros) by selecting for the analysis only genes that were methylated in more than 10% of the tumors. This selection reduced the initial 110 included CpGI to a final number of 59 regions. The statistical analyses of the 59 CpGI revealed that 18 of them (located within 16 different cancer-involved genes) were associated with TN tumors. The TN methylation profile was defined by the methylated status of CD44, MGMT, RB, p73 and 2 CpGI in CDKN2B plus the non-methylated status of MSH2, MLH1, DLC1, TWIST1, CACNA1G, CACNA1A, ID4, GSTP1, MSH6, MSH3 and 2 CpGI in PMS2 (P<0.05) (Figure 2). The strength of association of these 18 regions with TN tumors was assessed by Spearman's rho coefficient (Table 1). Note that the strongest association for the methylated status is given by the MGMT gene and the strongest association for the unmethylated status includes ID4 and the mismatch repair (MMR) genes.
Figure 2.
Diagram showing the 16 genes that define the epigenetic signature of TN tumors for FDR-corrected P values <0.05. Red represents methylated status and green represents unmethylated status of the analyzed CpGI.
Table 1. Epigenetic signature of TN tumors.
| Gene | P valuea | Spearman's rho coefficient | CpGI |
|---|---|---|---|
| CACNA1A | 0.042 | −0.389 | (+127 to +176 nt) |
| CACNA1G | 0.035 | −0.398 | (−75 to −20 nt) |
| CD44 | 0.037 | 0.277 | (+1 to +57 nt) |
| CDKN2B | 0.038 | 0.325 | (+88 to +144 nt exon 1) |
| CDKN2B | 0.041 | 0.331 | (−543 to −480 nt) |
| DLC1b | 0.034 | −0.379 | (−127 to −76 nt)b |
| GSTP1 | 0.03 | −0.313 | (+133 to +184 nt) |
| ID4 | 0.002 | −0.608 | (−993 to −942 nt) |
| MGMT | 0.026 | 0.385 | (−492 to −457 nt) |
| MLH1 | 0.030 | −0.423 | (−38 to +16 nt) |
| MSH2 | 0.024 | −0.423 | (−297 to −248 nt) |
| MSH3 | 0.038 | −0.306 | (+56 to +107 nt) |
| MSH6 | 0.037 | −0.381 | (−150 to−100 nt) |
| p73 | 0.04 | 0.265 | (+230 to +284 nt) |
| PMS2e | 0.04 | −0.341 | (−80 to −40 nt) |
| PMS2i | 0.05 | −0.502 | (+17 to +71 nt intron 1) |
| RB | 0.037 | 0.297 | (−487 to −432 nt) |
| TWIST | 0.033 | −0.389 | (+183 to +234 nt) |
Abbreviation: TN, tumor-negative.
P values corrected by the Benjamini and Hochberg false discovery rate.
DLC1 transcript variant 2.
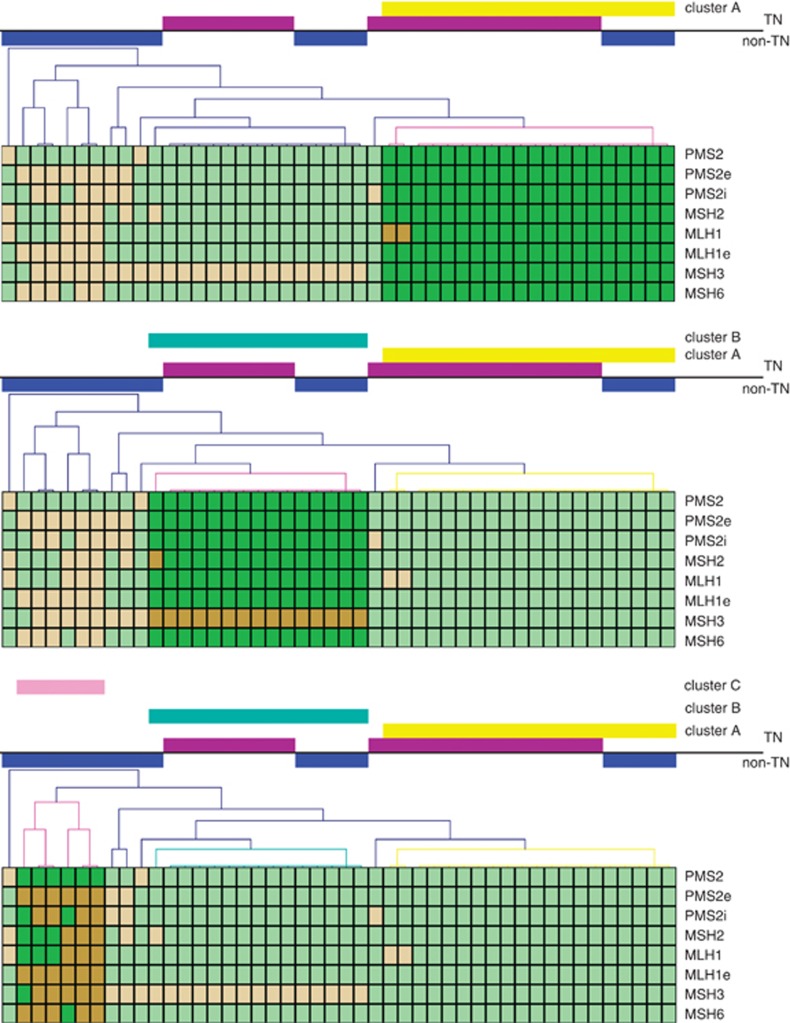
Given the genomic instability of TN tumors, we performed unsupervised hierarchical clustering analysis to examine if these tumors showed clusters based on the methylation profile of MMR genes. The analysis reveals three main groups. Cluster A is principally integrated by TN tumors lacking MMR methylation, cluster B is a mixed group composed by TN and non-TN tumors that share the methylation of MSH3, and cluster C is composed by non-TN tumors with methylation of several MMR genes (Figure 3).
Figure 3.
Unsupervised hierarchical clustering analysis among TN (represented by violet bars) and non-TN tumors (represented by blue bars), based on the methylation status of MMR genes. The map was generated using MeView software (TM 4 group, Dana Farber Cancer Institute, Boston, MA, USA). TN and non-TN tumors are arranged in columns, the genes are arranged in rows. Brown represents methylated status and green represents unmethylated status of the analyzed CpGI. Clusters are displayed as follows: cluster A in yellow, cluster B in light-blue and cluster C in pink.
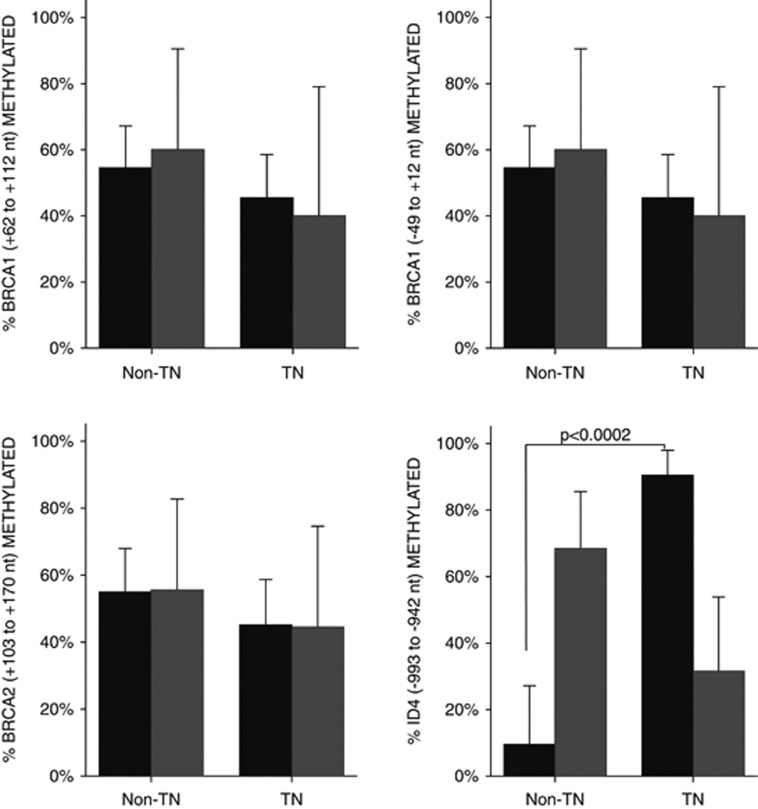
We consider worth mention that no significant association was detected between the methylation of either BRCA1 or BRCA2 and TN tumors. In this context the significant unmethylation of ID4, a negative regulator of BRCA1 pathways, is of high interest (Figure 4).
Figure 4.
Bar diagram representing the methylated (gray bars) and unmethylated (black bars) status of BRCA1, BRCA2 and ID4. Asterisk marks the significant difference in ID4 methylation.
Concurrent methylation in TN tumors
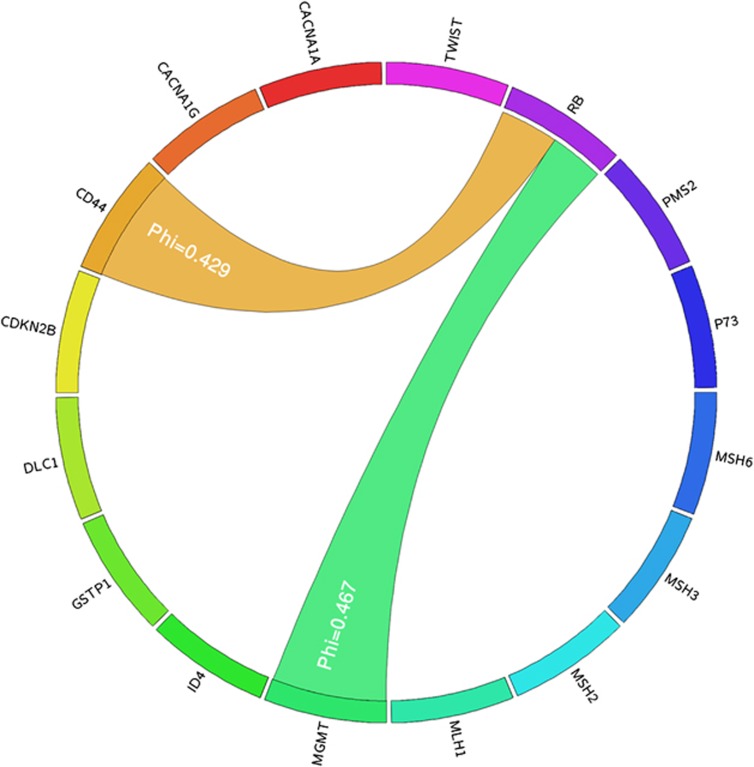
We tested whether the methylation of two regions occurred in a concurrent manner. To do so we analyzed by crosstab analysis if there was concurrent methylation between the 18 CpGI taken in pairs. Two concurrent methylations were detected: the methylation of CD44 with RB (Phi=0.429, P=0.007) and the methylation of MGMT with RB (Phi=0.467, P=0.002) (Table 2). This suggests that TN tumors that acquire the methylation of one of these two genes present a higher probability of acquiring the methylation of the other one (Figure 5).
Table 2. Concurrent methylation in TN tumors.
| Concurrent methylation | P valuea | Phi | OR (95% CI) | |
|---|---|---|---|---|
| MGMT → | RB | 0.002 | 0.467 | 16.4 (2.65–101.64) |
| CD44 → | RB | 0.007 | 0.429 | 12.8 (2.20–74.22) |
P value adjusted by Bonferroni correction.
Figure 5.
Circos diagram representing concurrent methylation in TN tumors. The outermost circle indicates the 16 genes that define the methylation profile of TN tumors. Only significant associations are shown. MGMT's concurrent methylation with RB is represented by green ribbons and CD44's concurrent methylation with RB is represented with orange ribbons. This figure was created using the Circos software.22
Discussion
The aim of this study was to evaluate the methylation profile of TN tumors. To do so we analyzed by MS-MLPA the methylation status of 110 GpGI located within 69 different cancer-involved genes, which had been previously found to be epigenetically altered in a variety of human neoplasms. We found that although the MI is similar in TN and non-TN breast tumors, the genes that are methylated are different. This indicates that the deregulation of the methylation mechanism occurs at a similar rate or intensity in both tumor subgroups, but with different targets.
The epigenetic profile of TN tumors is defined by 18 CpGI within 16 genes.
Among these 16 genes, those that showed strongest association coefficients are worth mention: the non-methylation of the MMR genes and the methylation of MGMT plus the non-methylation of ID4.
Although TN tumors present genomic instability, the methylation of MMR genes is not affected significantly. The genomic instability is probably acquired by other pathways rather than the methylation of MMR genes. We postulate that given the genomic instability of TN tumors, it is not an advantage to also inactivate the MMR system, which would probably be lethal. In addition to the repair of replication errors, MMR proteins also mediate the response to certain DNA-damaging agents that can modify the structure of bases such as cisplatin.16 Our result provides interesting data for studies focused on treatment targets in TN tumors. This new epigenetic evidence suggests that these tumors could be responsive to cisplatin-based drugs.
A second attractive result is that TN tumors present significant methylation of MGMT and RB. These two genes codify regulator proteins of the G1 phase of the cell cycle. It is thus possible that TN tumors acquire genomic instability because of the methylation of key genes of the cell cycle. Note that neither MMR nor BRCA1/2, which participate in different repair mechanisms, showed significant methylation in TN tumors.
The methylation of MGMT in particular is an interesting finding. This gene normally has a repair function removing alkyl groups from DNA. Our results are in line with what was recently published by Barbieris et al.17 The methylation of MGMT has also been described in glioblastomas.18 The presence of methylation on MGMT in these tumors is presently used as an indicator of the susceptibility to the drug temozolomide.18 This drug is currently being administered in combination with radiotherapy in patients with glioblastomas.19 Our results suggest that TN tumors could be responsive for the same drug. To our knowledge this is the first report presenting epigenetic evidence for two potential drug sensitivities in TN tumors.
It has been shown that in TN tumors the BRCA1 pathway may be dysfunctional, and in basal-like breast tumors BRCA1 protein levels are significantly lower.3, 20, 21 When we analyzed the methylation of two CpGI in the BRCA1 promoter and one CpGI in the BRCA2 promoter, we found no significant difference between TN and non-TN tumors. A study done by Turner's research group shows that the methylation of BRCA1, assessed by methylation-specific PCR, was the same between control and CK5/6-positive breast tumors. However, when they analyzed the expression of the BRCA1 messenger, they found it was lower in the CK5/6-positive breast tumors group. This partial silencing of BRCA1 expression was caused by the overexpression of the negative regulator of BRCA1, ID4. When we analyzed, by MS-MLPA, the methylation status of two CpGI in the ID4 promoter, we found that the CpGI of ID4 (−993 to −942 nt) is significantly not methylated in TN tumors (P<0.001). But an upstream CpGI in the same promoter (−379 to −316 nt) showed no statistical difference, and thus is probably not involved in ID4 expression regulation. To our knowledge this is the first report showing the methylation status of BRCA1, BRCA2 and ID4 in TN tumors. Our results can contribute to a better understanding of the causes of BRCA1 downregulation in TN tumors. Based on our observations, the absence of BRCA1 in TN tumors could be due to the activation of ID4 repressor, in line with the observations of Turner et al.21 and Matros et al.20
Our study provides possible epigenetic explanations for the genomic instability and BRCA-ness behavior of TN tumors and contributes to new epigenetic evidence of TN tumors with potential drug sensitivity. The unmethylated status of MMR and the methylation of MGMT open new perspectives for treatment of TN disease.
Materials and methods
Patients
Sixty-one women who had invasive ductal breast cancer were enrolled in the study. Ethical approval was obtained from the BioEthics Committee of the School of Medical Sciences, from the National University of Cuyo, Mendoza, Argentina.
Patients signed an informed consent based on the scientific and ethical principles of the World Medical Association's Declaration of Helsinki. Patients were treated in the Gineco-Mamario Institute or in the Italian Hospital of Mendoza, Argentina.
Histopathological analyses
All breast cancer specimens were reviewed by a single pathologist. Thirty-three non-TN and 28 TN tumors were analyzed and triple negativity was determined by means of immunohistochemistry by the same pathologist; tumors with expression of ER and/or PR and/or HER2 were included as non-TN. The clinical features of patients are presented in Table 3.
Table 3. Clinicopathological features of the patients included in the study.
| TN, n=28 (%) | Non-TN, n=33 (%) | |
|---|---|---|
| Age (mean) | 41.8 | 41.5 |
| ER | ||
| Positive | 0 (0) | 27 (82) |
| Negative | 28 (100) | 6 (18) |
| PR | ||
| Positive | 0 (0) | 21 (63) |
| Negative | 28 (100) | 12 (37) |
| HER2 | ||
| Negative | 28 (100) | 27 (81.8) |
| Positive moderate | 0 (0) | 3 (9.09) |
| Positive strong | 0 (0) | 3 (9.09) |
| Lymph node | ||
| Negative | 14 (51.8) | 19 (57.5) |
| Positive | 13 (48.1) | 14 (42.4) |
| Histological grade | ||
| I | 2 (7.4) | 2 (6.06) |
| II | 10 (37) | 14 (42.4) |
| III | 15 (55) | 14 (42.4) |
| Nuclear pleomorphism | ||
| I | 1 (3.7) | 3 (9.09) |
| II | 8 (29.6) | 17 (51.5) |
| III | 18 (66.6) | 10 (30.3) |
| Tumor grade | ||
| Low | 2 (7.4) | 6 (18.1) |
| Intermediate | 10 (37.0) | 17 (51.5) |
| High | 15 (55.5) | 10 (30.3) |
Abbreviation: TN, Triple-negative.
DNA extraction from tumor tissue
Tumor tissues were frozen at −80 °C and broken with a frozen mortar. The homogenate was collected and resuspended in T10E10 (10 mℳ Tris/HCl and 1 mℳ EDTA) buffer. All samples were frozen at least 24 h before DNA extraction to improve the efficiency of the process. DNA extraction was performed as described previously.15 Briefly, the homogenate was dissolved in 3 ml CTAB solution (2 g/l CTAB (Sigma-Aldrich, Bavaria, Germany), 100 mℳ Tris/HCl, 20 mℳ EDTA and 0.2% 2-mercaptoethanol) and incubated at 60 °C for 4 h for membrane lysis. Once the pellet was completely dissolved, 3 ml chloroform:isoamylic (24:1) solution was added, mixed gently for 5 min and centrifuged at 3000 r.p.m. for 5 min. The aqueous phase was collected and mixed with 6 ml of 100% ethanol. The precipitated DNA was dissolved in T10E buffer and stored at −20 °C.
DNA extraction from formalin-fixed paraffin-embedded tumor tissues
Fifty-micrometer paraffin sections were cut and deparaffinized with xylene. Tumor tissues were incubated in a pre-mix buffer containing 0.8 mg/ml proteinase K (Ambion, Austin, TX, USA) at 50 °C overnight. Proteinase K reaction was stopped by incubating at 70 °C for 15 min. Subsequently, DNA was isolated using phenol:chloroform:isoamyl alcohol (PCI) (25:24:1) pH 5.2. The aqueous phase was collected and mixed with 3 ℳ NaAc, which was vortexed and spinned down. Subsequently ethanol 100% was added and stored at −20 °C overnight. Finally, washing steps were performed using ethanol at different concentrations.
Methyl-specific multiplex ligation-dependent probe amplification assay
To assess the methylation status of 110 CpGI within 69 genes the MS-MLPA kits ME001B, ME002, ME003, ME011 and ME024 were used. The MS-MLPA assays were performed basically according to manufacturer's recommendations (MRC-Holland, Amsterdam, The Netherlands, www.mrc-holland.com),13 introducing subtle modifications (that is, extended restriction enzyme incubation time, separated ligation and restriction steps), to avoid background signals.15 The fluorescent-labeled PCR products were separated by capillary electrophoresis (ABI-3130 sequencer, Applied Biosystems, Foster City, CA, USA) and analyzed by GeneMarker (Softgenetics, State College, PA, USA) v1.75 software. This system normalizes the data by dividing the peak area of a single probe by the peak areas of all control probes. Then the normalized peaks from the analyzed samples are compared with the normalized peaks from the control reaction. We considered a dichotomized approach for the methylation status, given that we did not have information on the tumor cell content of our samples. Based on our previous HeLa cell line experiments, we established a cut-off threshold considering a promoter to show methylation if the methylation dosage ratio was >8%.
Statistical analyses
To decrease the influence of unmethylation events and the consequent excess of zeros in the data, we selected only genes that were methylated in more than 10% of tumors. The chi-square test and odds ratio analysis were applied to study the association between CpGI methylation status and tumor types. The strength of associations was assessed by the Spearman's rho coefficient. Pearson's correlation was used to compare relative MI among tumor types. We applied the Benjamini and Hochberg's approach (FDR) to adjust P-values for multiple testing. After correction P values <0.05 were considered statistically significant. Concurrent methylation was assessed by chi-square test, adjusting P values by Bonferroni. All statistical analyses were performed using the software SPSS v17 (SPSS Inc., Chicago, IL, USA). The hierarchical clustering analysis and heat maps were performed using the software Multi Experiment Viewer MeV v4.6 (TM 4 group, Dana Farber Cancer Institute, Boston, MA, USA).
Acknowledgments
This study was supported in part by the National Institute of Cancer (‘Instituto Nacional del Cancer') and the National University of Cuyo Argentina.
The authors declare no conflict of interest.
References
- Chacon RD, Costanzo MV. Triple-negative breast cancer. Breast Cancer Res. 2010;12 (Suppl 2:S3. doi: 10.1186/bcr2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JQ, Russo J. ERalpha-negative and triple negative breast cancer: molecular features and potential therapeutic approaches. Biochim Biophys Acta. 2009;1796:162–175. doi: 10.1016/j.bbcan.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy KB. Triple-negative breast cancers: an updated review on treatment options. Curr Oncol. 2011;18:e173–e179. doi: 10.3747/co.v18i4.738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey L, Winer E, Viale G, Cameron D, Gianni L. Triple-negative breast cancer: disease entity or title of convenience. Nat Rev Clin Oncol. 2010;7:683–692. doi: 10.1038/nrclinonc.2010.154. [DOI] [PubMed] [Google Scholar]
- Kreike B, van KM, Horlings H, Weigelt B, Peterse H, Bartelink H, et al. Gene expression profiling and histopathological characterization of triple-negative/basal-like breast carcinomas. Breast Cancer Res. 2007;9:R65. doi: 10.1186/bcr1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakha EA, El-Sayed ME, Green AR, Lee AH, Robertson JF, Ellis IO. Prognostic markers in triple-negative breast cancer. Cancer. 2007;109:25–32. doi: 10.1002/cncr.22381. [DOI] [PubMed] [Google Scholar]
- Kashiwagi S, Yashiro M, Takashima T, Nomura S, Noda S, Kawajiri H, et al. Significance of E-cadherin expression in triple-negative breast cancer. Br J Cancer. 2010;103:249–255. doi: 10.1038/sj.bjc.6605735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider BP, Winer EP, Foulkes WD, Garber J, Perou CM, Richardson A, et al. Triple-negative breast cancer: risk factors to potential targets. Clin Cancer Res. 2008;14:8010–8018. doi: 10.1158/1078-0432.CCR-08-1208. [DOI] [PubMed] [Google Scholar]
- Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12:R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey LA. Directed therapy of subtypes of triple-negative breast cancer. Oncologist. 2011;16 (Suppl 1:71–78. doi: 10.1634/theoncologist.2011-S1-71. [DOI] [PubMed] [Google Scholar]
- Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- Dworkin AM, Huang TH, Toland AE. Epigenetic alterations in the breast: Implications for breast cancer detection, prognosis and treatment. Semin Cancer Biol. 2009;19:165–171. doi: 10.1016/j.semcancer.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nygren AO, Ameziane N, Duarte HM, Vijzelaar RN, Waisfisz Q, Hess CJ, et al. Methylation-specific MLPA (MS-MLPA): simultaneous detection of CpG methylation and copy number changes of up to 40 sequences. Nucleic Acids Res. 2005;33:e128. doi: 10.1093/nar/gni127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schouten JP, McElgunn CJ, Waaijer R, Zwijnenburg D, Diepvens F, Pals G. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002;30:e57. doi: 10.1093/nar/gnf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzese DM, Gago FE, Vargas-Roig LM, Roque M. Simultaneous analysis of the methylation profile of 26 cancer related regions in invasive breast carcinomas by MS-MLPA and drMS-MLPA. Mol Cell Probes. 2010;24:271–280. doi: 10.1016/j.mcp.2010.05.002. [DOI] [PubMed] [Google Scholar]
- Martin SA, Lord CJ, Ashworth A. Therapeutic targeting of the DNA mismatch repair pathway. Clin Cancer Res. 2010;16:5107–5113. doi: 10.1158/1078-0432.CCR-10-0821. [DOI] [PubMed] [Google Scholar]
- Fumagalli C, Pruneri G, Possanzini P, Manzotti M, Barile M, Feroce I, et al. Methylation of O (6)-methylguanine-DNA methyltransferase (MGMT) promoter gene in triple-negative breast cancer patients Breast Cancer Res Treat 2012(e-pub ahead of print). [DOI] [PubMed]
- Hegi ME, Diserens AC, Gorlia T, Hamou MF, de TN, Weller M, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- Kim YS, Kim SH, Cho J, Kim JW, Chang JH, Kim DS, et al. MGMT gene promoter methylation as a potent prognostic factor in glioblastoma treated with temozolomide-based chemoradiotherapy: a single-institution study. Int J Radiat Oncol Biol Phys. 2012. [DOI] [PubMed]
- Matros E, Wang ZC, Lodeiro G, Miron A, Iglehart JD, Richardson AL. BRCA1 promoter methylation in sporadic breast tumors: relationship to gene expression profiles. Breast Cancer Res Treat. 2005;91:179–186. doi: 10.1007/s10549-004-7603-8. [DOI] [PubMed] [Google Scholar]
- Turner NC, Reis-Filho JS, Russell AM, Springall RJ, Ryder K, Steele D, et al. BRCA1 dysfunction in sporadic basal-like breast cancer. Oncogene. 2007;26:2126–2132. doi: 10.1038/sj.onc.1210014. [DOI] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]