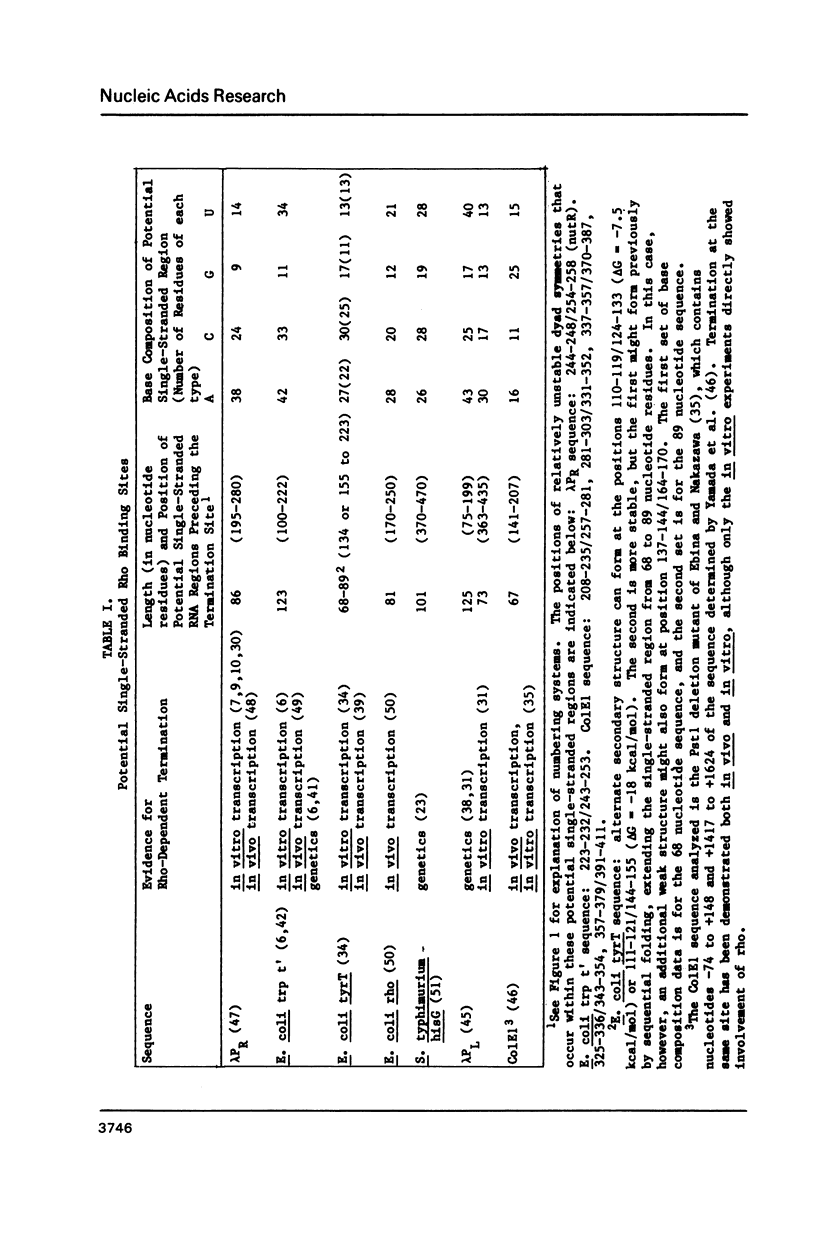
Abstract
The interaction of E. coli termination factor rho with the nascent RNA transcript appears to be a central feature of the rho-dependent transcription termination process. Based on in vitro studies of the rho-dependent termination of the transcript initiated at the PR promoter of bacteriophage lambda, and on earlier studies, Morgan, Bear and von Hippel (J. Biol. Chem. 258, 9565-9574, 1983) proposed a model defining the features of a potential binding site for rho protein on transcripts subject to rho-dependent termination. This model suggested that an effective rho binding site on a nascent RNA transcript should be: (i) greater than 70-80 nucleotide residues in length; (ii) essentially unencumbered with stable secondary structure; (iii) relatively sequence non-specific; and (iv) located within a few hundred nucleotide residues upstream of the potential rho-dependent terminus. In this paper we examine the sequences and secondary structures of several transcripts that exhibit rho-dependent termination to test this hypothesis further. Unstructured regions of approximately the expected size and location were found on all the transcripts examined. Though several short specific sequence elements were found to occur in a very similar arrangement on the lambda PR- and lambda PL-initiated transcripts of lambda phage, no such elements of sequence regularity were found on any of the other rho-dependent transcripts. The results of the sequence comparisons reported here strongly support the generality of the "unstructured binding site" hypothesis for rho-dependent termination.
Full text
PDF














Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Borer P. N., Dengler B., Tinoco I., Jr, Uhlenbeck O. C. Stability of ribonucleic acid double-stranded helices. J Mol Biol. 1974 Jul 15;86(4):843–853. doi: 10.1016/0022-2836(74)90357-x. [DOI] [PubMed] [Google Scholar]
- Brown S., Albrechtsen B., Pedersen S., Klemm P. Localization and regulation of the structural gene for transcription-termination factor rho of Escherichia coli. J Mol Biol. 1982 Dec 5;162(2):283–298. doi: 10.1016/0022-2836(82)90527-7. [DOI] [PubMed] [Google Scholar]
- Brutlag D. L., Clayton J., Friedland P., Kedes L. H. SEQ: a nucleotide sequence analysis and recombination system. Nucleic Acids Res. 1982 Jan 11;10(1):279–294. doi: 10.1093/nar/10.1.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calva E., Burgess R. R. Characterization of a rho-dependent termination site within the cro gene of bacteriophage lambda. J Biol Chem. 1980 Nov 25;255(22):11017–11022. [PubMed] [Google Scholar]
- Ciampi M. S., Schmid M. B., Roth J. R. Transposon Tn10 provides a promoter for transcription of adjacent sequences. Proc Natl Acad Sci U S A. 1982 Aug;79(16):5016–5020. doi: 10.1073/pnas.79.16.5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Court D., Brady C., Rosenberg M., Wulff D. L., Behr M., Mahoney M., Izumi S. U. Control of transcription termination: a rho-dependent termination site in bacteriophage lambda. J Mol Biol. 1980 Apr;138(2):231–254. doi: 10.1016/0022-2836(80)90285-5. [DOI] [PubMed] [Google Scholar]
- Drahos D., Szybalski W. Antitermination and termination functions of the cloned nutL, N, and tL1 modules of coliphage lambda. Gene. 1981 Dec;16(1-3):261–274. doi: 10.1016/0378-1119(81)90082-2. [DOI] [PubMed] [Google Scholar]
- Ebina Y., Nakazawa A. Cyclic AMP-dependent initiation and rho-dependent termination of colicin E1 gene transcription. J Biol Chem. 1983 Jun 10;258(11):7072–7078. [PubMed] [Google Scholar]
- Finger L. R., Richardson J. P. Stabilization of the hexameric form of Escherichia coli protein rho under ATP hydrolysis conditions. J Mol Biol. 1982 Mar 25;156(1):203–219. doi: 10.1016/0022-2836(82)90467-3. [DOI] [PubMed] [Google Scholar]
- Galluppi G. R., Richardson J. P. ATP-induced changes in the binding of RNA synthesis termination protein Rho to RNA. J Mol Biol. 1980 Apr 15;138(3):513–539. doi: 10.1016/s0022-2836(80)80016-7. [DOI] [PubMed] [Google Scholar]
- Grosschedl R., Schwarz E. Nucleotide sequence of the cro-cII-oop region of bacteriophage 434 DNA. Nucleic Acids Res. 1979 Mar;6(3):867–881. doi: 10.1093/nar/6.3.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L. P., Beckwth J., Wu A. M., Platt T. A mutation distal to the messenger RNA endpoint reduces transcription termination in the tryptophan operon in Escherichia coli. J Mol Biol. 1979 Sep 5;133(1):189–197. doi: 10.1016/0022-2836(79)90258-4. [DOI] [PubMed] [Google Scholar]
- Housley P. R., Leavitt A. D., Whitfield H. J. Genetic analysis of a temperature-sensitive Salmonella typhimurium rho mutant with an altered rho-associated polycytidylate-dependent adenosine triphosphatase activity. J Bacteriol. 1981 Jul;147(1):13–24. doi: 10.1128/jb.147.1.13-24.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassavetis G. A., Chamberlin M. J. Pausing and termination of transcription within the early region of bacteriophage T7 DNA in vitro. J Biol Chem. 1981 Mar 25;256(6):2777–2786. [PubMed] [Google Scholar]
- Kung H., Bekesi E., Guterman S. K., Gray J. E., Traub L., Calhoun D. H. Autoregulation of the rho gene of Escherichia coli K-12. Mol Gen Genet. 1984;193(2):210–213. doi: 10.1007/BF00330669. [DOI] [PubMed] [Google Scholar]
- Küpper H., Sekiya T., Rosenberg M., Egan J., Landy A. A rho-dependent termination site in the gene coding for tyrosine tRNA su3 of Escherichia coli. Nature. 1978 Mar 30;272(5652):423–428. doi: 10.1038/272423a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau L. F., Roberts J. W., Wu R., Georges F., Narang S. A. A potential stem-loop structure and the sequence CAAUCAA in the transcript are insufficient to signal rho-dependent transcription termination at lambda tR1. Nucleic Acids Res. 1984 Jan 25;12(2):1287–1299. doi: 10.1093/nar/12.2.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau L. F., Roberts J. W., Wu R. Transcription terminates at lambda tR1 in three clusters. Proc Natl Acad Sci U S A. 1982 Oct;79(20):6171–6175. doi: 10.1073/pnas.79.20.6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowery-Goldhammer C., Richardson J. P. An RNA-dependent nucleoside triphosphate phosphohydrolase (ATPase) associated with rho termination factor. Proc Natl Acad Sci U S A. 1974 May;71(5):2003–2007. doi: 10.1073/pnas.71.5.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowery C., Richardson J. P. Characterization of the nucleoside triphosphate phosphohydrolase (ATPase) activity of RNA synthesi termination factor p. I. Enzymatic properties and effects of inhibitors. J Biol Chem. 1977 Feb 25;252(4):1375–1380. [PubMed] [Google Scholar]
- Lowery C., Richardson J. P. Characterization of the nucleoside triphosphate phosphohydrolase (ATPase) activity of RNA synthesis termination factor p. II. Influence of synthetic RNA homopolymers and random copolymers on the reaction. J Biol Chem. 1977 Feb 25;252(4):1381–1385. [PubMed] [Google Scholar]
- Luk K. C., Szybalski W. Characterization of the cloned terminators tR1, tL3 and tI and the nut R antitermination site of coliphage lambda. Gene. 1982 Dec;20(2):127–134. doi: 10.1016/0378-1119(82)90030-0. [DOI] [PubMed] [Google Scholar]
- Morgan W. D., Bear D. G., von Hippel P. H. Rho-dependent termination of transcription. I. Identification and characterization of termination sites for transcription from the bacteriophage lambda PR promoter. J Biol Chem. 1983 Aug 10;258(15):9553–9564. [PubMed] [Google Scholar]
- Morgan W. D., Bear D. G., von Hippel P. H. Rho-dependent termination of transcription. II. Kinetics of mRNA elongation during transcription from the bacteriophage lambda PR promoter. J Biol Chem. 1983 Aug 10;258(15):9565–9574. [PubMed] [Google Scholar]
- Morgan W. D., Bear D. G., von Hippel P. H. Specificity of release by Escherichia coli transcription termination factor rho of nascent mRNA transcripts initiated at the lambda PR. J Biol Chem. 1984 Jul 10;259(13):8664–8671. [PubMed] [Google Scholar]
- Nussinov R., Tinoco I., Jr Sequential folding of a messenger RNA molecule. J Mol Biol. 1981 Sep 25;151(3):519–533. doi: 10.1016/0022-2836(81)90008-5. [DOI] [PubMed] [Google Scholar]
- Olson E. R., Flamm E. L., Friedman D. I. Analysis of nutR: a region of phage lambda required for antitermination of transcription. Cell. 1982 Nov;31(1):61–70. doi: 10.1016/0092-8674(82)90405-6. [DOI] [PubMed] [Google Scholar]
- Platt T. Termination of transcription and its regulation in the tryptophan operon of E. coli. Cell. 1981 Apr;24(1):10–23. doi: 10.1016/0092-8674(81)90496-7. [DOI] [PubMed] [Google Scholar]
- Reisbig R. R., Hearst J. E. Escherichia coli deoxyribonucleic acid dependent ribonucleic acid polymerase transcriptional pause sites on SV40 DNA F1. Biochemistry. 1981 Mar 31;20(7):1907–1918. doi: 10.1021/bi00510a029. [DOI] [PubMed] [Google Scholar]
- Richardson J. P., Conaway R. Ribonucleic acid release activity of transcription termination protein rho is dependent on the hydrolysis of nucleoside triphosphates. Biochemistry. 1980 Sep 2;19(18):4293–4299. doi: 10.1021/bi00559a022. [DOI] [PubMed] [Google Scholar]
- Richardson J. P., Macy M. R. Ribonucleic acid synthesis termination protein rho function: effects of conditions that destabilize ribonucleic acid secondary structure. Biochemistry. 1981 Mar 3;20(5):1133–1139. doi: 10.1021/bi00508a014. [DOI] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Rosenberg M., Court D., Shimatake H., Brady C., Wulff D. L. The relationship between function and DNA sequence in an intercistronic regulatory region in phage lambda. Nature. 1978 Mar 30;272(5652):414–423. doi: 10.1038/272414a0. [DOI] [PubMed] [Google Scholar]
- Rossi J., Egan J., Hudson L., Landy A. The tyrT locus: termination and processing of a complex transcript. Cell. 1981 Nov;26(3 Pt 1):305–314. doi: 10.1016/0092-8674(81)90199-9. [DOI] [PubMed] [Google Scholar]
- Salstrom J. S., Fiandt M., Szybalski W. N-independent leftward transcription in coliphage lambda: deletions, insertions and new promoters bypassing termination functions. Mol Gen Genet. 1979 Jan 10;168(2):211–230. doi: 10.1007/BF00431446. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Hong G. F., Hill D. F., Petersen G. B. Nucleotide sequence of bacteriophage lambda DNA. J Mol Biol. 1982 Dec 25;162(4):729–773. doi: 10.1016/0022-2836(82)90546-0. [DOI] [PubMed] [Google Scholar]
- Schwarz E., Scherer G., Hobom G., Kössel H. Nucleotide sequence of cro, cII and part of the O gene in phage lambda DNA. Nature. 1978 Mar 30;272(5652):410–414. doi: 10.1038/272410a0. [DOI] [PubMed] [Google Scholar]
- Sharp J. A., Platt T. Rho-dependent termination and concomitant NTPase activity requires a specific, intact RNA region. J Biol Chem. 1984 Feb 25;259(4):2268–2273. [PubMed] [Google Scholar]
- Shigesada K., Wu C. W. Studies of RNA release reaction catalyzed by E. coli transcription termination factor rho using isolated ternary transcription complexes. Nucleic Acids Res. 1980 Aug 11;8(15):3355–3369. doi: 10.1093/nar/8.15.3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somasekhar G., Drahos D., Salstrom J. S., Szybalski W. Sequence changes in coliphage lambda mutants affecting the nutL antitermination site and termination by tL1 and tL2. Gene. 1982 Dec;20(3):477–480. doi: 10.1016/0378-1119(82)90217-7. [DOI] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Wu A. M., Chapman A. B., Platt T., Guarente L. P., Beckwith J. Deletions of distal sequence after termination of transcription at the end of the tryptophan operon in E. coli. Cell. 1980 Apr;19(4):829–836. doi: 10.1016/0092-8674(80)90073-2. [DOI] [PubMed] [Google Scholar]
- Wu A. M., Christie G. E., Platt T. Tandem termination sites in the tryptophan operon of Escherichia coli. Proc Natl Acad Sci U S A. 1981 May;78(5):2913–2917. doi: 10.1073/pnas.78.5.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A. M., Platt T. Transcription termination: nucleotide sequence at 3' end of tryptophan operon in Escherichia coli. Proc Natl Acad Sci U S A. 1978 Nov;75(11):5442–5446. doi: 10.1073/pnas.75.11.5442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M., Ebina Y., Miyata T., Nakazawa T., Nakazawa A. Nucleotide sequence of the structural gene for colicin E1 and predicted structure of the protein. Proc Natl Acad Sci U S A. 1982 May;79(9):2827–2831. doi: 10.1073/pnas.79.9.2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Hippel P. H., Bear D. G., Morgan W. D., McSwiggen J. A. Protein-nucleic acid interactions in transcription: a molecular analysis. Annu Rev Biochem. 1984;53:389–446. doi: 10.1146/annurev.bi.53.070184.002133. [DOI] [PubMed] [Google Scholar]


