Abstract
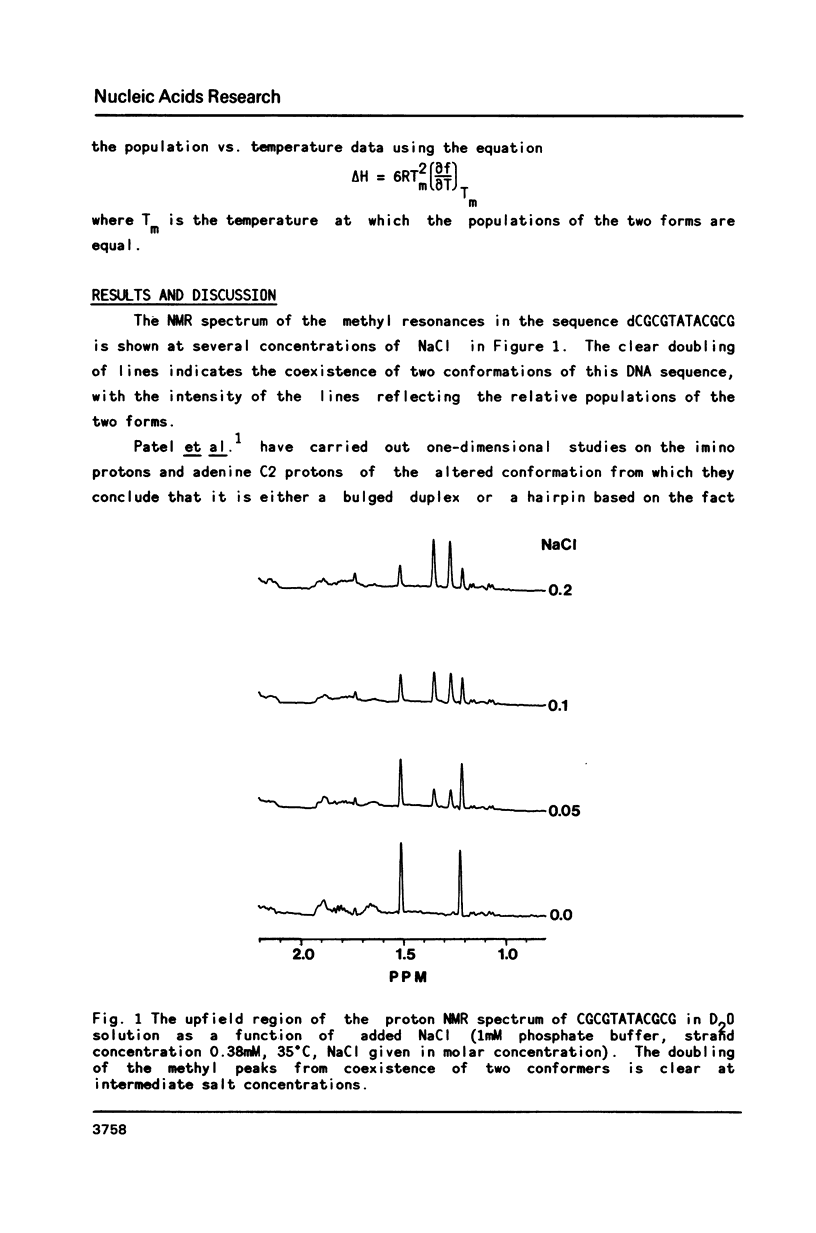
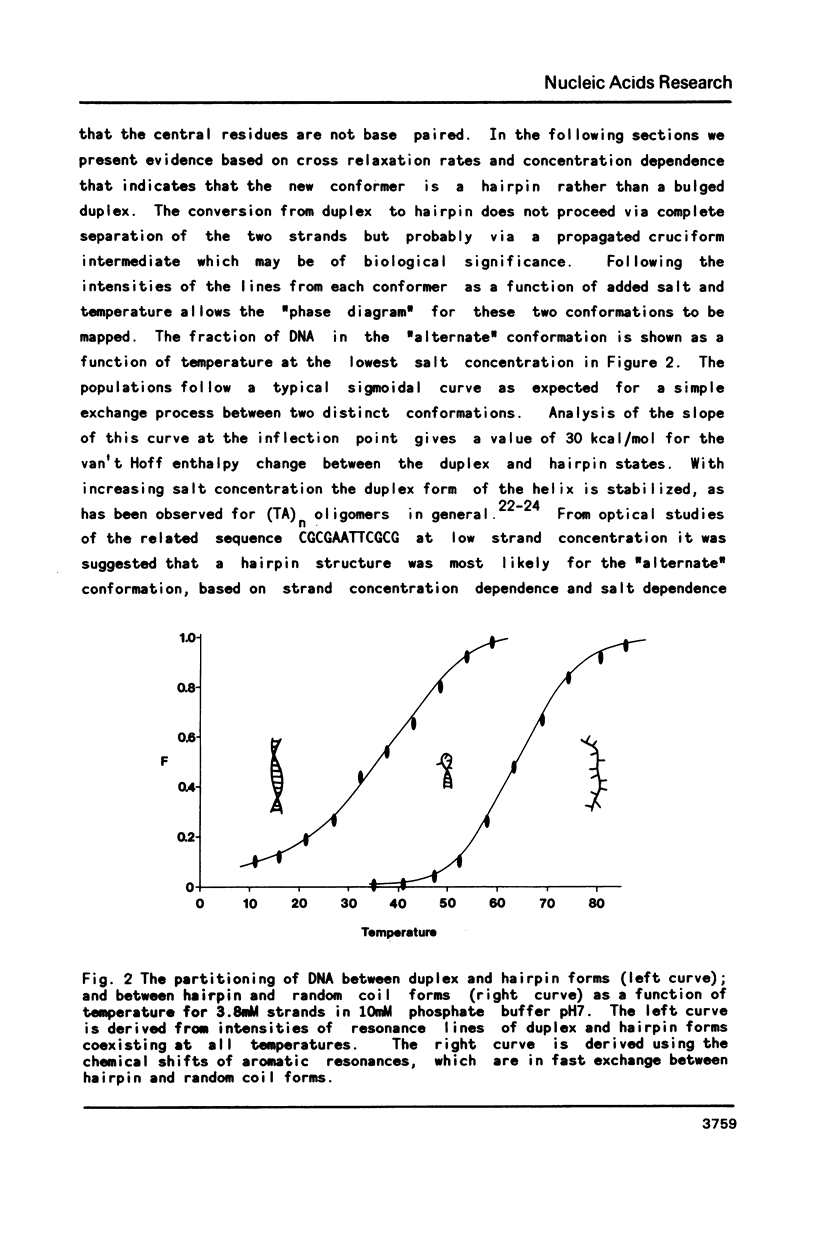
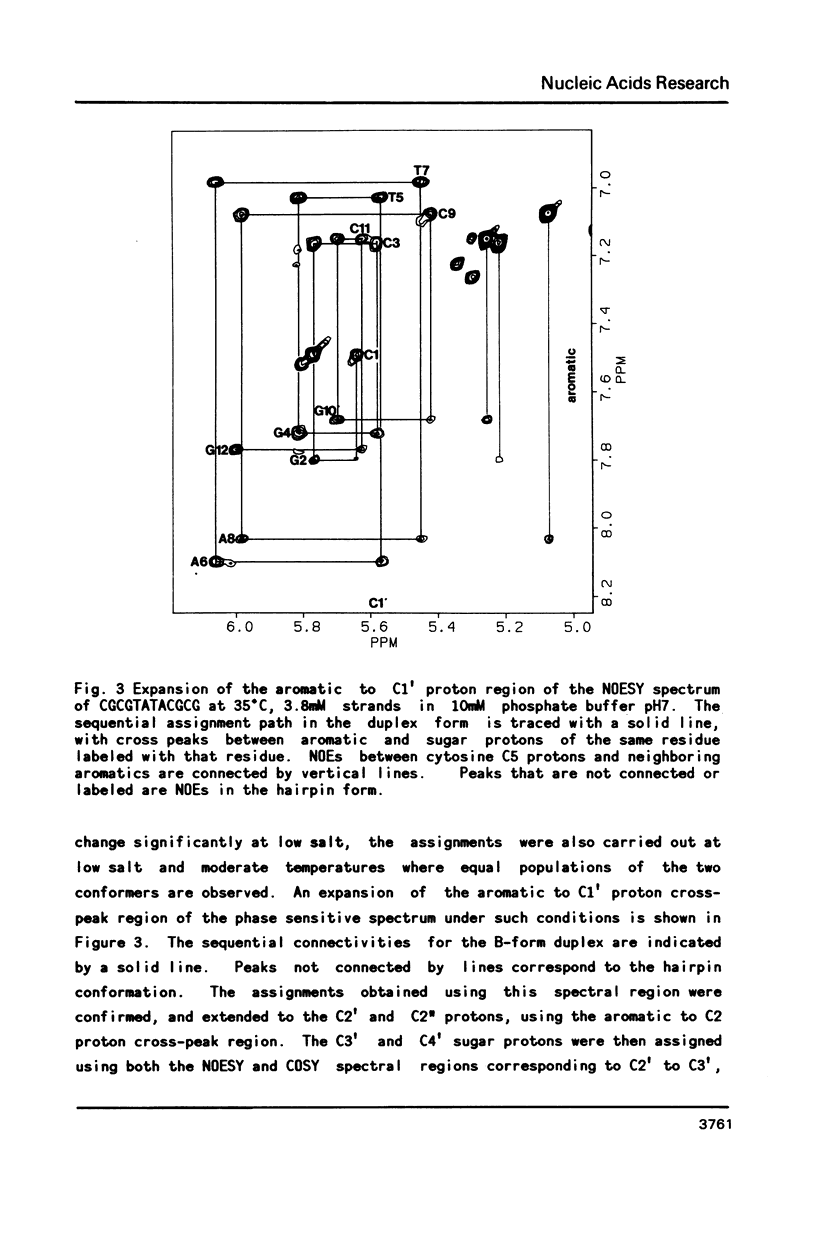
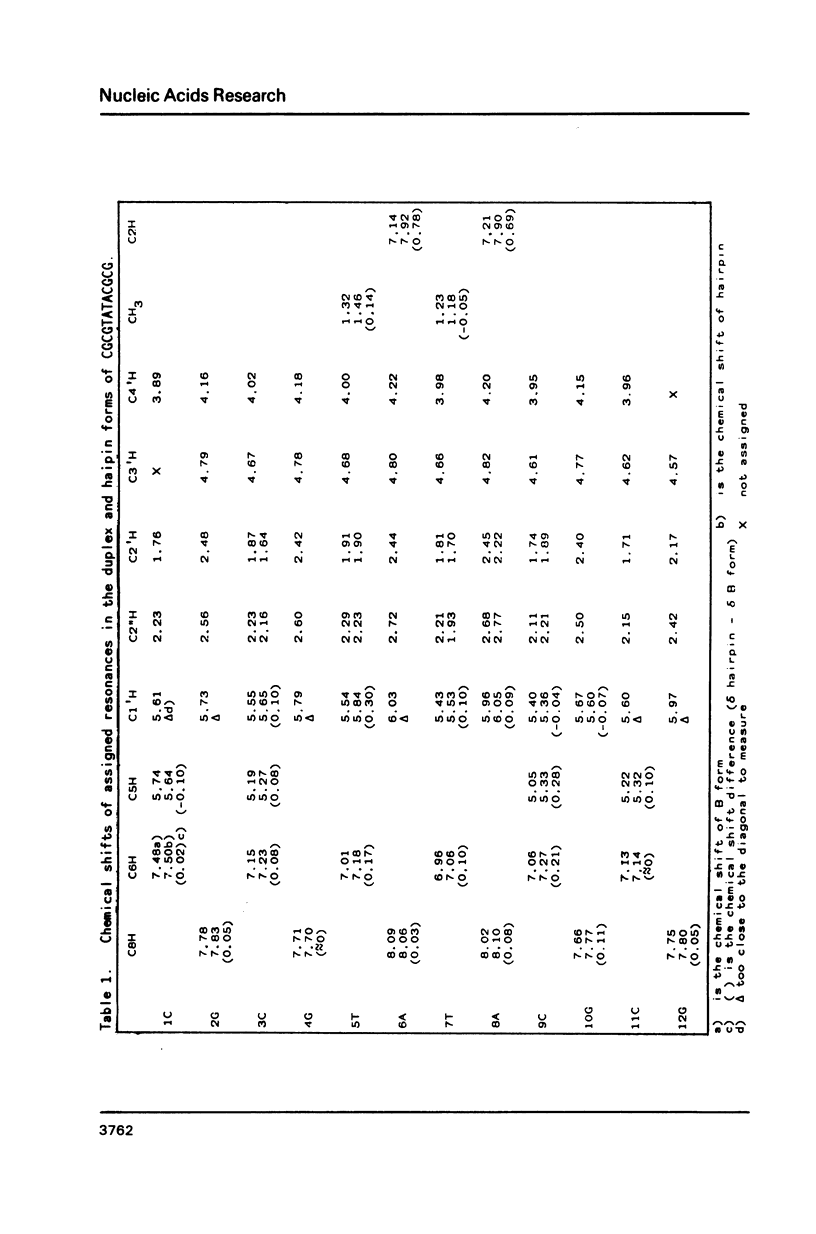
Two dimensional NMR methods have been used to assign proton resonances in the high salt (greater than or equal to 100mM Na+), low temperature duplex form of the self-complementary DNA dodecamer d(CGCGTATACGCG). At low salt (less than or equal to 10mM Na+) and higher temperature marked changes in the two-dimensional spectrum, and in the one-dimensional spectrum reported by others, indicate that the molecule converts to an alternate conformation. Using saturation transfer methods, many of the resonances of this new conformation have been assigned, and the kinetics of the interconversion of the two forms has been studied. The linewidth, correlation time, and concentration dependence of the formation of this alternate conformation support the idea that it is a unimolecular hairpin. Observation of chemical shifts and NOEs in the hairpin conformation allow some preliminary structural characterization. Examination of the energetics of the interconversion suggests that the exchange between forms does not proceed through a single stranded intermediate, but rather through another pathway, probably involving a cruciform structure.
Full text
PDF




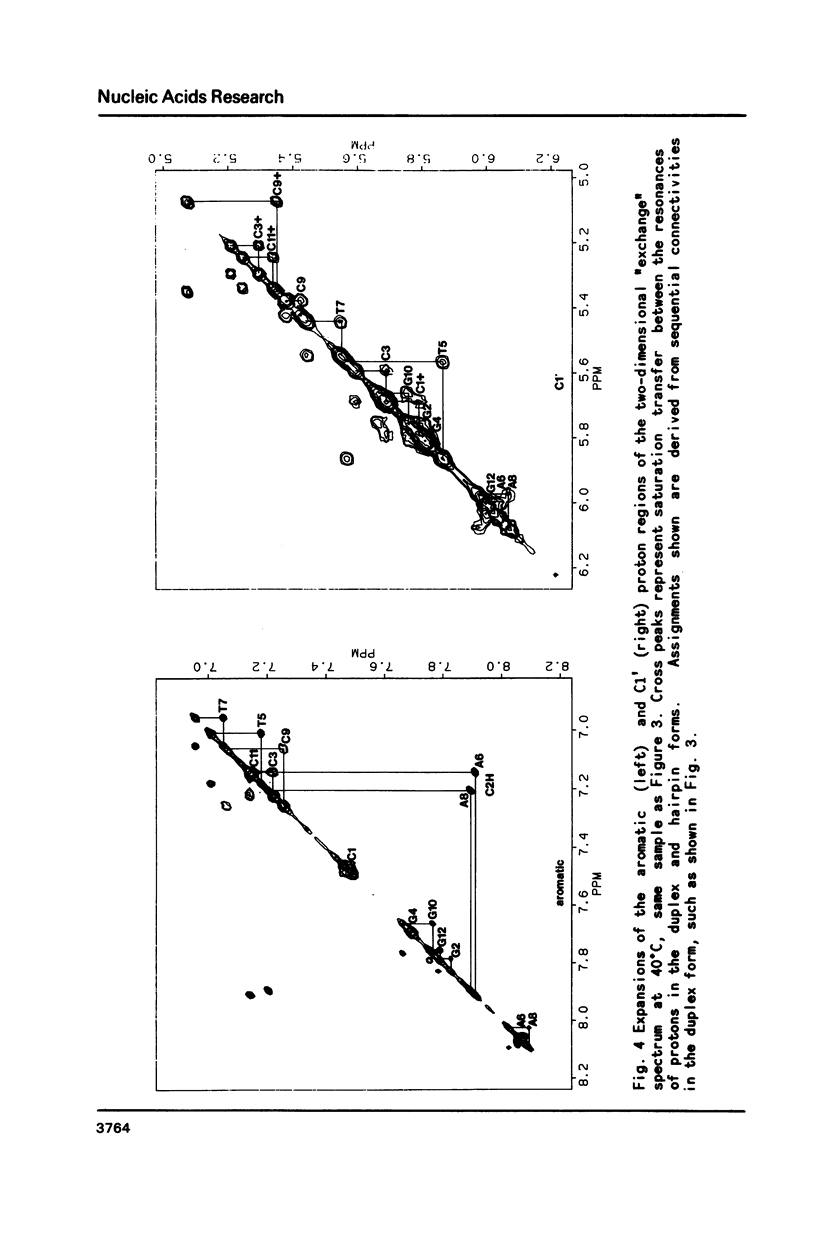

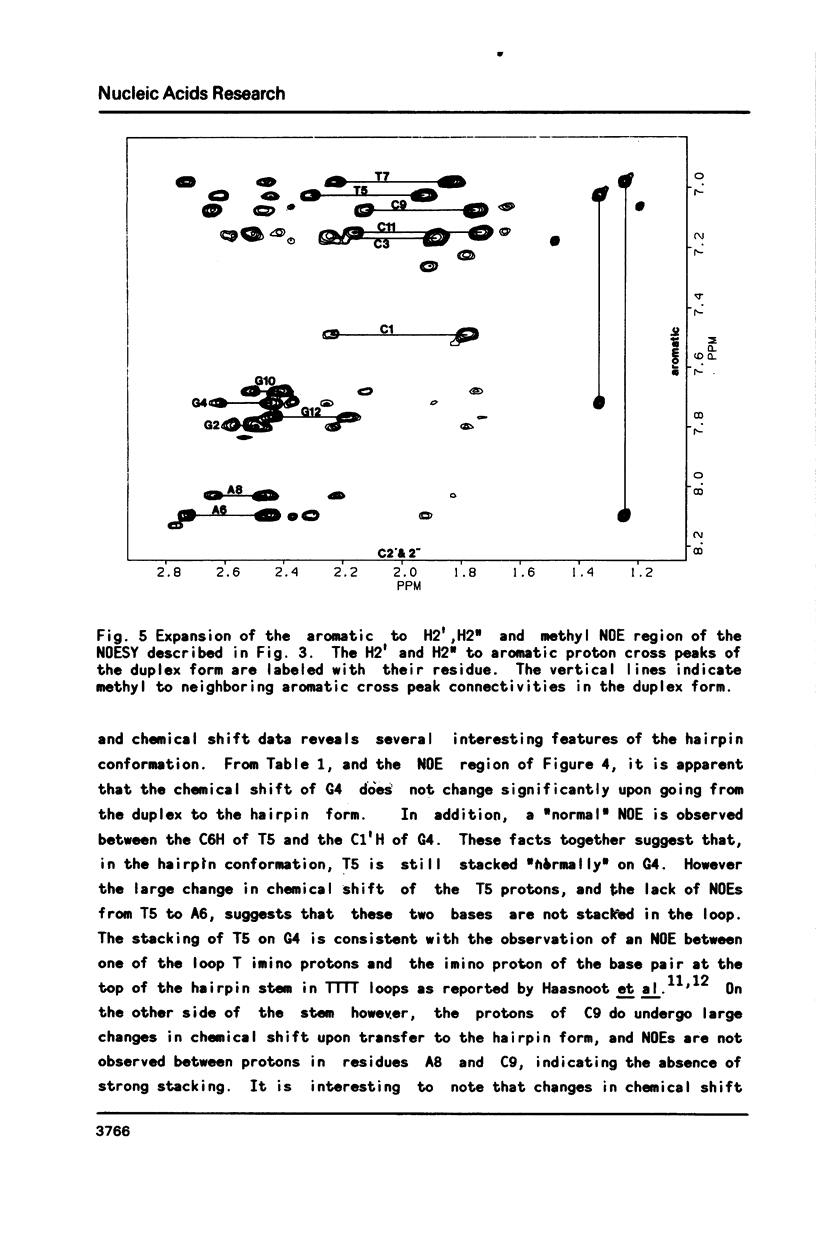


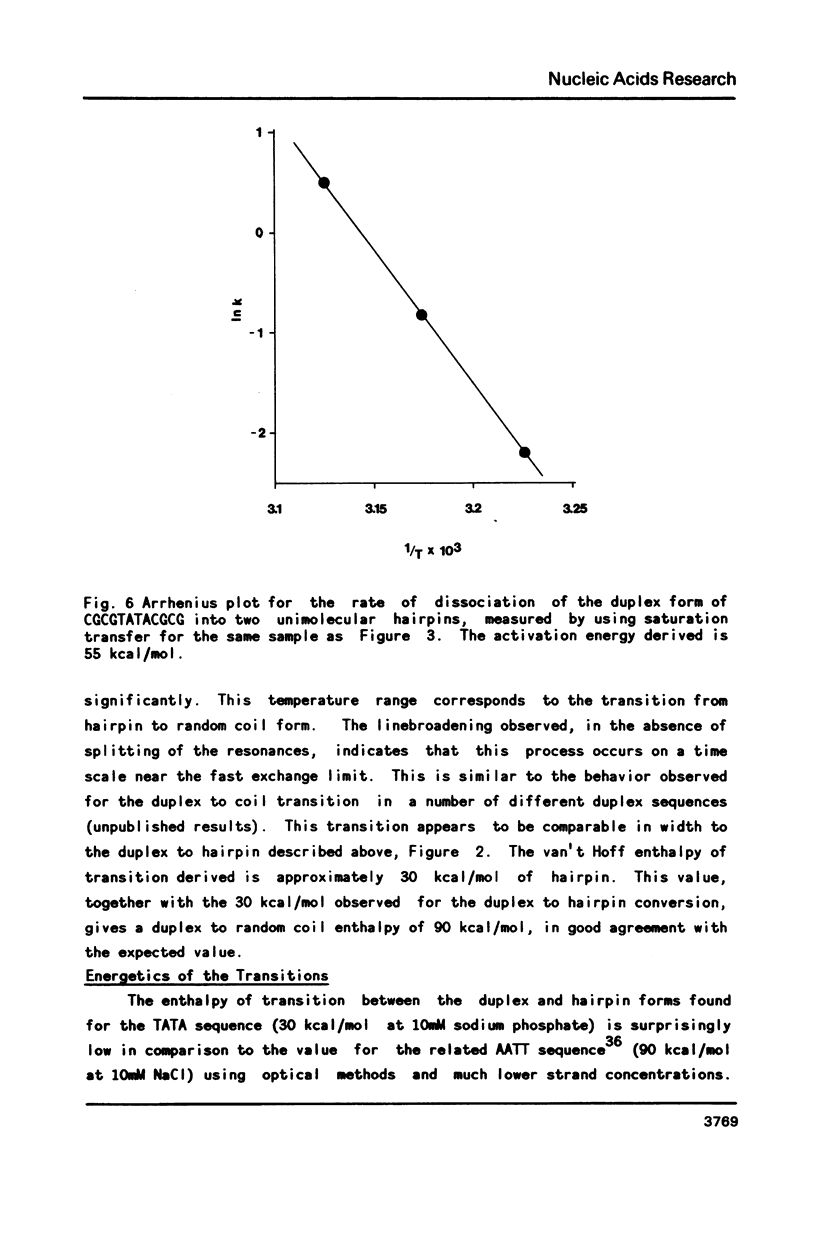



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Billeter M., Braun W., Wüthrich K. Sequential resonance assignments in protein 1H nuclear magnetic resonance spectra. Computation of sterically allowed proton-proton distances and statistical analysis of proton-proton distances in single crystal protein conformations. J Mol Biol. 1982 Mar 5;155(3):321–346. doi: 10.1016/0022-2836(82)90008-0. [DOI] [PubMed] [Google Scholar]
- Calladine C. R. Mechanics of sequence-dependent stacking of bases in B-DNA. J Mol Biol. 1982 Oct 25;161(2):343–352. doi: 10.1016/0022-2836(82)90157-7. [DOI] [PubMed] [Google Scholar]
- Dickerson R. E. Base sequence and helix structure variation in B and A DNA. J Mol Biol. 1983 May 25;166(3):419–441. doi: 10.1016/s0022-2836(83)80093-x. [DOI] [PubMed] [Google Scholar]
- Dickerson R. E., Drew H. R. Structure of a B-DNA dodecamer. II. Influence of base sequence on helix structure. J Mol Biol. 1981 Jul 15;149(4):761–786. doi: 10.1016/0022-2836(81)90357-0. [DOI] [PubMed] [Google Scholar]
- Drew H. R., Dickerson R. E. Conformation and dynamics in a Z-DNA tetramer. J Mol Biol. 1981 Nov 15;152(4):723–736. doi: 10.1016/0022-2836(81)90124-8. [DOI] [PubMed] [Google Scholar]
- Drew H. R., Wing R. M., Takano T., Broka C., Tanaka S., Itakura K., Dickerson R. E. Structure of a B-DNA dodecamer: conformation and dynamics. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2179–2183. doi: 10.1073/pnas.78.4.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feigon J., Leupin W., Denny W. A., Kearns D. R. Two-dimensional proton nuclear magnetic resonance investigation of the synthetic deoxyribonucleic acid decamer d(ATATCGATAT)2. Biochemistry. 1983 Dec 6;22(25):5943–5951. doi: 10.1021/bi00294a038. [DOI] [PubMed] [Google Scholar]
- Feigon J., Wang A. H., van der Marel G. A., Van Boom J. H., Rich A. A one- and two-dimensional NMR study of the B to Z transition of (m5dC-dG)3 in methanolic solution. Nucleic Acids Res. 1984 Jan 25;12(2):1243–1263. doi: 10.1093/nar/12.2.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haasnoot C. A., de Bruin S. H., Berendsen R. G., Janssen H. G., Binnendijk T. J., Hilbers C. W., van der Marel G. A., van Boom J. H. Structure, kinetics and thermodynamics of DNA hairpin fragments in solution. J Biomol Struct Dyn. 1983 Oct;1(1):115–129. doi: 10.1080/07391102.1983.10507429. [DOI] [PubMed] [Google Scholar]
- Haasnoot C. A., den Hartog J. H., de Rooij J. F., van Boom J. H., Altona C. Loopstructures in synthetic oligodeoxynucleotides. Nucleic Acids Res. 1980 Jan 11;8(1):169–181. doi: 10.1093/nar/8.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare D. R., Wemmer D. E., Chou S. H., Drobny G., Reid B. R. Assignment of the non-exchangeable proton resonances of d(C-G-C-G-A-A-T-T-C-G-C-G) using two-dimensional nuclear magnetic resonance methods. J Mol Biol. 1983 Dec 15;171(3):319–336. doi: 10.1016/0022-2836(83)90096-7. [DOI] [PubMed] [Google Scholar]
- Kallenbach N. R., Ma R. I., Wand A. J., Veeneman G. H., van Boom J. H., Seeman N. C. Fourth rank immobile nucleic acid junctions. J Biomol Struct Dyn. 1983 Oct;1(1):159–168. doi: 10.1080/07391102.1983.10507432. [DOI] [PubMed] [Google Scholar]
- Marky L. A., Blumenfeld K. S., Kozlowski S., Breslauer K. J. Salt-dependent conformational transitions in the self-complementary deoxydodecanucleotide d(CGCAATTCGCG): evidence for hairpin formation. Biopolymers. 1983 Apr;22(4):1247–1257. doi: 10.1002/bip.360220416. [DOI] [PubMed] [Google Scholar]
- Marky L. A., Breslauer K. J. Calorimetric determination of base-stacking enthalpies in double-helical DNA molecules. Biopolymers. 1982 Nov;21(11):2185–2194. doi: 10.1002/bip.360211107. [DOI] [PubMed] [Google Scholar]
- Ornstein R. L., Fresco J. R. Correlation of Tm and sequence of DNA duplexes with delta H computed by an improved empirical potential method. Biopolymers. 1983 Aug;22(8):1979–2000. doi: 10.1002/bip.360220811. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Kozlowski S. A., Ikuta S., Itakura K., Bhatt R., Hare D. R. NMR studies of DNA conformation and dynamics in solution. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):197–206. doi: 10.1101/sqb.1983.047.01.025. [DOI] [PubMed] [Google Scholar]
- Reid D. G., Salisbury S. A., Bellard S., Shakked Z., Williams D. H. Proton nuclear Overhauser effect study of the structure of a deoxyoligonucleotide duplex in aqueous solution. Biochemistry. 1983 Apr 12;22(8):2019–2025. doi: 10.1021/bi00277a044. [DOI] [PubMed] [Google Scholar]
- Sanderson M. R., Mellema J. R., van der Marel G. A., Wille G., van Boom J. H., Altona C. Assignment of non-exchangeable base proton and H1' resonances of a deoxyoctanucleoside heptaphosphate d(G-G-C*-C*-G-G-C-C) by using the nuclear Overhauser effect. Nucleic Acids Res. 1983 May 25;11(10):3333–3346. doi: 10.1093/nar/11.10.3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheek R. M., Boelens R., Russo N., van Boom J. H., Kaptein R. Sequential resonance assignments in 1H NMR spectra of oligonucleotides by two-dimensional NMR spectroscopy. Biochemistry. 1984 Mar 27;23(7):1371–1376. doi: 10.1021/bi00302a006. [DOI] [PubMed] [Google Scholar]
- Scheffler I. E., Elson E. L., Baldwin R. L. Helix formation by d(TA) oligomers. II. Analysis of the helix-coli transitions of linear and circular oligomers. J Mol Biol. 1970 Feb 28;48(1):145–171. doi: 10.1016/0022-2836(70)90225-1. [DOI] [PubMed] [Google Scholar]
- Shakked Z., Rabinovich D., Cruse W. B., Egert E., Kennard O., Sala G., Salisbury S. A., Viswamitra M. A. Crystalline A-dna: the X-ray analysis of the fragment d(G-G-T-A-T-A-C-C). Proc R Soc Lond B Biol Sci. 1981 Nov 24;213(1193):479–487. doi: 10.1098/rspb.1981.0076. [DOI] [PubMed] [Google Scholar]
- Sinden R. R., Broyles S. S., Pettijohn D. E. Perfect palindromic lac operator DNA sequence exists as a stable cruciform structure in supercoiled DNA in vitro but not in vivo. Proc Natl Acad Sci U S A. 1983 Apr;80(7):1797–1801. doi: 10.1073/pnas.80.7.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinden R. R., Pettijohn D. E. Cruciform transitions in DNA. J Biol Chem. 1984 May 25;259(10):6593–6600. [PubMed] [Google Scholar]
- Tran-Dinh S., Taboury J., Neumann J. M., Huynh-Dinh T., Genissel B., Langlois d'Estaintot B., Igolen J. 1H NMR and circular dichroism studies of the B and Z conformations of the self-complementary deoxyhexanucleotide d(m5C-G-C-G-m5-C-G): mechanism of the Z-B-coil transitions. Biochemistry. 1984 Mar 27;23(7):1362–1371. doi: 10.1021/bi00302a005. [DOI] [PubMed] [Google Scholar]
- Wagner G., Wüthrich K. Sequential resonance assignments in protein 1H nuclear magnetic resonance spectra. Basic pancreatic trypsin inhibitor. J Mol Biol. 1982 Mar 5;155(3):347–366. doi: 10.1016/0022-2836(82)90009-2. [DOI] [PubMed] [Google Scholar]
- Wang A. H., Quigley G. J., Kolpak F. J., Crawford J. L., van Boom J. H., van der Marel G., Rich A. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979 Dec 13;282(5740):680–686. doi: 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]
- Wang A. J., Quigley G. J., Kolpak F. J., van der Marel G., van Boom J. H., Rich A. Left-handed double helical DNA: variations in the backbone conformation. Science. 1981 Jan 9;211(4478):171–176. doi: 10.1126/science.7444458. [DOI] [PubMed] [Google Scholar]
- Weiss M. A., Patel D. J., Sauer R. T., Karplus M. Two-dimensional 1H NMR study of the lambda operator site OL1: a sequential assignment strategy and its application. Proc Natl Acad Sci U S A. 1984 Jan;81(1):130–134. doi: 10.1073/pnas.81.1.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wemmer D. E., Chou S. H., Reid B. R. Sequence-specific recognition of DNA. Nuclear magnetic resonance assignments and structural comparison of wild-type and mutant lambda OR3 operator DNA. J Mol Biol. 1984 Nov 25;180(1):41–60. doi: 10.1016/0022-2836(84)90429-7. [DOI] [PubMed] [Google Scholar]
- Wüthrich K., Wider G., Wagner G., Braun W. Sequential resonance assignments as a basis for determination of spatial protein structures by high resolution proton nuclear magnetic resonance. J Mol Biol. 1982 Mar 5;155(3):311–319. doi: 10.1016/0022-2836(82)90007-9. [DOI] [PubMed] [Google Scholar]



