Abstract
Background
The vast majority of the 1.1 million Alu elements are retrotranspositionally inactive, where only a few loci referred to as ‘source elements’ can generate new Alu insertions. The first step in identifying the active Alu sources is to determine the loci transcribed by RNA polymerase III (pol III). Previous genome-wide analyses from normal and transformed cell lines identified multiple Alu loci occupied by pol III factors, making them candidate source elements.
Findings
Analysis of the data from these genome-wide studies determined that the majority of pol III-bound Alus belonged to the older subfamilies Alu S and Alu J, which varied between cell lines from 62.5% to 98.7% of the identified loci. The pol III-bound Alus were further scored for estimated retrotransposition potential (ERP) based on the absence or presence of selected sequence features associated with Alu retrotransposition capability. Our analyses indicate that most of the pol III-bound Alu loci candidates identified lack the sequence characteristics important for retrotransposition.
Conclusions
These data suggest that Alu expression likely varies by cell type, growth conditions and transformation state. This variation could extend to where the same cell lines in different laboratories present different Alu expression patterns. The vast majority of Alu loci potentially transcribed by RNA pol III lack important sequence features for retrotransposition and the majority of potentially active Alu loci in the genome (scored high ERP) belong to young Alu subfamilies. Our observations suggest that in an in vivo scenario, the contribution of Alu activity on somatic genetic damage may significantly vary between individuals and tissues.
Keywords: Alu source elements, Alu expression, RT-PCR, ChIP-seq, Retrotransposition, SINE
Findings
Alu elements are major contributors to genomic instability [1] and genetic disease [2] due to their ability to generate new copies that randomly insert throughout the genome and to induce non-homologous recombination between different copies. When comparing copy numbers, Alu has been vastly more successful than other non-autonomous elements, such as the retropseudogenes and even the autonomous L1 element [3]. Alu-induced mutagenesis is responsible for the majority of the documented instances of human retroelement insertion-induced disease [2] and presents a retrotransposition rate estimated up to ten-fold higher than L1 [4,5]. The human genome contains over one million Alu inserts [3], which can be divided into subfamilies based on specific diagnostic nucleotides and their evolutionary period of activity [6,7]. About 80% of Alu elements belong to the older previously active Alu J and Alu S subfamilies [6]. Germline derived evidence supports the current activity of only the subsets of the younger Alu Y subfamilies (such asY, Ya, and Yb)[8], although recent data appear to indicate that Alu retrotransposition in germline and somatic tissues may show different distributions [9].
Only a few Alu elements, referred to as ‘source’ or ‘master’ elements, undergo retrotransposition. Identification of source Alu elements has been elusive, as bona fide Alu retrotransposition events never present 5′ or 3′ transductions that could help determine a parent locus. Because transcription by RNA polymerase III (pol III) is necessary for Alu retrotransposition, a first step to identify a source element is to determine the transcribing Alu loci. There are little available data on RNA pol III transcribed Alu loci. Current techniques using RT-PCR approaches are unable to distinguish bona fide pol III Alu transcripts from those pol II transcripts containing Alu sequences (see Figure 1 for details). One of the few sources of reliable information was generated using primer extension and C-tail RACE, which showed a limited amount of SINE expression ex vivo in some cell lines [10,11]. Recently, genome-wide chromatin immunoprecipitation (ChIP) analyses followed by parallel sequencing (ChIP-seq) performed by three different laboratories identified multiple Alu loci bound by RNA polymerase III factors [12-14]. These datasets (Table 1) were generated from a variety of cell lines including a relatively ‘normal’ cell line: IMR90 (a Tert-immortalized, untransformed human lung fibroblast) and the tumor-derived cell lines: HeLa (cervical adenocarcinoma), Jurkat (T-cell leukemia) and K562 (myelogenous leukemia). Although the binding by pol III factors is not synonymous with transcription, these Alu loci represent potential candidate source elements.
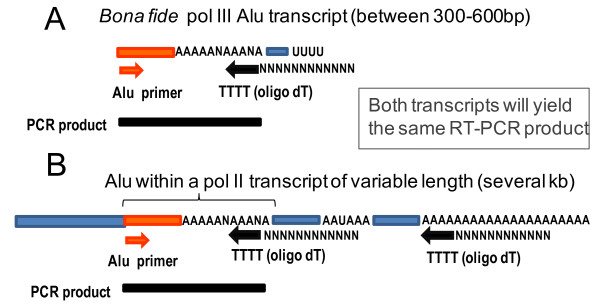
Figure 1.
RT-PCR assays are unable to distinguish between RNA pol III transcribed Alu transcripts and RNA pol II transcribed mRNAs containing Alu sequences. A. Schematic of a bona fide pol III Alu transcript. Transcripts of 300 to 600 bp in length contain an Alu body (orange) flanked at the 3′ end by a poly-A stretch and the unique region (blue) which is determined by the location of the pol III terminator (Us). The poly-A stretch may be either homogeneous or heterogeneous containing non-adenosine bases. B. Schematic of an mRNA containing an Alu sequence. The Alu (orange) present within the mRNA (blue) will also have an oligo dA stretch at its 3′ end. Most standard RT-PCR approaches, such as 3′ RACE, rely on generating a cDNA through the reverse transcription of the RNA using an oligo dT primer (black arrow). Because both types of transcripts (pol II versus pol III) contain Alu sequences flanked by a polyA stretch, both will be amplified during reverse transcription. PCR amplification of selected cDNAs can then be performed by using a gene specific primer (in this case Alu, shown as an orange arrow) and a primer to the 3′ sequence of the oligo dT (represented as ‘Ns’). The PCR products (shown as black bars) of the cDNAs generated by both types of transcripts will yield the same type of product, thus making it difficult to distinguish the data generated from the bona fide pol III Alu transcripts.
Table 1.
Data Source of Alu loci
| Study | Method | Cell lines |
|---|---|---|
| Canella et al. [13] |
ChIP-seqa for detection of sites bound by POLR3D |
IMR90 |
| (RPC4), TFIIIB subunits BDP1 and BRF1 |
|
|
| Oler et al. [12] |
ChIP-seqa and ChIP-arrayb for detection of sites bound by Pol III (RPC32 subunit), TFIIIC63 subunit, BRF1, BRF2 |
HeLa, Jurkat, HEK, 293 T |
| Moqtaderi et al. [14] | ||
| ChIP-seqa for detection of sites bound by TFIIIC-110 |
HeLa, K562 |
|
| subunit, TFIIIB subunits BDP1 and BRF1, Pol III (RPC155 subunit) and BRF2 |
aChromatin immunoprecipitation followed by massively parallel sequencing; bchromatin immunoprecipitation followed by complementary DNA microarray hybridization. ChIP, chromatin immunoprecipitation.
To evaluate these candidate elements, we retrieved the Alu-related sequences for those enriched with pol III initiation factors (pol III or TFIIIB) in the published datasets [12-14] including the ‘A-tail’ and ‘unique’ region at the 3′ flanking sequence (see schematic of an Alu in Figure 2). Each pol III bound Alu locus was assigned a name based on the dataset and/or cell line where it was identified. The 3′ flanking sequence included either 300 bp or up to the first pol III terminator (which was defined as four or more thymidine residues) of the downstream genomic flanking sequence (complete data set shown in Additional file 1: Tables S4-S9). We then selected only those that fit the standard dimeric Alu structure, eliminating any FLAMs, FRAMs and partial Alu elements. In addition, we eliminated any Alu sequences that contained an internal pol III terminator as these would generate truncated Alu transcripts.
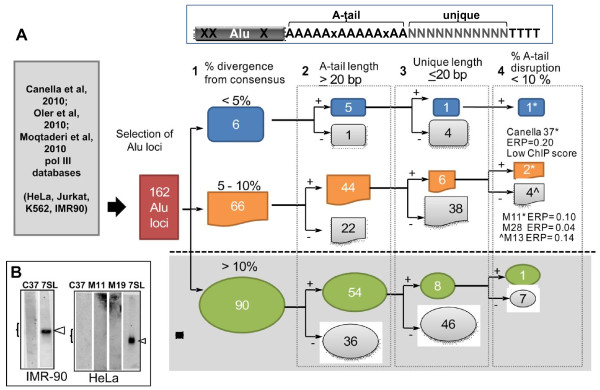
Figure 2.
Classification strategy of potentially active Alu loci. A schematic of an Alu element is on the top right, where the body is shown in gray. X represent sequences changes differing from the consensus or non-A residues in the A-tail. Ns represent the unique flanking sequence located downstream of the A-tail and the first RNA polymerase III terminator defined as four or more thymidine residues (TTTT). A. Flow chart used for the identification of potentially retrotransposition competent Alu elements. Datasets from previously published data sets were interrogated for Alu loci identified as bound by RNA polymerase III factors. A total of 162 Alu-candidate loci were retrieved and classified based on four parameters: 1- % divergence from the consensus sequence, which was subdivided into three categories based on the % sequence divergence from their Alu subfamily consensus (less than 5% in blue, 5 to 10% in orange and over 10% in green also classified as retrotranspositionally inactive). 2- A-tail length, where Alu elements with A-tail lengths of equal to or over 20 bp were included as potentially active (+). Alu elements not meeting the criteria were binned separately and not further evaluated (shown as gray symbols; -). 3- Length of the unique sequence, scoring positive Alu elements with 20 bp or less. 4- % disruption of the A-tail sequence was scored selecting for those with 10% or less disruption. The two Alu candidates evaluated for expression (show in B) are indicated by the asterisks in the flow diagram. ERP = estimated retrotranspositional potential. B. Expression of candidate Alu loci with retrotransposition potential. Total RNA extracts from HeLa or IMR90 cells were hybridized with radioactively endlabeled oligomers complementary to the unique region of the Alu loci (Additional file 1: Table S11) following a previously published protocol [24]. Results from Alu Yb8 Canella 37 (C37), Alu Sx Moq 11 h (M11), and Alu Sx Moq 19 k (M19) are shown. An oligo complementary to a region of the 7SL RNA that does not share sequence similarity to Alu RNA: 5′-CCGATCGGCATAGCGCACTA-3′ was used as a positive control (white arrowhead). Bracket approximates the expected location of the Alu transcript.
A total of 162 Alu elements fit our criteria (Additional file 1: Table S1). Several loci (24 out of 162, 14.8%) were identified in at least two separate cell lines (Additional file 1: Table S2), suggesting potential regions preferentially bound by RNA polymerase III factors. Each Alu locus is represented only once in our data set and analyses. Although the majority of Alu elements in the genome belong to the older Alu subfamilies (S + J) currently, only the ‘young’ Alu subfamilies appear to be retrotranspositionally active. Classification of the dataset of pol III-bound Alus revealed that the majority belong to the older subfamilies (Table 2) consistent with previously published expression data [11]. Although the Alu subfamily distribution from each individual cell line showed variation, the old Alu S + J subfamilies represented at least two thirds and up to 98% of the identified loci (Additional file 1: Table S3). When all Alus are considered together, a moderately significant association is found between pol III-binding and Alu J + S elements in at least one cell line but not Alu Y elements (odds ratio = 1.6, P = 0.098 in Fisher’s exact test), suggesting that pol III is approximately 1.6 times less likely to bind to an Alu Y element than to an Alu J or S element. Normalization of the collective dataset for Alu subfamily copy number differences (older Alus are vastly more abundant than younger elements), we observe that proportionally, there are more young Alu elements bound by pol III factors (Table 2); however, these differences are not significant (P = 0.21 and P = 0.44 for AluYa5 and AluYb8, respectively, in Fisher’s exact test).
Table 2.
Alu subfamily distribution of Alu elements bound by RNA polymerase III factors
|
Alu |
% Total |
# of disease |
% Alu |
% Alu loci |
Pol III |
# Pol III |
|---|---|---|---|---|---|---|
|
subfamilya |
full Alus (530,850) |
cases due to ade novo insertion |
transcriptsd |
bound by pol III |
bound Alu enrichment |
bound Alu with ERP |
| (%)c | factorse | ≥0.10 | ||||
|
S + J |
84.5 |
0 (0) |
66 |
88.4 |
1.0 |
2 |
|
Y |
15.5 |
13 (23) |
33 |
9.8 |
0.6 |
0 |
|
Ya5 |
0.63 |
24 (44) |
0.8 |
1.2 |
2.0 |
0 |
| Yb8 | 0.42 | 18 (33) | 0.5 | 0.6 | 1.5 | 1 |
aIncludes all subfamily variants: for example, AluYa5, AluYa5a2, AluYa8 are classified as Ya5; bnumber of Alu elements meeting full length criteria (Details in supplemental data); csubfamily distribution of the 55 de novo Alu elements reported to cause a disease [2,15]; danalysis was performed on transcripts isolated from the Ntera2 (teratocarcinoma) cell line [11]; eanalyses were performed on the cumulative data obtained from studies on IMR-90 (normal untransformed Tert-immortalized lung fibroblast) [13], HeLa (cervical adenocarcinoma), Jurkat (T-cell leukemia), HEK 293 T (T antigen-transformed kidney) [12] and K562 (myelogenous leukemia) cells [14] (raw data and detailed analysis for each cell line included in Additional file 1: Table S3); frepresents the increase in Alu loci bound by RNA polymerase III factors relative to copy number (that is, %AluY bound/%AluY copies in the genome, detailed analysis in Additional file 1: Table S3); gnumber of Alu loci bound by RNA polymerase III factors with estimated retrotransposition potential (ERP) scores of ≥0.10. An ‘ideal’ Alu will have ERP score of 1.00.
In addition to the ability to be transcribed, specific sequence features of Alu elements can influence retrotransposition efficiency [22]. Therefore, we proceeded to evaluate the individual pol III-bound Alu loci using our own designed dichotomous key based on the previously identified criteria known to affect retrotransposition rates: 1) sequence divergence from the consensus (loss of retrotransposition efficiency with higher divergence [22,16]); 2) A-tail length (a minimum length is required [17]); 3) length of the unique sequence (loss of efficiency with longer sequences [22]); and 4) A-tail homogeneity (loss of efficiency with higher % disruptions [22]). Our results are schematically represented in Figure 2A (details in Additional file 1: Table S1). We selected limits for our criteria parameters that have been shown to significantly reduce retrotransposition levels. We also separately assigned a numerical value of the impact on retrotransposition (‘R’) for each Alu feature variant relative to an Alu reference (Additional file 1: Tables S12-15) to roughly calculate the ERP of each individual Alu (Additional file 1: Table S1, column T). However, the ERP should not be taken as the sole defining criteria for in vivo predictions, as it is based on a limited amount of data generated from engineered Alus in a tissue culture system and does not include transcription status. This scoring system was applied to the pol III-bound subset as well as all Alus genome-wide using an algorithm that incorporates each of the scoring criteria (implemented in Perl, score_alus.pl; available upon request). As expected, young Alu elements had a higher score genome-wide than Alu J + S elements (median values of 0.0042 and 0.000001 for Y and J + S, respectively; P = 2.2e-16 in Wilcoxon test). While pol III-bound Alus had a higher ERP score in general than Alus not bound by pol III (median 0.000229 and 0.000001, respectively; P = 0.013 in Wilcoxon test), the ERP score for the vast majority of the pol III-bound Alus was considerably lower than the arbitrarily selected minimal threshold for retrotransposition competency of 0.20. Of the 162 pol III-bound Alu sequences only one AluYb8 (Canella 37 from IMR90 cells) was highly conserved relative to the consensus sequence, met the rest of the criteria and scored 0.20 ERP (an ‘ideal’ Alu will have a score of 1.00). In addition, it scored low in the pol III ChIP assay [13] and Canella 37 AluYb8 transcripts were undetectable in HeLa and IMR90 cells by northern blot probing with end-labeled oligonucleotides complementary to the unique sequence (Figure 2B). We opted not to use an RT-PCR approach, as it is unable to differentiate between RNA pol II and pol III transcripts (Figure 1). In contrast, when using a low ERP threshold to evaluate the reference genome, several thousands of Alus genome-wide were identified (6,103 and 1,818 Alus at ERP threshold of 0.10 or 0.20, respectively; Additional file 1: Table S16). Furthermore, a more conservative threshold (ERP scores of ≥0.50) yields only 163 of genome-wide Alus (all young elements), corroborating the previously proposed Alu source model that only a small portion of Alus in the genome are likely active [18].
The next ‘best’ candidates identified only partially met the criteria, corresponding to three Alu loci belonging to older S subfamilies: Moq 13 h (HeLa), Moq 11 h (HeLa) and Moq 28 k (K562) with 5.3%, 7.8% and 9.3% sequence divergence from consensus, respectively. Some of the sequence changes were within the RNA pol III A box and in the sequences predicted to bind the SRP9 and SRP14 proteins. Lower binding of SRP9/14 would likely reduce the retrotransposition capability of these elements, but further testing is required. Moq 28 k shows a very low ERP of 0.04. Interestingly, Moq 11 h and Moq 13 h present acceptable A-tail length with marginal ERP values of 0.10 and 0.14, respectively. Moq 13 h showed an A-tail with high % A-tail disruption (24.3%), which is not observed in de novo inserts [19]. The published work on Moq 11 h showed significant pol III binding by ChIP-seq [14]. If expressed, Moq 11 h could prove retrotranspositionally competent. However, the RNA-seq data showed only three sequence reads in HeLa and none in the K562 and a non-detectable transcript by northern blot analysis (Figure 2B). Evaluation of expression from five other randomly selected Alu loci, Moq 19 k (Figure 2B) and Canella 2 and 28, Oler 38 h, and 3c, (data not shown) by northern blot analysis also proved unsuccessful in the detection of pol III Alu transcripts. Due to the sensitivity limitation of our assay, we are unable to unambiguously confirm that these identified Alu candidate loci with the best retrotransposition potential (Canella 37 and Moq 11 h) are transcriptionally silent. Thus, we cannot eliminate the possibility that very low amounts of expression may occur, resulting in retrotransposition. Alternatively, these or other identified Alu loci may be more efficiently expressed in other cell types, tissues or under other conditions such as heat shock known to increase Alu expression [20].
Presently, we are unable to rule out that any of the other identified pol III-bound Alu candidates that partially fulfill our criteria or contain borderline attributes may undergo retrotransposition at very low rates. However, the limits of the criteria are based on the results using a tissue culture system [21] that significantly favors Alu activity through the overexpression of both a tagged Alu transcript and the enzymatic machinery required for retrotransposition(L1 ORF2 protein). This opens the possibility that an Alu locus identified as potentially active by the selected parameters may not be able to retrotranspose under natural cellular conditions. Thus, it is unlikely that the ‘less perfect’ Alu candidate elements (those with low ERP scores) contribute to retrotransposition in any significant manner.
Our findings indicate that up to now, most cells analyzed may support RNA pol III expression of a collection of Alu elements, although the vast majority lack sequence features associated with retrotransposition competence (Table 2). A striking observation is the overall low number of detected Alu loci (162), and even lower when considering retrotransposition potential (only three loci from all cell lines combined had ERPs above 0.10). So why is there little to no evidence of expression by pol III of the active younger Alu elements? Although speculative, these data could be indicative of a general mechanism, such as DNA methylation, that selectively limits Alu transcription of the retrotranspositionally competent elements. Also, it could be a reflection that younger, less mutated retroelements still maintain most of their CpGs making them good substrates for regulation by methylation [22]. In addition, the inability to detect transcripts from the candidates identified may reflect variability in Alu expression, where the same cell lines in different laboratories have different expression patterns. It is possible that Alu expression varies by cell type, growth conditions, epigenetic signals and transformation state. Our observations support the hypothesis that in an in vivo scenario, the contribution of Alu activity on somatic genetic damage may significantly vary between individuals and tissues.
Competing interests
The authors declare that they have no competing financial interests.
Authors’ contributions
AMR-E designed the experimental approach, directed, and performed analyses. ST-D mined some of the Alu sequences and performed the northern blot analyses, with the assistance of RSD. AJO mined the Alu sequences from the Moqtaderi data files, performed the reanalysis of ChIP-seq data, and wrote the algorithm for classification of Alus and ERP score. AJO and BRC provided RNA-seq data. AJO, BRC and DC provided scientific consultation on data interpretation and writing. All authors read and approved the final manuscript
Supplementary Material
Table S1. Final Alu data set. Table S2. Duplicate Alu loci (Alu locus was identified in more than one cell type). Table S3. Alu subfamily distribution. Table S4. Retrieved Alus from HeLa Table 1b Pol3 ChIP-chip in HeLa from Oler,A.J. et al. Nat Struct Mol Biol 17, 620–628 (2010). Table S5. Retrieved Alus from Table 1c HeLa ChIP-seq Knowns from Oler, A.J. et al. Nat Struct Mol Biol 17, 620–628 (2010). Table S6. Retrieved Alus from Table 1 g Pol3 Jurkat ChIP-seq from Oler, A.J. et al. Nat Struct Mol Biol 17, 620–628 (2010). Table S7. Retrieved Alus from Table 1 h Pol3 multiple cell types from Oler, A.J. et al. Nat Struct Mol Biol 17, 620–628 (2010). Table S8. Retrieved Alus from IMR90, Table S7 from Canella,D., et al. Genome Res. 20, 710–721 (2010). Table S9. Retrieved Alus from HeLa cells, Moqtaderi, Z, et al. Nat Struct Mol Biol, 20, 635–640 (2010). Table S10. Retrieved Alus from K562 cells, Moqtaderi, Z, et al. Nat Struct Mol Biol, 20, 635–640 (2010). Table S11. Probes for northern blot analysis. Table S12. Numerical value assigned base on the impact of Alu sequence divergence on retrotransposition potential. Table S13. Numerical value assigned base on the impact of A-tail length on retrotransposition potential. Table S14. Numerical value assigned base on the impact of length of unique sequence on retrotransposition potential. Table S15. Numerical value assigned base on the impact of A-tail disruptions on retrotransposition potential. Table S16. Output of genome-wide analysis of B box-containing Alus with ERP (Competence_score) > = 0.10.
Contributor Information
Andrew J Oler, Email: andrew.oler@nih.gov.
Stephen Traina-Dorge, Email: trainado@bc.edu.
Rebecca S Derbes, Email: rderbes@tulane.edu.
Donatella Canella, Email: Donatella.Canella@unil.ch.
Brad R Cairns, Email: Brad.Cairns@hci.utah.edu.
Astrid M Roy-Engel, Email: aengel@tulane.edu.
Acknowledgements
This publication was made possible by Grants Number GM45668 (PD) and P20GM103518/P20RR020152, plus R01GM079709A (to AMR-E) from the National Institutes of Health (NIH) and the Howard Hughes Medical Institute (AJO). An allocation of resources from the Center for High Performance Computing at the University of Utah is gratefully acknowledged. Huntsman Cancer Institute Biostatistics Core Facility supported by grant P30 CA042014 also participated in this work. The contents are solely the responsibility of the authors and do not necessarily represent the official views of NCRR or NIH. We would like to thank Prescott Deininger for his helpful comments on the manuscript.
References
- Xing J, Witherspoon DJ, Ray DA, Batzer MA, Jorde LB. Mobile DNA elements in primate and human evolution. Am J Phys Anthropol. 2007;45(Suppl):2–19. doi: 10.1002/ajpa.20722. [DOI] [PubMed] [Google Scholar]
- Belancio VP, Hedges DJ, Deininger P. Mammalian non-LTR retrotransposons: for better or worse, in sickness and in health. Genome Res. 2008;18:343–358. doi: 10.1101/gr.5558208. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C. et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Xing J, Zhang Y, Han K, Salem AH, Sen SK, Huff CD, Zhou Q, Kirkness EF, Levy S, Batzer MA, Jorde LB. Mobile elements create structural variation: analysis of a complete human genome. Genome Res. 2009;19:1516–1526. doi: 10.1101/gr.091827.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iskow RC, McCabe MT, Mills RE, Torene S, Pittard WS, Neuwald AF, Van Meir EG, Vertino PM, Devine SE. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell. 2010;141:1253–1261. doi: 10.1016/j.cell.2010.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen MR, Batzer MA, Deininger PL. Evolution of the master Alu gene(s) J Mol Evol. 1991;33:311–320. doi: 10.1007/BF02102862. [DOI] [PubMed] [Google Scholar]
- Batzer MA, Schmid CW, Deininger PL. Evolutionary analyses of repetitive DNA sequences. Methods Enzymol. 1993;224:213–232. doi: 10.1016/0076-6879(93)24017-o. [DOI] [PubMed] [Google Scholar]
- Stewart C, Kural D, Stromberg MP, Walker JA, Konkel MK, Stutz AM, Urban AE, Grubert F, Lam HY, Lee WP, Busby M, Indap AR, Garrison E, Huff C, Xing J, Snyder MP, Jorde LB, Batzer MA, Korbel JO, Marth GT. 1000 Genomes Project: A comprehensive map of mobile element insertion polymorphisms in humans. PLoS Genet. 2011;7:e1002236. doi: 10.1371/journal.pgen.1002236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baillie JK, Barnett MW, Upton KR, Gerhardt DJ, Richmond TA, De Sapio F, Brennan PM, Rizzu P, Smith S, Fell M, Talbot RT, Gustincich S, Freeman TC, Mattick JS, Hume DA, Heutink P, Carninci P, Jeddeloh JA, Faulkner GJ. Somatic retrotransposition alters the genetic landscape of the human brain. Nature. 2011;479:534–537. doi: 10.1038/nature10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson KE, Schmid CW. Transcriptional inactivity of Alu repeats in HeLa cells. Nucleic Acids Res. 1986;14:6145–6158. doi: 10.1093/nar/14.15.6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaikh TH, Roy AM, Kim J, Batzer MA, Deininger PL. cDNAs derived from primary and small cytoplasmic Alu (scAlu) transcripts. J Mol Biol. 1997;271:222–234. doi: 10.1006/jmbi.1997.1161. [DOI] [PubMed] [Google Scholar]
- Oler AJ, Alla RK, Roberts DN, Wong A, Hollenhorst PC, Chandler KJ, Cassiday PA, Nelson CA, Hagedorn CH, Graves BJ, Cairns BR. Human RNA polymerase III transcriptomes and relationships to Pol II promoter chromatin and enhancer-binding factors. Nat Struct Mol Biol. 2010;17:620–628. doi: 10.1038/nsmb.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canella D, Praz V, Reina JH, Cousin P, Hernandez N. Defining the RNA polymerase III transcriptome: genome-wide localization of the RNA polymerase III transcription machinery in human cells. Genome Res. 2010;20:710–721. doi: 10.1101/gr.101337.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moqtaderi Z, Wang J, Raha D, White RJ, Snyder M, Weng Z, Struhl K. Genomic binding profiles of functionally distinct RNA polymerase III transcription complexes in human cells. Nat Struct Mol Biol. 2010;17:635–640. doi: 10.1038/nsmb.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeaux MS, Roy-Engel AM, Hedges DJ, Deininger PL. Diverse cis factors controlling Alu retrotransposition: what causes Alu elements to die? Genome Res. 2009;19:545–555. doi: 10.1101/gr.089789.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett EA, Keller H, Mills RE, Schmidt S, Moran JV, Weichenrieder O, Devine SE. Active Alu retrotransposons in the human genome. Genome Res. 2008;18:1875–1883. doi: 10.1101/gr.081737.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewannieux M, Heidmann T. Role of poly(A) tail length in Alu retrotransposition. Genomics. 2005;86:378–381. doi: 10.1016/j.ygeno.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Deininger PL, Batzer MA, Hutchison CA, Edgell MH. Master genes in mammalian repetitive DNA amplification. Trends Genet. 1992;8:307–311. doi: 10.1016/0168-9525(92)90262-3. [DOI] [PubMed] [Google Scholar]
- Roy-Engel AM, Salem AH, Oyeniran OO, Deininger L, Hedges DJ, Kilroy GE. Active alu element "A-Tails": size does matter. Genome Res. 2002;12:1333–1344. doi: 10.1101/gr.384802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C, Rubin CM, Schmid CW. Genome-wide chromatin remodeling modulates the Alu heat shock response. Gene. 2001;276:127–133. doi: 10.1016/S0378-1119(01)00639-4. [DOI] [PubMed] [Google Scholar]
- Dewannieux M, Esnault C, Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nat Genet. 2003;35:41–48. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- Szpakowski S, Sun X, Lage JM, Dyer A, Rubinstein J, Kowalski D, Sasaki C, Costa J, Lizardi PM. Loss of epigenetic silencing in tumors preferentially affects primate-specific retroelements. Gene. 2009;448:151–167. doi: 10.1016/j.gene.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimmer K, Callens T, Wernstedt A, Messiaen L. The NF1 gene contains hotspots for L1 endonuclease-dependent de novo insertion. PLoS Genet. 2011;7:e1002371. doi: 10.1371/journal.pgen.1002371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy AM, West NC, Rao A, Adhikari P, Alemán C, Barnes AP, Deininger PL. Upstream flanking sequences and transcription of SINEs. J Mol Biol. 2000;302:17–25. doi: 10.1006/jmbi.2000.4027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Final Alu data set. Table S2. Duplicate Alu loci (Alu locus was identified in more than one cell type). Table S3. Alu subfamily distribution. Table S4. Retrieved Alus from HeLa Table 1b Pol3 ChIP-chip in HeLa from Oler,A.J. et al. Nat Struct Mol Biol 17, 620–628 (2010). Table S5. Retrieved Alus from Table 1c HeLa ChIP-seq Knowns from Oler, A.J. et al. Nat Struct Mol Biol 17, 620–628 (2010). Table S6. Retrieved Alus from Table 1 g Pol3 Jurkat ChIP-seq from Oler, A.J. et al. Nat Struct Mol Biol 17, 620–628 (2010). Table S7. Retrieved Alus from Table 1 h Pol3 multiple cell types from Oler, A.J. et al. Nat Struct Mol Biol 17, 620–628 (2010). Table S8. Retrieved Alus from IMR90, Table S7 from Canella,D., et al. Genome Res. 20, 710–721 (2010). Table S9. Retrieved Alus from HeLa cells, Moqtaderi, Z, et al. Nat Struct Mol Biol, 20, 635–640 (2010). Table S10. Retrieved Alus from K562 cells, Moqtaderi, Z, et al. Nat Struct Mol Biol, 20, 635–640 (2010). Table S11. Probes for northern blot analysis. Table S12. Numerical value assigned base on the impact of Alu sequence divergence on retrotransposition potential. Table S13. Numerical value assigned base on the impact of A-tail length on retrotransposition potential. Table S14. Numerical value assigned base on the impact of length of unique sequence on retrotransposition potential. Table S15. Numerical value assigned base on the impact of A-tail disruptions on retrotransposition potential. Table S16. Output of genome-wide analysis of B box-containing Alus with ERP (Competence_score) > = 0.10.