Abstract
Background
Plant-associated bacterial communities caught the attention of several investigators which study the relationships between plants and soil and the potential application of selected bacterial species in crop improvement and protection. Medicago sativa L. is a legume crop of high economic importance as forage in temperate areas and one of the most popular model plants for investigations on the symbiosis with nitrogen fixing rhizobia (mainly belonging to the alphaproteobacterial species Sinorhizobium meliloti). However, despite its importance, no studies have been carried out looking at the total bacterial community associated with the plant. In this work we explored for the first time the total bacterial community associated with M. sativa plants grown in mesocosms conditions, looking at a wide taxonomic spectrum, from the class to the single species (S. meliloti) level.
Results
Results, obtained by using Terminal-Restriction Fragment Length Polymorphism (T-RFLP) analysis, quantitative PCR and sequencing of 16 S rRNA gene libraries, showed a high taxonomic diversity as well as a dominance by members of the class Alphaproteobacteria in plant tissues. Within Alphaproteobacteria the families Sphingomonadaceae and Methylobacteriaceae were abundant inside plant tissues, while soil Alphaproteobacteria were represented by the families of Hyphomicrobiaceae, Methylocystaceae, Bradyirhizobiaceae and Caulobacteraceae. At the single species level, we were able to detect the presence of S. meliloti populations in aerial tissues, nodules and soil. An analysis of population diversity on nodules and soil showed a relatively low sharing of haplotypes (30-40%) between the two environments and between replicate mesocosms, suggesting drift as main force shaping S. meliloti population at least in this system.
Conclusions
In this work we shed some light on the bacterial communities associated with M. sativa plants, showing that Alphaproteobacteria may constitute an important part of biodiversity in this system, which includes also the well known symbiont S. meliloti. Interestingly, this last species was also found in plant aerial part, by applying cultivation-independent protocols, and a genetic diversity analysis suggested that population structure could be strongly influenced by random drift.
Background
Similar to the intensively studied animal microbioma, plants harbor a wide range of diverse bacteria forming a complex biological community, which includes pathogens, mutualists (symbionts), and commensals [1,2]. Depending on the colonized compartment, these bacteria are rhizospheric (root colonizers), endophytic (colonizing the endosphere, the bulk of internal tissues) and phyllospheric or epiphytic (leaf or stem surface). In recent years plant-associated bacteria (endophytic, epiphytic and rhizospheric) have been widely studied, mainly as promising tools for biotechnological applications [3-7], but investigations have also been carried out on the ecology and taxonomy of plant-associated bacterial communities [8-11]. Despite a high taxonomic diversity, only few bacterial taxa have been found characteristically associated to the majority of plant species, notably members of the Alphaproteobacteria class [2,7,8,12,13]. Consequently, the generally accepted idea is that the ability to colonize a plant is not a common, widespread feature present in the soil bacterial community, but preferentially resides in specific taxa which may be considered more ecologically versatile or genetically prone to the association with plants. This last hypothesis has recently been supported by the finding that, at least in the class of Alphaproteobacteria, a common gene repertoire seems to be present in all of its plant-associated members [14].
Medicago sativa L. (alfalfa) is one of the most important legume crop in temperate areas throughout the world, commonly used as forage or in crop rotation practices to contribute organic nitrogen to the soil via its symbiosis with the nitrogen fixing bacteria [15]. Moreover, it is important also for bioenergy production [16] and is one of the most suited plant species for land restoration [17]. Finally, this species, and the diploid relative M. truncatula Gaertn. (barrel medic), are among the most studied model species regarding the molecular aspects of plant-bacteria symbiosis, particularly in relation with the alphaproteobacterium Sinorhizobium (syn. Ensifer) meliloti[18-20]. Concerning S. meliloti, this species is present in most temperate soils, and, when conditions are suitable, it forms specialized structures, called nodules, in the roots of alfalfa plants where it differentiates into bacteroids [18]. It is assumed that a fraction of bacterial cells is released from dehiscent nodules to soil, giving rise to new free-living rhizobial clones [21]. In the last years S. meliloti has been found able to also endophytically colonize the aerial part of other plant species, as rice [22], suggesting the presence of several ecological niches for this species (soil, nodule, other plant tissues).
While the plant-associated bacterial flora of M. sativa has never been investigated at the community level, S. meliloti population genetics have been extensively studied in the past [23-28], but only on strains isolated from nodules, with a few early studies performed on bacteria directly recovered from soil [29,30], due to the lack of efficient selective culture media. No data have been reported on the presence in natural conditions of S. meliloti as endophytes in other plant compartments (such as leaves) and no comparison of soil vs. plant-associated populations has been done.
Based on the above mentioned considerations, there is a need to characterize the bacterial community associated with M. sativa in relation to both the potentially important role the class of Alphaproteobacteria seems to have as main component of a “core plant-associated bacterial community” in several different plant species [13,31-33], and to the relationships of soil vs. plant-associated populations of the symbiotic alphaproteobacterial partner S. meliloti.
In this work we investigated the bacterial communities associated with the legume M. sativa, focusing on both the total bacterial community composition and on the presence and populations structure of the symbiotic partner S. meliloti in soil and plant tissues.
The analysis was conducted by cultivation-independent techniques on alfalfa (M. sativa) plants grown in mesocosm pots. The bacterial community associated with M. sativa and that of the surrounding soil were analyzed at high (class, family) and low (single species, S. meliloti) taxonomic levels by employing Terminal-Restriction Fragment Length Polymorphism (T-RFLP) profiling [33], 16 S rRNA library screening and S. meliloti-specific markers [34,35]. These approaches allowed us to explore for the first time the bacterial community composition of such important plant species and the populations of S. meliloti without cultivation.
Results
Ribotype variability of the bacterial community
The ribotype variability of bacterial communities present in soil and associated to plant tissues (nodules, stems and leaves) was investigated by T-RFLP analysis. A total of 43 samples was analyzed: in particular one pooled soil sample for each one of the three pots, one pooled sample from all the nodules found in each pot and four plants per pot (one stem and 2–3 pools of leaves per plant). T-RFLP profiles on these samples produced 253 Terminal-Restriction Fragments (T-RFs) or ribotypes after the restriction digestion with two restriction enzymes, HinfI and TaqI. 16 S rRNA gene amplification and T-RFLP profiling was also performed on DNA extracted from surface-sterilized seeds, but no bands of 16 S rRNA gene amplification were recovered (data not shown), suggesting a very low bacterial titre in seeds.
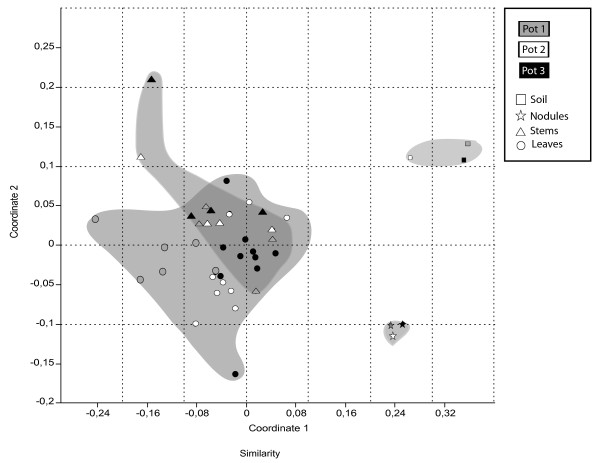
Figure 1 shows the pattern of similarity among T-RFLP profiles from total communities as Non-Metric Multidimensional Scaling (N-MDS). Soil and nodule bacterial communities were strongly differentiated from stem and leaf communities, forming relatively tight clusters. Large heterogeneity was detected in leaf and stem communities. To better evaluate the statistical significance of differentiation of communities we employed AMOVA. Most of the variation (71.75%) was due to intra-environment differences (Additional file 1: Table S1). However, significant differences between environments were found (P < 0.0001), in particular between a soil-nodule group and a stem-leaf group.
Figure 1.
Pattern of similarities of individual T-RFLP profiles from total community analysis. The pattern of similarity has been inspected by using Nonmetric Multidimensional scaling (N-MDS) based on Jaccard similarity matrix. Stress of N-MDS = 0.1896. Stars indicate nodules; squares, soils; circles, leaves; triangles, stems. Grey filling, pot 1; white, pot 2; black, pot 3. Samples of the same environment were grey shaded.
Interestingly, stem and leaf communities showed a significant (P < 0.0001), though small (pairwise FST = 0.05) separation (Additional file 2: Table S2). Moreover, AMOVA on stems and leaves community revealed a statistically significant differentiation between the three pots (P < 0.0001), irrespective of possible grouping (either plant genotype-related or unrelated), suggesting a pot-effect over the taxonomic shaping of the leaf-associated community and no effect of plant genotypes. These data confirmed a previous long-term experiment only addressing S. meliloti species [23].
Taxonomic composition of bacterial communities in soil, nodules and plant aerial parts
T-RFLP analysis has shown that bacterial communities clustered in three groups (soil, nodules and plant aerial parts). In order to elucidate which taxa are mainly represented in the bacterial communities of these assemblies, three 16 S rRNA gene clone libraries were constructed pooling together the DNAs extracted from the samples of each environment; additionally, leaves and stems samples were also pooled together due to their high similarities as mentioned above. Pooled samples did conceivably result in an enrichment of the more shared taxa possibly preventing the detection of taxa associated only to a few individual samples. DNA was used as template to construct three 16 S rRNA libraries; a total of 276 clones (from 78 to 116 per library) were sequenced. Sequence analysis revealed, as expected, that the soil community was the most diverse (Shannon H’ = 4.63; Chao1 = 168), while the nodule-associated community was less diverse (Shannon H’ = 1.98; Chao1 = 30), (Additional file 3: Table S3). As a consequence, the library of nodules showed a coverage (85.9%) higher than those of stems + leaves (74.1%) and soil (47.1%).
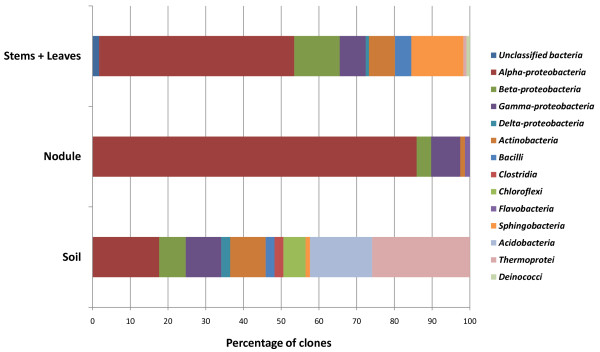
The percentages of taxonomic classes detected in the sequences of the clone libraries are reported in Figure 2. Seven classes were represented in both soil and stem + leaf communities, and 4 of them were also found in nodules. Alphaproteobacteria were dominant in nodules (as expected, due to the presence of high titres of the symbiotic alphaproteobacterium S. meliloti) and in stems + leaves. Also in soil Alphaproteobacteria were highly prevalent, but Acidobacteria and Crenarchaeota were also abundant. Flavobacteria were found only in nodules, however a low presence in the other environments cannot be excluded, especially in relation to the lower coverage of the respective libraries. Beta- and Gammaproteobacteria and Actinobacteria were found in all three libraries.
Figure 2.
Representation of bacterial divisions in the 16 S rRNA gene clone libraries. The percentage of clones accounting for each division with respect to its origin (nodule, stems + leaves, soil) is reported.
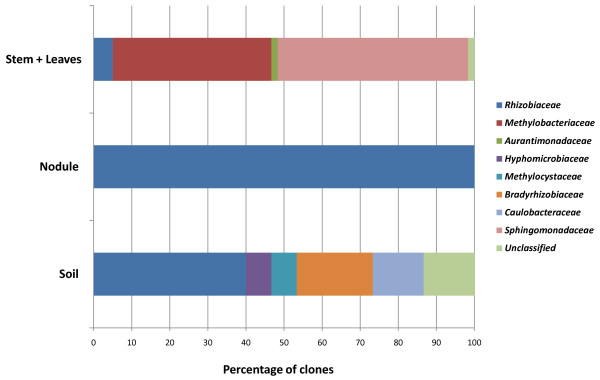
Concerning Alphaproteobacteria, only members of the Rhizobiaceae family were found in nodules, with all sequences assigned, as expected, to the Sinorhizobium/Ensifer genus (Figure 3). Alphaproteobacteria present in soil belonged to the Rhizobiaceae, Bradyrhizobiaceae, Methylocystaceae, Hypomicrobiaceae and Caulobacteraceae families. Rhizobiaceae, Aurantimonadaceae and Methylobacteriaceae, all belonging to the Rhizobiales, plus taxa of the order Sphingomonadales, were found in the stem + leaf library. The absence of sequences assigned to the Sinorhizobium/Ensifer genus from stem + leaves and soil libraries, though this species was found by qPCR in both these environments (see the following paragraph), could be due to its low abundance and to the relatively low coverage of clone libraries.
Figure 3.
Distribution of the recovered families inAlphaproteobacteriawith respect to their origin (nodule, stems + leaves, soil). The percentage of clones present in the libraries for each family is reported.
Detection and diversity of sinorhizobium meliloti in soil and plant tissues
Aiming to analyze presence and diversity of S. meliloti, we firstly estimated the population size by qPCR, using two species-specific primer pairs which amplify chromosomal (rpoE1) and megaplasmidic loci (nodC on pSymA), respectively [35]. The obtained results are reported in Table 1. Relatively higher titers of S. meliloti DNA were detected in root nodules, while lower values were obtained in soils, leaves and stems. Interestingly, nodule titers of S. meliloti DNA detected by rpoE marker were higher than those estimated by nodC marker (roughly one order of magnitude). The viable titers of S. meliloti cells from crushed nodules of M. sativa plants usually ranged from 2.1x108 to 5.0x108cells/g of fresh tissue (data not shown), suggesting that the titers from nodC marker are a better proxy of the number of bacteria involved in the symbiotic nitrogen-fixing process.
Table 1.
Titers ofS. melilotiin soil and plant tissues§
| Sample |
Titers |
|
|---|---|---|
| rpoE1-based | nodC-based | |
|
Pot 1 |
|
|
| Soil |
4.92 ± 2.82 x 104 |
2.78 ± 0.63 x 104 |
| Nodules |
3.07 ± 0.67 x 109 |
4.25 ±1.24 x 108 ** |
| Stems |
2.73 ± 1.21 x 104 |
3.22 ±2.4 x 103 * |
| Leaves |
8.65 ± 4.04 x 103 |
4.28 ± 1.23 x 103 |
|
Pot 2 |
|
|
| Soil |
1.16 ± 0.33 x 104 |
2.88 ± 1.09 x 104 |
| Nodules |
1.20 ± 0.50 x 1010 |
1.01 ± 0.10 x 109 ** |
| Stems |
2.37 ± 0.49 x 103 |
1.13 ± 0.15 x 103 |
| Leaves |
9.74 ± 5.08 x 102 |
2.34 ±0.78 x 102 |
|
Pot 3 |
|
|
| Soil |
2.70 ± 0.41 x 105 |
7.42 ±0.93 x 104 * |
| Nodules |
6.02 ± 1.45 x 109 |
2.02 ± 3.22 x 107 ** |
| Stems |
4.91 ± 0.95 x 105 |
1.07 ± 3.74 x 105 |
| Leaves | 5.54 ± 2.83 x 103 | 5.21 ± 3.01 x 103 |
§Titers were estimated by qPCR [35] with rpoE1 and nodC markers and are expressed as n. of gene copies/g of tissue or soil; ± standard deviation from triplicate experiments. Asterisks indicate significant differences between estimates based on rpoE1 and nodC markers (*, P < 0.05; **, P < 0.01).
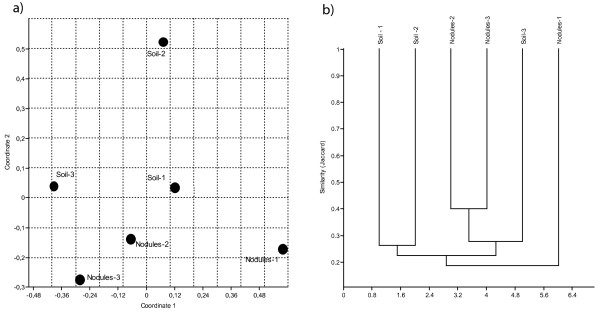
Then, to inspect the genetic diversity of S. meliloti populations present in the different environments, the amplification of the 1.3 kbp long 16 S-23 S ribosomal intergenic spacer (IGS) which proved to be an efficient marker for the study of S. meliloti populations [34], was attempted. Only DNAs from nodules and soil gave a PCR product, probably as a result of the low bacterial titers and high content in inhibitors present in DNA extracted from stems and leaves. Consequently, nodule tissue was taken as representative of the plant environment and was compared with soil. A total of 121 different IGS-T-RFs (16 S-23 S ribosomal intergenic spacer Terminal-Restriction Fragments) was detected after digestion with four restriction enzymes (AluI, MspI, HinfI, HhaI) in the six DNA samples (three from soil, three from nodules), after IGS amplification and T-RFLP profiling (Additional file 4: Figure S1a). Most of the 121 detected IGS-T-RFs (71.9%) were detected in one sample out of 6, while 8 (6.6%) IGS-T-RFs were present in all six samples (Additional file 4: Figure S1b). Moreover, from 25.5 to 53.3% of IGS-T-RFs present in soil were also detected in nodules and from 31.4 to 40.1% of IGS-T-RFs present in nodules were detected in the respective soil sample. Figure 4 shows the similarity relationships between IGS-T-RFLP profiles. Non-metric MDS plot of IGS-T-RFLP profiles (Figure 4a) showed a possible separation of nodule and soil populations on the second dimension. In particular, the nodule population in pot 1 was more separated from the soil population of the same pot and from the populations of the other pots. On the contrary, nodule populations of pots 2 and 3 were the closest ones, with soil population of pot 3 in the same cluster (Figure 4b), suggesting a possible effect of plant genotype as previously shown [23,36]. However, in agreement with the high number of single-sample haplotypes detected, an AMOVA carried out to evaluate the variance contribution to a hypothetical differentiation of soil and nodule S. meliloti population showed that 17.37% only of variance was attributed to a soil-nodule separation, the remaining 82.63% of variance being due to among-nodules and among-soil differences. Additionally, no statistical significant separation (P < 0.46) was detected for groupings based on the two plant genotypes present in the mesocosms.
Figure 4.
a) Non-metric MDS plot of similarities of IGS-T-RFLP profiles fromS. melilotipopulation analysis.a) The pattern of similarity of S. meliloti populations has been inspected by using Non-metric Multidimensional scaling (N-MDS) based on Jaccard similarity matrix. Stress = 0.0898. b) Cluster analysis based on Jaccard similarity matrix. Scale bar represents Jaccard similarity coefficient
Discussion
In recent years there has been an increasing interest in exploring the bacterial flora associated with plants [37-41]. A recent field survey indicates [8] that plant aerial parts (leaves) harbor complex, but highly variable, bacterial communities, and that only a small number of bacterial taxa (mainly belonging to Alphaproteobacteria) are plant-specific. In the experiments reported here, as in the majority of the reports on endophytic microflora, we refer to endophytic and epiphytic bacteria indicating all those that are inside the plant tissue or strongly adhering to the plant surface, such as they resist washing and sterilization (or their DNA is retained by plant tissue), therefore a more correct definition could be “plant-associated bacteria”.
The present study shows that root nodules and aerial parts of Medicago sativa plants grown in mesocosm conditions, harbor distinct bacterial communities with specific signatures at the class, family and species levels and that these communities do not mirror soil bacterial communities.
Initially, T-RFLP profiles allowed us to show that bacterial communities present in the different environments (soil, nodules, stems and leaves) were strongly differentiated and in particular that a large heterogeneity was present between leaves of individual plants, though soil profiles were highly similar. Moreover a clear separation between above-ground (stem and leaves) and below-ground environments (soil and nodules) was detected. An analysis of the clone libraries, prepared from above-ground and below-ground pooled samples, revealed an uneven distribution of bacterial classes, with a marked pattern highlighting the class of Alphaproteobacteria as the more abundant in plant tissues (this class represented half of the clones in the stem + leaf library). The same uneven pattern was then observed, at lower taxonomic ranks, within the Alphaproteobacteria, with sequences of clones belonging to members of the Methylobacteriaceae and Sphingomonadaceae families being more abundant in stem than in soil and nodules. Methylobacteria and Sphingomonadaceae have been found as endophytes in a number of plants [8,12,31,33,42-45] and it is believed that this group of bacteria may take advantage from living as plant-associated, thanks to its ability to utilize the one-carbon alcohol methanol discharged by wall-associated pectin metabolism of growing plant cells.
Concerning root nodule bacterial communities, obtained data indicated that very diverse bacterial taxa are associated with nodules, the most represented being the specific rhizobial host of M. sativa, the alphaproteobacterium S. meliloti. However, additional taxa have been found, including members of ActinobacteriaFlavobacteriaGammaproteobacteria and Betaproteobacteria, which may have some additional plant growth-promoting activities (see for instance [46,47]).
In soil, Acidobacteria was one of the most important divisions (in terms of number of clones in the library) and was present exclusively in the soil clone library, in agreement with many previous observations [48,49]. A relatively high presence of Archaea (Thermoprotei) was also found. Checking the 16 S rRNA gene sequences present in the Ribosomal Database for 799f/pHr primer annealing, we found that PCR amplification from Thermoprotei was theoretically possible with this primer pair (data not shown). The presence of Archaea in the soil is not unexpected [50] and could be linked also to the anoxic or nearly anoxic conditions present in the bottom of the pot. However, since the low coverage of soil clone library, the presence of many other additional taxa, as well of different proportions of those found here cannot be excluded. In addition, it should be mentioned that differences between soil and plant-tissues bacterial communities could also be ascribed to the different DNA extraction protocols we were obliged to use, since a unique protocol (bead-beading protocol for both soil DNA and plant DNA) failed in a successful extraction of DNA from both soil and plant tissues (data not shown). A similar technical need was encountered by other authors also [33], which renders the study of the relationships between plant-associated and soil bacterial communities still at its beginning.
At the lowest taxonomic rank here investigated, within the species S. meliloti, we detected the presence of this species in all environment analyzed (soil, nodules and plant aerial tissues). This finding is confirming earlier reports on the ability of S. meliloti to behave as an endophytic strain, colonizing all plant compartments, besides being a root symbiont of legumes [22], and suggest a potential higher genetic variability of S. meliloti population, and, from the other side, potential new ecological and functional roles for this species, not investigated so far[29,51,52]. Unfortunately, the low population size of S. meliloti in stems and leaves and the possible presence of PCR inhibitors (plant DNA or phenolic compounds, for instance) did not permit the amplification of 16 S-23 S rRNA intergenic region from plant aerial parts to obtain information about the genetic diversity and structure of S. meliloti population resident in plant aerial part. No hypothesis could then be drawn about the relationships between this population and those of soil and nodules. Concerning S. meliloti populations present in soil and nodules, similar values for diversity were detected in nodules and in soil, suggesting that both environments harbor a consistent fraction of the population’s genetic diversity. Interestingly, most of the T-RFs were detected in one sample only, and a very small fraction of T-RFs was shared among all samples, though the original soil material was homogeneous and should, in theory, contain the same S. meliloti haplotypes. Therefore, S. meliloti populations from all the three mesocosms investigated were highly differentiated between each other and, as expected from previous studies on S. meliloti[23] and on Bradyrhizobium[53], no statistically significant plant genotype- related haplotypes were detected.
A possible explanation of such findings could be linked to the relatively low titers of S. meliloti in soil (104-105 cells/g), which is roughly 1/10,000 of the total bacterial community of soil (estimated at ~109 16 S rRNA gene copies/g of soil by qPCR, data not shown). Such estimated S. meliloti titers were similar to those previously observed in other soil and plant tissues [35] and in line with those normally found in soil with viable (Most Probable Number, MPN) estimates [26,54]. As a consequence of this low population size, founder effect and genetic drift are likely to be among the main shaping forces of S. meliloti population in this experimental set-up, perhaps permitting the fixation of sample-specific haplotypes by simple chance [55]. Regarding the nodule-soil relationships, though our experiments did not directly address this issue, the reported S. meliloti population analysis suggests the presence of somewhat nonoverlapping soil and nodule population fractions, even if no specific patterns of soil and nodule populations were detected. The presence of different rhizobial haplotypes in nodules and soil was previously found in chickpea [51] and clover [52], though no simple conclusion could be drawn, because of limited sampling. However, as for total bacterial community analysis, it should be mentioned that the use of two different DNA extraction protocols for soil and plant DNA may have produced some bias in the proportion of the different haplotyes detected.
Conclusion
In conclusion, we show on M. sativa that its associated microflora, though highly variable, is mainly related to the presence of Alphaproteobacteria. This class has an uneven presence of families in stems + leaves, nodules and soil. We then speculated that a sort of “pan-plant-associated bacterial community” may be composed of a large plethora of “accessory” taxa, which are occasionally associated with plants, and a small number of “core” taxa (e.g. Alphaproteobacteria families) which, on the contrary, are consistently found in the plants. Moreover, within Alphaproteobacteria the specific alfalfa symbiotic species S. meliloti, abundant as symbiont in root nodules, was also detected in soil and in leaves, with potentially different populations, suggesting a more complex interplay of colonization of multiple environments (soil, root nodules, other plant tissues) by this species.
Methods
Experimental design and sampling procedure
A controlled experiment was set-up in mesocosms composed of three pots (numbered 1, 2, 3) containing Medicago sativa (alfalfa) plants grown at CRA-FLC Lodi, Italy, in outdoor conditions. Two of the three pots were planted with the same line of alfalfa (1x5) while the third pot was planted with a different line (5x7). The pots (cylinders of 25 cm diameter x 80 cm depth) with a drainage layer on the bottom, were filled with a sandy loam non-calcareous soil (57.8% sand, 32% silt, 10.2% clay, 1.7% organic matter and 0.09% total N; pH 6.7) in which alfalfa has never been grown. Phosphorus and potassium equivalent to 120 Kg ha-1 of P2O5 and 180 Kg ha-1 of K2O were distributed into the soil, while no mineral N was added; irrigation was not limiting. Twenty plants/pot (density equivalent to 400 plants m-2) were transplanted in March 2008 and allowed to grow until the 2nd year (the end of September 2009), when plant aerial parts of 12 plants were harvested and the pots were opened to allow sampling of the whole eye-detectable nodules present (approximately 80–100 of various sizes per pot) and of bulk soil. Roots were excluded from the analysis since the presence of small nodules or nodule primordia could not be excluded, possibly inducing a strong bias in the estimation of “non-nodule-associated root colonizers”. The plant sample size was chosen on the basis of a previous analysis of plant-by-plant variation in which the overall diversity of communities did not change from 2 to 30 plants (unpublished data and [8]). Stems, leaves (pools of around 10 leaves per plant) and nodules were washed with water and with 10 mM MgSO4 twice to remove most soil and dust particles and eliminate bacteria loosely adhering to the surface and then surface sterilized with 1% HClO for 1 min. Samples of soil, nodules, stem and leaves were then stored at −80°C from 1–2 weeks before DNA extraction.
A control of seed-borne bacteria was also prepared with seeds of M. sativa surface sterilized with 1% HgCl2.
S. meliloti viable titres in sterilized nodules have been estimated by serial dilution of crushed nodules as previously reported [54].
DNA extraction real-time PCR and T-RFLP profiling
DNA was extracted from soil by using a commercial kit (Fast DNA Spin kit for soil, QBiogene, Cambridge, UK) following the manufacturer’s instructions. DNA extraction from plant tissues and surface sterilized control seeds was performed by a 2X CTAB protocol as previously described [56]. The 16 S rRNA gene pool of total bacterial community was amplified from the extracted DNA with primer pairs 799f (labeled with HEX) and pHr which allow the amplification of most bacterial groups without targeting chloroplast DNA [33]. PCR conditions and Terminal-Restriction Fragment Length Polymorphism (T-RFLP) profiling were as previously reported [8], by using HinfI and TaqI restriction enzymes. For sinorhizobial populations, T-RFLP was carried out on 16 S-23 S ribosomal intergenic spacer amplified from total DNA (IGS-T-RFLP) with S. meliloti specific primers and AluI and HhaII restriction enzymes, as already reported [34]. Real-Time PCR (qPCR) for quantification of S. meliloti DNA was carried out on rpoE1 and nodC loci, as previously reported [35]; two different calibration curves were constructed, one for soil samples and the other one for plant samples, by using as template DNA extracted from sterile soil (without presence of S. meliloti) and from sterile plant (grown in petri dishes), both spiked with serial dilutions of known titres of S. meliloti cells, as previously reported [35]. Controls with S. medicae WSM419 DNA were included in both IGS-T-RFLP and qPCR, for S. meliloti species-specificity check [35].
Library construction and sequencing
Amplified (with 799f and pHr primer pair) 16 S rRNA genes from DNA extracted from soil, nodules, pooled stems and leaves of a 1:1:1 mix of all pots were inserted into a pGemT vector (Promega, Fitchburg, WI, USA) and cloned in E. coli JM109 cells. Positive clones were initially screened by white/blue coloring and the inserted amplified 16SrRNA genes sequenced. Plasmid purification and sequencing reactions were performed by Macrogen Europe Inc. (Amsterdam, The Netherlands). The nucleotide sequences obtained were deposited in Gen- Bank/DDBJ/EMBL databases under accession numbers from HQ834968 to HQ835246.
Data processing and statistical analyses
For qPCR data, 1-way ANOVA with Tukey post hoc test was employed. Analyse-it 2.0 software (Analyse-It, Ldt., Leeds, UK) was used for both tests. For T-RFLP, chromatogram files from automated sequencer sizing were imported into GeneMarker ver. 1.71 software (SoftGenetics LLC, State College, PA, USA) by filtering with the default options of the module for AFLP analysis. Peaks above 100 fluorescence units and whose size ranged from 35 to 500 nt were considered for profile analysis. Only the presence/absence of peaks was considered as informative data from the chromatograms. Statistical analyses were performed on a binary matrix obtained as previously reported [8]. Past 2.02 [57] software package was used to compute Non-Metric Multidimensional Scaling (N-MDS). To test the distribution of the variance of T-RFLP profiles within plant tissues and among pots, Analysis of Molecular Variance (AMOVA [58]) was applied using Arlequin 3.5.1.2 software (http://cmpg.unibe.ch/software/arlequin3/). Although developed for population genetic analysis, the general procedure implemented by AMOVA is flexible enough to estimate the statistical significance of groups of bacterial communities as reported previously [13,42,59]. Pairwise FST distances [60] between T-RFLP profiles of plant tissues and soils were used to infer a Neighbor-Joining dendrogram with the MEGA4 software [61].
Partial 16 S rRNA sequences were manually inspected for quality, then aligned and clustered with the furthest neighboring algorithm with the module present in Mothur v.1.12.3 [62]. Diversity indices (Shannon H’ and Chao-1) were calculated with the same software. Library coverage was estimated with the formula C = 1-(n/N)[63], where n is the number of singletons (defined at 97% sequence identity in Mothur) that are encountered only once in the library and N is the total number of sequenced clones. Taxonomic assignment was performed with the Classifier module present in Ribosomal Database Project 10 website [64] (http://rdp.cme.msu.edu/) at 80% confidence threshold. Sequences with 97% similarity were treated as a single Operational Taxonomic Unit (OTU). Sequences (one for each OTU) were aligned with the 16 S rRNA gene sequences of the closest match retrieved from NCBI databases, using MUSCLE [65] and a Neighbor-Joining dendrogram was constructed using MEGA4 [61]. Phylogenetic inference and evolutionary distance calculations were generated using the Maximum Composite Likelihood; 1000 bootstrap replicates were used to obtain confidence estimates for the phylogenetic trees.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
FP performed most of the analyses, prepared the figures and contributed in writing the draft of the manuscript. This work is part of FP PhD thesis. AF and LS gave technical assistance to the experimental work during the preparation of their master degree theses. CS settled the mesocosm experiment and assisted in the samplings. EGB, MB, FP and AM conceived the idea and contributed in performing part of the analyses and in drafting the manuscript. All authors have given final approval of the version to be published.
Supplementary Material
Table S1. Hierarchical analysis of differentiation between bacterial communities. AMOVA was performed with T-RFLP profiles from samples of the four different environments (soil, nodules, stems and leaves). Data show the degrees of freedom (d.f.), the sum of squared deviation, the variance component estimate, the percentage of total variance contributed by each component, and the probability (P) of obtaining a more extreme component estimate by chance alone, estimated from 10,000 permutations.
Table S2. Matrix of pairwise FST values. Statistical significance (p < 0.05) has been computed after 1000 random permutation; n.s., not significant. Only below diagonal values are reported.
Table S3. Statistical analysis of 16SrRNA gene clone libraries. OTUs were arbitrarily defined at 97% sequence identity based on Mothur clustering. Confidence intervals at 95% are given in parentheses. Coverage is defined C = [1 − (n/N)] × 100, where n is the number of unique clones, and N is the total number of clones examined.
Figure S1. S. meliloti IGS-T-RFLP profiling of nodule and soil samples. A), the schematic representation of the binary matrix of IGS-T-RF presence (black) and absence (empty cell); the IGS-T-RF number is reported on the right side of each row. B) The occurrence of “private” and “public” IGS-T-RFs. The percentage of total number of scored IGS-T-RFs is reported for T-RFs present from 1 to all 6 samples analyzed.
Contributor Information
Francesco Pini, Email: francesco.pini@unifi.it.
Arcangela Frascella, Email: affabula@gmail.com.
Luisa Santopolo, Email: luisasantop@hotmail.it.
Marco Bazzicalupo, Email: marco.bazzicalupo@unifi.it.
Emanuele G Biondi, Email: emanuele.biondi@iri.univ-lille1.fr.
Carla Scotti, Email: carla.scotti@entecra.it.
Alessio Mengoni, Email: alessio.mengoni@unifi.it.
Acknowledgements
This work was partially supported by intramural funding of the University of Florence to AM and MB and by Soil-Sink (FISR-MIUR) project funding to MB. We are grateful to Mary Forrest, Professor of Scientific English, Department of Pharmacology, Faculty of Medicine and Surgery, University of Florence for editing English language. We acknowledge two anonymous reviewers for helpful suggestions for the improvement of the manuscript.
References
- Ryan RP, Germaine K, Franks A, Ryan DJ, Dowling DN. Bacterial endophytes: recent developments and applications. FEMS Microbiol Lett. 2008;278(1):1–9. doi: 10.1111/j.1574-6968.2007.00918.x. [DOI] [PubMed] [Google Scholar]
- Mengoni A, Schat H, Vangronsveld J. Plants as extreme environments? Ni-resistant bacteria and Ni-hyperaccumulators of serpentine flora. Plant and Soil. 2010;331:5–16. doi: 10.1007/s11104-009-0242-4. [DOI] [Google Scholar]
- Danhorn T, Fuqua C. Biofilm formation by plant-associated bacteria. Annu Rev Microbiol. 2007;61:401–422. doi: 10.1146/annurev.micro.61.080706.093316. [DOI] [PubMed] [Google Scholar]
- Lodewyckx C, Vangronsveld J, Porteous F, Moore ERB, Taghavi S, Mezgeay M, van der Lelie D. Endophytic bacteria and their potential applications. Crit Rev Plant Sci. 2002;21(6):583–606. doi: 10.1080/0735-260291044377. [DOI] [Google Scholar]
- Rajkumar M, Ae N, Freitas H. Endophytic bacteria and their potential to enhance heavy metal phytoextraction. Chemosphere. 2009;77(2):153–160. doi: 10.1016/j.chemosphere.2009.06.047. [DOI] [PubMed] [Google Scholar]
- Mastretta C, Taghavi S, Van der Lelie D, Mengoni A, Galardi F, Gonnelli C, Barac T, Boulet J, Weyens N, Vangronsveld J. Endophytic bacteria from seeds of Nicotiana tabacum can reduce cadmium phytotoxicity. Int J Phytoremediation. 2009;11:251–267. doi: 10.1080/15226510802432678. [DOI] [Google Scholar]
- Ikeda S, Okubo T, Anda M, Nakashita H, Yasuda M, Sato S, Kaneko T, Tabata S, Eda S, Momiyama A. et al. Community- and Genome-Based Views of Plant-Associated Bacteria: Plant-Bacterial Interactions in Soybean and Rice. Plant Cell Physiol. 2010;51(9):1398–1410. doi: 10.1093/pcp/pcq119. [DOI] [PubMed] [Google Scholar]
- Mengoni A, Pini F, Huang L-N, Shu W-S, Bazzicalupo M. Plant-by-plant variations of bacterial communities associated with leaves of the nickel-hyperaccumulator Alyssum bertolonii Desv. Microbial Ecol. 2009;58:660–667. doi: 10.1007/s00248-009-9537-5. [DOI] [PubMed] [Google Scholar]
- Manter DK, Delgado JA, Holm DG, Stong RA. Pyrosequencing Reveals a Highly Diverse and Cultivar-Specific Bacterial Endophyte Community in Potato Roots. Microbial Ecol. 2010;60:157–166. doi: 10.1007/s00248-010-9658-x. [DOI] [PubMed] [Google Scholar]
- Bulgari D, Casati P, Brusetti L, Quaglino F, Brasca M, Daffonchio D, Bianco PA. Endophytic Bacterial Diversity in Grapevine (Vitis vinifera L.) Leaves Described by 16S rRNA Gene Sequence Analysis and Length Heterogeneity-PCR. J Microbiol. 2009;47(4):393–401. doi: 10.1007/s12275-009-0082-1. [DOI] [PubMed] [Google Scholar]
- Prakamhang J, Minamisawa K, Teamtaisong K, Boonkerd N, Teaumroong N. The communities of endophytic diazotrophic bacteria in cultivated rice (Oryza sativa L.) Applied Soil Ecol. 2009;42(2):141–149. doi: 10.1016/j.apsoil.2009.02.008. [DOI] [Google Scholar]
- Idris R, Kuffner M, Bodrossy L, Puschenreiter M, Monchy S, Wenzel WW, Sessitsch A. Characterization of Ni-tolerant methylobacteria associated with the hyperaccumulating plant Thlaspi goesingense and description of Methylobacterium goesingense sp nov. Syst Applied Microbiol. 2006;29(8):634–644. doi: 10.1016/j.syapm.2006.01.011. [DOI] [PubMed] [Google Scholar]
- Okubo T, Ikeda S, Kaneko T, Eda S, Mitsui H, Sato S, Tabata S, Minamisawa K. Nodulation-dependent communities of culturable bacterial endophytes from stems of field-grown soybeans. Microbes Environ. 2009;24(3):253–258. doi: 10.1264/jsme2.ME09125. [DOI] [PubMed] [Google Scholar]
- Pini F, Galardini M, Bazzicalupo M, Mengoni A. Plant-bacteria association and symbiosis: are there common genomic traits in Alphaproteobacteria? Genes. 2011;2:1017–1032. doi: 10.3390/genes2041017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprent JI. Nodulation in legumes. Royal Botanic Gardens, Kew, London; 2001. [Google Scholar]
- Sanderson MA, Adler PR. Perennial forages as second generation bioenergy crops. Int J Mol Sci. 2008;9(5):768–788. doi: 10.3390/ijms9050768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw AD, Chadwick MJ. The restoration of land: the ecology and reclamation of derelict and degraded land. University of California Press, Berkley; 1980. [Google Scholar]
- Gibson KE, Kobayashi H, Walker GC. Molecular Determinants of a Symbiotic Chronic Infection. Annual Rev Genet. 2008;42:413–441. doi: 10.1146/annurev.genet.42.110807.091427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd GED, Downie JM. Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu Rev Plant Biol. 2008;59:519–546. doi: 10.1146/annurev.arplant.59.032607.092839. [DOI] [PubMed] [Google Scholar]
- Downie JA. The roles of extracellular proteins, polysaccharides and signals in the interactions of rhizobia with legume roots. Fems Microbiol Rev. 2010;34(2):150–170. doi: 10.1111/j.1574-6976.2009.00205.x. [DOI] [PubMed] [Google Scholar]
- Oono R, Denison RF, Kiers ET. Controlling the reproductive fate of rhizobia: how universal are legume sanctions? New Phytologist. 2009;183(4):967–979. doi: 10.1111/j.1469-8137.2009.02941.x. [DOI] [PubMed] [Google Scholar]
- Chi F, Shen SH, Cheng HP, Jing YX, Yanni YG, Dazzo FB. Ascending migration of endophytic rhizobia, from roots to leaves, inside rice plants and assessment of benefits to rice growth physiology. Appl Environ Microbiol. 2005;71(11):7271–7278. doi: 10.1128/AEM.71.11.7271-7278.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carelli M, Gnocchi S, Fancelli S, Mengoni A, Paffetti D, Scotti C, Bazzicalupo M. Genetic diversity and dinamics of Sinorhizobium meliloti populations nodulating different alfalfa varieties in Italian soils. Applied Environ Microbiol. 2000;66:4785–4789. doi: 10.1128/AEM.66.11.4785-4789.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jebara M, Mhamdi R, Aouani ME, Ghrir R, Mars M. Genetic diversity of Sinorhizobium populations recovered from different Medicago varieties cultivated in Tunisian soils. Can J Microbiol. 2001;47(2):139–147. doi: 10.1139/w00-135. [DOI] [PubMed] [Google Scholar]
- Bailly X, Olivieri I, De Mita S, Cleyet-Marel JC, Bena G. Recombination and selection shape the molecular diversity pattern of nitrogen-fixing Sinorhizobium sp. associated to Medicago. Mol Ecol. 2006;15(10):2719–2734. doi: 10.1111/j.1365-294X.2006.02969.x. [DOI] [PubMed] [Google Scholar]
- Trabelsi D, Mengoni A, Aouani ME, Bazzicalupo M, Mhamdi R. Genetic diversity and salt tolerance of Sinorhizobium populations from two Tunisian soils. Annals of Microbiol. 2010;60(3):541–547. doi: 10.1007/s13213-010-0084-6. [DOI] [Google Scholar]
- Roumiantseva ML, Andronov EE, Sharypova LA, Dammann-Kalinowski T, Keller M, Young JPW, Simarov BV. Diversity of Sinorhizobium meliloti from the central Asian alfalfa gene center. Applied Environ Microbiol. 2002;68(9):4694–4697. doi: 10.1128/AEM.68.9.4694-4697.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biondi EG, Pilli E, Giuntini E, Roumiantseva ML, Andronov EE, Onichtchouk OP, Kurchak ON, Simarov BV, Dzyubenko NI, Mengoni A. et al. Genetic relationship of Sinorhizobium meliloti and Sinorhizobium medicae strains isolated from Caucasian region. FEMS Microbiol Lett. 2003;220(2):207–213. doi: 10.1016/S0378-1097(03)00098-3. [DOI] [PubMed] [Google Scholar]
- Bromfield ESP, Barran LR, Wheatcroft R. Relattive genetic structure of a population of Rhizobium meliloti isolated directly from soil and from nodules of alfalfa (Medicago sativa) and sweet clover (Melilotus alba) Mol Ecol. 1995;4(2):183–188. doi: 10.1111/j.1365-294X.1995.tb00207.x. [DOI] [Google Scholar]
- Hartmann A, Giraud JJ, Catroux G. Genotypic diversity of Sinorhizobium (formerly Rhizobium) meliloti strains isolated directly from a soil and from nodules of alfalfa (Medicago sativa) grown in the same soil. Fems Microbiol Ecol. 1998;25(2):107–116. [Google Scholar]
- Ikeda S, Okubo T, Kaneko T, Inaba S, Maekawa T, Eda S, Sato S, Tabata S, Mitsui H, Minamisawa K. Community shifts of soybean stem-associated bacteria responding to different nodulation phenotypes and N levels. ISME J. 2010;4(3):315–326. doi: 10.1038/ismej.2009.119. [DOI] [PubMed] [Google Scholar]
- Ikeda S, Rallos LEE, Okubo T, Eda S, Inaba S, Mitsui H, Minamisawa K. Microbial Community Analysis of Field-Grown Soybeans with Different Nodulation Phenotypes. Appl Environ Microbiol. 2008;74(18):5704–5709. doi: 10.1128/AEM.00833-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idris R, Trifonova R, Puschenreiter M, Wenzel WW, Sessitsch A. Bacterial communities associated with flowering plants of the Ni hyperaccumulator Thlaspi goesingense. Appl Environ Microbiol. 2004;70(5):2667–2677. doi: 10.1128/AEM.70.5.2667-2677.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabelsi D, Pini F, Bazzicalupo M, Biondi EG, Aouani ME, Mengoni A. Development of a cultivation-independent approach for the study of genetic diversity of Sinorhizobium meliloti populations. Mol Ecol Res. 2010;10(1):170–172. doi: 10.1111/j.1755-0998.2009.02697.x. [DOI] [PubMed] [Google Scholar]
- Trabelsi D, Pini F, Aouani ME, Bazzicalupo M, Mengoni A. Development of real-time PCR assay for detection and quantification of Sinorhizobium meliloti in soil and plant tissue. Letters in Applied Microbiol. 2009;48(3):355–361. doi: 10.1111/j.1472-765X.2008.02532.x. [DOI] [PubMed] [Google Scholar]
- Paffetti D, Daguin F, Fancelli S, Gnocchi S, Lippi F, Scotti C, Bazzicalupo M. Influence of plant genotype on the selection of nodulating Sinorhizobium meliloti strains by Medicago sativa. Antonie Van Leeuwenhoek. 1998;73(1):3–8. doi: 10.1023/A:1000591719287. [DOI] [PubMed] [Google Scholar]
- Hardoim PR, van Overbeek LS, Elsas JDv. Properties of bacterial endophytes and their proposed role in plant growth. Trends in Microbiol. 2008;16(10):463–471. doi: 10.1016/j.tim.2008.07.008. [DOI] [PubMed] [Google Scholar]
- Rosenblueth M, Martinez-Romero E. Bacterial endophytes and their interactions with hosts. Mol Plant Microbe Interact. 2006;19(8):827–837. doi: 10.1094/MPMI-19-0827. [DOI] [PubMed] [Google Scholar]
- Sessitsch A, Puschenreiter M. In: Microbiology of Extreme Soils Soil Biology 1. Dion P, Nautiyal CS, editor. Springer, Berlin Heidelberg; 2008. Endophytes and Rhizosphere Bacteria of Plants Growing in Heavy Metal-Containing Soils. [Google Scholar]
- Hartmann A, Stoffels M, Eckert B, Kirchhof G, Schloter M. In: Prokaryotic nitrogen fixation: a model system for the analysis of a biological process. Triplett EW, editor. Horizon Scientific Press, Wymondham; 2000. Analysis of the presence and diversity of diazotrophic endophytes; pp. 727–736. [Google Scholar]
- Weyens N, van der Lelie D, Taghavi S, Newman L, Vangronsveld J. Exploiting plant-microbe partnerships to improve biomass production and remediation. Trends in Biotechnology. 2009;27(10):591–598. doi: 10.1016/j.tibtech.2009.07.006. [DOI] [PubMed] [Google Scholar]
- Mengoni A, Mocali S, Surico G, Tegli S, Fani R. Fluctuation of endophytic bacteria and phytoplasmosis in elm trees. Microbiol Res. 2003;158(4):363–369. doi: 10.1078/0944-5013-00216. [DOI] [PubMed] [Google Scholar]
- Van Aken B, Peres CM, Doty SL, Yoon JM, Schnoor JL. Methylobacterium populi sp. nov., a novel aerobic, pink-pigmented, facultatively methylotrophic, methane-utilizing bacterium isolated from poplar trees (Populus deltoides x nigra DN34) Int J Syst Evol Microbiol. 2004;54(Pt 4):1191–1196. doi: 10.1099/ijs.0.02796-0. [DOI] [PubMed] [Google Scholar]
- Ulrich K, Ulrich A, Ewald D. Diversity of endophytic bacterial communities in poplar grown under field conditions. Fems Microbiol Ecol. 2008;63:169–180. doi: 10.1111/j.1574-6941.2007.00419.x. [DOI] [PubMed] [Google Scholar]
- López-López A, Rogel MA, Ormeño-Orrillo E, Martínez-Romero J, Martínez-Romero E. Phaseolus vulgaris seed-borne endophytic community with novel bacterial species such as Rhizobium endophyticum sp. nov. Syst Applied Microbiol. 2010;33(6):322–327. doi: 10.1016/j.syapm.2010.07.005. [DOI] [PubMed] [Google Scholar]
- Hayat R, Ali S, Amara U, Khalid R, Ahmed I. Soil beneficial bacteria and their role in plant growth promotion: a review. Annals of Microbiol. 2010;60(4):579–584. doi: 10.1007/s13213-010-0117-1. [DOI] [Google Scholar]
- Lugtenberg B, Kamilova F. Plant-growth-promoting rhizobacteria. Ann Rev Microbiol. 2009. pp. 541–556. [DOI] [PubMed]
- Nunes da Rocha U, Van Overbeek L, Van Elsas JD. Exploration of hitherto-uncultured bacteria from the rhizosphere. Fems Microbiol Ecol. 2009;69(3):313–328. doi: 10.1111/j.1574-6941.2009.00702.x. [DOI] [PubMed] [Google Scholar]
- Kielak A, Pijl AS, van Veen JA, Kowalchuk GA. Phylogenetic diversity of Acidobacteria in a former agricultural soil. ISME J. 2008;3(3):378–382. doi: 10.1038/ismej.2008.113. [DOI] [PubMed] [Google Scholar]
- Gubry-Rangin C, Nicol GW, Prosser JI. Archaea rather than bacteria control nitrification in two agricultural acidic soils. Fems Microbiol Ecol. 2010;74(3):566–574. doi: 10.1111/j.1574-6941.2010.00971.x. [DOI] [PubMed] [Google Scholar]
- Sarita S, Sharma PK, Priefer UB, Prell J. Direct amplification of rhizobial nodC sequences from soil total DNA and comparison to nodC diversity of root nodule isolates. Fems Microbiol Ecol. 2005;54(1):1–11. doi: 10.1016/j.femsec.2005.02.015. [DOI] [PubMed] [Google Scholar]
- Zézé A, Mutch LA, Young JPW. Direct amplification of nodD from community DNA reveals the genetic diversity of Rhizobium leguminosarum in soil. Environ Microbiol. 2001;3(6):363–370. doi: 10.1046/j.1462-2920.2001.00202.x. [DOI] [PubMed] [Google Scholar]
- Sachs JL, Kembel SW, Lau AH, Simms EL. In Situ Phylogenetic Structure and Diversity of Wild Bradyrhizobium Communities. Appl Environ Microbiol. 2009;75(14):4727–4735. doi: 10.1128/AEM.00667-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thies JE, Singleton PW, Bohlool BB. Influence of the Size of Indigenous Rhizobial Populations on Establishment and Symbiotic Performance of Introduced Rhizobia on Field-Grown Legumes. Appl Environ Microbiol. 1991;57(1):19–28. doi: 10.1128/aem.57.1.19-28.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Aravind L, Kondrashov AS. The impact of comparative genomics on our understanding of evolution. Cell. 2000;101:573–576. doi: 10.1016/S0092-8674(00)80867-3. [DOI] [PubMed] [Google Scholar]
- Mengoni A, Barabesi C, Gonnelli C, Galardi F, Bazzicalupo M. Genetic diversity of heavy metal-tolerant populations in Silene paradoxa L. (Caryophyllaceae): a chloroplast microsatellite analysis. Mol Ecol. 2001;10(8):1909–1916. doi: 10.1046/j.0962-1083.2001.01336.x. [DOI] [PubMed] [Google Scholar]
- Hammer Ø, Harper DAT, Ryan PD. PAST: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontologia Electronica. 2001;41(1):9. [Google Scholar]
- Excoffier L, Smouse PE, Quattro M. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genet. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocali S, Bertelli E, Di Cello F, Mengoni A, Sfalanga A, Viliani F, Caciotti A, Tegli S, Surico G, Fani R. Fluctuation of bacteria isolated from elm tissues during different seasons and from different plant organs. Res Microbiol. 2003;154(2):105–114. doi: 10.1016/S0923-2508(03)00031-7. [DOI] [PubMed] [Google Scholar]
- Slatkin M. A measure of population subdivision based on microsatellite allele frequencies. Genet. 1995;139:457–462. doi: 10.1093/genetics/139.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ. et al. Introducing mothur: Open Source, Platform-independent, Community-supported Software for Describing and Comparing Microbial Communities. Appl Environ Microbiol. 2009;75(23):7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good IJ. The population frequencies of species and the estimation to the population parameters. Biometrika. 1953;40:237–264. [Google Scholar]
- Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM. et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucl Acids Res. 2009;37(suppl_1):D141–145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Hierarchical analysis of differentiation between bacterial communities. AMOVA was performed with T-RFLP profiles from samples of the four different environments (soil, nodules, stems and leaves). Data show the degrees of freedom (d.f.), the sum of squared deviation, the variance component estimate, the percentage of total variance contributed by each component, and the probability (P) of obtaining a more extreme component estimate by chance alone, estimated from 10,000 permutations.
Table S2. Matrix of pairwise FST values. Statistical significance (p < 0.05) has been computed after 1000 random permutation; n.s., not significant. Only below diagonal values are reported.
Table S3. Statistical analysis of 16SrRNA gene clone libraries. OTUs were arbitrarily defined at 97% sequence identity based on Mothur clustering. Confidence intervals at 95% are given in parentheses. Coverage is defined C = [1 − (n/N)] × 100, where n is the number of unique clones, and N is the total number of clones examined.
Figure S1. S. meliloti IGS-T-RFLP profiling of nodule and soil samples. A), the schematic representation of the binary matrix of IGS-T-RF presence (black) and absence (empty cell); the IGS-T-RF number is reported on the right side of each row. B) The occurrence of “private” and “public” IGS-T-RFs. The percentage of total number of scored IGS-T-RFs is reported for T-RFs present from 1 to all 6 samples analyzed.