The two recombinant apo subunits H1 and H2 from H. americanus have been structurally characterized. Reconstitution studies with astaxanthin reproduced the bathochromic shift of 85–95 nm typical of the natural crustacyanin subunits.
Keywords: bathochromic shift, astaxanthin, colouration, recombinant carotenoproteins, lobster carapace
Abstract
Crustacean crustacyanin proteins are linked to the production and modification of carapace colour, with direct implications for fitness and survival. Here, the structural and functional properties of the two recombinant crustacyanin subunits H1 and H2 from the American lobster Homarus americanus are reported. The two subunits are structurally highly similar to the corresponding natural apo crustacyanin CRTC and CRTA subunits from the European lobster H. gammarus. Reconstitution studies of the recombinant crustacyanin proteins H1 and H2 with astaxanthin reproduced the bathochromic shift of 85–95 nm typical of the natural crustacyanin subunits from H. gammarus in complex with astaxanthin. Moreover, correlations between the presence of crustacyanin genes in crustacean species and the resulting carapace colours with the spectral properties of the subunits in complex with astaxanthin confirmed this genotype–phenotype linkage.
1. Introduction
The typical slate-grey/blue colour of the lobster carapace is generated by a number of distinct carotenoproteins, with the predominant one being the multimeric α-crustacyanin in complex with astaxanthin (3,3′-dihydroxy-β,β-carotene-4,4′-dione), which is characterized by a λmax of 632 nm (Zagalsky, 1985 ▶). In contrast, the absorption spectrum λmax in the visible region of astaxanthin in organic solvent is drastically blue-shifted (λmax of 478 nm in acetone). The change in colour of α-crustacyanin from blue to red upon protein denaturation and from red to blue upon astaxanthin-complex reconstitution has been a subject of investigation for over 60 years (Wald et al., 1948 ▶; Zagalsky, 1985 ▶; Buchwald & Jenks, 1968 ▶).
The multimeric α-crustacyanin, a protein complex consisting of 16 crustacyanin protein subunits, dissociates reversibly in low ionic strength solution, initially forming α′-crustacyanin (λmax = 595 nm) and then with further incubation the heterodimeric β-crustacyanin (λmax = 580–590 nm) (Cheeseman et al., 1966 ▶). Chemical removal of the carotenoid from β-crustacyanin results in reversible dissociation into two types of apoproteins with molecular weights of 21 kDa (type I; CRTC) and 19 kDa (type II; CRTA), respectively (Buchwald & Jenks, 1968 ▶; Quarmby et al., 1977 ▶). β-Crustacyanin is in fact a heterodimer formed by one CRTC lipocalin subunit, one CRTA lipocalin subunit and two noncovalently bound astaxanthin molecules (Zagalsky, 1985 ▶; Cianci et al., 2002 ▶). The crustacyanins of the lobsters Homarus americanus and H. gammarus exhibit identical absorption spectra but behave differently in ion-exchange chromatography and polyacrylamide gel electrophoresis (Zagalsky & Tidmarsh, 1985 ▶). In the case of H. gammarus five distinct subunits are evident on 6 M urea–PAGE gels, namely A1, C1 and C2 (type I) and A2 and A3 (type II). The H. americanus crustacyanin subunits appear to consist of only two major subunits, namely H1 (type I; CRTC) and H2 (type II; CRTA), both of which behave like Ax subunits on a 6 M urea–PAGE gel (Fig. 1 ▶ a). Only two genes encoding crustacyanin subunits have so far been identified in lobsters (Wade et al., 2009 ▶), one for each group, suggesting that the differences between protein members of the same group could arise from post-translational modifications.
Figure 1.
(a) 6 M urea–PAGE of crustacyanin pigments of H. gammarus (left) and H. americanus (right). Copyright Elsevier (1985), reproduced with permission from Zagalsky & Tidmarsh (1985 ▶). Sequence alignments of (b) CRTC and (c) CRTA crustacyanin proteins from H. americanus (HomAm) using the H. gammarus (HomGa) sequences as a reference. Amino-acid alignment was performed via the ClustalW2 algorithm (http://www.ebi.ac.uk/Tools/msa/clustalw2/; Altschul et al., 1997 ▶).
The presence of CRTC and/or CRTA might be linked to the ability to produce or modify carapace colour, with direct implications for the fitness and survival of many crustaceans via camouflage or mate selection (Wade et al., 2009 ▶).
Although the crystal structure of β-crustacyanin has been solved at 3.2 Å resolution (Cianci et al., 2002 ▶) and the structural architecture of α-crustacyanin from H. gammarus has been investigated by EM/SAXS (Rhys et al., 2011 ▶), there is still debate on the origin of the bathochromic shift of astaxanthin upon binding to carotenoproteins. The contributions of coplanarization, exciton interaction and polarization to the bathochromic shift of astaxanthin have also been investigated using chemical crystallography of astaxanthin and its derivatives (Bartalucci et al., 2007 ▶, 2009 ▶; Helliwell, 2008 ▶) or by using halogenated canthaxanthins in crustacyanin-reconstitution studies (Liu et al., 2002 ▶). These contributions have also been the subject of several theoretical reports (Durbeej & Eriksson, 2004 ▶; Ilagan et al., 2005 ▶; Wijk et al., 2005 ▶; Strambi & Durbeej, 2009 ▶; Helliwell, 2010 ▶; Polívka et al., 2010 ▶; Neugebauer et al., 2011 ▶).
The present study reports the cloning, heterologous expression and structural characterization of the two apo subunits H1 and H2 from H. americanus and the results of the reconstitution of their complexes with astaxanthin.
2. Materials and methods
2.1. Sequence analyses
A BLAST search (Altschul et al., 1997 ▶) for sequences similar to the CRA1_HOMGA sequence (type I subunit from H. gammarus; GenBank accession No. P58989) identified EST sequence DV771534 (Fig. 1 ▶ b) among 5184 H. americanus olfactory-organ 5′-end sequences obtained from a subtracted directional cDNA library prepared in pBluescript (Stepanyan et al., 2006 ▶). A second BLAST search obtained using the CRA2_HOMGA sequence (type II subunit from H. gammarus; GenBank accession No. P80007) as the query identified a second EST sequence of interest from the same source: DV774018 (Fig. 1 ▶ c). Clones containing these putative crustacyanin sequences were rescued from frozen stocks of this library by ampicillin selection on agar plates.
The EST sequence DV771534 (H1 from H. americanus) encoded 181 amino acids, the same as CRTC_HOMGA, with an overall protein sequence identity of 97%. The EST sequence DV774018 (H2 from H. americanus) encoded 159 amino acids with a sequence identity of 95%. Based on this high degree of identity, the 15 N-terminal codons of A3 from H. gammarus were added to cDNA clone DV774018 to produce what is predicted to be a full-length CRTA sequence (Fig. 2 ▶ b).
Figure 2.
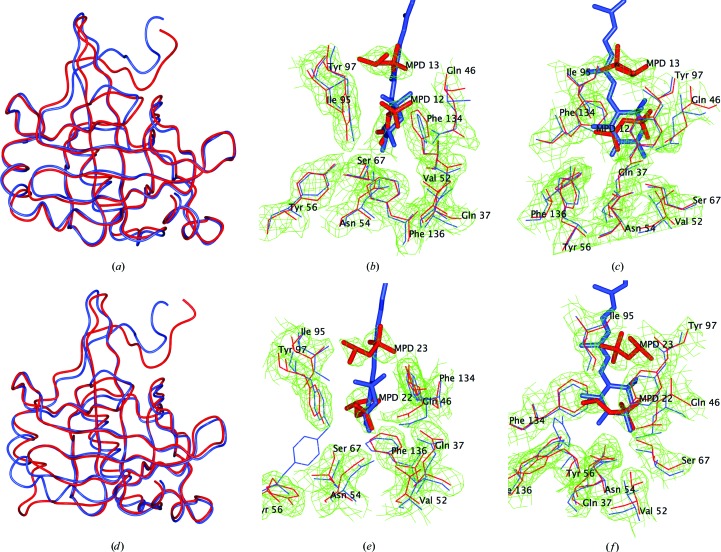
Worm-type diagram of the H1 subunit (red) superimposed on the A1 subunit from β-crustacyanin (blue; PDB entry 1gka): (a) monomer A, (d) monomer B. Superimposition of the H1 subunit binding pocket (red) superimposed on the A1 subunit binding pocket from β-crustacyanin (blue), resulting in the astaxanthin molecule AXT1 from 1gka superimposed on the MPD-binding sites: (b) monomer A, (c) monomer A rotated by 90°, (e) monomer B, (f) monomer B rotated by 90°. 2F o − F c maps contoured at 1σ r.m.s. are shown.
2.2. Cloning, expression and purification of the H1 subunit
The region of the cDNA clone DV771534 corresponding to the mature H1 subunit was amplified by PCR using an NdeI-tailed upstream primer 5′-CATATGGACAAAATCCCAGACTTC-3′ and the downstream primer 5′-CTAGAGTGTCTTCTGAGTATCGTA-3′. The product was then cloned into pGEM-T Easy (Promega) using the T/A cloning method and then transferred into the NdeI site of the expression plasmid pET11b (Novagen). Escherichia coli BL21 Rosetta-gami cells (Novagen) were transformed with the expression plasmid. The expression of the H1 subunit was induced overnight at 310 K by adding 1 mM IPTG and the cells were lysed by sonication. The recombinant CRTC_HOMAM, which was present in the insoluble fraction, was dissolved in a denaturing medium [50 mM Tris–HCl pH 7.5, 2 mM EDTA, 2 mM PMSF, 0.1%(v/v) Triton X-100, 8 M urea] and then refolded by dialysis against 50 mM Tris–HCl pH 9.3. The soluble fraction at pH 7 was centrifuged and concentrated by ultrafiltration. The H1 subunit was then purified to homogeneity using Ultrogel AcA54 gel-filtration chromatography. The final yield of purified protein was 2–2.5 mg per litre of culture.
2.3. Cloning, expression and purification of the H2 subunit
The cDNA clone DV774018 lacked a complete N-terminus. To produce a full-length protein, albeit chimeric, for expression studies, three PCR reactions were performed sequentially to generate a cDNA encoding the first 15 N-terminal residues of CRA_HOMGA followed in-frame by the DV774018 sequence. For the 5′ elongation of the coding sequence, three forward partially overlapping primers and a reverse primer were employed (Table 1 ▶). The sequence encoding the full-length chimeric protein was amplified using an NcoI-tailed upstream primer (Table 1 ▶), cloned into pGEM-T Easy (Promega) using the T/A cloning method and transferred into the NcoI site of the expression plasmid pET28b (Novagen). E. coli BL21 Rosetta-gami cells (Novagen) were transformed with the expression plasmid. Protein expression and purification was carried out as previously described for the H. americanus H1 subunit. The final yield of purified protein was 1.5–2 mg per litre of culture.
Table 1. Primers used for cloning of the H. americanus H2 subunit.
| CRTA1 | Forward, 5′-AAATGTGCCTCCGTAGCAAACCAGGCCAAC-3′ |
| CRTA2 | Forward, 5′-TTTGTAACTGCAGGAAAATGTGCCTCCGTAGCAAAC-3′ |
| CRTA3 | Forward, 5′-GATGGAATTCCTTCATTTGTAACTGCAGGAAAATGTG-3′ |
| CRTA | Reverse, 5′-TTAAGCTCTGTAGACACACTC-3′ |
2.4. Crystallization, data collection and analysis of the H1 subunit
Recombinant H1 subunit was concentrated to a final concentration of 10.7 mg ml−1 in 1 mM EDTA, 0.1 M Tris–HCl pH 7.0 by ultrafiltration using an Amicon cell with a 10 kDa cutoff membrane and used in crystallization experiments. Crystallization was performed at room temperature using the sitting-drop vapour-diffusion method at the EMBL Hamburg high-throughput crystallization facility (Mueller-Dieckmann, 2006 ▶). The best crystals were grown by mixing equal volumes (1 µl) of protein solution and 1 mM EDTA, 2.4 M ammonium sulfate, 5%(v/v) 2-methyl-2,4-pentanediol (MPD), 0.1 M Tris–HCl pH 8.0 buffer. Crystals grew to dimensions of 100 × 30 × 30 µm in a week.
X-ray diffraction data were collected from H1 crystals using synchrotron radiation on the BW6-MPG beamline at the DORIS storage ring, c/o Deutsches Elektronen Synchrotron (DESY), Hamburg, Germany. Each single crystal was exposed to a cold nitrogen stream at 100 K without further cryoprotection. The data were processed with HKL-2000 (Otwinowski & Minor, 1997 ▶). The crystals diffracted to 2.38 Å resolution and belonged to space group P212121, with unit-cell parameters a = 40.37, b = 78.42, c = 105.63 Å.
The structure was solved by molecular replacement with AMoRe (Winn et al., 2011 ▶) using the crystal structure of apo A1 from H. gammarus (PDB entry 1h91; Cianci et al., 2001 ▶) as the search model. The structure was refined using REFMAC5 (Winn et al., 2011 ▶; Murshudov et al., 2011 ▶). Model building and water assignments were performed using Coot (Emsley et al., 2010 ▶). General criteria for water assignment were B factor lower than 80 Å2, 2F o − F c map σ level greater than 1 and interatomic contacts of between 2.3 and 3.5 Å.
Refinement using isotropic temperature factors converged to final R-factor and R free (5% of data) values of 20.2% and 26.8%, respectively.
The stereochemistry of the final model was checked using PROCHECK (Winn et al., 2011 ▶). Structural superpositions were performed using LSQKAB (Winn et al., 2011 ▶). Figures were prepared using CCP4mg (McNicholas et al., 2011 ▶). Data-collection and refinement statistics are reported in Table 2 ▶.
Table 2. Data-collection and model-refinement statistics.
Values in parentheses are for the highest resolution bin.
| Data collection | |
| Wavelength (Å) | 1.05 |
| Detector | MAR 165 CCD |
| Oscillation angle (°) | 0.35 |
| No. of images | 250 |
| Space group | P212121 |
| Unit-cell parameters (Å) | a = 40.38, b = 78.46, c = 105.63 |
| Resolution range (Å) | 20.0–2.38 (2.46–2.38) |
| Total No. of reflections | 48851 |
| Unique reflections | 13980 |
| Multiplicity | 3.5 (3.5) |
| Completeness (%) | 98.9 (99.0) |
| R merge † (%) | 7.5 (34.1) |
| Mean I/σ(I) | 14.0 (2.5) |
| Refinement statistics | |
| No. of molecules in the asymmetric unit | 2 |
| No. of residues‡ | 362 |
| R factor‡ (%) | 22.3 |
| R free ‡ (%) | 26.6 |
| Cruickshank’s DPI for coordinate error based on R factor‡ (Å) | 0.98 |
| B factors (Å2) | |
| Average all-atom§ | 31.5 |
| Main-chain atoms¶ | 35.0 |
| Side-chain atoms and waters¶ | 35.9 |
| Average r.m.s. B factor (Å2) | |
| Main-chain atoms¶ | 0.4 |
| Side-chain atoms¶ | 0.9 |
| Total No. of atoms | 3083 |
| Total No. of water molecules | 97 |
| Solvent content (%) | 40.5 |
| Matthews coefficient (Å3 Da−1) | 2.06 |
| Ramachandran plot†† | |
| Most favoured region | 286 [88.8%] |
| Additionally allowed region | 34 [10.6%] |
| Generously allowed region | 1 [0.3%] |
| Disallowed region | 1 [0.3%] |
R
merge = 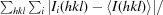
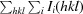 , where Ii(hkl) is the intensity of a reflection and 〈I(hkl)〉 is the mean intensity of all symmetry-related reflections i.
, where Ii(hkl) is the intensity of a reflection and 〈I(hkl)〉 is the mean intensity of all symmetry-related reflections i.
DPI = [N atoms/(N refl − N params)]1/2 × R factor × D max × compl−1/3, where N atoms is the number of atoms included in the refinement, N refl is the number of reflections included in the refinement, D max is the maximum resolution of the reflections included in the refinement, compl is the completeness of the observed data and, in isotropic refinement, N params ≃ 4N atoms (Cruickshank, 1999 ▶).
Taken from BAVERAGE (CCP4; Winn et al., 2011 ▶).
Taken from PROCHECK (CCP4; Winn et al., 2011 ▶).
2.5. SAXS data collection and analysis of the H2 subunit from H. americanus
The synchrotron-radiation X-ray scattering data of the H2 subunit were collected following standard procedures on SAXS beamline X33 (Roessle et al., 2007 ▶) at the DORIS III storage ring, DESY, Hamburg. The scattering patterns from the H2 subunit were measured using a sample-to-detector distance of 2.4 m, covering the momentum-transfer range 0.01 < s < 0.5 Å−1 [s = 4πsin(θ)/λ, where θ is the scattering angle and λ = 1.5 Å is the X-ray wavelength]. In order to check for interprotein interactions, measurements were made at three protein concentrations: 15, 7.5 and 3.8 mg ml−1. Repetitive measurements (120 s) of the same protein solution were performed in order to check for radiation damage, and no aggregation was found during the initial 120 s exposure. This initial exposure frame was taken as a reference for further analysis. The data were normalized to the intensity of the incident beam; the scattering of the buffer was subtracted and the difference curves were scaled for concentration. Data sets collected at different concentrations were merged to improve the data quality. All data-processing steps were performed using the PRIMUS package (Konarev et al., 2003 ▶). The forward scattering I(0) and the radius of gyration R g were evaluated using the Guinier approximation (Guinier & Fournet, 1955 ▶) assuming that at very small angles (s < 1.3/R g) the intensity was represented by I(s) = I(0)exp[−(sR g)2/3]. These parameters were computed from the entire scattering patterns using the indirect transform package GNOM (Semenyuk & Svergun, 1991 ▶), which provides the distance distribution function p(r) of the particle. The molecular mass of the proteins was calculated by comparison with the forward scattering from a reference solution of bovine serum albumin (BSA).
Initial ab initio modelling trials using DAMMIN or GASBOR from the ATSAS program suite (Svergun, 1997 ▶; Svergun et al., 2001 ▶) did not yield unique models (high χ values) and could not be used for structural interpretation. The multimeric state of the protein in solution was tested by the program OLIGOMER (Konarev et al., 2003 ▶), which calculates the theoretical volume fractions of different protein oligomers, and this was compared with experimental SAXS data. For this analysis the A3 model from the heterodimeric structure of β-crustacyanin (PDB entry 1gka; Cianci et al., 2002 ▶) was used to create a pseudodimer of H2. The trimeric H2 model was derived from the crystal packing of β-crustacyanin (Fig. 3 ▶ c).
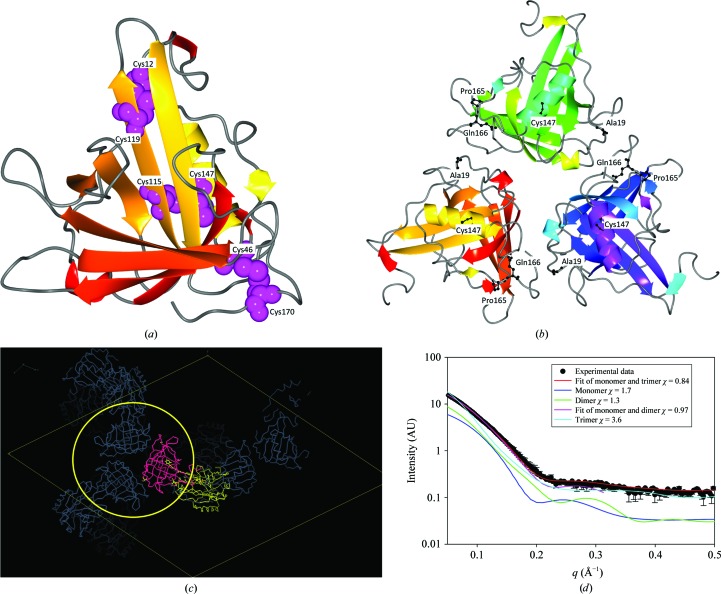
Figure 3.
(a) Ribbon diagram of an H2 monomer, with cysteine residues highlighted. (b) H2 trimer generated using the 1gka crystal packing, with point mutations shown in ball-and-stick representation. (c) 1gka crystal packing viewed down the threefold axis, showing the H2 trimer. (d) Profile-fitting results of SAXS data using different models.
2.6. Complex reconstitution with astaxanthin
The complex reconstitution of either H1 or H2 or a mixture of both subunits in the presence of astaxanthin was probed using the acetone method (Zagalsky, 1985 ▶). The protein (about 5 mM) dissolved in a mixture of equal volumes of 40 mM Tris–HCl pH 7, 0.2 M ammonium sulfate and acetone was incubated with astaxanthin dissolved in acetone (about 12 mM). The mixture was dialyzed overnight at 277 K in the dark against phosphate buffer pH 7. Protein solutions were analyzed for absorption spectra in the 240 and 700 nm range.
3. Results and discussion
3.1. H1 subunit structure
The crystal structure of H1 apo crustacyanin from H. americanus contains a homodimer in the crystallographic asymmetric unit.
The H1 protein has 181 residues with the typical lipocalin folding of crustacyanins A1 (Cianci et al., 2001 ▶, 2002 ▶), C1 (Gordon et al., 2001 ▶) and C2 (Habash et al., 2004 ▶) (Figs. 2 ▶ a–2 ▶ d), with a β-barrel made up of two distinct β-sheets. Homodimerization is achieved via a close contact between the two β-strands of each subunit, similarly to the A1 (Cianci et al., 2001 ▶), C1 (Gordon et al., 2001 ▶) and C2 (Habash et al., 2004 ▶) crystal structures (see Fig. 4 of Gordon et al., 2001 ▶). All the residues that differed between species were located on the protein surface in the crystal structure. Tyr16, Glu61 and Met99 are located on hairpins, Met30 on the first α-helix, Try66 on β-strand C and Gln145 on an α-helix.
Superimposition of the complete H1 apo crustacyanin homodimer with its analogues leads to an r.m.s. deviation for Cα atoms of 1.39 Å from the crystal structure of the apo A1 subunit (PDB entry 1h91; Cianci et al., 2001 ▶), of 1.41 Å from that of apo C1 (PDB entry 14u1; Gordon et al., 2001 ▶) and of 1.37 Å from that of apo C2 (PDB code: 1sp2; Habash et al., 2004 ▶). The comparison shows a good overall fit, in agreement with the high sequence homology.
The statistics of the Ramachandran plot for the apo H1 subunit are reported in Table 2 ▶. The outlier residues are Tyr112 in both chains A and B, which are in a generously allowed region, similar to the previously reported apo CRTC subunits from H. gammarus (Cianci et al., 2001 ▶). Other members of the lipocalin family have, as a feature, the torsion angles of this residue in a disallowed region of the Ramachandran plot (Cowan et al., 1990 ▶; Zanotti et al., 1993 ▶).
Apo subunits A1 (Cianci et al., 2001 ▶) and C1 (Gordon et al., 2001 ▶) from H. gammarus were crystallized in the presence of MPD, which could be found in the astaxanthin binding site of both proteins. Comparison of the MPD positions in the apo crustacyanin H1 (this work), A1 (Cianci et al., 2001 ▶), C1 (Gordon et al., 2001 ▶) and C2 (Habash et al., 2004 ▶) subunits revealed that the MPD-binding mode was preserved throughout the CRTC subunits from both H. gammarus and H. americanus given that no mutations were observed for the residues defining the MPD-binding pocket.
Moreover, the astaxanthin-binding site in the β-crustacyanin A1 monomer corresponds to that of MPD in each H1 monomer of the apo crustacyanin H1 dimer. The superimposition of the three-dimensional structure of apo crustacyanin H1 monomer A, for instance, onto the structure of the A1 monomer of β-crustacyanin (PDB entry 1gka; Cianci et al., 2002 ▶) results in the superimposition of an astaxanthin molecule onto the positions of the pair of MPD molecules (Figs. 2 ▶ b, 2 ▶ c, 2 ▶ e and 2 ▶ f). MPD molecules are therefore mimicking the binding of parts of a carotenoid ring and the C14 atom. This is also evident for apo crustacyanin C1, C2 and A1 dimers. The superimposition of the residues of the apo H1 crustacyanin that define the MPD-binding pocket (Gln37, Gln46, Val52, Asn54, Tyr56, Ser67, Ile95, Tyr97, Phe134 and Phe136) with their analogues in the A1 monomer of β-crustacyanin (PDB entry 1gka; Cianci et al., 2002 ▶) results in r.m.s. deviations of 0.9 Å for monomer A and 1.7 Å for monomer B. The larger r.m.s. deviation for monomer B is owing to Tyr56, which appears to be able to adopt a different conformation from monomer A (Fig. 2 ▶ e) without affecting MPD binding (the r.m.s. deviation of Tyr56 in monomer B versus monomer A is 4.8 Å). The overall residue arrangement of the binding site of apo crustacyanin H1 for the astaxanthin molecule is preserved.
3.2. H2 subunit structure
The H2 subunit shares 95% sequence identity with the A3 subunit from H. gammarus (Cianci et al., 2002 ▶). It is therefore highly likely that the H2 subunit and the A3 monomers adopt the same three-dimensional structure. The structure of the H2 subunit from H. americanus was modelled on the known structure of the A3 subunit from H. gammarus (PDB entry 1gka; Cianci et al., 2002 ▶) and was thus predicted to have a lipocalin topology with a β-barrel made up of two distinct β-sheets (Fig. 3 ▶ a).
The H2 subunit structure model was then confirmed by SAXS. No conditions suitable for the production of protein crystals were found for this protein. A Kratky analysis of the SAXS data gave clear evidence for a folded H2 subunit in solution. Further analysis of the SAXS data revealed that the H2 subunit in solution is present in a dynamic equilibrium between monomeric and multimeric forms. Analysis using standard ab initio or rigid-body SAXS modelling did not give good results because of the polydispersity of the H2 subunit in solution.
Several possible protein-oligomerization states were considered: monomer–dimer, monomer–dimer–trimer, monomer–trimer and dimer–trimer mixtures.
Optimum scattering curve fitting (χ = 0.84) resulted from combination of the H2 monomeric subunit (volume fractions of 65%) and its trimeric form (35%) predicted from the A3 trimer in the crystal structure of β-crustacyanin (Cianci et al., 2002 ▶), where it generates a crystallographic threefold axis (Figs. 3 ▶ b and 3 ▶ c).
Other combinations of monomeric and multimeric forms of the H2 subunit tested with OLIGOMER resulted in no improvement in fit (Fig. 3 ▶ d). The fact that only a mixture of the monomer and the trimer based on the crystallographic studies of the A3 subunit could explain the scattering profile suggests not only that the apo crustacyanin H2 subunit structure is conserved compared with apo crustacyanin A3, but also that the subunit preserves the same oligomerization properties in solution as observed in the crystal lattice. All of the residues that differed between species were located on the protein surface: Ala19, Pro165 and Gln166 are located on hairpins and Cys147 on an α-helix. They were not involved in generating intermolecular contacts (Fig. 3 ▶ b). Within the putative apo H2 subunit structure the Cys147 residue (associated with the T147C mutation) is directly opposite the Cys115 residue, thus potentially generating an intramolecular Cys115–Cys147 disulfide bridge, structurally similar to the observed Cys117–Cys150 disulfide bridge present in the H1 subunit (Fig. 3 ▶ a).
3.3. Complex reconstitution with astaxanthin
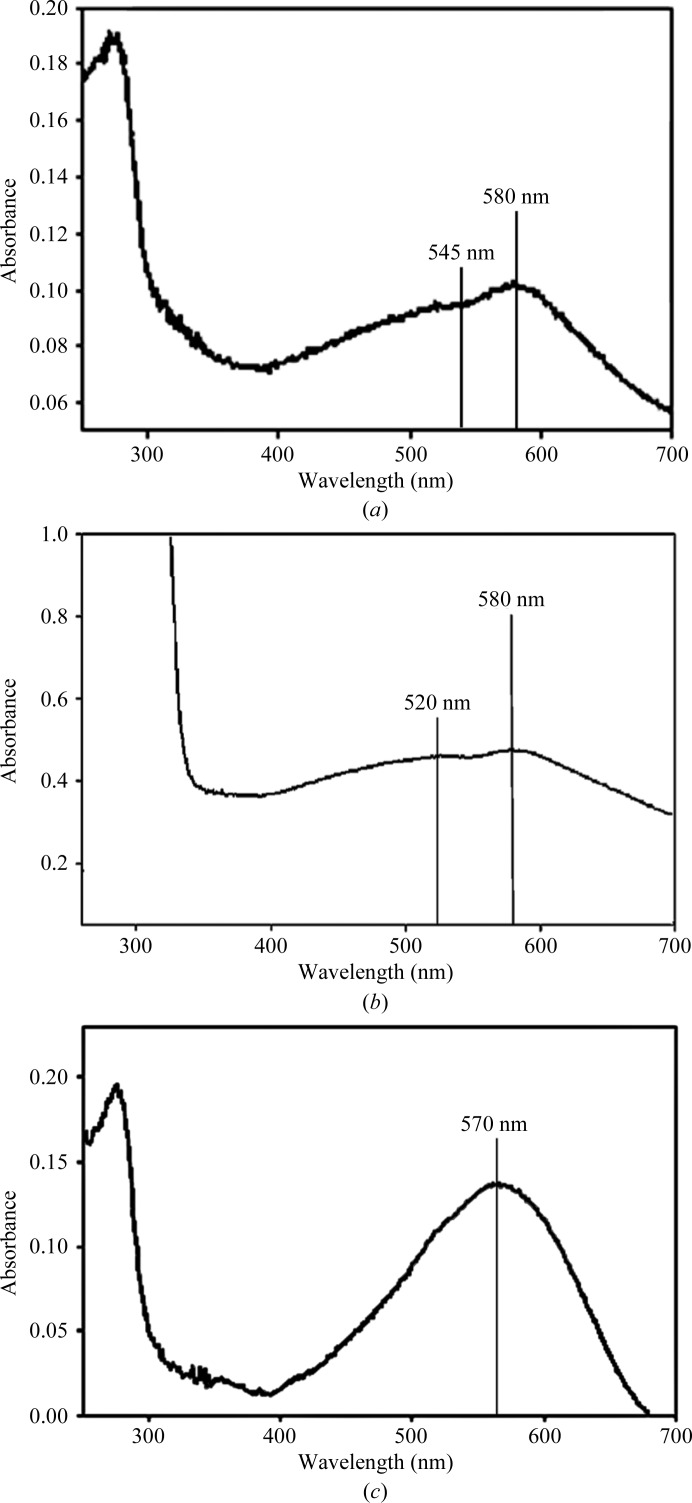
Recombination of apo H1 protein singly with astaxanthin produced a bathochromic shift with λmax = 580 nm (Fig. 4 ▶ a), albeit with a low yield (OD580/OD285 = 0.6). A shoulder is also present with a peak at λmax ≃ 545 nm (OD545/OD285 = 0.5), suggesting the presence of a second species in solution. For comparison, recombination of freshly prepared natural apo subunits from H. gammarus carapace singly with astaxanthin gives a dimeric-like product with λmax = 585 nm in high yields when testing C1 and C2 apo proteins or a monomeric-like product with λmax = 565 nm in low yields when testing A1, A2 and A3 apo proteins (Quarmby et al., 1977 ▶).
Figure 4.
Reconstitution studies: (a) H1 subunit incubated with astaxanthin, (b) H2 subunit incubated with astaxanthin, (c) a solution containing both H1 and H2 subunits incubated with astaxanthin.
On the other hand, combination of astaxanthin with mixtures of the individual apo protein units separated by gel filtration or ion exchange gave only monomeric-like or dimeric-like products with λmax = 565 nm (Zagalsky, 1985 ▶). The apo H1 protein from H. americanus therefore has the ability to produce a bathochromic shift and to generate two recombination products with distinguishable λmax at 580 and 545 nm, analogous to CRTC apo subunits from H. gammarus.
Recombination of apo H2 subunit from H. americanus singly with astaxanthin gave a bathochromic shift with λmax = 580 nm (Fig. 4 ▶ b), albeit with a low yield. A shoulder is also present with a peak at λmax ≃ 520 nm, suggesting the presence of a second species in solution. Previous reconstitution studies using natural apo H2 subunit from H. americanus singly with astaxanthin showed a single peak at λmax ≃ 540 nm after chromatofocusing (Milicua et al., 1986 ▶).
The recombination of a mixture of H1 and H2 subunits with astaxanthin gave rise to a new single product attributable to β-crustacyanin and characterized by a single absorption maximum at λmax = 570 nm (Fig. 4 ▶ c) and with a yield (OD570/OD285 = 0.75) larger than those determined in the cases of the individual recombinant subunits in the presence of astaxanthin. For comparison, recombination of both types of freshly prepared natural apo subunits from H. gammarus with astaxanthin gives β-crustacyanin with variable but nevertheless good yields (OD580/OD285 > 2.5) and absorption maxima (565–585 nm) that differed according to the combination of apoproteins selected (Zagalsky, 1985 ▶; Quarmby et al., 1977 ▶).
3.4. Genotype–phenotype correlation
Examination of the spectral properties of the subunits in complex with astaxanthin shed light on carapace coloration resulting from expression of CRTC and/or CRTA subunits (Table 3 ▶). In crustaceans the carapace colour is generated by subtractive mixing of the colours absorbed by the adducts. In crustaceans expressing only CRTC subunits the colours absorbed would be yellow (580 nm) and yellow–green (545 nm) similar to the H1 subunit. By using Munsell’s colour wheel (Munsell, 1912 ▶; Supplementary Fig. 11) it can be determined that the complementary colours are violet and purple, respectively, thus generating a visible purple–violet colour. In the carapace of the shrimp Alpheus alpheopsides only CRTC subunits were found and the colour was reported to be blue (Wade et al., 2009 ▶).
Table 3. Type of crustacyanin (CRTN) proteins present in Decapoda crustacea and their related colours.
| Sample animal | Tissue | CRTN | Colour | Reference |
|---|---|---|---|---|
| Panulirus cygnus | Epithelium/carapace | A1/A2 | Red | Wade et al. (2009 ▶) |
| Panulirus ornatus | Epithelium/carapace | — | Blue/orange | Wade et al. (2009 ▶) |
| Panulirus versicolour | Epithelium/carapace | A/C | Blue/green | Wade et al. (2009 ▶) |
| Penaeus monodon | Epithelium | A/C | Blue | Wade et al. (2009 ▶) |
| Marsupenaeus japonicus | Epithelium/EST | A/C | Red | Wade et al. (2009 ▶) |
| Homarus americanus | Carapace | A/C | Blue/green | Zagalsky (1985 ▶) |
| Homarus gammarus | Carapace | A/C | Blue/green | Zagalsky (1985 ▶) |
| Cherax quadricanatus | Epithelium/carapace/EST | A/C | Blue | Wade et al. (2009 ▶) |
| Alpheus sp. | Whole animal | C | Blue | Wade et al. (2009 ▶) |
| Macrobranchium rosenbergii | Carapace | A(?)/C | Blue | Yang et al. (2011 ▶) |
| Dardanus megistos | Epithelium/carapace | A | Red | Wade et al. (2009 ▶) |
| Gonadactylus smithii | Epithelium/carapace | A | Green | Wade et al. (2009 ▶) |
In crustaceans expressing only CRTA subunits the colours absorbed would be yellow (580 nm) and bluish–green (520 nm), similarly to the H2 subunit, thus generating a visible purple–red colour. In the carapaces of Palinurus cygnus and Dardanus megistos only CRTA subunits are expressed and the reported colour is red (Wade et al., 2009 ▶). Expression of the CRTA subunit alone may then result in red hues and expression of the CRTC subunit alone results in blue hues.
In crustaceans expressing both CRTC and CRTA subunits the colours absorbed would be yellow (580 nm, β-crustacyanin-like complexes) to red (630 nm, α-crustacyanin-like complexes), similar to lobsters with visible blue to bluish–green colours. In the carapaces of Palinurus versicolor, Penaeus monodon and Cherax quadricarinatus both CRTC and CRTA subunits are expressed and their reported colours are blue or blue–green (Wade et al., 2009 ▶). In Macrobrachium rosenbergii the MrLC gene encodes a lipocalin protein which has been shown to specifically bind astaxanthin (Yang et al., 2011 ▶). Knockdown of the MrLC gene by RNA interference (RNAi) resulted in a shift in body colour from blue to orangish–red over the entire carapace (Yang et al., 2011 ▶).
Apparent discrepancies in the correlation between genotype (the presence or absence of CRTC and/or CRTA crustacyanin genes) and phenotype (carapace colouration or hue) in crustaceans can potentially be explained either by different levels of protein expression, post-translational modifications, oligomerization (such as α-crustacyanin) or environmental and dietary factors (Wade et al., 2009 ▶). The correlations between the presence of CRTC and/or CRTA crustacyanin genes in crustacean species and carapace colours, as reported in the literature, with the spectral properties of the subunit in complex with astaxanthin confirm this genotype–phenotype linkage.
Heterologous expression and purification of the two major recombinant crustacyanin subunits from H. americanus produce two apo subunits that are highly structurally similar to the corresponding subunits A1, C1 and C2 from H. gammarus characterized in previous studies (Cianci et al., 2002 ▶; Gordon et al., 2001 ▶; Habash et al., 2004 ▶). Moreover, complex-reconstitution studies of the recombinant proteins H1 and H2 with astaxanthin show the bathochromic shift of 85–95 nm typical of natural crustacyanin proteins from H. gammarus. Preparations of recombinant crustacyanin subunits provide several opportunities to further dissect the molecular basis of crustacean carapace coloration and its biotechnological applications.
The crystallographic coordinates (PDB entry 4alo) and structure-factor amplitudes for the H1 subunit have been deposited in the Protein Data Bank. The theoretical coordinates for the H2 subunit validated by SAXS are available from the corresponding author upon request.
Supplementary Material
PDB reference: 4alo
Supplementary material file. DOI: 10.1107/S1744309112026103/hv5216sup1.pdf
Acknowledgments
MC thanks Dr G. Bourenkov for support during data collection. MPG is thanked for the provision of beam time at DORIS, DESY, Germany. Dr T. R. Schneider (EMBL Hamburg) is thanked for general support. MC is grateful to Dr P. Zagalsky, Professor J. R. Helliwell (University of Manchester, England) and Dr I. Manolaridis (EMBL Hamburg) for helpful discussions.
Footnotes
Supplementary material has been deposited in the IUCr electronic archive (Reference: HV5216).
References
- Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997). Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed]
- Bartalucci, G., Coppin, J., Fisher, S., Hall, G., Helliwell, J. R., Helliwell, M. & Liaaen-Jensen, S. (2007). Acta Cryst. B63, 328–337. [DOI] [PubMed]
- Bartalucci, G., Fisher, S., Helliwell, J. R., Helliwell, M., Liaaen-Jensen, S., Warren, J. E. & Wilkinson, J. (2009). Acta Cryst. B65, 238–247. [DOI] [PubMed]
- Buchwald, M. & Jenks, W. (1968). Biochemistry, 7, 844–859. [DOI] [PubMed]
- Cheeseman, D. F., Zagalsky, P. F. & Ceccaldi, J. H. (1966). Proc. R. Soc. Lond. B, 164, 130–151.
- Cianci, M., Rizkallah, P. J., Olczak, A., Raftery, J., Chayen, N. E., Zagalsky, P. F. & Helliwell, J. R. (2001). Acta Cryst. D57, 1219–1229. [DOI] [PubMed]
- Cianci, M., Rizkallah, P. J., Olczak, A., Raftery, J., Chayen, N. E., Zagalsky, P. F. & Helliwell, J. R. (2002). Proc. Natl Acad. Sci USA, 99, 9795–9800. [DOI] [PMC free article] [PubMed]
- Cowan, S. W., Newcomer, M. E. & Jones, T. A. (1990). Proteins, 8, 44–61. [DOI] [PubMed]
- Cruickshank, D. W. J. (1999). Acta Cryst. D55, 583–601. [DOI] [PubMed]
- Durbeej, B. & Eriksson, L. A. (2004). Phys. Chem. Chem. Phys. 6, 4190–4198.
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Gordon, E. J., Leonard, G. A., McSweeney, S. & Zagalsky, P. F. (2001). Acta Cryst. D57, 1230–1237. [DOI] [PubMed]
- Guinier, A. & Fournet, G. (1955). Small Angle Scattering of X-rays New York: Wiley.
- Habash, J., Helliwell, J. R., Raftery, J., Cianci, M., Rizkallah, P. J., Chayen, N. E., Nneji, G. A. & Zagalsky, P. F. (2004). Acta Cryst. D60, 493–498. [DOI] [PubMed]
- Helliwell, J. R. (2010). Crystallogr. Rev. 16, 231–242.
- Helliwell, M. (2008). Carotenoids, Vol. 4, Natural Functions, edited by G. Britton, S. Liaaen-Jensen & H. Pfander, pp. 37–52. Basel: Birkhäuser Verlag.
- Ilagan, R. P., Christensen, R. L., Chapp, T. W., Gibson, G. N., Pascher, T., Polívka, T. & Frank, H. A. (2005). J. Phys. Chem. A, 109, 3120–3127. [DOI] [PubMed]
- Konarev, P. V., Volkov, V. V., Sokolova, A. V., Koch, M. H. J. & Svergun, D. I. (2003). J. Appl. Cryst. 36, 1277–1282.
- Liu, J., Shelton, N. L. & Liu, R. S. (2002). Org. Lett. 4, 2521–2524. [DOI] [PubMed]
- McNicholas, S., Potterton, E., Wilson, K. S. & Noble, M. E. M. (2011). Acta Cryst. D67, 386–394. [DOI] [PMC free article] [PubMed]
- Milicua, J. C. G., Gárate, A. M., Barbon, P. G. & Gomez, R. (1986). Comp. Biochem. Physiol. B Comp. Biol. 85, 621–626.
- Mueller-Dieckmann, J. (2006). Acta Cryst. D62, 1446–1452. [DOI] [PubMed]
- Munsell, A. H. (1912). Am. J. Psychol. 23, 236–244.
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Neugebauer, J., Veldstra, J. & Buda, F. (2011). J. Phys. Chem. B, 115, 3216–3225. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Polívka, T., Frank, H. A., Enriquez, M. M., Niedzwiedzki, D. M., Liaaen-Jensen, S., Hemming, J., Helliwell, J. R. & Helliwell, M. (2010). J. Phys. Chem. B, 114, 8760–8769. [DOI] [PubMed]
- Quarmby, R., Norden, D. A., Zagalsky, P. F., Ceccaldi, H. J. & Daumas, R. (1977). Comp. Biochem. Physiol. B Comp. Biol. 56, 55–61. [DOI] [PubMed]
- Rhys, N. H., Wang, M.-C., Jowitt, T. A., Helliwell, J. R., Grossmann, J. G. & Baldock, C. (2011). J. Synchrotron Rad. 18, 79–83. [DOI] [PMC free article] [PubMed]
- Roessle, M. W., Klaering, R., Ristau, U., Robrahn, B., Jahn, D., Gehrmann, T., Konarev, P., Round, A., Fiedler, S., Hermes, C. & Svergun, D. (2007). J. Appl. Cryst. 40, s190–s194.
- Semenyuk, A. V. & Svergun, D. I. (1991). J. Appl. Cryst. 24, 537–540.
- Stepanyan, R., Day, K., Urban, J., Hardin, D. L., Shetty, R. S., Derby, C. D., Ache, B. W. & McClintock, T. S. (2006). Physiol. Genomics, 25, 224–233. [DOI] [PubMed]
- Strambi, A. & Durbeej, B. (2009). J. Phys. Chem. B, 113, 5311–5317. [DOI] [PubMed]
- Svergun, D. I. (1997). J. Appl. Cryst. 30, 792–797.
- Svergun, D. I., Petoukhov, M. V. & Koch, M. H. J. (2001). Biophys. J. 80, 2946–2953. [DOI] [PMC free article] [PubMed]
- Wade, N. M., Tollenaere, A., Hall, M. R. & Degnan, B. M. (2009). Mol. Biol. Evol. 26, 1851–1864. [DOI] [PubMed]
- Wald, G., Nathanson, N., Jenks, W. & Tarr, E. (1948). Biol. Bull. Mar. Biol. 95, 249–250. [PubMed]
- Wijk, A. A. van, Spaans, A., Uzunbajakava, N., Otto, C., de Groot, H. J., Lugtenburg, J. & Buda, F. (2005). J. Am. Chem. Soc. 127, 1438–1445. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Yang, F., Wang, M.-R., Ma, Y.-G., Ma, W.-M. & Yang, W.-J. (2011). J. Exp. Zool. A Ecol. Genet. Physiol. 315, 562–571. [DOI] [PubMed]
- Zagalsky, P. F. (1985). Methods Enzymol, 111, 216–247. [DOI] [PubMed]
- Zagalsky, P. F. & Tidmarsh, M.-L. (1985). Comp. Biochem. Physiol. B Comp. Biol. 80, 599–601.
- Zanotti, G., Ottonello, S., Berni, R. & Monaco, H. L. (1993). J. Mol. Biol. 230, 613–624. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: 4alo
Supplementary material file. DOI: 10.1107/S1744309112026103/hv5216sup1.pdf