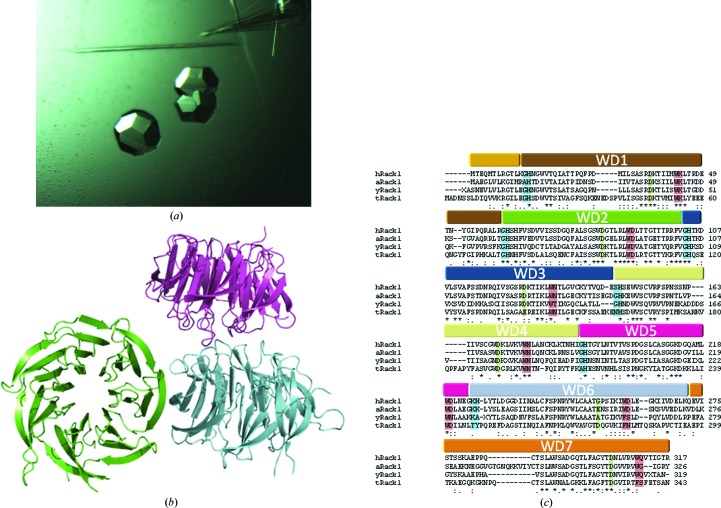
The crystal structure of human Rack1 protein is reported at 2.45 Å resolution.
Keywords: Rack1, WD repeat, β-propeller, protein kinase C
Abstract
The crystal structure of human receptor for activated C-kinase 1 (hRack1) protein is reported at 2.45 Å resolution. The crystals belongs to space group P41212, with three molecules per asymmetric unit. The hRack1 structure features a sevenfold β-propeller, with each blade housing a sequence motif that contains a strictly conserved Trp, the indole group of which is embedded between adjacent blades. In blades 1–5 the imidazole group of a His residue is wedged between the side chains of a Ser residue and an Asp residue through two hydrogen bonds. The hRack1 crystal structure forms a starting basis for understanding the remarkable scaffolding properties of this protein.
1. Introduction
Receptor for activated C-kinase 1 (Rack1) is a 37 kDa protein (UniProt P63244) that is ubiquitously expressed in mammals (Chou et al., 1999 ▶) and is involved in cell signalling (Ron et al., 1999 ▶), growth (Hermanto et al., 2002 ▶) and development (McLeod et al., 2000 ▶). Rack1 has a cytosolic (Yarwood et al., 1999 ▶) and nuclear (Ron et al., 2000 ▶) localization and is devoid of enzymatic activity. Rack1 has drawn considerable attention since its first description as a binding partner of the active form of protein kinase C (PKC). Indeed, the main feature of Rack1 is its scaffolding ability, which promotes the formation of supra-macromolecular assemblies (Adams et al., 2011 ▶). The anchoring capacity of the Rack1 protein has been extensively documented and, apart from PKC (Stebbins & Mochly Rosen, 2001 ▶), its list of more than 80 binding partners includes the cyclic AMP-specific phosphodiesterase PDE4D5 (Yarwood et al., 1999 ▶) and the Src tyrosine kinase (Yaka et al., 2003 ▶; Mamidipudi et al., 2004 ▶). In addition, Rack1 can act as an enzymatic cofactor and as a cellular shuttle for its partners (Ron et al., 1999 ▶).
Rack1 belongs to the family of tryptophan–aspartate (WD)-repeat-containing proteins (Xu & Min, 2011 ▶; Steele et al., 2001 ▶). These proteins comprise polypeptide stretches of 40–60 amino acids in which each repeat folds into a four-stranded antiparallel β-blade (Xu & Min, 2011 ▶). The WD-repeat sequence includes a Gly-His dipeptide at its N-terminal end and a Trp-Asp dipeptide at its C-terminus. However, the repeat signature sequence is not strictly conserved and the Trp residue can be substituted by tyrosine or phenylalanine (Xu & Min, 2011 ▶).
The seven WD repeats that are present in hRack1 (Fig. 1 ▶ c) have been predicted to build up a β-propeller tertiary structure (Steele et al., 2001 ▶). β-Propeller proteins are found in all organisms and may or may not be endowed with enzymatic activity (Paoli, 2001 ▶; Xu & Min, 2011 ▶). Their shape resembles a cylinder with a channel that traverses the molecule in a direction parallel to its symmetry axis and their structural stability arises from extensive hydrophobic intramolecular interactions that are established between adjacent β-blades. In some β-propellers additional stability is provided by the presence of a ‘velcro’ motif that tethers the toroidal structure by joining the N- and C-terminal ends of the protein in the same β-blade.
Figure 1.
(a) Crystals of hRack1. (b) Content of the asymmetric unit, with the three different hRack1 molecules coloured cyan, green and purple. (c) Sequence alignment of the Rack1 orthologues for which structures have been reported: hRack1, human (this work); yRack1, yeast; aRack1, Arabidopsis thaliana; tRack1, Tetrahymena thermophila. See text for details. The seven WD domains found in hRack1 are highlighted in different colours. Conserved residues that typically define the primary sequence of WD domains are shown in red for the WD dipeptide, in blue for the GH dipeptide, in green for aspartate residues and in yellow for serine residues.
The crystal structures of several orthologues of Rack1 have been reported, including those from Saccharomyces cerevisiae (yRack1; Yatime et al., 2011 ▶; Coyle et al., 2009 ▶), Arabidopsis thaliana (aRack1; Ullah et al., 2008 ▶) and Tetrahymena thermophila (tRack1; Rabl et al., 2011 ▶). In addition, structural information for Rack1 embedded in the eukaryotic ribosome from the plant Triticum aestivum has been reported using electron microscopy (Armache et al., 2010 ▶; Chandramouli et al., 2008 ▶). Interestingly, yRack1 lacks the stabilizing ‘velcro’ motif and its molecular dimerization is achieved via the formation of an intermolecular β-sheet that involves the N- and C-termini of the protein. The structure of tRack1 has been solved in the context of the eukaryotic 40S ribosome subunit, shedding light on how Rack1 can play a significant role in protein translation and also how it can interact with its binding partners (Rabl et al., 2011 ▶). With a view to determining the molecular basis of the ability of hRack1 to act as a scaffolding protein, we report its crystal structure at a resolution of 2.45 Å.
2. Materials and methods
2.1. Cloning, expression and purification
The cDNA of hRack1 was cloned into the pNIC28-Bsa4 vector with forward primer TACTTCCAATCCATGATGACTGAGCAGATGACCCT and reverse primer TATCCACCTTTACTGTCAGCGTGTGCCAATGGTCACC using ligation-independent cloning (Gräslund et al., 2008 ▶), yielding a construct with an N-terminal hexahistidine tag followed by a TEV protease cleavage site. Protein expression conducted in Escherichia coli BL21 (DE3) Rosetta T1R cells in the presence of kanamycin and chloramphenicol was initiated at 310 K by inoculating 1 l Terrific broth with a 2%(v/v) inoculum until an OD600 of 0.6–0.8 was reached, at which point the temperature was decreased to 289 K. After 30 min, isopropyl β-d-1-thiogalactopyranoside (Affymetrix) was added to the culture to a final concentration of 500 µM. After overnight incubation, cells were harvested at 4000g for 10 min and resuspended in 100 mM HEPES, 500 mM NaCl, 10 mM imidazole, 10%(v/v) glycerol, 0.5 mM tris(2-carboxyethyl)phosphine (TCEP) pH 8.0. Pellets stored at 193 K were thawed in an iced-water bath, and 25 µl EDTA-free protease-inhibitor cocktail (Calbiochem) and 10 mg lysozyme were added to the thawed cell resuspension. Sonication was performed for 10 min in cycles of 5 s pulse followed by 5 s delay at 30% amplitude (Sonics Vibra-cell). Cells debris was discarded following centrifugation at 47 000g for 30 min at 277 K. The supernatant was filtered with a 1.2 µm pore syringe filter and subsequently loaded onto a 1 ml Ni–NTA Superflow column (Qiagen). Proteins were eluted using a 20–500 mM imidazole gradient in a buffer consisting of 20 mM HEPES, 500 mM NaCl, 10%(v/v) glycerol, 0.5 mM TCEP pH 7.5. The eluted fractions were pooled, concentrated using a 30 kDa cutoff concentrator (Vivascience) and loaded onto a preparative gel-filtration column (HiLoad 16/60 Superdex 200 prep-grade; GE Healthcare) pre-equilibrated with 20 mM HEPES, 300 mM NaCl, 10%(v/v) glycerol, 0.5 mM TCEP pH 7.5. hRack1 eluted as a single peak of apparent molecular mass 37 kDa and the identity of the protein was confirmed by mass spectrometry and SDS–PAGE. The protein, which still contained the hexahistidine tag, was concentrated to 8 mg ml−1, flash-cooled in liquid nitrogen and stored at 193 K until use.
2.2. Crystallization
Initial crystal hits were found using the JCSG+ screen from Qiagen. After optimization, crystals of about 100–300 µm in size (Fig. 1 ▶ a) were grown at 291 K using the hanging-drop technique in 100 mM CHES pH 9.5, 20%(w/v) PEG 8000 by mixing 1 µl reservoir buffer with 2 µl protein solution at a concentration of 8 mg ml−1. Crystals appeared after overnight incubation. Crystals were mounted in a nylon loop, briefly soaked in a cryoprotecting solution consisting of the reservoir solution with 25%(v/v) glycerol and flash-cooled in liquid nitrogen.
2.3. Data collection and processing and model building
Data were collected on the PXIII beamline at the Swiss Light Source at a wavelength of 1.00 Å using 2 s exposure time. Images were collected over a total angular range of 120° using a 1° oscillation range with a 220 mm distance between the crystal and the detector. The crystal mosaicity was 0.29°. The data were processed using the CCP4 suite (Winn et al., 2011 ▶) and XDS (Kabsch, 2010 ▶). The structure was solved by molecular replacement with Phaser using the structure of aRack1 (PDB entry 3dm0; Ullah et al., 2008 ▶) as the search probe after the sequence of hRack1 had been inserted using the program CHAINSAW. The molecular-replacement solution was used for iterative cycles of model building and structure refinement using Coot (Emsley & Cowtan, 2004 ▶), REFMAC5 (Murshudov et al., 2011 ▶) and autoBUSTER (Bricogne et al., 2011 ▶). A total of 5% of the structure-factor amplitudes were excluded from refinement for R free calculations. Throughout refinement, noncrystallographic restraints were applied between the three independent molecules within the asymmetric unit using tight restraints for the main-chain atoms and medium restraints for the side-chains atoms. In the last stages of refinement, translation/libration/screw motion (TLS) refinement was introduced, breaking each of the three molecules into two groups: residues Glu3–Thr141 and Val142–Ile314 for molecules A and B and residues Glu3–Ser146 and His147–Ile314 for molecule C. These groups were assigned using the TLS Motion Determination server (Painter & Merritt, 2006 ▶). The model quality was assessed using PROCHECK (Winn et al., 2011 ▶). Electrostatic surfaces were calculated and displayed with Coot (Emsley & Cowtan, 2004 ▶). The coordinates and structure factors have been deposited in the PDB with access code 4aow.
3. Results and discussion
3.1. Overall structure
Data-collection and refinement statistics are given in Table 1 ▶. The model (Fig. 2 ▶) includes residues Glu3–Ile314 of hRack1 from a total of 317 amino acids. The electron-density map is well defined except for a short stretch between residues Gln273 and Glu282 and several flexible exposed side chains.
Table 1. Data-collection and refinement statistics.
| Source | PXIII beamline, Swiss Light Source |
| Wavelength (Å) | 1.00 |
| Space group | P41212 |
| Unit-cell parameters (Å) | a = b = 134.3, c = 135.2 |
| Molecules in the asymmetric unit | 3 |
| Resolution range (Å) | 47.47–2.45 (2.59–2.45) |
| No. of observed reflections | 421161 (47700) |
| No. of unique reflections | 45262 (6031) |
| Completeness (%) | 98.8 (92.1) |
| Multiplicity | 9.3 (7.9) |
| Wilson B factor (Å2) | 37 |
| 〈I/σ(I)〉 | 15.4 (2.5) |
| R sym † | 0.108 (0.751) |
| R work/R free ‡ | 0.1934/0.2156 |
| R.m.s.d. bonds (Å) | 0.010 |
| R.m.s.d. angles (°) | 1.18 |
| Overall B factors (Å2) | |
| Protein | |
| Chain A | 41.4 |
| Chain B | 46.5 |
| Chain C | 49.2 |
| Water molecules (185) | 43.7 |
| Glycerol (2) | 57.2, 50.8 |
| Ramachandran plot (%) | |
| Favoured | 96.8 |
| Allowed | 3.2 |
| Disallowed | 0.0 |
| PDB code | 4aow |
R
sym = 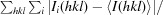
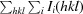 , where I
i(hkl) is the intensity of the ith observation of reflection hkl and 〈I(hkl)〉 is the average over all observations of reflection hkl.
, where I
i(hkl) is the intensity of the ith observation of reflection hkl and 〈I(hkl)〉 is the average over all observations of reflection hkl.
R
work = 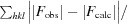
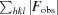 , where F
obs and F
calc are the observed structure-factor amplitudes and the calculated structure-factor amplitudes of the model, respectively. R
free is the same as R
work but for 5% of reflections not included in the model refinement.
, where F
obs and F
calc are the observed structure-factor amplitudes and the calculated structure-factor amplitudes of the model, respectively. R
free is the same as R
work but for 5% of reflections not included in the model refinement.
Figure 2.
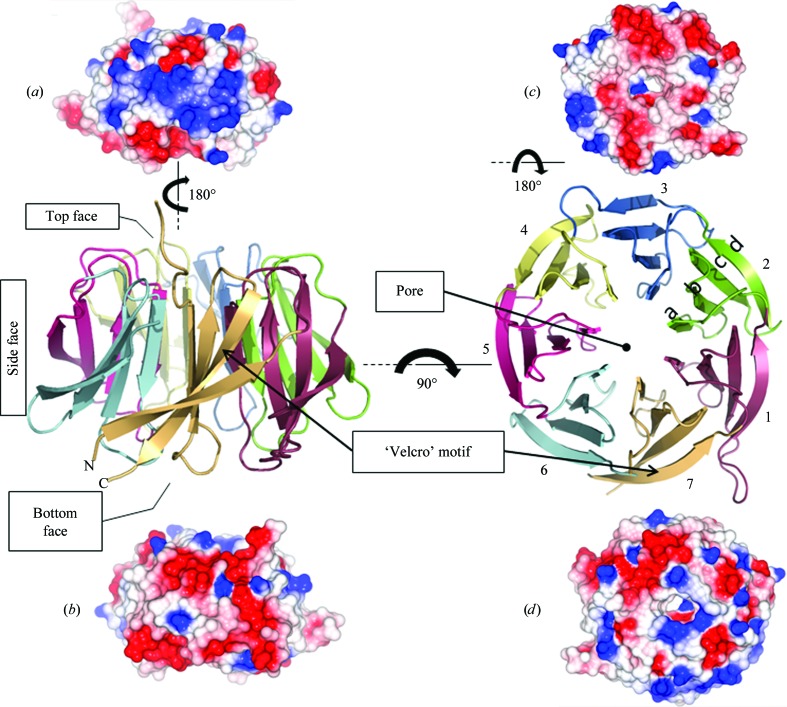
Views of the hRack1 crystal structure. Central panel: cartoon representations of hRack1 viewed from the side (left) and from the top (right). The locations of the protein pore and the ‘velcro’ motif are indicated. Each β-sheet or blade is numbered sequentially from the N-terminus of the protein and their β-strands are labelled a, b, c, and d starting from the inside of the propeller near the pore. The electrostatic surface of hRack1 is shown from four different angles. Side surfaces, bottom and top views are displayed in (a) (blades 1–3), (b) (blades 5–7), (c) and (d), respectively.
The structure features the expected sevenfold β-propeller architecture, with an overall shape that resembles a cylinder (Fig. 2 ▶) of approximate dimensions 50 Å in diameter and 30 Å in width. One face comprises the N- and C-termini (‘bottom face’). The seven β-sheets are arranged around a central axis and pack against one another such that a channel with a diameter of ∼7 Å runs through the propeller. The asymmetric unit contains three hRack1 molecules (Fig. 1 ▶ b). Two hRack1 molecules are stacked on top of each other with their central axes parallel but offset by approximately 15 Å and a rotation of ∼90°. The ‘bottom face’ of one hRack1 molecule makes contact with the ‘top’ surface of the adjacent molecule and this interaction buries a surface area of 571 Å2. In contrast, the axis of the third independent hRack monomer runs approximately perpendicular to the other two molecules (Fig. 1 ▶ b). Each β-sheet (1–7) shows the same basic architecture comprising four antiparallel strands labelled a, b, c and d from the inner side of the propeller to the outer side. The residues of the inwards-facing a β-strands display the lowest temperature factors, while the residues located on exposed loops, especially the loop spanning residues 266–268, show the highest values. The β-sheet is twisted, with an angle of approximately 75° between the inner and outer β-strands (Figs. 2 ▶ and 3 ▶ a). The canonical ‘velcro’ motif is present in the seventh blade and connects the N- and C-termini of the polypeptide chain. Here, unlike in the structure of yRack1 (Yatime et al., 2011 ▶), the ‘velcro’ motif does not participate in intermolecular interactions but stabilizes the overall structure as an interlocking device. The electrostatic surface of the hRack1 monomer is predominantly acidic on one side of the propeller (Fig. 2 ▶ c), while the opposite ‘top’ side is more basic, particularly in the vicinity of the entrance to the central pore (Fig. 2 ▶ d). Furthermore, electrostatic clustering is observed on the side surfaces of the molecule, with either predominantly acidic or basic areas. Thus, an acidic patch is located between blades 5 and 7 (Fig. 2 ▶ b) and a basic patch is located between blades 1 and 4 (Fig. 2 ▶ a).
Figure 3.
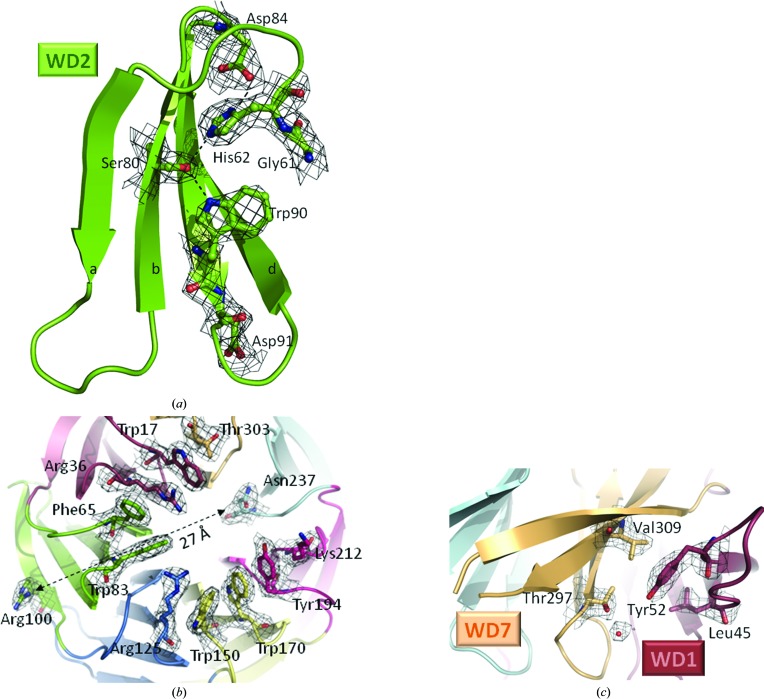
(a) Close-up view of the WD2 motif shown in cartoon representation, with the conserved residues Gly61, His62, Ser80, Asp84, Trp90 and Asp91 shown as sticks and spheres and with the electron-density map overlaid. (b) Close-up view of the ‘top side’ of hRack1 shown as a cartoon representation and as a stick representation for residues involved in the hydrophobic ring with the electron-density map overlaid. The arrow indicates the separation between two residues (Arg100 and Asn237) involved in the binding of the same substrate, PKCβ. (c) Close-up view showing the Tyr52 phosphorylation site wedged between WD7 (pale brown) and WD1 (red) overlaid with the electron-density map. Maps were calculated with 2F o − F c coefficients and phases from the refined model at a level of 1.2σ.
3.2. The WD repeats
The indole side chains of the strictly conserved Trp residues from the WD repeat (Fig. 1 ▶ c) are embedded in the hydrophobic interstices between blades, making contacts with the side chains of a leucine and an isoleucine residue from the preceding blade and with hydrophobic residues projecting from the same blade (Figs. 1 ▶ c and 3 ▶ a). In addition, in blades 1–5 the Trp side chain is stabilized by a hydrogen bond to a serine residue situated at position −10 compared with the Trp, whilst the Trp side-chain orientation differs in blades 6 and 7, in which the serine is absent (Figs. 1 ▶ c and 3 ▶ a). In contrast, the Asp residue from the WD motif is not conserved: it is substituted by Asn in blades 3 and 4, by Lys in the first blade and Gln in the seventh blade (Fig. 1 ▶ c). The corresponding side chains are exposed to the solvent and their main-chain carbonyl accepts a hydrogen bond from the nitrogen amide of the adjacent β-strand. The GH motif present in blades 1, 2, 3 and 5 is substituted by SH in blade 4 and by KH in blade 6 (Fig. 1 ▶ c). This dipeptide is located in the loop that connects two adjacent blades. The imidazole ring makes two hydrogen bonds to the carboxylic moiety of a conserved Asp residue located six residues upstream of the WD motif and to the serine residue situated at position −10 compared with the Trp (Figs. 1 ▶ c and 3 ▶ a).
3.3. Comparison with Rack1 homologues
The structures of Rack1 from S. cerevisiae (yRack1; Yatime et al., 2011 ▶), A. thaliana (aRack1; Ullah et al., 2008 ▶) and T. thermophila (tRack1; Rabl et al., 2011 ▶) show good agreement with that of hRack1. Nevertheless, significant differences exist, mainly in the conformation of the interconnecting loops between the a and b strands of the first and fourth blades. The loop that connects blades 6 and 7, which is variable in length and sequence between the various Rack orthologues (Fig. 1 ▶ c), is not visible in the electron-density map of hRack1. This structural flexibility is probably used by Rack1 to bind different protein partners. The structures of hRack1 and aRack1 show the highest similarity, with an r.m.s.d. of 0.88 Å for 300 superimposed Cα atoms. The comparison with the yRack1 structure stresses the fact that the mechanism used by the yeast protein to achieve dimerization is not found in the human protein. It has been shown that phosphorylation of Ser146 leads to hRack1 dimerization (Liu et al., 2007 ▶), suggesting that a dimeric form of hRack1 may involve blades 3 and 4 (Fig. 2 ▶) at the interface.
3.4. Protein sites prone to macromolecular interactions
Yeast two-hybrid screening, mutagenesis and peptide-mapping studies helped to draw a possible map of the binding sites of Rack1 partners. The extraordinary scaffolding attributes of proteins containing the WD repeat allow them to recruit various substrates using either overlapping or distinct binding modes (Xu & Min, 2011 ▶). Thus, while tRack1 establishes an extensive network of contacts with the rpS17e ribosome subunit (PDB entries 2xzm and 2xzn; Rabl et al., 2011 ▶) via its ‘top side’, its interaction with the rpS3e ribosome subunit is mainly formed with the side surface of the propeller in the fifth blade region. Interestingly, 60% of the Trp residues of hRack1 that do not belong to WD repeats are clustered on the top face of the propeller, thus forming a possible binding site for protein ligands (Fig. 3 ▶ b). This feature is also present in other Rack1 orthologues, e.g. those from S. cerevisiae (Yatime et al., 2011 ▶; PDB entry 3rfg) and A. thaliana (Ullah et al., 2008 ▶; PDB entry 3dm0). This binding surface is characterized by a hydrophobic ring lined by Trp17, Trp83, Trp150 and Trp170 together with Phe67 and Tyr194, with the side chains of Arg36, Arg125 and Lys212 forming cation–π interactions with the hydrophobic residues (Zacharias & Dougherty, 2002 ▶). The peptide 234-DIINALCF-241 is able to compete with the complete Rack1 protein for PKCβ binding and can activate PKCβ both in vivo and in vitro (Ron & Mochly Rosen, 1994 ▶; Ron et al., 1995 ▶). The peptide 234-DIINALCF-241 encompasses β-strand a of blade 6 as well as the preceding inter-blade connecting loop. In the hRack1 structure residues Asp234, Ile235 and Asn237 are exposed to the solvent and thus accessible for protein–protein interactions, while residues 236 and 238–241 are buried. Likewise, the peptide 99-RRFVGHTKDV-108 has been shown to disrupt the association between Rack1 and PKCβ (Grosso et al., 2008 ▶). In this case, this polypeptide stretch belongs to β-strand d from blade 2, with two arginine residues largely exposed, and the inter-blade loop that joins blades 2 and 3. Regions 99–100 and 234–237 both belong to the exposed ‘top side’ of the β-propeller of hRack1 (Fig. 3 ▶ b), indicating that this surface may be involved in the association between hRack1 and PKCβ. However, these two binding sites for PKCβ predicted from the peptide-mapping studies are distant from each other on the surface of hRack1 (Fig. 3 ▶ b), suggesting the existence of multiple discontinuous contact sites.
Other known binding partners of Rack1 are PDE4D5 (Yarwood et al., 1999 ▶) and the cytoplasmic tails of β-integrins. Interestingly, both share WD repeats 5, 6 and 7 as their Rack1 binding site (McCahill et al., 2002 ▶). The loop connecting blades 6 and 7 of hRack1 is not visible, indicating a certain degree of flexibility that may be required to allow attachment to its different binding partners. Indeed, Rack1 is able to simultaneously bind proteins that target overlapping binding sites (Thornton et al., 2004 ▶; Yaka et al., 2002 ▶) and flexible loops appear as a solution to establish promiscuous intermolecular contacts. Moreover, post-translational modifications of Rack1, including its phosphorylation, are known to influence substrate binding. Tyr246 and Tyr228 are phosphorylated by the Src kinase (Chang et al., 2001 ▶, 2002 ▶). These two residues are next to each other at the edge of blades 5 and 6 and are exposed to the solvent. Phosphorylation of Tyr246 is required for Src binding, suggesting that Src attachment and Rack1 phosphorylation are concomitant events. In this case, Tyr246 phosphorylation is expected to alter the conformation of the ab loop of blade 6. Likewise, phosphorylation of Tyr52 mediates the interaction between Rack1 and FAK (Kiely et al., 2009 ▶), and modification of Tyr302 of Rack1 mediates the binding to PP2A and β-integrins (Kiely et al., 2006 ▶, 2008 ▶). Interestingly, in the nonphosphorylated hRack1 protein residue Tyr52 is buried between blades 1 and 7, making hydrophobic contacts with Val309 and Leu45 (Fig. 3 ▶ c). Moreover, Arg8 and Arg47 shield access of Tyr52 to the solvent. Thus, phosphorylation of Tyr52 is likely to involve local alterations in the protein conformation that could propagate to the cd and ab loops of blades 1 and 7.
Supplementary Material
PDB reference: human Rack1, 4aow
Acknowledgments
We would like to thank Drs Vincent Olieric and Meitian Wang for their expert support during data acquisition at the Swiss Light Source. The laboratory of JL is supported by grants (08/1/22/19/589) from the BMRC and NRF CRP4-20082.
References
- Adams, D. R., Ron, D. & Kiely, P. A. (2011). Cell Commun. Signal. 9, 22. [DOI] [PMC free article] [PubMed]
- Armache, J.-P. et al. (2010). Proc. Natl Acad. Sci. USA, 107, 19754–19759.
- Bricogne, G., Blanc, E., Brandl, M., Flensburg, C., Keller, P., Paciorek, W., Roversi, P, Sharff, A., Smart, O. S., Vonrhein, C. & Womack, T. O. (2011). BUSTER v.2.8.0. Cambridge: Global Phasing Ltd.
- Chandramouli, P., Topf, M., Ménétret, J.-F., Eswar, N., Cannone, J. J., Gutell, R. R., Sali, A. & Akey, C. W. (2008). Structure, 16, 535–548. [DOI] [PMC free article] [PubMed]
- Chang, B. Y., Chiang, M. & Cartwright, C. A. (2001). J. Biol. Chem. 276, 20346–20356. [DOI] [PubMed]
- Chang, B. Y., Harte, R. A. & Cartwright, C. A. (2002). Oncogene, 21, 7619–7629. [DOI] [PubMed]
- Chou, Y.-C., Chou, C.-C., Chen, Y.-K., Tsai, S., Hsieh, F. M. J., Liu, H. J. & Hseu, T.-H. (1999). Biochim. Biophys. Acta, 1489, 315–322. [DOI] [PubMed]
- Coyle, S. M., Gilbert, W. V. & Doudna, J. A. (2009). Mol. Cell. Biol. 29, 1626–1634. [DOI] [PMC free article] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Gräslund, S. et al. (2008). Nature Methods, 5, 135–146.
- Grosso, S., Volta, V., Sala, L. A., Vietri, M., Marchisio, P. C., Ron, D. & Biffo, S. (2008). Biochem. J. 415, 77–85. [DOI] [PubMed]
- Hermanto, U., Zong, C. S., Li, W. & Wang, L.-H. (2002). Mol. Cell. Biol. 22, 2345–2365. [DOI] [PMC free article] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 133–144. [DOI] [PMC free article] [PubMed]
- Kiely, P. A., Baillie, G. S., Barrett, R., Buckley, D. A., Adams, D. R., Houslay, M. D. & O’Connor, R. (2009). J. Biol. Chem. 284, 20263–20274. [DOI] [PMC free article] [PubMed]
- Kiely, P. A., Baillie, G. S., Lynch, M. J., Houslay, M. D. & O’Connor, R. (2008). J. Biol. Chem. 283, 22952–22961. [DOI] [PubMed]
- Kiely, P. A., O’Gorman, D., Luong, K., Ron, D. & O’Connor, R. (2006). Mol. Cell. Biol. 26, 4041–4051. [DOI] [PMC free article] [PubMed]
- Liu, Y. V., Hubbi, M. E., Pan, F., McDonald, K. R., Mansharamani, M., Cole, R. N., Liu, J. O. & Semenza, G. L. (2007). J. Biol. Chem. 282, 37064–37073. [DOI] [PMC free article] [PubMed]
- Mamidipudi, V., Zhang, J., Lee, K. C. & Cartwright, C. A. (2004). Mol. Cell. Biol. 24, 6788–6798. [DOI] [PMC free article] [PubMed]
- McCahill, A., Warwicker, J., Bolger, G. B., Houslay, M. D. & Yarwood, S. J. (2002). Mol. Pharmacol. 62, 1261–1273. [DOI] [PubMed]
- McLeod, M., Shor, B., Caporaso, A., Wang, W., Chen, H. & Hu, L. (2000). Mol. Cell. Biol. 20, 4016–4027. [DOI] [PMC free article] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Painter, J. & Merritt, E. A. (2006). Acta Cryst. D62, 439–450. [DOI] [PubMed]
- Paoli, M. (2001). Prog. Biophys. Mol. Biol. 76, 103–130. [DOI] [PubMed]
- Rabl, J., Leibundgut, M., Ataide, S. F., Haag, A. & Ban, N. (2011). Science, 331, 730–736. [DOI] [PubMed]
- Ron, D., Jiang, Z., Yao, L., Vagts, A., Diamond, I. & Gordon, A. (1999). J. Biol. Chem. 274, 27039–27046. [DOI] [PubMed]
- Ron, D., Luo, J. & Mochly Rosen, D. (1995). J. Biol. Chem. 270, 24180–24187. [DOI] [PubMed]
- Ron, D. & Mochly Rosen, D. (1994). J. Biol. Chem. 269, 21395–21398. [PubMed]
- Ron, D., Vagts, A. J., Dohrman, D. P., Yaka, R., Jiang, Z., Yao, L., Crabbe, J., Grisel, J. E. & Diamond, I. (2000). FASEB J. 14, 2303–2314. [DOI] [PubMed]
- Stebbins, E. G. & Mochly Rosen, D. (2001). J. Biol. Chem. 276, 29644–29650. [DOI] [PubMed]
- Steele, M. R., McCahill, A., Thompson, D. S., MacKenzie, C., Isaacs, N. W., Houslay, M. D. & Bolger, G. B. (2001). Cell. Signal. 13, 507–513. [DOI] [PubMed]
- Thornton, C., Tang, K.-C., Phamluong, K., Luong, K., Vagts, A., Nikanjam, D., Yaka, R. & Ron, D. (2004). J. Biol. Chem. 279, 31357–31364. [DOI] [PubMed]
- Ullah, H., Scappini, E. L., Moon, A. F., Williams, L. V., Armstrong, D. L. & Pedersen, L. C. (2008). Protein Sci. 17, 1771–1780. [DOI] [PMC free article] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Xu, C. & Min, J. (2011). Protein Cell, 2, 202–214. [DOI] [PMC free article] [PubMed]
- Yaka, R., Phamluong, K. & Ron, D. (2003). J. Neurosci. 23, 3623–3632. [DOI] [PMC free article] [PubMed]
- Yaka, R., Thornton, C., Vagts, A. J., Phamluong, K., Bonci, A. & Ron, D. (2002). Proc. Natl Acad. Sci. USA, 99, 5710–5715. [DOI] [PMC free article] [PubMed]
- Yarwood, S. J., Steele, M. R., Scotland, G., Houslay, M. D. & Bolger, G. B. (1999). J. Biol. Chem. 274, 14909–14917. [DOI] [PubMed]
- Yatime, L., Hein, K. L., Nilsson, J. & Nissen, P. (2011). J. Mol. Biol. 411, 486–498. [DOI] [PubMed]
- Zacharias, N. & Dougherty, D. A. (2002). Trends Pharmacol. Sci. 23, 281–287. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: human Rack1, 4aow