Xylosidase C from Thermoanaerobacterium saccharolyticum, the first enzyme classified as GH120 family, has been successfully crystallized. The crystal diffracted to a resolution of 2.2 Å.
Keywords: thermophilic enzymes, xylan, β-xylosidases
Abstract
Xylosidases hydrolyze xylopolymers at the nonreducing end to free xylose units. The β-xylosidase (XylC) from Thermoanaerobacterium saccharolyticum JW/SL-YS485 was expressed in Escherichia coli and the recombinant protein was purified and crystallized. A BLASTP search with the XylC protein sequence showed that no similar structure had previously been solved. XylC was classified as a member of the new glycoside hydrolase family GH120 according to the CAZy website (http://www.cazy.org/). Crystals belonging to the monoclinic space group P21, with unit-cell parameters a = 88.36, b = 202.20, c = 99.87 Å, β = 99.04°, were obtained by the sitting-drop vapour-diffusion method and diffracted to 2.2 Å resolution. Structure determination using MIR and MAD methods is in progress.
1. Introduction
Xylans are the most abundant heteropolymeric hemicellulose polysaccharides in the plant cell wall and can be efficiently enzymatically hydrolyzed into their sugar constituents (e.g. xylose; Saha, 2003 ▶). The complete degradation of xylan requires a number of xylanolytic enzymes with various activities because of the structural complexity of xylan. In the last step, β-d-xylosidase (EC 3.2.1.37) hydrolyzes short xylooligosaccharides which are produced by other xylanolytic enzymes and releases xylose as the final product (Biely, 1985 ▶).
Three xylosidases (XylA, XylB and XylC) from Thermoanaerobacterium saccharolyticum JW/SL-YS485 have been purified and characterized (Lorenz & Wiegel, 1997 ▶; Shao et al., 2011 ▶). T. saccharolyticum is a highly xylanolytic anaerobic thermophile that grows over a broad range of pH (pH 3.85–6.35) and temperature (303–339 K) and that can utilize xylan as a sole carbon source. XylC has drawn much attention because it shows weaker product (xylose) inhibition compared with other enzymes (Shao et al., 2011 ▶) and also because no similar structure has been solved. BLASTP searches of XylC against the nonredundant protein-sequence database showed that XylC is totally different from any other corresponding glycosyl hydrolases in the CAZy database. Thus, XylC has recently been classified into a new glycohydrolase family (GH120; http://www.cazy.org/). Consequently, it is important to solve its crystal structure in order to better understand this novel GH family. Furthermore, structural analysis will provide additional information for modifying the enzyme for use in biotechnological applications such as the remediation of waste streams, bioethanol production and the pulp and paper industries.
2. Materials and methods
2.1. Protein preparation
Expression and purification of XylC protein followed the procedure described previously by Shao et al. (2011 ▶). Briefly, the XylC gene (GenBank accession No. EF193646) was constructed in pHsh (Wu et al., 2010 ▶), which was electrotransformed into Escherichia coli JM109 cells using a Gene Pulser (Bio-Rad). For overexpression of the recombinant xylosidase, E. coli JM109 cells harbouring pHsh-xylC were grown with vigorous aeration at 303 K in LB broth containing ampicillin (100 µg ml−1) until an optical density at 600 nm of 0.6–0.8 was reached. XylC expression was induced at a higher temperature of 315 K for 8 h. The recombinant XylC protein was purified using the following procedures: (i) the E. coli JM109 lysate was heated at 343 K in 50 mM sodium phosphate buffer pH 6.8 for 40 min, (ii) the supernatant was applied onto a DEAE-Sepharose column (2.5 × 56 cm; GE Heathcare Biosciences) for ion-exchange chromatography and (iii) the XylC protein was precipitated by adding ammonium sulfate to 70% saturation and then dissolved in and dialyzed against 25 mM Tris–HCl buffer pH 7.2. The yield was about 6 mg per litre of culture after purification and the purity was checked by SDS–PAGE analysis (>95% purity; Shao et al., 2011 ▶). The tag-free XylC was detected as a single band with a molecular mass of about 73 kDa. Based on the results of native gel electrophoresis (265 kDa), the native XylC is probably a tetramer (Shao et al., 2011 ▶). Expression of selenomethionine-substituted (SeMet) XylC was conducted based on the previous method of Van Duyne et al. (1993 ▶) and the purification procedures were the same as those used for the wild-type enzyme. The yield of SeMet XylC was about 2 mg per litre of culture after purification. The proteins (73 kDa; amino acids 1–638) were concentrated to 15 mg ml−1 in 25 mM Tris–HCl, 150 mM NaCl pH 7.5 using an Amicon Ultra-15 Centricon (Millipore).
2.2. Crystallization and data collection
Initial crystallization screening was performed manually using 768 different reservoir conditions from Hampton Research (Laguna Niguel, California, USA) kits, including Crystal Screen, Crystal Screen 2, Crystal Screen Cryo, Crystal Screen Lite, MembFac, Natrix, Index, SaltRx, SaltRx 2, PEG/Ion, PEG/Ion 2, Quick Screen and Grid Screens (Ammonium Sulfate, MPD, Sodium Chloride, Sodium Malonate, PEG 6000 and PEG/LiCl), and the sitting-drop vapour-diffusion method. 2 µl XylC solution (in 25 mM Tris–HCl, 150 mM NaCl pH 7.5) was mixed with 2 µl reservoir solution in 24-well Cryschem plates (Hampton Research) and equilibrated against 300 µl reservoir solution at 295 K. Initial crystals of XylC were obtained within 2 d using Index condition No. 94 [0.2 M sodium citrate tribasic dehydrate pH 5.6, 20%(w/v) polyethylene glycol 3350]. The optimized crystallization condition used to obtain the crystals described here was 0.2 M sodium citrate tribasic dehydrate pH 5.6, 15–17%(w/v) polyethylene glycol 3350. The crystals reached dimensions of about 0.45 × 0.1 × 0.1 mm within 2–3 d. An X-ray diffraction data set was collected to 2.2 Å resolution on beamline BL13B1 at the National Synchrotron Radiation Research Center (NSRRC), Hsinchu, Taiwan. The diffraction images were processed using the program HKL-2000 (Otwinowski & Minor, 1997 ▶). Data-collection statistics are shown in Table 1 ▶.
Table 1. Data-collection statistics for the XylC crystal.
Values in parentheses are for the highest resolution shell.
| Beamline | BL13B1, NSRRC |
| Wavelength (Å) | 1.0 |
| Resolution (Å) | 25–2.2 (2.28–2.20) |
| Space group | P21 |
| Unit-cell parameters | |
| a (Å) | 88.36 |
| b (Å) | 202.20 |
| c (Å) | 99.87 |
| β (°) | 99.04 |
| No. of measured reflections | 669429 (55187) |
| No. of unique reflections | 173739 (17246) |
| Completeness (%) | 99.8 (99.2) |
| R merge † (%) | 14.6 (41.9) |
| 〈I/σ(I)〉 | 11.6 (3.1) |
| Multiplicity | 3.9 (3.2) |
| Detector | ADSC Q315r |
| Oscillation range (°) | 1 |
| Exposure time (s) | 1 |
| Crystal-to-detector distance (mm) | 280 |
R
merge = 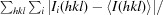
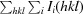 .
.
3. Results and discussion
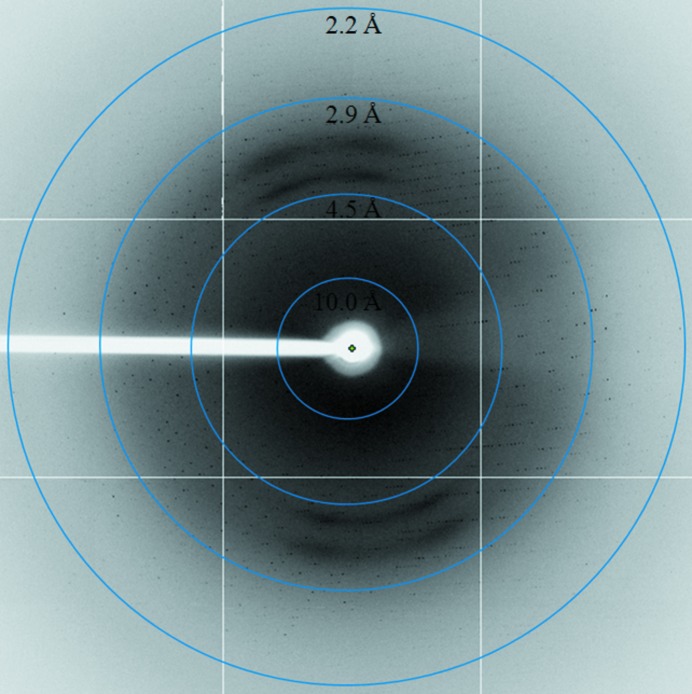
As shown in Fig. 1 ▶, large single XylC crystals were obtained using 0.2 M sodium citrate tribasic dehydrate pH 5.6, 15–17%(w/v) polyethylene glycol 3350. Prior to data collection at 100 K, the crystal was mounted in a cryoloop and flash-cooled in liquid nitrogen with a slightly modified cryoprotectant consisting of 0.2 M sodium citrate pH 5.6, 18%(w/v) polyethylene glycol 3350 and 15%(v/v) glycerol. Based on the diffraction pattern (Fig. 2 ▶), the XylC crystals belonged to the monoclinic space group P21, with unit-cell parameters a = 88.36, b = 202.20, c = 99.87 Å, β = 99.04°. Assuming that there are four molecules in the asymmetric unit, the Matthews coefficient V M (Matthews, 1968 ▶) is 3.08 Å3 Da−1 and the estimated solvent content is 60%.
Figure 1.
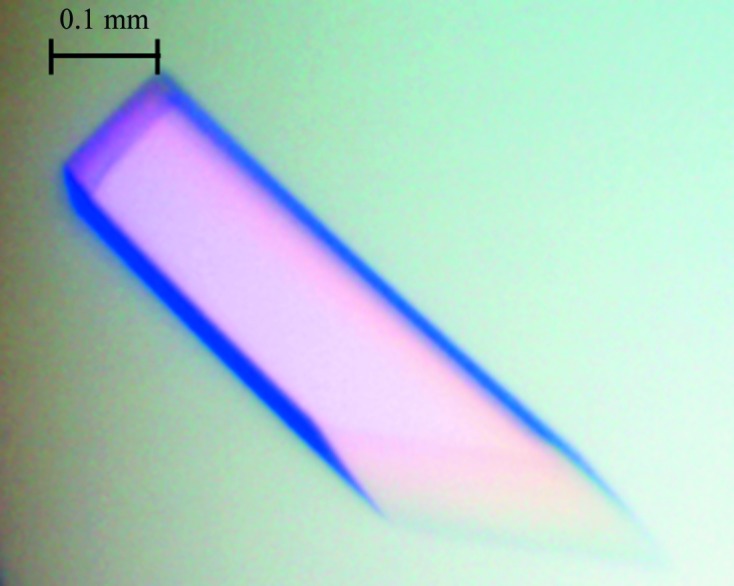
A crystal of β-xylosidase (XylC). The crystal reached approximate dimensions of 0.45 × 0.1 × 0.1 mm in 2–3 d.
Figure 2.
A diffraction pattern of the XylC crystal.
No similar structure to XylC has previously been solved. BLASTP searches of the PDB using XylC failed to identify any similar structure (Shao et al., 2011 ▶). Consequently, there is no structure that can serve as a template for molecular replacement. We are currently working on preparing crystals of heavy-atom derivatives and attempting to solve the structure using the multiple isomorphous replacement method. The preparation of SeMet protein for multiple-wavelength anomalous diffraction is also in progress.
Acknowledgments
The synchrotron data collection was conducted on beamline BL13B1 at NSRRC (National Synchrotron Radiation Research Center, Taiwan) supported by the National Science Council (NSC). This work was supported by grants from the National Basic Research Program of China (2011CB710800) and Tianjin Municipal Science and Technology Commission (10ZCKFSY06000).
References
- Biely, P. (1985). Trends Biotechnol. 3, 286–290.
- Lorenz, W. W. & Wiegel, J. (1997). J. Bacteriol. 179, 5436–5441. [DOI] [PMC free article] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Saha, B. C. (2003). J. Ind. Microbiol. Biotechnol. 30, 279–291. [DOI] [PubMed]
- Shao, W., Xue, Y., Wu, A., Kataeva, I., Pei, J., Wu, H. & Wiegel, J. (2011). Appl. Environ. Microbiol. 77, 719–726. [DOI] [PMC free article] [PubMed]
- Van Duyne, G. D., Standaert, R. F., Karplus, P. A., Schreiber, S. L. & Clardy, J. (1993). J. Mol. Biol. 229, 105–124. [DOI] [PubMed]
- Wu, H., Pei, J., Jiang, Y., Song, X. & Shao, W. (2010). Biotechnol. Lett. 32, 795–801. [DOI] [PubMed]