A putative lipase, LipS, from the soil metagenome of an environmental sample taken at the Biocenter Klein Flottbek, Hamburg, Germany was cloned, expressed and purified. Two different constructs of the protein were crystallized in a search for well diffracting crystals. Preliminary X-ray analyses indicated that crystals in space groups P42212 and P4 diffracted X-ray radiation to 2.8 and 2.0 Å resolution, respectively.
Keywords: lipases, metagenome
Abstract
LipS is a novel thermostable putative lipase that was isolated from a metagenomic library using functional screening methods. The corresponding gene shows high similarity to that encoding a putative but uncharacterized esterase from Symbiobacterium thermophilum IAM14863 (99% nucleotide-sequence similarity). Two different constructs of the recombinant lipase were crystallized. Crystals belonging to space group P42212 diffracted X-ray radiation to 2.8 Å resolution and crystals belonging to space group P4 diffracted to 2.0 Å resolution. The most probable content of their asymmetric units were two molecules (P42212) and four or five molecules (P4), respectively.
1. Introduction
Lipases (EC 3.1.1.3) are used in numerous biotechnological processes, e.g. in the pharmaceutical, cosmetics, food, paper, detergent and biofuel industries (Jaeger & Eggert, 2002 ▶). They are also applied in esterification reactions for the synthesis of organic compounds such as sugar esters (Kobayashi, 2011 ▶) and polymers (Jiang, 2008 ▶; Kobayashi, 2010 ▶). As an important characteristic, lipases usually display high stereoselectivity and regioselectivity as well as a high stability towards solvents (Gupta et al., 2004 ▶). This group of enzymes is therefore highly coveted in many industrially significant processes. In order to address this demand, the development of novel and improved lipases through directed evolution or the exploitation of natural sources, such as the metagenomic pool, is on the rise.
The bacterial metagenome is a vast source of a virtually unlimited number of enzymes and proteins with unknown functions and/or properties (Streit & Schmitz, 2004 ▶). By definition, these microorganisms are difficult or impossible to cultivate under standard laboratory conditions. The identification and isolation of proteins from this source therefore requires specialized setups and methods. In our search for novel thermostable lipases, we constructed a metagenomic library using DNA isolated from soil and water samples collected in the Botanical Garden Klein Flottbek (University Hamburg, Germany). The enrichment media were kept under moderately aerobic conditions at 338 K to preselect for thermophilic bacteria. Details of the DNA isolation, cosmid library construction and functional screening for lipolytic clones will be described elsewhere.
One positive clone with lipolytic activity contained a 26.5 kb insert. The insert encodes an enzyme with 99% identity to a putative esterase from Symbiobacterium thermophilum IAM14863 (Ueda et al., 2004 ▶; Fig. 1 ▶). The ability of this microorganism to undergo horizontal gene acquisition is supposed to be the highest amongst known bacteria (Nishida & Yun, 2011 ▶). Further characterization of the source organism of the putative lipase will proceed by 16S rRNA analysis of the metagenomic DNA. The enzyme encoded by the 26.5 kb insert was named LipS. It consisted of 280 amino acids with a molecular weight of 30 180 Da.
Figure 1.
Alignment of LipS (top line) with a putative esterase from S. thermophilum IAM14863 (bottom line). The single-amino-acid difference at position 81 is marked by an asterisk.
Carboxyl esterases and lipases have been classified into eight families according to amino-acid similarity and physiological function (Arpigny & Jaeger, 1999 ▶). Interestingly, LipS could not be unequivocally grouped into any of these families. Therefore, LipS is the first metagenomic member of a new group of lipolytic enzymes. Further biochemical analysis together with the crystallographic structure will help to characterize this lipase from a novel family.
2. Materials and methods
2.1. Cloning, expression and purification
Lips was isolated from the metagenome of soil and water samples taken at the Biocenter Klein Flottbek, University of Hamburg. The enrichment of cultures, DNA isolation and library construction, as well as the functional screening for clones with lipolytic activity, will be described elsewhere. A sequence search with BLASTX against the NCBI database identified a predicted esterase from S. thermophilum, an uncultivable bacterium (Ueda et al., 2004 ▶), as the closest homologue (Fig. 1 ▶).
The gene was first amplified by PCR from a pre-selected cosmid. The sequences of the forward and reverse primers were 5′-CATATGAGCCGGAAAAGCAGG-3′ and 3′-CAAGTAGGCCTTCGTGTCGTTCGAA-5′, respectively. The primers contained NdeI and HindIII restriction sites (bold) for directed cloning into the expression vector pET21a (5.44 kb; Novagen, Darmstadt, Germany), which adds a noncleavable His6 tag at the C-terminus. The final construct starts with its natural N-terminus and ends with KLAAALEHHHHHH immediately after the regular C-terminus. The molecular weight of this construct is 31 701 Da
LipS with a TEV-cleavable His6 tag at the N-terminus was amplified for cloning into vector pETM11 (6.03 kb; courtesy of A. Geerlof, EMBL Hamburg Outstation, Hamburg, Germany). Two different forward primers were used in order to obtain one construct with the native N-terminus and another which lacks 26 N-terminal residues. These residues were predicted to be unstructured and may constitute a localization signal. The sequences of the forward primer (PscI restriction site shown in bold) and the reverse primer for the TEV-cleavable full-length construct were 5′-TACACATGTGCCGGAAAAGCAGGAACTG-3′ and 3′-GCCTTCGTGACTTTCGAACGCCGGCGTG-5′, respectively. After removal of the His tag, this construct additionally contained the amino-acid sequence GA at the N-terminus before MCRKSRN. Note the replacement of serine at position 2 with a cysteine residue. The construct ends with His279. The molecular weight of the final product after cleavage is thus 30 239 Da.
The sequences of the forward and the reverse primers for the N-terminally truncated construct were 5′-AACATGTCCGGTATGTCGACGACGCCCCTTC-3′ and 3′-GCCTTCGTGACTTTCGAACGCCGGCGTG-5′, respectively. This construct adds GAM to the truncated N-terminus beginning with Ser27 after His-tag removal. It also terminates with His279.
All final inserts were verified by DNA sequencing. All plasmids were transformed into Escherichia coli strain BL21 (DE3) (Novagen, Darmstadt, Germany) by heat shock. Transformants were selected on LB (Luria–Bertani medium) plates containing 100 µg ml−1 ampicillin (pET21a) or 25 µg ml−1 kanamycin (pETM11). 10 ml overnight cultures were grown at 290 K in LB medium containing the same concentrations of the appropriate antibiotics. Erlenmeyer flasks containing 750 ml LB medium were inoculated with 1%(v/v) overnight cultures and incubated aerobically at 290 K until the medium reached an optical density of 0.8 at 600 nm. Overexpression of the constructs was induced by the addition of 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG). After incubation for 16 h at 290 K, the cells were harvested by centrifugation at 8000 rev min−1 for 20 min at 277 K (Sorvall RC6+ centrifuge, F10S-6x500y rotor; Thermo Scientific, Braunschweig, Germany). Cell pellets were immediately processed and resuspended in lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole) augmented with EDTA-free protease-inhibitor cocktail (Roche) and 1 mg ml−1 DNAse. The suspension was lysed by ultrasonication (5 × 1 min on ice with 1 min interruptions; ultrasonication processor UP 200S, 24 kHz, 200 W, Dr Hielscher GmbH, Teltow, Germany). Cell debris was removed by centrifugation for 30 min at 13 000 rev min−1 and 277 K (Sorvall RC6+ centrifuge, SS-34 rotor; Thermo Scientific, Braunschweig, Germany). The crude extract was incubated at 277 K with 1 ml Ni–NTA resin (Ni–NTA coupled to Sepharose; Qiagen, Hilden, Germany) per 4 ml lysate on a rotary shaker (200 rev min−1) for at least 2 h or overnight. The sample was loaded onto a column and the flowthrough was collected for analysis. Unbound material was eluted with 16 ml (four column volumes) of wash buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole). His6-tagged protein was eluted with 6 ml elution buffer (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole). The fractions were analyzed on SDS–PAGE gels stained with Coomassie Brilliant Blue. Fractions that contained LipS were combined and concentrated in a Vivaspin column (molecular-weight cutoff 10 kDa) to a final volume of 3 ml. Tagged protein was digested with 2%(w/w) TEV protease overnight and then subjected to another round of Ni–NTA chromatography to remove uncleaved LipS.
Size-exclusion chromatography (HiLoad 16/60 Superdex 75 prep grade, Amersham Biosciences) was used as a final step of purification. The column was equilibrated with 50 mM HEPES pH 8.0, 100 mM NaCl, 0.5 mM TCEP. The samples eluted as a single peak with an elution volume of 61 ml, corresponding to LipS in a dimeric state. Peak fractions were analyzed by SDS–PAGE and pooled. The purity of the combined protein fractions was assessed by 12% SDS–PAGE stained with Coomassie Brilliant Blue.
2.2. Crystallization
On the basis of precipitation results using the Hampton Pre-Crystallization Test (Hampton Research, California, USA), initial crystallization screens were performed at 4 mg ml−1 by the sitting-drop vapour-diffusion method at 292 K. The sample concentration was determined from the absorption at 280 nm using a NanoDrop ND-1000 spectrophotometer. The calculated absorption coefficients (obtained using http://web.expasy.org/protparam) for a concentration of 1 mg ml−1 were 0.929 for His-tagged LipS and 0.974 for tag-free LipS. Initial crystallization trials were carried out with four different 96-well screens from Qiagen (The Classics, Classics II, PEGs and PEGs II Suites) at the EMBL Hamburg high-throughput crystallization facility (Mueller-Dieckmann, 2006 ▶). 300 nl sample solution was mixed with 300 nl reservoir solution and equilibrated against 50 µl reservoir solution. Crystallization trials with the His-tagged construct resulted in a single lead in condition C9 of The Classics II Suite screen. Optimization of this condition was carried out with The Opti-Salts Suite from Qiagen, which improved the size of the crystals through the addition of 3.5 M NaBr. The final crystallization condition thus consisted of 1 M sodium malonate, 0.1 M Tris pH 8.5, 0.5%(v/v) Jeffamine ED-2001 and 3.5 M NaBr.
Tag-free LipS with a full-length N-terminus was found to form crystals in condition A8 of The Classics II Suite screen. In the course of optimization trials the addition of 0.01 M spermidine (Additive Screen, Hampton Research) improved the crystal size and the quality of the observed diffraction spots. The final crystallization condition thus consisted of 0.1 M sodium acetate pH 4.5, 3 M NaCl and 0.01 M spermidine. The tag-free and N-terminally truncated version of LipS could not be crystallized.
2.3. X-ray diffraction data collection and processing
Prior to data collection, crystals of LipS were flash-cooled to 77 K in liquid nitrogen. Crystals of His-tagged LipS did not require cryoprotection and were taken straight from the mother liquor. Crystals of tag-free LipS were soaked in a cryoprotectant solution comprising reservoir solution plus 20%(v/v) glycerol. Complete X-ray diffraction data sets were collected on beamlines ID23-1 and ID23-2 at the ESRF, Grenoble, France using ADSC Q315R and MAR 225 Mosaic detectors, respectively.
The data were indexed and integrated using iMOSFLM v.1.0.4 (Battye et al., 2011 ▶) and scaled with SCALA (Winn et al., 2011 ▶). Table 1 ▶ summarizes the data-collection and processing statistics. Self-rotation functions were computed with MOLREP (Vagin & Teplyakov, 2010 ▶).
Table 1. X-ray data-collection and processing statistics.
Values in parentheses are for the highest resolution shell.
| Protein | His-tagged LipS | Tag-free LipS |
|---|---|---|
| X-ray source | ID23-1, ESRF | ID23-2, ESRF |
| Wavelength (Å) | 0.87260 | 0.87260 |
| Temperature (K) | 100 | 100 |
| Crystal-to-detector distance (mm) | 310 | 240 |
| Rotation range per image (°) | 0.45 | 1.0 |
| Total rotation range (°) | 54 | 120 |
| Space group | P42212 | P4 |
| Unit-cell parameters (Å) | a = b = 170.7, c = 46.8 | a = b = 105.3, c = 121.0 |
| Resolution range (Å) | 46.3–2.8 (3.2–2.8) | 40.3–2.0 (2.1–2.0) |
| Observed reflections | 76113 (11247) | 463607 (65626) |
| Unique reflections | 16386 (2243) | 90469 (13179) |
| R merge (%) | 18.9 (69.0) | 11.7 (80.1) |
| R p.i.m. (%) | 9.1 (33.7) | 5.7 (39.9) |
| Completeness (%) | 93.7 (95.9) | 100 (100) |
| 〈I/σ(I)〉 | 7.9 (2.6) | 9.8 (2.0) |
| Multiplicity | 4.6 (5.0) | 5.1 (5.0) |
| Wilson B factor (Å2) | 36 | 29 |
| Optical resolution (Å) | 2.0 | 1.6 |
3. Results and discussion
A novel metagenome-derived putative lipase was expressed in E. coli. The enzyme, LipS, has 99% sequence identity to a putative and uncharacterized lipase from S. thermophilum IAM14863 (NCBI accession No. NC_006177; Fig. 1 ▶). Further classification of the possible host organism will proceed via 16S rRNA analysis of the metagenomic DNA. Expression of three different constructs of LipS resulted in close to 100% soluble protein and yielded 3–4 mg pure protein per litre of cell culture. Purification via Ni–NTA affinity chromatography and affinity-tag removal where applicable followed by size-exclusion chromatography resulted in protein samples that were >95% pure as estimated by SDS–PAGE. The apparent molecular weights from size-exclusion chromatography correspond to dimers.
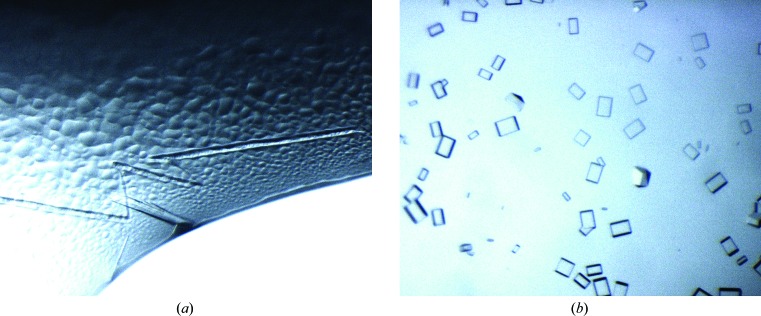
The initial full-length construct contained a noncleavable His6 tag at the C-terminus. Crystallization trials produced a large number of microcrystals from a single lead within 2 d. Overnucleation was reduced through the addition of 3.5 M NaBr. The use of seeds from these crystals eventually produced single crystals of sufficient size (Fig. 2 ▶ a). However, these crystals did not diffract X-ray radiation beyond 2.8 Å resolution. Assuming interference from the affinity tag, two constructs with TEV-cleavable His6 tags at the N-termini were produced. One of the constructs was truncated at the N-terminus to remove the first 26 amino acids, which were predicted to be unstructured. The other construct extended to the native N-terminus. Both constructs were tested in initial crystallization experiments, but only LipS with the full-length N-terminus yielded crystals. The addition of 0.01 M spermidine, identified in an additive screen, helped to improve the size of crystals, which grew as cuboids (Fig. 2 ▶ b).
Figure 2.
Crystals of LipS. (a) Large single needle-like crystals of full-length LipS with an uncleaved His6 tag as obtained after addition of NaBr and seeding. The largest needle is about 300 µm in length. (b) Single crystals grown from LipS without an affinity tag. The largest cuboids measure about 80 × 50 µm.
Data from crystals of the initial affinity-tagged construct were indexed in space group P42212, with unit-cell parameters a = 170.65, c = 46.83 Å. During data collection, it became apparent that the crystal morphology strongly correlated with the overall data quality as well as the scattering isotropy. The best crystals had precise edges in all directions and diffracted to 2.8 Å resolution (Table 1 ▶). Two molecules per asymmetric unit are most likely for these crystals, based on a Matthews parameter (Matthews, 1968 ▶) of 2.7 Å3 Da−1, corresponding to a solvent content of 54%. The self-rotation function did not indicate any peaks corresponding to noncrystallographic symmetry at χ = 60, 90, 120 or 180°.
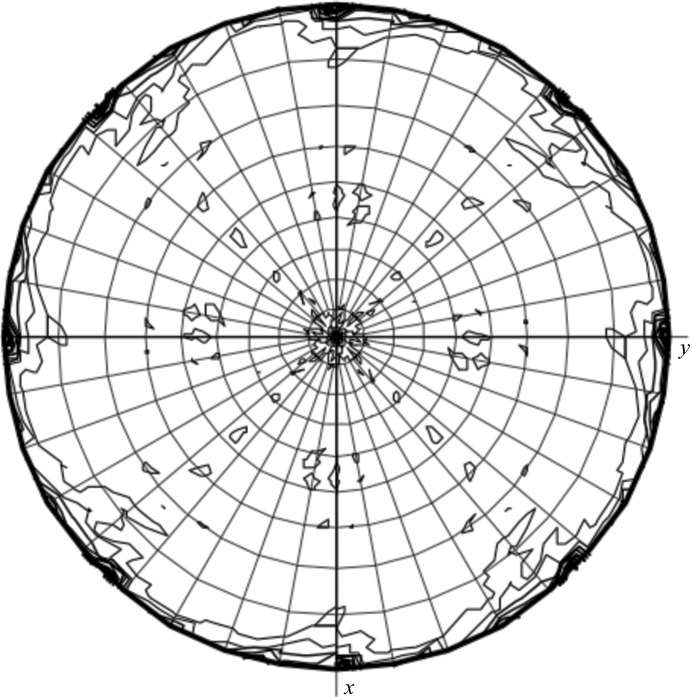
Diffraction data for the tag-free construct were indexed in space group P4, with unit-cell parameters a = 105.27, c = 120.98 Å (Table 1 ▶). Five molecules per asymmetric unit are most likely for these crystals in space group P4, with a Matthews parameter of 2.2 Å3 Da−1 and a solvent content of 45%. The self-rotation function showed small peaks in the χ = 180° section for θ = 90° and ϕ = 45° and 90° (Fig. 3 ▶). This indicates a packing arrangement which is similar to that of the His6-tagged LipS in space group P42212. Such an arrangement points to the possibility of only four molecules per asymmetric unit in space group P4, with a Matthews parameter of 2.8 Å3 Da−1 and a solvent content of 56%. A complete data set to just below 2 Å resolution was collected. Data from this construct will be used for phasing via molecular replacement using the structure of the carboxylesterase from Geobacillus stearothermophilus (27% sequence identity; PDB entry 1tqh; Liu et al., 2004 ▶) as a model.
Figure 3.
Plot of the self-rotation function of tag-free LipS in space group P4. The χ = 180° section is shown. The aditional peaks at θ = 90° and ϕ = 45° and 90° emulate a P422 arrangement as observed for the packing of uncleaved LipS.
Acknowledgments
The research leading to these results received funding from the European Community’s Seventh Framework Programme (FP7/2007–2013) under grant agreement No. 227764 (P-CUBE).
References
- Arpigny, J. L. & Jaeger, K.-E. (1999). Biochem. J. 343, 177–183. [PMC free article] [PubMed]
- Battye, T. G. G., Kontogiannis, L., Johnson, O., Powell, H. R. & Leslie, A. G. W. (2011). Acta Cryst. D67, 271–281. [DOI] [PMC free article] [PubMed]
- Gupta, R., Gupta, N. & Rathi, P. (2004). Appl. Microbiol. Biotechnol. 64, 763–781. [DOI] [PubMed]
- Jaeger, K.-E. & Eggert, T. (2002). Curr. Opin. Biotechnol. 13, 390–397. [DOI] [PubMed]
- Jiang, Z. (2008). Biomacromolecules, 9, 3246–3251. [DOI] [PubMed]
- Kobayashi, S. (2010). Proc. Jpn Acad. Ser. B Phys. Biol. Sci. 86, 338–365. [DOI] [PMC free article] [PubMed]
- Kobayashi, T. (2011). Biotechnol. Lett. 33, 1911–1919. [DOI] [PubMed]
- Liu, P., Wang, Y.-F., Ewis, H. E., Abdelal, A. T., Lu, C.-D., Harrison, R. W. & Weber, I. T. (2004). J. Mol. Biol. 342, 551–561. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Mueller-Dieckmann, J. (2006). Acta Cryst. D62, 1446–1452. [DOI] [PubMed]
- Nishida, H. & Yun, C.-S. (2011). Int. J. Evol. Biol. 2011, 34505. [DOI] [PMC free article] [PubMed]
- Streit, W. R. & Schmitz, R. A. (2004). Curr. Opin. Microbiol. 7, 492–498. [DOI] [PubMed]
- Ueda, K., Yamashita, A., Ishikawa, J., Shimada, M., Watsuji, T., Morimura, K., Ikeda, H., Hattori, M. & Beppu, T. (2004). Nucleic Acids Res. 32, 4937–4944. [DOI] [PMC free article] [PubMed]
- Vagin, A. & Teplyakov, A. (2010). Acta Cryst. D66, 22–25. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.