The C-terminal domain of the cation diffusion facilitator MamM has been successfully constructed, purified and crystallized. Diffraction data was collected to to a resolution of 2.0 Å.
Keywords: MamM, magnetosome-associated proteins, Magnetospirillum gryphiswaldense MSR-1
Abstract
MamM is a unique magnetosome-associated protein that shares substantial homology with cation diffusion facilitator (CDF) proteins, a group of heavy-metal-ion efflux transporters that participate in metal-ion homeostasis in all domains of life. Magnetotactic bacteria utilize CDF proteins in iron-oxide biomineralization and in magnetosome formation. Here, the crystallization and preliminary X-ray analysis of recombinant Magnetospirillum gryphiswaldense MamM is reported. The C-terminal domain of MamM was crystallized in the orthorhombic space group C2221, with unit-cell parameters a = 37.1, b = 94.0, c = 53.3 Å. X-ray diffraction data were collected to a resolution of 2.0 Å.
1. Introduction
All living organisms require divalent metal cations for proper cellular development and function (Alberts et al., 1998 ▶). Although divalent metal cations are essential, their cellular concentrations are tightly regulated as their excess accumulation can be highly cytotoxic (Outten & O’Halloran, 2001 ▶). Cells thus rely on diverse export and sensory systems to avoid the excess accumulation of metal ions and to sustain cellular homeostasis. Cation diffusion facilitator (CDF) proteins are one group of heavy-metal-ion efflux transporters that participate in metal-ion homeostasis and can be found in all domains of life (Paulsen & Saier, 1997 ▶). For example, CDF proteins are found in the bacterial cell membrane and the vacuolar membranes of plants and yeast, as well as in the Golgi apparatus of animal cells (Haney et al., 2005 ▶). Members of the CDF family transport cytoplasmic divalent metal cations, including Cd, Co, Fe, Mn, Ni and Zn, by exploiting the proton motive force (Anton et al., 1999 ▶; Persans et al., 2001 ▶; Delhaize et al., 2003 ▶; Munkelt et al., 2004 ▶; Grass et al., 2005 ▶). According to bioinformatics analysis, CDF proteins can be phylogenetically divided into three substrate-specific branches, namely the Zn-CDF, Fe/Zn-CDF and Mn-CDF subgroups (Montanini et al., 2007 ▶). All CDF transporters share a common two-domain architecture consisting of a transmembrane domain (TMD) and a cytoplasmic C-terminal domain (CTD) (Paulsen & Saier, 1997 ▶). Despite the broad distribution of CDFs, the Escherichia coli FieF (YiiP) protein is the only standalone structure of the entire transporter deposited in the Protein Data Bank to date (Lu et al., 2009 ▶). FieF (YiiP), which belongs to the Fe/Zn-CDF subgroup, is thus the best characterized CDF. The FieF (YiiP) homodimeric structure was determined in a zinc-bound state and presented a CTD with a metallochaperone-like fold, despite a lack of sequence homology to such proteins. Indeed, given the difficulty of transmembrane-protein purification and crystallization, structural studies of CDFs have mainly focused on the cytosolic CTD. Such efforts have determined CTD structures of the Zn-bound and apo forms of the Thermus thermophilus CzrB protein as well as the apo form of the Thermotoga maritima TM0876 protein (Cherezov et al., 2008 ▶; Higuchi et al., 2009 ▶). These CTDs structures share the same metallochaperone-like fold as FieF (YiiP), with CzrB displaying a flexible structural shift upon zinc binding. This conformational shift is considered to be significant for transport-activity regulation.
A unique bacterial system that utilizes CDF proteins for iron transport and biomineralization is the magnetosome of magnetotactic bacteria (Uebe et al., 2011 ▶). Magnetotactic bacteria (MTB) are a diverse phylogenic group of Gram-negative bacteria that share the unique ability of being able to navigate along geomagnetic fields, a behaviour referred to as magnetotaxis (Bazylinski & Frankel, 2004 ▶; Schüler, 2008 ▶; Komeili, 2012 ▶). MTBs can be found in transition environments, such as the oxic/anoxic transition zone, where they employ magnetotaxis to simplify their environmental search (Faivre & Schüler, 2008 ▶). The MTB geomagnetic field sensor is the magnetosome chain, which allows the biomineralization of iron oxides. Each magnetosome biomineralizes magnetite or greigite nanocrystals, which are enclosed by a phospholipid membrane that originates from the plasma membrane. Several magnetosomes are arranged into a linear chain along actin-like filaments, thus promoting the formation of a single and stable magnetic dipole (Klumpp & Faivre, 2012 ▶).
Magnetosome formation and magnetite biomineralization are genetically controlled by a large set of genes that encode both soluble and integral proteins (Schüler, 2004 ▶; Murat et al., 2010 ▶). The genes encoding magnetosome proteins are situated on a single genomic island that contains four main operons: mamAB, mamCD, mms6 and mamXY (Jogler et al., 2009 ▶). Deletion of the magnetosome-related genomic island results in the loss of magnetic orientation (Bazylinski & Frankel, 2004 ▶; Komeili et al., 2004 ▶; Murat et al., 2010 ▶; Lohsse et al., 2011 ▶). Studies of magnetosome-forming genes revealed that the biomineralization process is controlled by these four operons, with the mamAB operon thought to be involved in iron transport and magnetosome vesicle alignment (Amemiya et al., 2007 ▶; Komeili, 2007 ▶; Scheffel & Schüler, 2007 ▶; Faivre & Schüler, 2008 ▶; Jogler et al., 2011 ▶; Murat et al., 2010 ▶; Lefèvre et al., 2011 ▶; Lohsse et al., 2011 ▶).
MamM and MamB are two conserved and abundant integral membrane magnetosome-associated proteins. Both MamM and MamB share high similarity to CDF proteins and are presumed to play a role in the iron transport and biomineralization stages of magnetosome formation (Uebe et al., 2011 ▶). Bioinformatics analysis of MTB CDF protein sequences has shown that these proteins form a distinct and coherent branch within the Fe/Zn-CDF subfamily. Recent in vivo studies have demonstrated the ability of MamM to form dimers, as well as to stabilize MamB by forming heterodimeric complexes. In addition, these studies revealed a role for MamM in proper magnetite but not hematite crystal formation.
To analyze the structure–function relationship of MamM, a CDF protein that is presumed to transport iron, we initiated crystallographic studies of the cytosolic domain of the Magnetospirillum gryphiswaldense MSR-1 MamM protein. Here, we report the crystallization and preliminary X-ray analysis of a truncated version of MamM (MamM-CTD).
2. Materials and methods
2.1. Expression of M. gryphiswaldense MSR-1 MamM in E. coli
The truncated mamM gene (UniProt Q6NE57, residues 215–318) was amplified by polymerase chain reaction (PCR) using M. gryphiswaldense MSR-1 genomic DNA as template, together with primers Fw, 5′-GCATTACGCATATGGATACCGAACTGTTGCAGACGGCC-3′, and Rev, 5′-CGTATGCGGATCCTCAGACCCGGACCGTCACGGC-3′. The primers were designed to introduce an NdeI site at the initiation codon, ATG, and a BamHI site after the termination codon. The DNA fragments were digested with NdeI and BamHI and cloned into the respective sites of plasmid pET28a(+), giving rise to plasmid pET28aMamM-CTD-MSR1. In this construct, the mamM gene was fused in-frame to express a six-His tag at the N-terminus of the protein followed by a thrombin proteolysis site.
E. coli Rosetta strain cells harbouring plasmid pET28aMamM-CTD-MSR1 were grown in auto-induction medium (Studier, 2005 ▶) containing kanamycin (100 mg ml−1) and chloramphenicol (30 mg ml−1) at 310 K for 8 h. The cultivation temperature was then reduced from 310 to 300 K for a further 48 h. The cells were harvested by centrifugation at 7438g for 8 min at 277 K.
2.2. Purification of MamM-CTD
MamM-CTD-expressing cells were suspended in buffer A (50 mM Tris–HCl pH 8, 300 mM NaCl, 20 mM imidazole, 5 mM β-mercaptoethanol, 0.1% Triton X-100) and incubated with DNase I (10 mg ml−1) and protease-inhibitor cocktail [100 µM phenylmethylsulfonyl fluoride (PMSF), 1.2 µg ml−1 leupeptin and 1 µM pepstatin A] for 30 min at 277 K. The cells were then disrupted by two cycles in a French press pressure cell at 172 MPa. Cell debris was separated by centrifugation at 19 000g for 2 h at 277 K and the soluble fraction was applied onto a home-made gravity Ni–NTA column (4 ml bed volume, 2.5 cm diameter; Bio-Rad Econo-Column chromatography column, Thermo Scientific HisPur Ni–NTA resin) pre-equilibrated with buffer A. The protein was washed with 50 ml buffer B (20 mM Tris–HCl pH 8, 1 M NaCl, 40 mM imidazole, 5 mM β-mercaptoethanol) and eluted with buffer C (20 mM Tris–HCl pH 8, 150 mM NaCl, 500 mM imidazole, 5 mM β-mercaptoethanol). To remove the six-His tag, bovine thrombin (10 U ml−1; t4648-10KU, Sigma–Aldrich) was added to the eluted protein and the mixture was dialyzed against buffer D (10 mM Tris–HCl pH 8, 150 mM NaCl, 5 mM β-mercaptoethanol) for 16 h at 277 K. The protein was concentrated to 8 mg ml−1 using a Vivaspin-4 (3000 molecular-weight cutoff; Sartorius Stedim Biotech) and applied onto a size-exclusion column (HiLoad 26/60 Superdex 75, GE Healthcare Biosciences) pre-equilibrated with buffer D. Purified MamM-CTD was then concentrated to 31 mg ml−1 for crystallization, flash-cooled in liquid nitrogen and stored at 193 K. Throughout the purification process, protein concentrations were determined by protein spectrometric absorption at 280 nm using a calculated extinction coefficient of 0.588 M −1 cm−1. Sample purity at this stage was analyzed by SDS–PAGE and protein identification was confirmed by tandem mass spectroscopy.
2.3. Crystallization
MamM-CTD was crystallized using the sitting-drop vapour-diffusion method at 293 K. 0.2 µl MamM-CTD solution and 0.2 µl reservoir solution were mixed to form the drop. The initial crystallization conditions were examined using commercial screening kits from Hampton Research (Crystal Screen, Crystal Screen 2 and Index) and Molecular Dimensions (Structure Screens I and II).
2.4. Diffraction data collection
Crystals were harvested and flash-cooled in liquid nitrogen without the addition of cryoprotecting solution. Diffraction data were collected using an image-plate detector system (MAR 345 mm; MAR Research, Germany) on a marμX X-ray system home source (MAR Research, Germany). Data collection was performed at 100 K. For the MamM-CTD data set, a total of 192 frames were collected with an oscillation range of 1° and an exposure time of 2 min per image. The crystal-to-detector distance was 160 mm. The data were processed using HKL-2000 (Otwinowski & Minor, 1997 ▶) and POINTLESS (Evans, 2006 ▶) from the CCP4 program suite (Winn et al., 2011 ▶).
3. Results and discussion
The M. gryphiswaldense MamM amino-acid sequence (residues 215–318) with an additional four N-terminal residues (NGSHMC) that remained after thrombin proteolysis predicts a protein that contains 108 residues with a molecular mass of 11 895 Da and a theoretical pI of 4.97. MamM-CTD was expressed in E. coli and purified to homogeneity, with a yield of approximately 50 mg purified protein from 20 g bacterial culture. During MamM-CTD purification, we performed size-exclusion chromatography at a flow rate of 4.4 ml min−1 and demonstrated that the protein elutes at a volume of 170 ml. According to the size-exclusion calibration curve generated using standard proteins (ferritin, 440 kDa; conalbumin, 75 kDa; carbonic anhydrase, 29 kDa; ribonuclease A, 13.7 kDa) and the calculated molecular weight of the MamM-CTD monomer, it was concluded that the chromatography elution volume corresponds to ∼25 kDa, suggesting a protein functional unit consisting of a dimer. Such a molecular size is similar to the dimeric form presented by the CzrB C-terminal domain in solution (Cherezov et al., 2008 ▶). SDS–PAGE revealed that the histidine tag was fully removed from MamM-CTD by thrombin and a 99% correlation was observed between the mass reported by tandem mass spectroscopy and the pre-calculated protein mass: 11 878 and 11 895 Da, respectively. By screening MamM against several crystallization screens, we were able to detect crystals which appeared in Index condition No. 62 (0.2 M ammonium sulfate, 0.1 M bis-Tris pH 5.5, 25% polyethylene glycol 3350). This crystal was harvested directly from the 96-well plate screen and flash-cooled in liquid nitrogen without addition of cryoprotecting solution. Crystal-harbouring conditions were further optimized by performing minor alternations to both the polyethylene glycol 3350 concentration (20–35% PEG 3350) and the bis-Tris pH value (pH 5–6). These optimized conditions yielded similarly diffracting crystals.
Crystals of MamM-CTD were reproducibly obtained. The diffraction data to a resolution of 2.0 Å obtained from a crystal harvested from Index screen condition No. 62 (Fig. 1 ▶) fulfilled the systematic absence rules of space group C2221. Further validation of the chosen space group was performed by POINTLESS (Evans, 2006 ▶), which indicated that the space-group choice was correct, with a symmetry-absence probability of 0.735. The unit-cell parameters are a = 37.1, b = 94.0, c = 53.3 Å (Table 1 ▶). We believe that this crystal should be adequate for structural determination, as it diffracted to high resolution and yielded a data set with a low R merge and a high multiplicity. Matthews coefficient calculations of the scaled data set only permitted the presence of a single monomer in the asymmetric unit. Assuming the presence of this single monomer, the calculated V M value (Matthews, 1968 ▶) and solvent content are 2.11 Å3 Da−1 and 41.83%, respectively, which are both within the normal range of values observed for soluble protein crystals. The V M value suggesting the presence of a single monomer in the asymmetric unit is surprising, as MamM-CTD was found to be a stable dimer in solution. In-depth examination of the asymmetric unit compositions of other previously determined CDF proteins revealed some differences with respect to MamM-CTD (see Table 2 ▶ for sequence identities and similarities). FieF (PDB entries 2qfi and 3ho9) contains two or four monomers in the symmetric unit (Lu & Fu, 2007 ▶; Lu et al., 2009 ▶), while CzrB (PDB entries 3byp and 3byr) contains two monomers in the asymmetric unit (Cherezov et al., 2008 ▶). Since CDFs are functional in their dimeric form, we presume that the crystal contacts of MamM-CTD will exhibit a similar V-shape-like packing as appears in the crystal structure of the Thermotoga maritima TM0876 CDF cytosolic domain owing to the twofold symmetry axis in the C2221 space group. In the TM0876 structure the asymmetric unit also contains a single monomer as the crystallographic twofold symmetry axis overlaps with the homodimer twofold axis (PDB entry 2zzt; Higuchi et al., 2009 ▶). However, we cannot fully rule out the possibility of a MamM-CTD monomeric form within the crystal owing to dimer disruption by crystallization conditions and/or crystal contacts.
Figure 1.
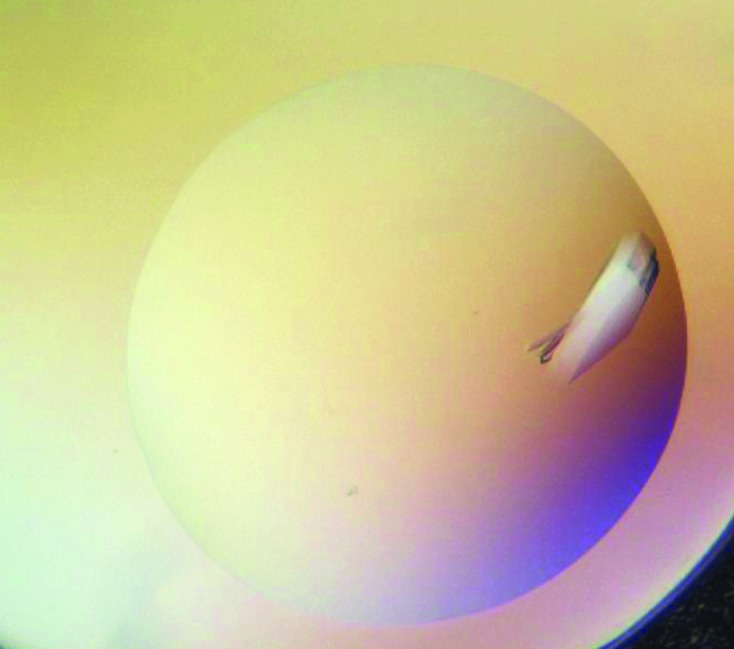
Micrograph of a crystal of MamM-CTD from M. gryphiswaldense MSR-1. Crystal dimensions are 98 × 142 × 310 µm.
Table 1. Diffraction data-collection and processing statistics for MamM-CTD.
Values in parentheses are for the highest resolution shell.
| Space group | C2221 |
| Unit-cell parameters (Å, °) | a = 37.13, b = 93.99, c = 53.29, α = β = γ = 90 |
| Resolution (Å) | 20.00–2.00 (2.03–2.00) |
| Reflections, total | 48043 |
| Reflections, unique | 6474 |
| R merge † (%) | 4.7 (33.0) |
| 〈I/σ(I)〉 | 48.20 (3.69) |
| Completeness (%) | 98.1 (93.7) |
| Multiplicity | 7.4 |
| Wavelength (Å) | 1.5417 |
R
merge = 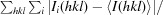
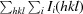 , where I
i(hkl) is the observed intensity of an individual reflection and 〈I(hkl)〉 is the mean intensity of that reflection.
, where I
i(hkl) is the observed intensity of an individual reflection and 〈I(hkl)〉 is the mean intensity of that reflection.
Table 2. Sequence identity and similarity of MamM and structurally determined CDF proteins.
MamM-CTD is one of the few magnetosome-associated proteins and the first CDF/CTD protein from a magnetotactic bacterium to be crystallized. Moreover, MamM-CTD is the first protein to be crystallized that is derived from a CDF protein that transports high amounts of iron and participates in the biomineralization process. To obtain phases and determine the MamM-CTD structure, molecular-replacement methodologies will be applied. For this protocol, the previously determined structures of FieF (YiiP), CzrB or TM0876 proteins will be consulted. Such molecular-replacement experiments are now in progress.
References
- Alberts, I. L., Nadassy, K. & Wodak, S. J. (1998). Protein Sci. 7, 1700–1716. [DOI] [PMC free article] [PubMed]
- Amemiya, Y., Arakaki, A., Staniland, S. S., Tanaka, T. & Matsunaga, T. (2007). Biomaterials, 28, 5381–5389. [DOI] [PubMed]
- Anton, A., Grosse, C., Reissmann, J., Pribyl, T. & Nies, D. H. (1999). J. Bacteriol. 181, 6876–6881. [DOI] [PMC free article] [PubMed]
- Bazylinski, D. A. & Frankel, R. B. (2004). Nature Rev. Microbiol. 2, 217–230. [DOI] [PubMed]
- Cherezov, V., Höfer, N., Szebenyi, D. M., Kolaj, O., Wall, J. G., Gillilan, R., Srinivasan, V., Jaroniec, C. P. & Caffrey, M. (2008). Structure, 16, 1378–1388. [DOI] [PMC free article] [PubMed]
- Delhaize, E., Kataoka, T., Hebb, D. M., White, R. G. & Ryan, P. R. (2003). Plant Cell, 15, 1131–1142. [DOI] [PMC free article] [PubMed]
- Evans, P. (2006). Acta Cryst. D62, 72–82. [DOI] [PubMed]
- Faivre, D. & Schüler, D. (2008). Chem. Rev. 108, 4875–4898. [DOI] [PubMed]
- Grass, G., Otto, M., Fricke, B., Haney, C. J., Rensing, C., Nies, D. H. & Munkelt, D. (2005). Arch. Microbiol. 183, 9–18. [DOI] [PubMed]
- Haney, C. J., Grass, G., Franke, S. & Rensing, C. (2005). J. Ind. Microbiol. Biotechnol. 32, 215–226. [DOI] [PubMed]
- Higuchi, T., Hattori, M., Tanaka, Y., Ishitani, R. & Nureki, O. (2009). Proteins, 76, 768–771. [DOI] [PubMed]
- Jogler, C., Kube, M., Schübbe, S., Ullrich, S., Teeling, H., Bazylinski, D. A., Reinhardt, R. & Schüler, D. (2009). Environ. Microbiol. 11, 1267–1277. [DOI] [PubMed]
- Jogler, C., Wanner, G., Kolinko, S., Niebler, M., Amann, R., Petersen, N., Kube, M., Reinhardt, R. & Schüler, D. (2011). Proc. Natl Acad. Sci. USA, 108, 1134–1139. [DOI] [PMC free article] [PubMed]
- Klumpp, S. & Faivre, D. (2012). PLoS One, 7, e33562. [DOI] [PMC free article] [PubMed]
- Komeili, A. (2007). Annu. Rev. Biochem. 76, 351–366. [DOI] [PubMed]
- Komeili, A. (2012). FEMS Microbiol. Rev. 36, 232–255. [DOI] [PMC free article] [PubMed]
- Komeili, A., Vali, H., Beveridge, T. J. & Newman, D. K. (2004). Proc. Natl Acad. Sci. USA, 101, 3839–3844. [DOI] [PMC free article] [PubMed]
- Lefèvre, C. T., Menguy, N., Abreu, F., Lins, U., Pósfai, M., Prozorov, T., Pignol, D., Frankel, R. B. & Bazylinski, D. A. (2011). Science, 334, 1720–1723. [DOI] [PubMed]
- Lohsse, A., Ullrich, S., Katzmann, E., Borg, S., Wanner, G., Richter, M., Voigt, B., Schweder, T. & Schüler, D. (2011). PLoS One, 6, e25561. [DOI] [PMC free article] [PubMed]
- Lu, M., Chai, J. & Fu, D. (2009). Nature Struct. Mol. Biol. 16, 1063–1067. [DOI] [PMC free article] [PubMed]
- Lu, M. & Fu, D. (2007). Science, 317, 1746–1748. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Montanini, B., Blaudez, D., Jeandroz, S., Sanders, D. & Chalot, M. (2007). BMC Genomics, 8, 107. [DOI] [PMC free article] [PubMed]
- Munkelt, D., Grass, G. & Nies, D. H. (2004). J. Bacteriol. 186, 8036–8043. [DOI] [PMC free article] [PubMed]
- Murat, D., Quinlan, A., Vali, H. & Komeili, A. (2010). Proc. Natl Acad. Sci. USA, 107, 5593–5598. [DOI] [PMC free article] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Outten, C. E. & O’Halloran, T. V. (2001). Science, 292, 2488–2492. [DOI] [PubMed]
- Paulsen, I. T. & Saier, M. H. (1997). J. Membr. Biol. 156, 99–103. [DOI] [PubMed]
- Persans, M. W., Nieman, K. & Salt, D. E. (2001). Proc. Natl Acad. Sci. USA, 98, 9995–10000. [DOI] [PMC free article] [PubMed]
- Scheffel, A. & Schüler, D. (2007). J. Bacteriol. 189, 6437–6446. [DOI] [PMC free article] [PubMed]
- Schüler, D. (2004). Arch. Microbiol. 181, 1–7. [DOI] [PubMed]
- Schüler, D. (2008). FEMS Microbiol. Rev. 32, 654–672. [DOI] [PubMed]
- Studier, F. W. (2005). Protein Expr. Purif. 41, 207–234. [DOI] [PubMed]
- Uebe, R., Junge, K., Henn, V., Poxleitner, G., Katzmann, E., Plitzko, J. M., Zarivach, R., Kasama, T., Wanner, G., Pósfai, M., Böttger, L., Matzanke, B. & Schüler, D. (2011). Mol. Microbiol. 82, 818–835. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.


