Nectin-4 belongs to a family of immunoglobulin-like cell adhesion molecules and is highly expressed in cancer cells. Recently, nectin-4 was found to be a receptor of measles virus and the IgV domain sustains strong binding to measles virus H protein. In this study, the successful expression and purification of human nectin-4 V domain (nectin-4v) is reported
Keywords: nectin-4, IgV domain, cell adhesion molecules
Abstract
Nectin-4 belongs to a family of immunoglobulin-like cell adhesion molecules and is highly expressed in cancer cells. Recently, nectin-4 was found to be a receptor of measles virus and the IgV domain sustains strong binding to measles virus H protein. In this study, the successful expression and purification of human nectin-4 V domain (nectin-4v) is reported. The purified protein was crystallized using the sitting-drop vapour-diffusion method. The crystals diffracted to 1.8 Å resolution and belonged to space group P21, with unit-cell parameters a = 33.1, b = 51.7, c = 56.9 Å, β = 94.7°. Preliminary analysis of the diffraction data was also performed.
1. Introduction
Nectins are cell adhesion molecules (CAMs) that belong to the immunoglobulin superfamily and play essential roles in Ca2+-independent intracellular interactions (Takahashi et al., 1999 ▶). The nectin family has four members: nectin-1, nectin-2, nectin-3 and nectin-4 (Takai & Nakanishi, 2003 ▶). In addition, five molecules named nectin-like (Necl) with domain structures similar to those of nectins have also been discovered. Both nectin and nectin-like molecules have an extracellular region with three immunoglobulin-like (Ig-like) domains (one V domain and two C2 domains; VCC), a single transmembrane region and a cytoplasmic tail region (Takai et al., 2003 ▶). The major difference between nectins and Necls is that the former can associate with afadin, an F-actin-binding protein, through their conserved C-terminal motif, while the latter cannot form any such association (Fuchs & Colonna, 2006 ▶). All members of the nectin family have two or three splicing variants, i.e. nectin-1α, nectin-1β, nectin-1γ, nectin-2α, nectin-2δ, nectin-3α, nectin-3c and nectin-3γ isoforms exist. Nectin-1, nectin-2 and nectin-3 are widely expressed in a variety of cells, including epithelial cells, neurons and fibroblasts. Nectin-2 and nectin-3 are also expressed in cells that lack cadherins, such as monocytes, spermatids and B cells (Takai & Nakanishi, 2003 ▶).
While nectin-1, nectin-2 and nectin-3 have been intensively studied, work on nectin-4 has been relatively rare. Based on the results of Takai et al. (2008 ▶), two splicing variants of nectin-4 exist, with the shorter one lacking amino acids 412–436. Like the other members of the nectin family, nectin-4 is involved in cell–cell adhesion by forming a homodimer or a heterodimer with nectin-1. Initially, human nectin-4 was mainly detected in the placenta (Reymond et al., 2001 ▶), but recent research has shown that nectin-4 is also highly expressed in cancer cells such as lung cancer, breast cancer and ovarian cancer cells (Fabre-Lafay et al., 2007 ▶; Takano et al., 2009 ▶; Derycke et al., 2010 ▶). As early detection of cancer is difficult owing to a lack of specific and effective tests, the potential use of nectin-4 as a cellular marker of cancer should be considered (Derycke et al., 2010 ▶). In addition, a recent report showed that nectin-4 can support measles virus entry and lateral spread of the virus in well differentiated primary human airway epithelial sheets (Mühlebach et al., 2011 ▶). The high expression of nectin-4 in cancer cells and its strong specificity for measles virus are also suggestive of a potential function of nectin-4 in oncolysis (Noyce et al., 2011 ▶). Therefore, as an important member of the nectin family, nectin-4 deserves further study of its structural features and cellular functions.
Several studies have demonstrated that viruses utilize the IgV domain of nectin or Necl molecules as the authentic binding entity (Zhang et al., 2008 ▶, 2011 ▶). The IgV domain of nectin-4 plays a critical role in cell adhesion and measle virus entry. Accordingly, the work of Mühlebach et al. (2011 ▶) has shown that the IgV domain of nectin-4 sustains strong binding to measles virus H protein. Thus, a high-resolution crystal structure of the V domain of nectin-4 (nectin-4v) should supply valuable information on the possible interaction between nectin-4 and measles virus, thereby facilitating future modelling studies and structure-directed drug design. In the present study, we report the successful expression, purification and crystallization of the nectin-4v protein. A data set was collected to 1.8 Å resolution using synchrotron radiation. Preliminary analysis of the diffraction data revealed a Matthews coefficient of 1.89 Å3 Da−1 and an estimated solvent content of about 35.0% for the crystal, with two nectin-4v molecules per asymmetric unit.
2. Materials and methods
2.1. Subcloning
The construction of the expression plasmid followed the canonical subcloning strategy. In brief, the DNA fragment encoding the V domain of human nectin-4 (nectin-4v; amino acids 32–145; gene accession No. NM_030916) was generated by PCR from PVRL4-pENTR221 (purchased from GeneCopoeia) using the following specific primer pair: 5′-GGAATTCCATATGGGCGAACTGGAAACCTCAGACGTG-3′ (forward) and 5′-CCGCTCGAGCACTCGGAGCCGCAGCCGCGC-3′ (reverse). The amplified product was then inserted into the NdeI and XhoI restriction sites of the pET-21a expression vector (Novagen, Darmstadt, Germany). The recombinant protein expressed from this vector contains an extra methionine residue (from the initial codon) at the N-terminus and additional leucine and glutamic acid residues (encoded by the XhoI sequence) and a hexahistidine tag at the C-terminus. The recombinant plasmid was transformed into DH5α (Novagen, Darmstadt, Germany) for plasmid amplification and was then verified by PCR, double enzyme digestion and direct DNA sequencing (GENEWIZ, Beijing, People’s Republic of China).
2.2. Expression and purification
The verified recombinant plasmid was transformed into Escherichia coli BL21 (DE3) cells and grown at 310 K overnight on a Luria–Bertani (LB) agar plate containing 100 µg ml−1 ampicillin. A single colony was inoculated into 50 ml LB medium containing 100 µg ml−1 ampicillin and incubated overnight. 20 ml of the preculture was then transferred into 2 l of fresh LB medium supplemented with ampicillin and grown at 310 K. When the culture density (OD600) reached approximately 0.6, isopropyl β-d-1-thiogalactopyranoside (IPTG; Sigma, Beijing, People’s Republic of China) was added to the cell culture to a final concentration of 0.1 mM and the cells were induced for 18 h at 289 K. The nectin-4v protein was expressed in soluble form with a C-terminal hexahistidine tag. The cells were harvested by centrifugation at 5000g for 10 min at 277 K and were resuspended in ice-cold PBS buffer consisting of 10 mM Na2HPO4, 2 mM KH2PO4, 2.7 mM KCl, 137 mM NaCl pH 7.4. The cell pellet was disrupted using an ultrasonic cell crusher.
Cell debris was removed by centrifugation at 15 000g for 30 min at 277 K. The crude protein extract was filtered through a 0.22 µm low-protein-binding membrane and loaded onto a 5 ml HisTrap HP column (GE Healthcare, Beijing, People’s Republic of China) equilibrated with PBS buffer at 277 K. The column was washed with five column volumes of buffer A (20 mM Tris–HCl pH 8.0, 50 mM NaCl) and the protein was eluted at room temperature with a gradient of 0–100% buffer B (20 mM Tris–HCl pH 8.0, 50 mM NaCl, 400 mM imidazole) using an ÄKTAexplorer system (GE Healthcare, Beijing, People’s Republic of China). Fractions containing the target protein were collected and applied onto a HiLoad 16/60 Superdex 75 column (GE Healthcare) at room temperature to further remove aggregates and impurities. For molecular-weight (MW) calculations, the Superdex column was calibrated in advance using the Low Molecular Weight (LMW) calibration kit (GE Healthcare) containing five standard proteins of MWs in the range 6500–75 000 Da. The purified nectin-4v protein was concentrated by centrifugation at 277 K using an Amicon Ultra-15 centrifugal filter device (3 kDa cutoff; Millipore, Beijing, People’s Republic of China). The protein buffer was then exchanged to 10 mM Tris–HCl, 10 mM NaCl pH 8.0. The concentration of the protein was measured using the Pierce BCA Protein Assay Kit (Thermo Scientific, Beijing, People’s Republic of China). The final protein preparation was immediately used for crystallization screening at concentrations of 5 and 10 mg ml−1.
2.3. Protein crystallization and data collection
Screening for crystallization conditions was carried out by mixing 1 µl protein solution with 1 µl reservoir solution and equilibrating against 100 µl reservoir solution in 48-well double-sample sitting-drop crystallization plates (XtalQuest, Beijing, People’s Republic of China). All crystallization attempts were performed at 291 K using the sitting-drop vapour-diffusion method. Crystal Screen, Crystal Screen 2 and Index (Hampton Research, California, USA) were used for crystallization trials. Crystals of good quality were obtained in a condition consisting of 5%(v/v) Tacsimate, 0.1 M HEPES pH 7.0, 10%(w/v) polyethylene glycol monomethyl ether 5000.
For data collection, a single crystal was picked up using a nylon loop, immersed into a cryoprotectant solution consisting of the reservoir solution with 20% glycerol for about 30 s and flash-cooled at 100 K. Diffraction data were collected using synchrotron radiation on beamline BL-5A (wavelength 1.0000 Å) at the Photon Factory, KEK, Japan. A total of 360 diffraction images were collected with an oscillation angle of 1°. Data were indexed, integrated and scaled using DENZO and SCALEPACK as implemented in HKL-2000 (Otwinowski & Minor, 1997 ▶). Detailed statistics are summarized in Table 1 ▶.
Table 1. Data-collection statistics.
Values in parentheses are for the outermost resolution shell.
| Space group | P21 |
| Unit-cell parameters | |
| a (Å) | 33.1 |
| b (Å) | 51.7 |
| c (Å) | 56.9 |
| β (°) | 94.7 |
| Wavelength (Å) | 1.00000 |
| Resolution range (Å) | 50.00–1.80 (1.86–1.80) |
| No. of observed reflections | 128137 |
| No. of unique reflections | 17741 |
| Completeness (%) | 100.0 (100.0) |
| Multiplicity | 7.2 (7.2) |
| R merge † (%) | 0.085 (0.406) |
| Average I/σ(I) | 23.2 (5.1) |
R
merge = 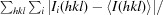
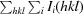 , where 〈I(hkl)〉 is the mean intensity of reflection I(hkl) and I
i(hkl) is the intensity of an individual measurement of reflection I(hkl).
, where 〈I(hkl)〉 is the mean intensity of reflection I(hkl) and I
i(hkl) is the intensity of an individual measurement of reflection I(hkl).
3. Results and discussion
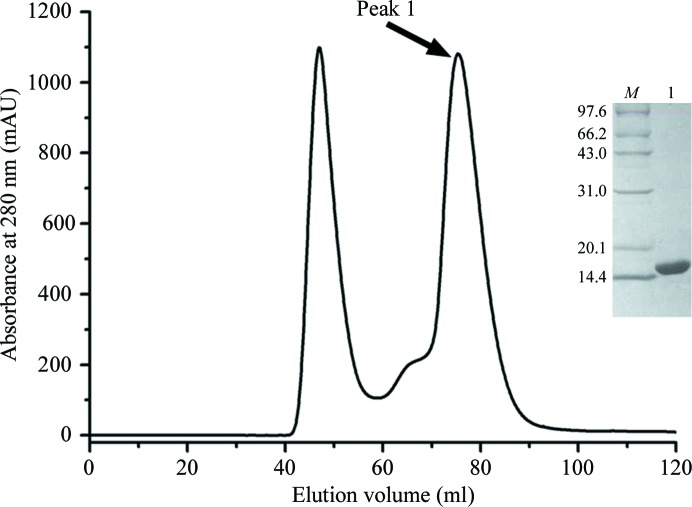
The human nectin-4 protein contains one IgV-like and two IgC-like domains in its extracellular region. The IgV-like domain of nectin-4 (nectin-4v) roughly spans Glu32–Pro145. This V-domain protein was expressed as a recombinant protein with a C-terminal hexahistidine tag in E. coli and was further purified by affinity and gel-filtration chromatography. As shown in Fig. 1 ▶, nectin-4v eluted as a symmetric peak at about 75.4 ml on a Superdex 75 column, corresponding to a protein with a molecular weight of about 15 kDa. Therefore, nectin-4v exists mainly as a monomer in solution. The final purity of nectin-4v was demonstrated to be greater than 99% on 15% SDS–PAGE (Fig. 1 ▶). The yield of pure nectin-4v protein was about 15 mg per litre of cell culture.
Figure 1.
Purification of the nectin-4v protein by gel filtration. A typical separation profile of nectin-4v on a Superdex 75 (HiLoad 16/60) column is shown. Proteins with an elution volume of 75.4 ml were collected and analyzed by 15% SDS–PAGE (inset). Lane M, standard protein molecular-weight markers (labelled in kDa); lane 1, peak 1.
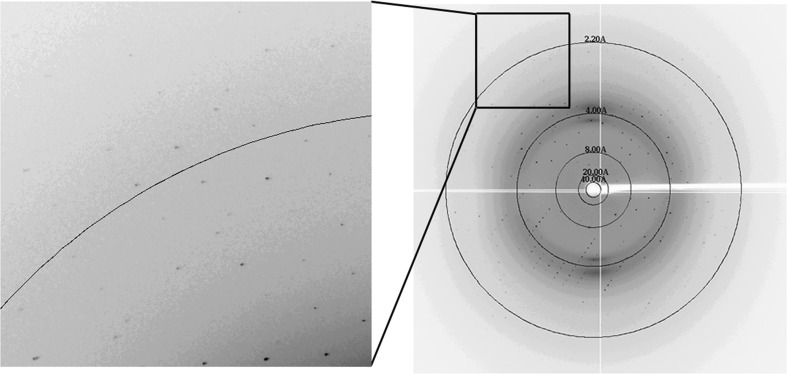
The initial crystallization screening trials for nectin-4v were carried out using kits from Hampton Research. The purified protein could easily be crystallized and protein crystals were obtained within a week at 291 K under several conditions. Good-quality crystals were finally selected from a condition consisting of 5%(v/v) Tacsimate, 0.1 M HEPES pH 7.0, 10%(w/v) polyethylene glycol monomethyl ether 5000. A crystal of about 0.2 × 0.1 × 0.04 mm in size (Fig. 2 ▶) diffracted to 1.8 Å resolution (Fig. 3 ▶). The data-processing statistics are summarized in Table 1 ▶. The space group of the crystal was P21 and the unit-cell parameters were a = 33.1, b = 51.7, c = 56.9 Å, β = 94.7°. Overall, the diffraction data were of good quality, with an R merge of only about 0.085 for all resolution shells from 50.0 to 1.80 Å. The Matthews coefficient (Matthews, 1968 ▶) for the crystal was calculated to be 1.89 Å3 Da−1, with an estimated solvent content of about 35.0%, which corresponds to two nectin-4v molecules in the asymmetric unit. Nevertheless, the self-rotation function [generated by POLARRFN (Winn et al., 2011 ▶) using data from 12 to 5 Å resolution with an integration radius of 30 Å] did not show any sign of twofold noncrystallographic symmetry. The largest off-origin peak in the native Patterson was 8.65% of the height of the origin peak, which suggested no significant pseudo-translation. We managed to determine the structure of nectin-4v with MOLREP (Vagin & Teplyakov, 2010 ▶) using the coordinates of nectin-1 residues 35–144 (PDB entry 3u83; Zhang et al., 2011 ▶) as the search model and we indeed found two molecules in the asymmetric unit. The R factor after the translation function was 0.618. The final nectin-4v model has been refined to R work = 0.1915 and R free = 0.2398. Further refinement and structural analysis are currently under way.
Figure 2.
Representative crystal of the nectin-4v protein. The dimensions of the crystal are about 0.2 × 0.1 × 0.04 mm.
Figure 3.
A representative diffraction image collected from a nectin-4v crystal. The resolution shells are shown.
Acknowledgments
This work was completed in the laboratory of Professor George F. Gao at the Institute of Microbiology, Chinese Academy of Sciences (IMCAS). We thank Dr Jianxun Qi for help with the collection and processing of the X-ray data.
References
- Derycke, M. S., Pambuccian, S. E., Gilks, C. B., Kalloger, S. E., Ghidouche, A., Lopez, M., Bliss, R. L., Geller, M. A., Argenta, P. A., Harrington, K. M. & Skubitz, A. P. (2010). Am. J. Clin. Pathol. 134, 835–845. [DOI] [PMC free article] [PubMed]
- Fabre-Lafay, S. et al. (2007). BMC Cancer, 7, 73. [DOI] [PMC free article] [PubMed]
- Fuchs, A. & Colonna, M. (2006). Semin. Cancer Biol. 16, 359–366. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Mühlebach, M. D. et al. (2011). Nature (London), 480, 530–533.
- Noyce, R. S., Bondre, D. G., Ha, M. N., Lin, L.-T., Sisson, G., Tsao, M.-S. & Richardson, C. D. (2011). PLoS Pathog. 7, e1002240. [DOI] [PMC free article] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 267, 307–326. [DOI] [PubMed]
- Reymond, N., Fabre, S., Lecocq, E., Adelaïde, J., Dubreuil, P. & Lopez, M. (2001). J. Biol. Chem. 276, 43205–43215. [DOI] [PubMed]
- Takahashi, K., Nakanishi, H., Miyahara, M., Mandai, K., Satoh, K., Satoh, A., Nishioka, H., Aoki, J., Nomoto, A., Mizoguchi, A. & Takai, Y. (1999). J. Cell Biol. 145, 539–549. [DOI] [PMC free article] [PubMed]
- Takai, Y., Ikeda, W., Ogita, H. & Rikitake, Y. (2008). Annu. Rev. Cell Dev. Biol. 24, 309–342. [DOI] [PubMed]
- Takai, Y., Irie, K., Shimizu, K., Sakisaka, T. & Ikeda, W. (2003). Cancer Sci. 94, 655–667. [DOI] [PMC free article] [PubMed]
- Takai, Y. & Nakanishi, H. (2003). J. Cell Sci. 116, 17–27. [DOI] [PubMed]
- Takano, A., Ishikawa, N., Nishino, R., Masuda, K., Yasui, W., Inai, K., Nishimura, H., Ito, H., Nakayama, H., Miyagi, Y., Tsuchiya, E., Kohno, N., Nakamura, Y. & Daigo, Y. (2009). Cancer Res. 69, 6694–6703. [DOI] [PubMed]
- Vagin, A. & Teplyakov, A. (2010). Acta Cryst. D66, 22–25. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Zhang, P., Mueller, S., Morais, M. C., Bator, C. M., Bowman, V. D., Hafenstein, S., Wimmer, E. & Rossmann, M. G. (2008). Proc. Natl Acad. Sci. USA, 105, 18284–18289. [DOI] [PMC free article] [PubMed]
- Zhang, N., Yan, J., Lu, G., Guo, Z., Fan, Z., Wang, J., Shi, Y., Qi, J. & Gao, G. F. (2011). Nature Commun. 2, 577. [DOI] [PubMed]