Crystallization of the complex of the transcription factor AmrZ with DNA was accomplished through the combination of established and newly developed methods. Here, a general method to obtain and optimize crystals of protein–DNA complexes consisting of these combined procedures is described.
Keywords: protein–DNA complexes, crystallization screens, Pseudomonas aeruginosa, transcription factors, AmrZ
Abstract
The AmrZ protein from the pathogenic bacterium Pseudomonas aeruginosa is a transcription factor that activates and represses the genes for several potent virulence factors, which gives the bacteria a selective advantage in infection. AmrZ was crystallized in complex with DNA containing the amrZ1 repressor binding site. Obtaining crystals of the complex required the integration of a number of well known techniques along with the development of new methods. Here, these processes are organized and combined into a comprehensive method which yielded diffraction-quality crystals. Part of this method included thorough data mining of the crystallization conditions of protein–DNA complexes to create a new directed crystallization screen. An optimized technique for the verification of protein–DNA complexes in crystals is also presented. Taken together, the methods described in this article attempt to streamline the difficult process of obtaining diffraction-quality crystals of protein–DNA complexes through the organization of older methods combined with the introduction of new techniques.
1. Introduction
The transcription factor AmrZ from the pathogenic bacterium Pseudomonas aeruginosa has been shown to both transcriptionally activate and repress the genes for many potent virulence factors which give the bacteria a selective advantage in infection (Baynham et al., 1999 ▶, 2006 ▶; Baynham & Wozniak, 1996 ▶; Tart et al., 2005 ▶; Ramsey et al., 2005 ▶). P. aeruginosa is a common, opportunistic, Gram-negative bacillus which is responsible for bacteremia in burn patients, endocarditis and nosocomial infections (Regules et al., 2008 ▶; Carruthers & Kanokvechayant, 1973 ▶; Richards et al., 1999 ▶). Perhaps most importantly, chronic P. aeruginosa lung infection leads to respiratory failure in 80–90% of patients with the autosomal recessive disorder cystic fibrosis (CF; Lyczak et al., 2002 ▶). Initial P. aeruginosa infection in CF patients is by planktonic bacteria which cause minor, acute infections; however, as time progresses complex bacterial biofilms form which mark the transition from an acute infection to a severe chronic infection (Costerton et al., 1999 ▶). One major component of P. aeruginosa biofilms is the exopolysaccharide alginate. This virulence factor gives P. aeruginosa a selective advantage in the CF lung by protecting the bacteria from antibiotic penetration and host defenses (Govan & Deretic, 1996 ▶). The enzymes involved in alginate biosynthesis are encoded in the algD operon, which is tightly regulated by the transcription factor AmrZ (Baynham & Wozniak, 1996 ▶). AmrZ is also responsible for the repression of flagellum biosynthesis through regulation of the fleQ gene, as well as being necessary for the proper surface localization and twitching motility of type IV pili (Tart et al., 2006 ▶; Baynham et al., 2006 ▶). In addition to regulating these three virulence factors of P. aeruginosa, AmrZ has also been shown to bind two sites on its own promoter, causing repression of its own transcription (Ramsey et al., 2005 ▶).
AmrZ is a 12.3 kDa transcription factor and a member of the ribbon–helix–helix family of DNA-binding proteins. Amino-acid sequence analysis of AmrZ reveals three domains: an extended N-terminal domain, a putative ribbon–helix–helix DNA-binding domain and a C-terminal domain. Both the N-terminal domain and the C-terminal domain lack any significant sequence similarity to other proteins; however, the putative ribbon–helix–helix domain has sequence similarity to the Arc and Mnt repressors from bacteriophage P22, with 36.6 and 28.2% identity, respectively (Huang & Miller, 1998 ▶). A critical element in understanding the transcriptional control of P. aeruginosa virulence factors is to determine the mechanism by which AmrZ interacts with its various binding sites, ultimately leading to different phenotypes within the bacteria. The structures of other ribbon–helix–helix proteins have previously been determined; however, these proteins lack the unique features which make AmrZ a novel ribbon–helix–helix DNA-binding protein (Schreiter & Drennan, 2007 ▶). These novel features of AmrZ include the extended N-terminal domain and a different consensus sequence in the DNA-binding β-sheet (Waligora et al., 2010 ▶). Additionally, with the exception of Helicobacter pylori NikR (Schreiter et al., 2003 ▶), AmrZ is the only ribbon–helix–helix protein which has been shown to have dual roles as both an activator and a repressor of transcription. Understanding the specific protein–nucleic acid interactions between AmrZ and its many binding sites on the P. aeruginosa chromosome is paramount in advancing future therapeutic strategies to eliminate these potent virulence factors.
Our first attempts to crystallize the AmrZ protein with and without DNA were unsuccessful. In these initial experiments, we started by using the 14 bp binding site from the algD operon determined by DNA footprinting (Baynham & Wozniak, 1996 ▶). AmrZ–algD complexes were formed by mixing the full-length protein and DNA in a 2:1 molar ratio. This ratio was chosen based upon biological information and the hypothesized protein:DNA stoichiometry. This complex was used in crystallization experiments with several commercially available screens. In each of these crystallization experiments we observed aggregation and precipitation in the majority of the conditions; however, no initial crystals were ever obtained. Even employing some of the standard methods of protein–DNA crystallography, such as varying the length and ends of the algD site, as performed in the structure of the lambda repressor in complex with the high-affinity OL1 binding site (Jordan et al., 1985 ▶), as well as limited proteolysis of the AmrZ protein, which was successful for the Max DNA-binding protein (Cohen et al., 1995 ▶), did not yield crystals. It became clear that varying individual parameters alone would not yield crystals and that a more systematic approach would be required. In this article, we describe the methods used to obtain diffraction-quality crystals of the AmrZ protein in complex with the amrZ1 repressor binding site. To obtain these crystals, biochemical and biophysical information about the biology of AmrZ was tied together with existing methods of protein–DNA crystallography. The methods used to obtain these crystals include the creation of a new directed crystallization screen based on a comprehensive review of protein–DNA complex structures, as well as the utilization of a gel-based method for the verification of protein and DNA substrate in crystals. Finally, we summarize these techniques into a comprehensive method which can be applied to the crystallization of many other protein–DNA complexes.
2. Materials and methods
2.1. Cloning and expression of the P. aeruginosa amrZ gene
The P. aeruginosa amrZ gene was amplified by PCR from the genomic strain PAO1 with the primers amrZ_F (5′-CGCCATCACATATGCGCCCACTGAAACAGGC) and amrZ_wt_R (5′-CGCCATCAGGATCCTCAGGCCTGGGCCAGCTC). The resulting gene product was ligated into a modified pET-19b (Novagen) expression vector which expresses the AmrZ protein fused with an N-terminal polyhistidine affinity tag. The affinity tag was linked by a sequence encoding the rhinovirus 3C protease recognition site which permits the removal of the polyhistidine tag by treatment with PreScission Protease (GE Healthcare). Δ42, Δ22 and Δ7 C-terminal truncation mutants of AmrZ were created by designing primers that encoded a stop codon after the codon encoding the 66th, 86th and 101st amino acid, respectively. The Δ42 full C-terminal truncation of AmrZ was created by amplifying the amrZ gene from the P. aeruginosa genomic strain PAO1 with the primers amrZ_F and amrZ_Δ42_R (5′-CGCCATCAGGATCCTCAAACACCGAGATTGTCTTG). The amplified product was then purified, ligated into a modified pET-19b vector and expressed as described above. The Δ22 and Δ7 constructs were created using the same methods, but with the reverse primers amrZ_Δ22_R (5′-CGCCATCAGGATCCTCACTGGCGGAAACGCTGC) and amrZ_Δ7_R (5′-CGCCATCAGGATCCTCAGTGTGCGATCAGGGCG) being used in the PCR reaction.
For overexpression of the AmrZ protein, Escherichia coli C41 (DE3) cells transformed with the pET-19b-amrZ vector were incubated at 310 K in 1 l Luria–Bertani broth supplemented with 50 µg ml−1 ampicillin until they reached an OD600 of 0.5. The cells were then cooled to 293 K on ice and induced with isopropyl β-d-1-thiogalactopyranoside to a final concentration of 1 mM. The cells were then incubated at 289 K for 20 h followed by harvesting by centrifugation at 5000g for 20 min at 277 K. The harvested cells were resuspended in lysis buffer (100 mM KH2PO4 pH 7.5, 500 mM NaCl, 10% glycerol, 4 M urea) and lysed using an EmusiFlex-C5 cell homogenizer (Avestin); the cell debris was pelleted by centrifugation at 30 000g. The cell-free extract was immediately loaded onto a 10 ml Ni–NTA column (Qiagen) pre-equilibrated with lysis buffer. The column was washed with ten column volumes of lysis buffer followed by 20 column volumes of wash buffer 1 (100 mM KH2PO4 pH 7.5, 500 mM NaCl, 10% glycerol, 35 mM imidazole, 3 M urea) and ten column volumes of wash buffer 2 (100 mM KH2PO4 pH 7.5, 500 mM NaCl, 10% glycerol, 50 mM imidazole, 2 M urea). To recover bound AmrZ protein, elution buffer (100 mM KH2PO4 pH 7.5, 500 mM NaCl, 10% glycerol, 500 mM imidazole, 1 M urea) was then passed over the column and fractions containing the AmrZ protein were pooled. The partial denaturing conditions introduced by the 4 M urea are necessary for protein solubility and column affinity during the first step of purification. Circular dichroism was utilized to ensure that the final purified protein was properly folded (data not shown). These fractions were treated with PreScission Protease (GE Healthcare) according to the manufacturer’s directions and dialyzed at 277 K for 16 h against 100 mM bis-Tris pH 5.5, 100 mM NaCl, 5% glycerol, 2 mM DTT, 0.5 mM EDTA. Cleaved AmrZ protein was then passed over a MonoS cation-exchange column (GE Healthcare) and eluted with a 100 mM–1 M gradient of NaCl. Peak fractions were analyzed for purity via SDS–PAGE and fractions containing pure AmrZ protein were pooled and dialyzed into buffer consisting of 100 mM bis-Tris pH 5.5, 100 mM NaCl, 2% glycerol. The protein was then concentrated to 20 mg ml−1, flash-cooled in liquid nitrogen and stored at 193 K. The molecular weight of each protein construct was verified by MALDI-TOF mass spectrometry (data not shown). Selenomethionine-derivatized Δ42 AmrZ was prepared using previously published methods (Doublié, 1997 ▶) and was purified using the same methods as described above, with the only change being the addition of 5 mM DTT to the final dialysis buffer.
2.2. Preparation of DNA oligonucleotides for crystallization
There are currently three known binding sites for the AmrZ protein on the P. aeruginosa genome. AmrZ binding to the algD promoter is necessary for the transcriptional activation of the genes required for alginate production, while AmrZ binding to the two binding sites on the amrZ promoter, amrZ1 and amrZ2, is required for transcriptional repression of AmrZ. In order to limit the total experimental search space, one of these three binding sites was chosen for the initial crystallographic experiments by utilizing information from protein–DNA affinity measurements. DNA-binding experiments employing fluorescence anisotropy were performed at each of the three binding sites. AmrZ binding to the amrZ1 site had the highest affinity, while binding to the amrZ2 and algD sites was approximately 14-fold and 25-fold weaker, respectively (Waligora et al., 2010 ▶). Using this information, the amrZ1 binding site was chosen for the first round of crystallographic experiments. The initial library of oligonucleotides for crystallization was created using the 14 bp amrZ1 recognition sequence and sequentially adding and removing base pairs to each end to change the DNA length. The library contained oligonucleotides ranging in size from 12 to 24 bp (Table 1 ▶).
Table 1. DNA oligonucleotides used in initial crystallographic experiments.
| Name | Sequence |
|---|---|
| amrZ1_12bp | GGCAAAACGCCG |
| CCGTTTTGCGGC | |
| amrZ1_14bp | TGGCAAAACGCCGG |
| ACCGTTTTGCGGCC | |
| amrZ1_16bp | CTGGCAAAACGCCGGC |
| GACCGTTTTGCGGCCG | |
| amrZ1_18bp | ACTGGCAAAACGCCGGCA |
| TGACCGTTTTGCGGCCGT | |
| amrZ1_20bp | TACTGGCAAAACGCCGGCAC |
| ATGACCGTTTTGCGGCCGTG | |
| amrZ1_22bp | GTACTGGCAAAACGCCGGCACG |
| CATGACCGTTTTGCGGCCGTGC | |
| amrZ1_24bp | GGTACTGGCAAAACGCCGGCACGC |
| CCATGACCGTTTTGCGGCCGTGCG |
DNA oligonucleotides were created by chemically synthesizing the forward and reverse strands (Integrated DNA Technologies). These oligonucleotides were resuspended at a concentration of 10 mM and the forward and reverse strands were annealed by mixing equal volumes of each strand with 10× annealing buffer (25 mM MES pH 5.5, 100 mM NaCl, 25 mM MgCl2) to give a final concentration of 4.55 mM. The DNA was then placed in a 373 K water bath and allowed to cool to room temperature.
2.3. Determination of protein constructs to use for crystallization
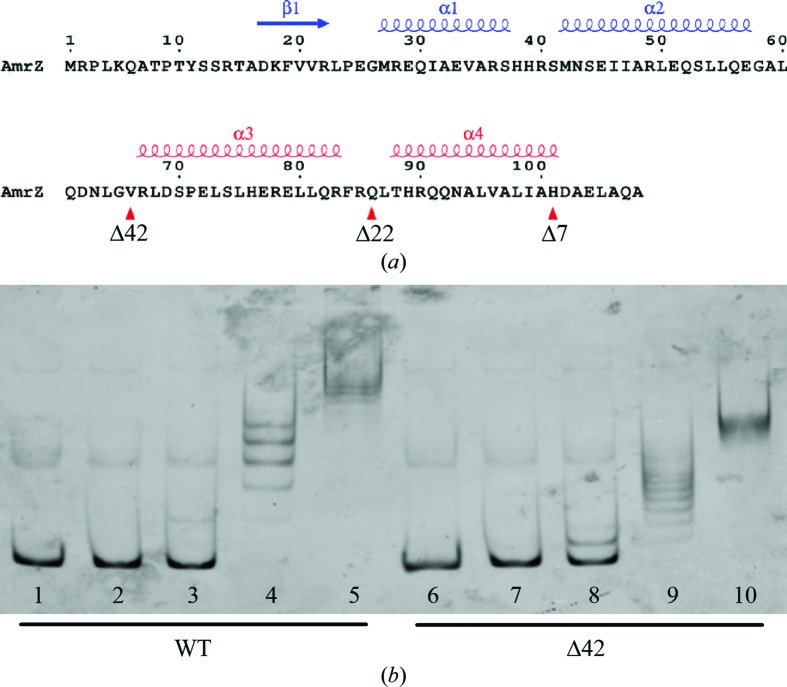
In addition to variations in DNA length, the protein constructs used in optimization were also optimized in order to increase the probability of successfully crystallizing the complex of AmrZ bound to its operator DNA. Previous electrophoretic mobility-shift assay (EMSA) experiments (Baynham & Wozniak, 1996 ▶) and glutaraldehyde cross-linking experiments (Waligora et al., 2010 ▶) showed that AmrZ forms higher order oligomers beyond the dimeric state through interactions of the C-terminal region. The dimeric form of the protein is necessary for DNA binding and these higher order and possibly irregular oligomeric structures could potentially interfere with crystal packing of the protein–DNA complex and may have contributed to the aggregation observed in the initial crystallization experiments. In an attempt to determine the region of AmrZ that is responsible for higher order protein oligomerization, we utilized the secondary-structure program JPred (Cuff et al., 1998 ▶) and the structure-modeling software Phyre (Kelley et al., 2000 ▶) to determine the regions of the various domains of AmrZ. The results from these programs show that the putative ribbon–helix–helix DNA-binding domain of AmrZ is located from residues 17 to 66 (Fig. 1 ▶ a). Using this prediction, we surmised that the C-terminal domain of AmrZ, located from residues 67 to 108, is responsible for the higher order oligomerization of the protein. Since our main goal in determining the structure of the AmrZ–DNA complex was to understand the specific protein–nucleic acid interactions responsible for AmrZ binding, we decided to create three additional protein constructs that truncate parts of the C-terminal domain of AmrZ but leave the ribbon–helix–helix DNA-binding domain intact. The C-terminal domain from residues 67 to 108 is predicted to be composed of two α-helices separated by a disordered region. The first construct, Δ42, is the complete C-terminal truncation mutant of the AmrZ protein. The Δ22 construct of AmrZ contains the putative ribbon–helix–helix domain but also includes the first predicted α-helix, while the final AmrZ construct, Δ7, contains the putative ribbon–helix–helix domain and the two predicted α-helices in the C-terminal domain, but truncates the last seven residues of the protein predicted to have aperiodic structure. After expression and purification, EMSA assays using an algD promoter fragment were performed as described previously (Waligora et al., 2010 ▶). The results of these assays confirm that each of these constructs retains wild-type DNA binding (Fig. 1 ▶ b).
Figure 1.
(a) AmrZ sequence and secondary structure. Deletion constructs were made for the AmrZ protein to reduce protein aggregation during crystallization experiments. The deletions were directed by secondary-structure prediction and identification of the ribbon–helix–helix domain (shown in blue) of the protein necessary for DNA binding. Deletions of 42 (Δ42), 22 (Δ22) and seven (Δ7) amino acids at the C-terminal end of the protein were made (positions indicated by red triangles) to truncate the protein between secondary-structural elements (shown in red). (b) Activity of the wild-type and Δ42 AmrZ constructs on a 300 bp fluorescently labeled algD promoter region. Lanes 1 and 6 contain no protein, lanes 2 and 7 contain 39 nM protein, lanes 3 and 8 contain 156.25 nM protein, lanes 4 and 9 contain 625 nM protein and lanes 5 and 10 contain 2500 nM protein.
2.4. Formation of the protein–DNA complex
After creating the DNA oligonucleotide library and determining the protein constructs to be used in the crystallization experiments, the next step was to optimize the conditions for forming a stable and homogeneous protein–DNA complex. The initial AmrZ–amrZ1 complex was formed by mixing AmrZ and a double-stranded oligonucleotide in a 2:1 molar ratio. The conditions for this reaction consisted of 810 µM (10 mg ml−1) AmrZ, 405 µM double-stranded DNA, 20 mM bis-tris pH 5.5, 100 mM NaCl and 2 mM DTT; however, upon the addition of the DNA the reaction became turbid and cloudy. Previously, the addition of a few microlitres of NaCl or centrifugation of the reaction was used to clear the precipitated components of the reaction. To avoid the precipitation of macromolecules in the reaction, a method was developed to screen many types of salts to determine which ones could be used in the reaction to keep the components soluble. The protein–DNA complex was formed as described above and the reaction was kept in the precipitated state. 2 µl of precipitated complex was then mixed with an equal volume of various salt solutions at concentrations of 0, 10, 50 and 100 mM. These salts contained many of the common cations found in crystallization conditions, including Li+, Na+, K+, Mg2+ and Ca2+, and anions such as Cl−, sulfate, acetate and formate. The reactions were sealed to prevent evaporation and incubated at 277 K for 2 h. Each condition was then evaluated for precipitate. The conditions with the lowest amount of precipitate contained MgSO4. Further refinement revealed that 50 mM MgSO4 was the minimum concentration required to prevent precipitation of the protein–DNA complex. The final conditions for forming the AmrZ–amrZ1 complex were 810 µM AmrZ, 405 µM amrZ1, 20 mM bis-tris pH 5.5, 100 mM NaCl, 50 mM MgSO4, 2 mM DTT.
2.5. Creation of the PEG–DNA crystallization screen
The success of this project relied on finding appropriate crystallization conditions for the AmrZ protein bound to DNA. Currently, many macromolecular crystallization screens are commercially available; however, relatively few are specifically designed for protein–nucleic acid complexes. The most common commercially available screens are the Natrix and Natrix 2 screens offered by Hampton Research, the Nucleix Suite created by Qiagen and the JBScreen Nuc-Pro screen offered by Jena Biosystems. Although useful, these three screens contain many conditions that are also designed to crystallize only nucleic acids, which limit the number of conditions specifically formulated for protein–nucleic acid complexes. We have created a 48-condition screen, called the PEG–DNA screen, with conditions that are specifically directed towards the crystallization of protein–DNA complexes (Table 2 ▶). To create this screen, the Biological Macromolecule Crystallization Database (http://xpdb.nist.gov:8060/BMCD4) was searched to identify the crystallization conditions of many protein–DNA complexes (Tung & Gallagher, 2009 ▶). By using the search term macromol:‘protein’ AND macromol:‘DNA’ AND Content:‘complex’, crystallization conditions for 264 deposited structures of protein–DNA complexes were returned. Individual searching of each entry led to the removal of six entries that only contained protein, leaving the final count of crystallization conditions for protein–DNA complexes at 258. Various conditions from these entries were analyzed, such as pH, buffer, precipitant and salt.
Table 2. Directed crystallization screen for protein–DNA complexes.
| No. | Precipitant | Buffer | Salt |
|---|---|---|---|
| 1 | 6% PEG 8K | 0.1 M sodium acetate pH 4.6 | None |
| 2 | 16% PEG 8K | 0.1 M MES pH 6.0 | 0.1 M CaCl2, 0.1 M NaCl |
| 3 | 11% PEG 4K | 0.1 M HEPES pH 7.5 | 0.05 M MgCl2 |
| 4 | 20% PEG 8K | 0.1 M HEPES pH 7.5 | 0.1 M CaCl2 |
| 5 | 6% PEG 4K | 0.1 M sodium acetate pH 4.6 | 0.05 M CaCl2 |
| 6 | 16% PEG 4K | 0.1 M HEPES pH 7.5 | 0.05 M MgCl2, 0.1 M NaCl |
| 7 | 11% PEG 2K | 0.1 M MES pH 6.0 | 0.1 M NaCl |
| 8 | 20% PEG 2K | 0.1 M MES pH 6.0 | 0.05 M CaCl2, 0.1 M NaCl |
| 9 | 16% PEG 2K | 0.1 M sodium acetate pH 4.6 | 0.1 M CaCl2, 0.1 M NaCl |
| 10 | 25% PEG 2K | 0.1 M HEPES pH 7.5 | 0.1 M CaCl2, 0.1 M NaCl |
| 11 | 20% PEG 4K | 0.1 M MES pH 6.0 | 0.1 M MgCl2 |
| 12 | 11% PEG 8K | 0.1 M sodium acetate pH 4.6 | None |
| 13 | 25% PEG 4K | 0.1 M MES pH 6.0 | 0.1 M MgCl2, 0.1 M NaCl |
| 14 | 20% PEG 2K | 0.1 M HEPES pH 7.5 | 0.1 M NaCl |
| 15 | 11% PEG 8K | 0.1 M sodium acetate pH 4.6 | 0.1 M CaCl2 |
| 16 | 6% PEG 2K | 0.1 M MES pH 6.0 | None |
| 17 | 11% PEG 2K | 0.1 M sodium acetate pH 4.6 | 0.05 M MgCl2, 0.1 M NaCl |
| 18 | 20% PEG 4K | 0.1 M sodium acetate pH 4.6 | 0.1 M NaCl |
| 19 | 16% PEG 4K | 0.1 M MES pH 6.0 | 0.05 M MgCl2 |
| 20 | 6% PEG 8K | 0.1 M MES pH 6.0 | 0.1 M CaCl2, 0.1 M NaCl |
| 21 | 20% PEG 8K | 0.1 M sodium acetate pH 4.6 | 0.1 M MgCl2, 0.1 M NaCl |
| 22 | 11% PEG 4K | 0.1 M HEPES pH 7.5 | 0.1 M CaCl2 |
| 23 | 16% PEG 2K | 0.1 M HEPES pH 7.5 | None |
| 24 | 6% PEG 4K | 0.1 M sodium acetate pH 4.6 | 0.1 M NaCl |
| 25 | 16% PEG 2K | 0.1 M sodium acetate pH 4.6 | 0.05 M CaCl2, 0.1 M NaCl |
| 26 | 20% PEG 4K | 0.1 M MES pH 6.0 | 0.05 M CaCl2, 0.1 M NaCl |
| 27 | 6% PEG 2K | 0.1 M MES pH 6.0 | 0.1 M CaCl2, 0.1 M NaCl |
| 28 | 11% PEG 8K | 0.1 M HEPES pH 7.5 | 0.1 M CaCl2 |
| 29 | 20% PEG 8K | 0.1 M HEPES pH 7.5 | 0.05 M CaCl2 |
| 30 | 16% PEG 4K | 0.1 M HEPES pH 7.5 | 0.1 M CaCl2 |
| 31 | 11% PEG 8K | 0.1 M MES pH 6.0 | None |
| 32 | 6% PEG 8K | 0.1 M HEPES pH 7.5 | None |
| 33 | 16% PEG 8K | 0.1 M MES pH 6.0 | 0.1 M MgCl2, 0.1 M NaCl |
| 34 | 20% PEG 2K | 0.1 M sodium acetate pH 4.6 | 0.1 M CaCl2, 0.1 M NaCl |
| 35 | 11% PEG 4K | 0.1 M sodium acetate pH 4.6 | 0.1 M NaCl |
| 36 | 6% PEG 4K | 0.1 M MES pH 6.0 | 0.05 M CaCl2, 0.1 M NaCl |
| 37 | 6% PEG 4K | 0.1 M HEPES pH 7.5 | 0.05 M CaCl2 |
| 38 | 16% PEG 8K | 0.1 M sodium acetate pH 4.6 | 0.05 M CaCl2, 0.1 M NaCl |
| 39 | 20% PEG 2K | 0.1 M MES pH 6.0 | 0.1 M CaCl2 |
| 40 | 11% PEG 2K | 0.1 M sodium acetate pH 4.6 | 0.1 M MgCl2, 0.1 M NaCl |
| 41 | 11% PEG 4K | 0.1 M sodium acetate pH 4.6 | 0.1 M CaCl2, 0.1 M NaCl |
| 42 | 6% PEG 2K | 0.1 M MES pH 6.0 | None |
| 43 | 20% PEG 8K | 0.1 M HEPES pH 7.5 | 0.1 M CaCl2, 0.1 M NaCl |
| 44 | 16% PEG 4K | 0.1 M HEPES pH 7.5 | 0.05 M MgCl2, 0.1 M NaCl |
| 45 | 6% PEG 2K | 0.1 M HEPES pH 7.5 | 0.05 M CaCl2 |
| 46 | 6% PEG 8K | 0.1 M sodium acetate pH 4.6 | 0.05 M MgCl2 |
| 47 | 11% PEG 8K | 0.1 M MES pH 6.0 | 0.05 M CaCl2, 0.1 M NaCl |
| 48 | 25% PEG 4K | 0.1 M HEPES pH 7.5 | 0.1 M CaCl2 |
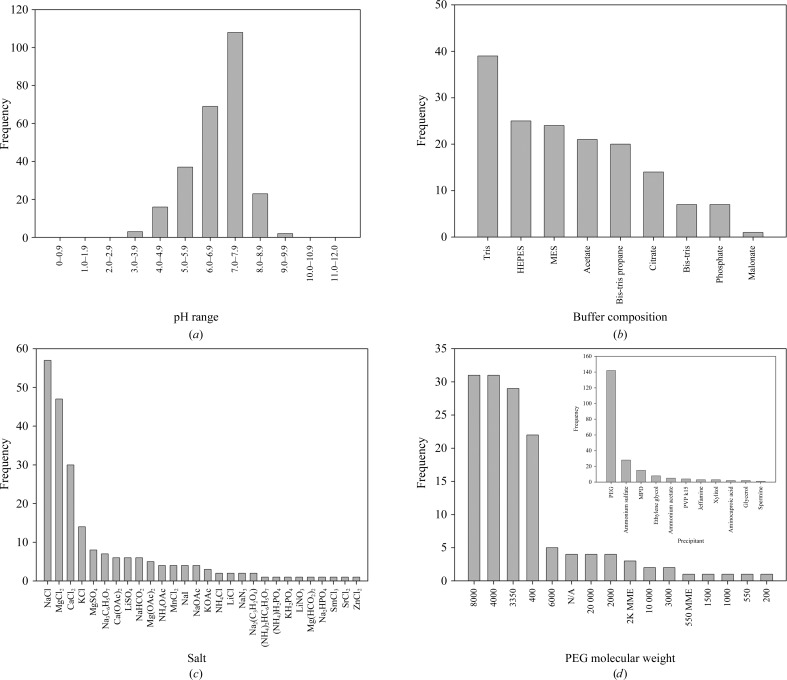
Our search shows that protein–DNA complexes crystallize in a wide pH range from 3.8 to 9.1 (Fig. 2 ▶ a). The majority of protein–DNA crystals are obtained between pH 7.0 and 7.9 (41.9%), with the next highest range from pH 6.0 to 6.9 (26.7%). The largest number of protein–DNA complexes crystallized in Tris buffer (Fig. 2 ▶ b). The pH of Tris buffer is dependent on temperature; we wanted buffers that could be used in a general screen at different temperatures. HEPES, MES and acetate buffer were the next three commonest buffers used to obtain crystals; combining these data with the pH data, three buffer/pH combinations were selected for use in the directed screen. These three buffers are sodium acetate pH 4.6, MES pH 6.5 and HEPES pH 7.5. The lower pH for sodium acetate was selected in an attempt to vary the search space and explore a wider range of pH conditions.
Figure 2.
Crystallization conditions for protein–DNA complexes. Data mining of the Biomolecular Crystallization Database (BMCD; Tung & Gallagher, 2009 ▶) yielded conditions used in the crystallization of many protein–DNA complexes. (a) The pH used in 258 crystallization conditions shows that the majority of protein–DNA complexes crystallize between pH 7.0 and 7.9, with an overall preference for slightly acidic pH. (b) Tris was used as the predominate buffer for crystallization, but was excluded from the directed screen owing to the variation in pH at different temperatures. The next three most commonly used buffers were HEPES, MES and sodium acetate. (c) In the 155 conditions that reported salts a total of 28 different salts were used, with the largest majority of conditions containing sodium chloride, magnesium chloride and calcium chloride. (d) Of the 193 conditions that reported precipitant composition, 74% contained some variation of PEG (inset). Of these conditions, there is a strong preference for PEG 8000, PEG 4000 and PEG 3350 in protein–DNA crystals.
Out of the 258 crystallization conditions identified for protein–DNA complexes, 155 conditions reported the salts used to crystallize the complex (Fig. 2 ▶ c). Although 28 different salts were used in these crystallization conditions, a large majority (36.8%) of these conditions contained sodium chloride. Magnesium chloride and calcium chloride were the next two commonest salts used and were seen in 30.3 and 19.4% of the crystallization conditions, respectively. These data led us to select sodium chloride, magnesium chloride and calcium chloride for the directed screen.
The final component of the crystallization conditions that we analyzed was the precipitant, which was provided for 193 of the 258 conditions. Out of the 11 different precipitants used, various molecular weights of polyethylene glycol (PEG) were used in 73.6% of the conditions (Fig. 2 ▶ d). The next commonest used precipitant was ammonium sulfate, which was used in only 14.5% of the conditions. When these results are compared with the crystallization conditions for all of the proteins in the Biological Macromolecule Crystallization Database, where PEG is used 46.5% of the time and ammonium sulfate is used 20.4% of the time, it can be inferred that there is a strong preference for PEG in the crystallization conditions for protein–DNA complexes. Looking in more depth at the 142 conditions containing PEG (Fig. 2 ▶ d), PEG 8000 and PEG 4000 were both used the most often (21.9% of conditions). In an attempt to screen across a wide range of PEG molecular weights, PEG 2000 was chosen as the third precipitant in the directed screen.
To create the PEG–DNA screen, five concentrations (6, 11, 16, 20 and 25%) of each molecular weight PEG (2K, 4K and 8K) were chosen, along with 0.1 M of each of the three buffers (sodium acetate pH 4.6, MES pH 6.0 and HEPES pH 7.5). Each condition contains either no salt, 0.1 M NaCl, 0.05/0.1 M MgCl2 or 0.05/0.1 M CaCl2. Conditions with MgCl2 and CaCl2 could also contain 0.1 M NaCl. Since exploring all combinations of these conditions would yield 450 unique conditions (three PEGs × five PEG concentrations × three buffers × ten salt combinations), incomplete factorial methods were utilized to narrow the search space. Following methods described previously, each crystallization experiment had one value for each variable and combinations were chosen randomly, but were balanced so that each value was represented approximately the same number of times to ensure that the effect of each variable could be assessed from the larger group of conditions (Carter & Carter, 1979 ▶; Abergel et al., 1991 ▶; Yin & Carter, 1996 ▶). The final PEG–DNA screen contains 48 unique conditions specifically directed towards the crystallization of protein–DNA complexes. Interestingly, PEG 3350 was used in a large number (20.4%) of the crystallization conditions (Fig. 2 ▶ d). From this observation, we paired the PEG–DNA screen with the 48-condition PEG/Ion screen (Hampton Research), which only uses PEG 3350 as a precipitant, offering a 96-condition screen.
2.6. Crystallization and optimization of AmrZ–amrZ1 crystals
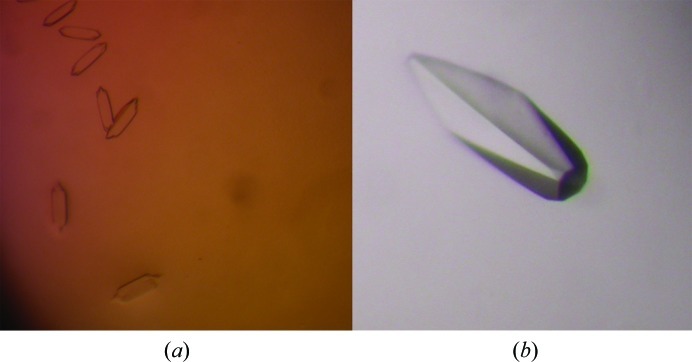
After determining the optimal AmrZ binding site for crystallization, creating the oligonucleotide library, choosing the four protein constructs and optimizing the conditions to form the protein–DNA complex, crystallization experiments were performed. Each of the four protein constructs was combined with one of the seven double-stranded oligonucleotides from the library in a pairwise fashion. Each AmrZ construct was also screened in the absence of DNA to give a total of 32 unique complexes to be screened. Each complex was screened through the 48 conditions of the newly created PEG–DNA screen and the 48 conditions from the PEG/Ion Screen as well as a number of other commercially available screens at room temperature. Initial crystals were observed in many conditions of the PEG–DNA screen using the Δ42 AmrZ–18 bp amrZ1 complex. Of these initial crystals, the best crystals in terms of size and morphology were found in condition No. 20 of the PEG–DNA screen (6% PEG 8K, 0.1 M MES pH 6.0, 0.1 M CaCl2, 0.1 M NaCl), which measured approximately 2.5 × 6.25 µm (Fig. 3 ▶ a). This condition was further optimized by varying the PEG, salt and protein concentrations. During optimization, a switch from native Δ42 AmrZ to selenomethionine-derivatized Δ42 AmrZ was made to facilitate phasing. Various additives were added to the crystallization conditions, with 2 mM TCEP pH 8.0 having the greatest effect on crystal growth. Finally, the greatest improvement in crystal size was achieved by screening different protein:DNA ratios in the complex. As seen in both the crystallization of the Trp and lac repressor proteins bound to DNA (Joachimiak et al., 1987 ▶; Cohen et al., 1995 ▶), a molar excess of DNA in the crystallization experiment, beyond the biological stochiometric ratio, was needed to obtain crystals. By using a 2:1.5 ratio of Δ42 AmrZ to 18 bp amrZ1 DNA, the crystals increased in size to 375 × 100 µm (Fig. 3 ▶ b). The final crystallization conditions for the Δ42 AmrZ–18 bp amrZ1 complex were 3% PEG 8K, 0.1 M MES pH 6.0, 0.15 M NaCl, 2 mM TCEP pH 8.0. Crystallization experiments were carried out at 298 K and crystals appeared after approximately one week. For cryoprotection, the effects of many different additives were tested both by growing the crystals in the presence of the additive and by soaking. The best results were obtained by soaking the crystals in a solution of 5% PEG 8K, 0.1 M MES pH 6.0, 0.15 M NaCl, 20% 2-methyl-1,3-propanediol (MPD) for 1 min before immediately cooling the crystal in liquid nitrogen. These crystals were screened at our home facility, with the best crystals diffracting to approximately 3.2 Å resolution.
Figure 3.
Crystals of the Δ42 AmrZ–18 bp amrZ1 complex. (a) Initial crystals of the Δ42 AmrZ–18 bp amrZ1 complex from condition No. 20 of the directed PEG–DNA screen. These crystals measured approximately 2.5 × 6.25 µm. (b) Systematic variations of the crystallization conditions yielded optimized crystals of the AmrZ–amrZ1 complex which measured approximately 375 × 100 µm. Additionally, selenomethionine-derivatized Δ42 AmrZ was used in crystallization to obtain experimental phase information.
2.7. Verification of protein–DNA complex in crystals
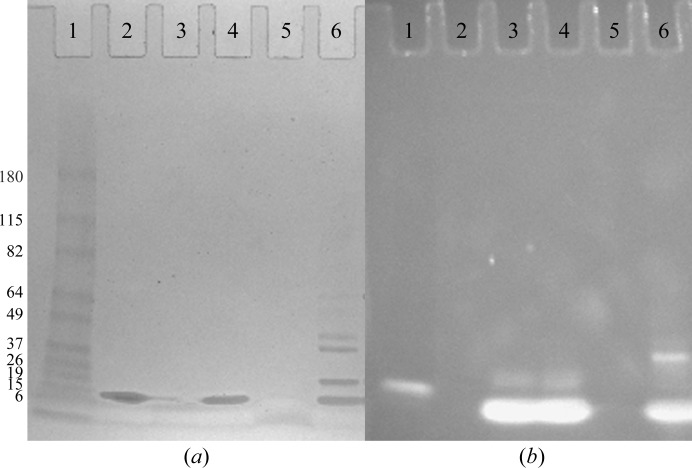
One major obstacle in the crystallization of protein–DNA complexes is verifying that the crystals contain both the protein and the DNA. Most often, this is verified at the end of the structure-determination process when diffraction data are collected and electron density for the DNA is present. We have optimized a gel-based method which can be used to determine whether both the protein and DNA components are present in the initial crystals before optimization. There are several advantages of this gel-based method over other methods, such as UV absorbance (Joachimiak et al., 1987 ▶), to determine the crystal composition of protein–DNA complexes. This gel-based method can be performed with smaller amounts of sample material, it can detect either single- or double-stranded nucleic acids and the method is not dependent on the extinction coefficients of the protein and nucleic acid. Approximately 10–15 crystals which were too small for diffraction experiments were harvested and washed in three separate drops containing the well solution to remove any dissolved macromolecules in the crystal solution. After the third wash, the crystals were transferred to a 10 µl drop containing 1% SDS and 1 M urea. 3 µl of 6× SDS loading buffer was added to the drop and the crystals were dissolved by a combination of vortexing and boiling. The sample was resolved on a 4–15% gradient SDS–PAGE gel (Fig. 4 ▶, lane 6) along with approximately 2.5 µg purified Δ42 AmrZ (lane 2), 1 µg 18 bp amrZ1 DNA (lane 3) and a lane containing a mixture of 2.5 µg purified Δ42 AmrZ and 1 µg 18 bp amrZ1 DNA (lane 4) as controls. Lane 5 of the gel contained a sample from the third wash drop to ensure that the bands observed from dissolving the crystals came from the crystals and not the buffer in the crystallization reaction. After resolving the samples, the gel was first washed for 15 min in water followed by staining for protein with GelCode Blue Stain Reagent (Thermo Scientific) for 1 h. The gel was then washed with water for 45 min, followed by staining with 1× SYBR Gold Nucleic Acid Gel Stain (Invitrogen) dissolved in TBE buffer for 45 min. Protein bands on the gel were visualized with visible light (Fig. 4 ▶ a), while the DNA bands were visualized with ultraviolet light (302 nm wavelength; Fig. 4 ▶ b). Lane 6 of the gel shows the presence of protein consistent with the molecular weight of the purified Δ42 AmrZ protein control, in addition to a DNA band consistent with the molecular weight of the 18 bp amrZ1 DNA. The additional bands observed in lane 6 are likely to be the result of the fact that AmrZ forms very stable oligomeric structures in solution (dimers and tetramers) and we often observe these species on gels, even under denaturing conditions. The three control experiments (lanes 2–4) show that there is no cross reactivity between the GelCode Blue Stain Reagent and the SYBR Gold Nucleic Acid Gel Stain. The DNA band seen in the molecular-weight standards (lane 1) could possibly arise from an interaction between the SYBR Gold stain and one of the protein standards; however, the composition of these standards is not available from the manufacturer (BenchMark Pre-Stained Protein Ladder, Invitrogen). Compared with other methods of staining acrylamide gels containing both protein and nucleic acid, such as ethidium bromide and silver staining, this dual stain method allows independent visualization of the protein and nucleic acid species, and can also visualize both single- and double-stranded nucleic acids. Additionally, this method is useful if the protein and DNA species migrate to the same position on a gel, as both species can be observed independently. This gel confirmed that these crystals contained both the AmrZ protein and the amrZ1 DNA.
Figure 4.
Gel-based method to verify that crystals contained protein–DNA complex. Crystals were harvested, washed and dissolved before resolving via SDS–PAGE. The gel was stained for protein using GelCode Blue Stain Reagent (Thermo Scientific) prior to being stained for DNA with SYBR Gold Nucleic Acid Gel Stain (Invitrogen). Visible-light (a) and ultraviolet-light (b) exposures of the gel were used to visualize protein and DNA bands, respectively. Lane 1, molecular-weight markers (labeled in kDa); lane 2, 2.5 ng purified Δ42 AmrZ protein; lane 3, 1 ng 18 bp amrZ1 DNA; lane 4, mixture of 2.5 ng Δ42 AmrZ protein and 1 ng 18 bp amrZ1 DNA; lane 5, smple from crystal wash drop; lane 6, sample from dissolved crystals.
2.8. Data collection/verification of DNA in the structure
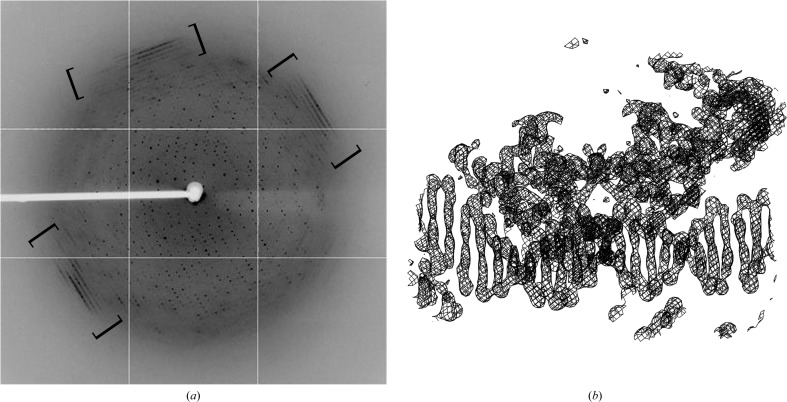
X-ray diffraction data from selenomethionine-derivatized Δ42 AmrZ–DNA crystals were collected to 3.1 Å resolution at 100 K on beamline X25 of the NSLS at Brookhaven National Laboratory (Fig. 5 ▶ a). The data were collected at the selenium peak, with an X-ray wavelength of 0.9793 nm. The data were indexed in space group I422. The diffraction images of the Δ42 AmrZ–18 bp amrZ1 crystals showed diffuse diffraction located at 3.4 Å resolution similar to that observed for the 434 repressor in complex with the 14 bp OL2 binding site (Anderson et al., 1984 ▶; Fig. 5 ▶ a, bracketed region). This diffuse diffraction was our second indication, in addition to visualizing both protein and DNA species on a gel, that these crystals consisted of a protein–DNA complex. X-ray diffraction data were scaled and integrated using HKL-2000 (Otwinowski & Minor, 1997 ▶). The expected complex in the crystal is two dimers of AmrZ bound to the DNA. The calculated Matthews coefficient (V M) is 3.94 Å3 Da−1 for a single complex or 1.97 Å3 Da−1 for two complexes within the asymmetric unit; both of these are within the probability curve for all protein–DNA complexes in the PDB. Single-wavelength anomalous dispersion (SAD) was used to phase the data and was performed using SOLVE (Terwilliger & Berendzen, 1999 ▶) and RESOLVE (Terwilliger, 2000 ▶). The initial electron-density maps obtained from this data set confirmed the presence of DNA, which exists as long continuous helices throughout the crystals (Fig. 5 ▶ b).
Figure 5.
Data-collection image from a crystal containing the Δ42 AmrZ–18 bp amrZ1 complex. (a) Data were collected on beamline X25 at the NSLS, Brookhaven, New York, USA. Crystals diffracted to 3.1 Å resolution and were indexed in space group I422. Diffuse diffraction believed to be caused by long helical segments of DNA was observed (bracketed regions), providing further evidence that these crystals contained a protein–DNA complex. (b) The presence of DNA in these crystals was verified in the electron density, in which a clear double helix can be observed. The 2F o − F c map contoured at 2σ is shown.
3. Results and discussion
DNA-binding proteins play an important role in many areas of biological homeostasis and are tasked with important processes critical to the survival of all organisms. Understanding these biological mechanisms, as well as advancing progress in structure-based drug-design initiatives, hinges on elucidating the specific protein–DNA interactions at an atomic level. As of the 1 January 2012 release of the Protein Data Bank (PDB; Berman et al., 2002 ▶), a total of 78 207 structures of biological macromolecules have been deposited, the majority of which (69 470) were determined by X-ray crystallography. Of these, DNA-binding proteins in complex with their substrate account for 3.3% of the deposited structures. In humans, E. coli and yeast, proteins containing DNA-binding domains account for 8.1, 6.2 and 3.9% of the total genomic transcripts, respectively (Babu et al., 2004 ▶). In this paper, using the AmrZ protein as an example, we discuss a systematic approach for obtaining diffraction-quality crystals of this protein in complex with its DNA substrate which can further be generalized to any protein–nucleic acid complex. An advantage of a general approach is that it can be further optimized for any individual case by taking into account the specific considerations for that case. For example, if a specific nucleic acid-binding protein is known to be especially redox-sensitive then various reducing reagents could be further included to optimize crystallization. Likewise, if the formation of a protein–nucleic acid complex is temperature-sensitive then the screening procedures can be carried out at the desired temperature.
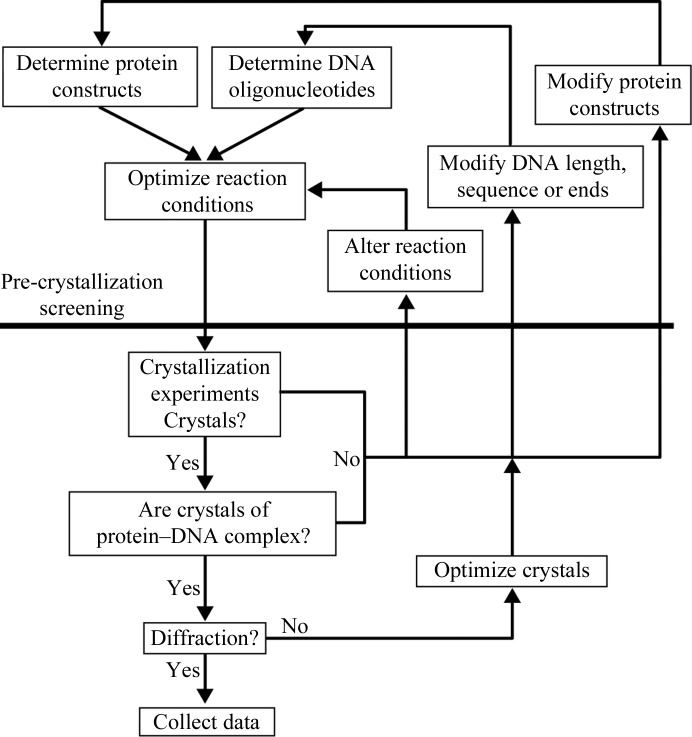
The methods used to obtain diffraction-quality crystals of the Δ42 AmrZ–18 bp amrZ1 complex can be used as a general method for obtaining crystals of most protein–DNA complexes. These methods are summarized in Fig. 6 ▶. Careful attention must be paid to the choice and design of both the initial protein constructs and the initial DNA constructs used in crystallization by utilizing existing knowledge about the biology of the system in the pre-crystallization stages to limit the total search space explored during the crystallographic experiments. The PEG–DNA screen, which was created by in-depth analysis of the crystallographic conditions of many existing structures of protein–DNA complexes, is a directed screen that is especially useful when a limited amount of starting material is available. One way to alter the crystallographic conditions is through modifications of the DNA library, which can be performed by screening additional lengths of DNA and adding both 3′ and 5′ overhangs to the DNA, as well as more drastic measures such as changing the target binding site. Once initial crystals have been obtained, optimization can be carried out by multiple methods such as the screening of various additives and altering the protein:DNA ratio used to form the complex, as well as varying the initial crystallization conditions to vary the pH, the temperature and the concentration of salt and PEG and protein.
Figure 6.
Flowchart of the comprehensive method for protein–DNA crystallography. This article stresses the point that important considerations in pre-crystallization screening are vital to the success of obtaining crystals of protein–DNA complexes. Combining information from multiple sources, such as biochemical experiments and secondary-structural prediction, provides fruitful information to carefully design protein and DNA constructs for crystallization and optimal conditions to form the protein–DNA complex.
Crystallization of protein–nucleic acid complexes can be a very time-consuming endeavor, so ensuring that the crystals are composed of both protein and nucleic acid at an early stage is a critical piece of information. In the crystallization of the Δ42 AmrZ–18 bp amrZ1 complex, we employ multiple methods throughout the crystallization process to ensure that the composition of the crystals includes both protein and nucleic acid, which is ultimately verified by observing the electron density. We present a method for assessing crystals for the presence of protein and DNA through gel electrophoresis with Coomassie Blue staining to visualize the band for the protein and a nucleic acid-specific stain, SYBR-Gold, to visualize the DNA.
Acknowledgments
This work was supported by a Cystic Fibrosis Foundation Student Traineeship, PRYOR10H0 (EEP), and Public Health Service grants AI061396 and HL58334 (DJW). We would like to thank Annie Heroux and the staff at the National Synchrotron Light Source (Brookhaven National Laboratory, Upton, New York, USA) for their generous help in the collection of X-ray data.
References
- Abergel, C., Moulard, M., Moreau, H., Loret, E., Cambillau, C. & Fontecilla-Camps, J. C. (1991). J. Biol. Chem. 266, 20131–20138. [PubMed]
- Anderson, J., Ptashne, M. & Harrison, S. C. (1984). Proc. Natl Acad. Sci. USA, 81, 1307–1311. [DOI] [PMC free article] [PubMed]
- Babu, M. M., Luscombe, N. M., Aravind, L., Gerstein, M. & Teichmann, S. A. (2004). Curr. Opin. Struct. Biol. 14, 283–291. [DOI] [PubMed]
- Baynham, P. J., Brown, A. L., Hall, L. L. & Wozniak, D. J. (1999). Mol. Microbiol. 33, 1069–1080. [DOI] [PubMed]
- Baynham, P. J., Ramsey, D. M., Gvozdyev, B. V., Cordonnier, E. M. & Wozniak, D. J. (2006). J. Bacteriol. 188, 132–140. [DOI] [PMC free article] [PubMed]
- Baynham, P. J. & Wozniak, D. J. (1996). Mol. Microbiol. 22, 97–108. [DOI] [PubMed]
- Berman, H. M. et al. (2002). Acta Cryst. D58, 899–907. [DOI] [PubMed]
- Carruthers, M. M. & Kanokvechayant, R. (1973). Am. J. Med. 55, 811–818. [DOI] [PubMed]
- Carter, C. W. Jr & Carter, C. W. (1979). J. Biol. Chem. 254, 12219–12223. [PubMed]
- Cohen, S. L., Ferré-D’Amaré, A. R., Burley, S. K. & Chait, B. T. (1995). Protein Sci. 4, 1088–1099. [DOI] [PMC free article] [PubMed]
- Costerton, J. W., Stewart, P. S. & Greenberg, E. P. (1999). Science, 284, 1318–1322. [DOI] [PubMed]
- Cuff, J. A., Clamp, M. E., Siddiqui, A. S., Finlay, M. & Barton, G. J. (1998). Bioinformatics, 14, 892–893. [DOI] [PubMed]
- Doublié, S. (1997). Methods Enzymol. 276, 523–530. [PubMed]
- Govan, J. R. & Deretic, V. (1996). Microbiol. Rev. 60, 539–574. [DOI] [PMC free article] [PubMed]
- Huang, X. & Miller, W. (1998). Adv. Appl. Math. 12, 337–357.
- Joachimiak, A., Marmorstein, R. Q., Schevitz, R. W., Mandecki, W., Fox, J. L. & Sigler, P. B. (1987). J. Biol. Chem. 262, 4917–4921. [PubMed]
- Jordan, S. R., Whitcombe, T. V., Berg, J. M. & Pabo, C. O. (1985). Science, 230, 1383–1385. [DOI] [PubMed]
- Kelley, L. A., MacCallum, R. M. & Sternberg, M. J. (2000). J. Mol. Biol. 299, 499–520. [DOI] [PubMed]
- Lyczak, J. B., Cannon, C. L. & Pier, G. B. (2002). Clin. Microbiol. Rev. 15, 194–222. [DOI] [PMC free article] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Ramsey, D. M., Baynham, P. J. & Wozniak, D. J. (2005). J. Bacteriol. 187, 4430–4443. [DOI] [PMC free article] [PubMed]
- Regules, J. A., Glasser, J. S., Wolf, S. E., Hospenthal, D. R. & Murray, C. K. (2008). Burns, 34, 610–616. [DOI] [PubMed]
- Richards, M. J., Edwards, J. R., Culver, D. H. & Gaynes, R. P. (1999). Crit. Care Med. 27, 887–892. [DOI] [PubMed]
- Schreiter, E. R. & Drennan, C. L. (2007). Nature Rev. Microbiol. 5, 710–720. [DOI] [PubMed]
- Schreiter, E. R., Sintchak, M. D., Guo, Y., Chivers, P. T., Sauer, R. T. & Drennan, C. L. (2003). Nature Struct. Biol. 10, 794–799. [DOI] [PubMed]
- Tart, A. H., Blanks, M. J. & Wozniak, D. J. (2006). J. Bacteriol. 188, 6483–6489. [DOI] [PMC free article] [PubMed]
- Tart, A. H., Wolfgang, M. C. & Wozniak, D. J. (2005). J. Bacteriol. 187, 7955–7962. [DOI] [PMC free article] [PubMed]
- Terwilliger, T. C. (2000). Acta Cryst. D56, 965–972. [DOI] [PMC free article] [PubMed]
- Terwilliger, T. C. & Berendzen, J. (1999). Acta Cryst. D55, 849–861. [DOI] [PMC free article] [PubMed]
- Tung, M. & Gallagher, D. T. (2009). Acta Cryst. D65, 18–23. [DOI] [PubMed]
- Waligora, E. A., Ramsey, D. M., Pryor, E. E., Lu, H., Hollis, T., Sloan, G. P., Deora, R. & Wozniak, D. J. (2010). J. Bacteriol. 192, 5390–5401. [DOI] [PMC free article] [PubMed]
- Yin, Y. & Carter, C. W. Jr (1996). Nucleic Acids Res. 24, 1279–1286. [DOI] [PMC free article] [PubMed]