Abstract
Hearing loss, which is genetically heterogeneous, can be caused by mutations in the mitochondrial DNA (mtDNA). The A1555G mutation of the 12S ribosomal RNA (rRNA) gene in the mtDNA has been associated with both aminoglycoside-induced and non-syndromic hearing loss in many ethnic populations. Here, we report for the first time the clinical and genetic characterization of nine Korean pedigrees with aminoglycoside-induced and non-syndromic hearing loss. These Korean families carry in the A1555G mutation of 12S rRNA gene and exhibit variable penetrance and expressivity of hearing loss. Specifically, the penetrance of hearing loss in these families ranged between 28.6% and 75%, with an average of 60.8%. These results were higher than the 29.8% penetrance that was previously reported in a Chinese population but similar to the 65.4% and 54.1% penetrance observed in a large Arab-Israeli population and nineteen Spanish pedigrees, respectively. The mutational analysis of the complete mtDNA genome in these families showed that the haplogroups of the Korean population, which belongs to the eastern Asian population, were similar to those of the Chinese population but different from the Spanish population, which belongs to the European-Caucasian population. The mtDNA variants that may act as modifier factors were also found to be similar to the Chinese population. Although the mtDNA haplogroups and variants were similar to the eastern Asian population, we did find some differing phenotypes, although some subjects had the same variants. This result suggests that both the ethnic background and environmental factors lead to a variable phenotype of the A1555G mutation.
Introduction
Hearing loss is the most common sensorineural disorder in humans, affecting one in 1000 newborns and 10% and 50% of people aged 65 and 80 years or older, respectively [1]. It is genetically heterogeneous and can be caused by mitochondrial DNA (mtDNA) mutations [2]. MtDNA mutations have been reported in both non-syndromic and syndromic hearing loss such as Kearns-Sayre Syndrome (KSS) [3], myoclonic epilepsy and ragged red fibers (MERRF) [4], mitochondrial encephalopathy, lactic acidosis and stroke-like episodes (MELAS) [5], maternally inherited diabetes and deafness (MIDD) [6] and are associated with presbycusis [7], [8].
The A1555G mutation of the 12S ribosomal RNA (rRNA) gene in the mtDNA is associated with both aminoglycoside-induced and non-syndromic hearing loss in many ethnic populations [9], [10], [11], [12]. Transition of A to G results in an additional G–C pair in the 12S rRNA gene, which has been predicted to encode an aminoglycoside binding based on sequence similarity to the bacterium Escherichia coli [13], [14]. In addition, sporadic aminoglycoside-induced hearing loss has been reported [15], and even without the use of antibiotics, non-syndromic hearing loss also occurs in ethnic families. Families with hearing loss caused by the A1555G mutation in the 12S rRNA gene have variable phenotypes, including varying severity, age of onset, and penetrance [2], [9],[16],[17],[18]. Penetrance appears to differ with the use of aminoglycosides, even within the same pedigree [19]. The variable phenotypes of hearing loss in persons carrying the A1555G mutation are difficult to explain because it is a single point mutation. Therefore, additional modifying factors, including the mtDNA haplogroup, nuclear DNA or mtDNA variations and aminoglycoside antibiotics, have been proposed to be associated with the variable phenotypic expression [18], [20], [21]. A nuclear modifier gene, tRNA 5-methylaminomethyl-2-thiouridylate methyltransferase (TRMU), has been identified, and this gene encodes a highly conserved mitochondrial protein related to transfer RNA (tRNA) modification [22]. The mtDNA variations that have been shown to influence the variable phenotype of hearing loss associated with the A1555G mutation are tRNALys T5802C [18] and G5821A [19], tRNASer(UCN) G7444A [23], tRNAArg T10454C [24], tRNAGlu A14693G [24], [25], tRNAThr T15908C [24] and G15927A [18], and T12338C [18] in the ND5 gene. Ten eastern Asian haplogroups, including A, B, C, D, F, G, M, N, R and Y, have been detected in Chinese pedigrees carrying the A1555G mutation [26]. Seven European haplogroups have been detected in Spanish pedigrees, including H, I, J, K, T, U and K [27]. Notably, the frequency of the A1555G mutation is much higher in haplogroups D and H in the Chinese and Spanish populations, respectively, than the other haplogroups [26], [27]. However, the A1555G mutation has been detected in all of the haplogroups, suggesting that the A1555G mutation in the mtDNA occurred sporadically and persisted over generations. Chinese pedigrees carrying the haplogroups C, Y and F2 have been shown to have higher penetrance than the pedigrees carrying the other haplogroups [26].
In the present study, we investigated the association of modifier factors and variable phenotypes of hearing loss in Korean pedigrees carrying the A1555G mutation. We performed clinical, molecular, and genetic characterizations of the pedigrees, including a sequence analysis of the complete mtDNA genome.
Subjects and Methods
Subjects and Audiological Evaluation
A total of 281 unrelated Korean subjects with non-syndromic hearing loss participated for the mtDNA A1555G mutation screening. All the subjects were subjected to appropriate audiological examinations, including pure-tone audiometry (PTA) and/or auditory brainstem response (ABR). The average of pure-tone audiometry (PTA) was calculated from the average of the audiometric thresholds at 500, 1000, 2000, and 3000 Hz. The severity of hearing loss was classified as follows: normal <26 decibels (dB), mild; 26–40 dB, moderate; 41–70 dB, severe; 71–90 dB, and profound; >90 dB. The subjects with the mtDNA A1555G mutation were subjected to a comprehensive history interview and physical examination to identify other symptoms and their history of aminoglycoside use. All subjects provided written informed consent according to the protocol approved by the Ethics Committee of Kyungpook National University Hospital prior to the study.
Mutation and Haplogroup Analysis of the mtDNA Genome
Genomic DNA was extracted from the peripheral blood of subjects using the Qiagen Flexigene DNA Extraction Kit (Qiagen, Hilden, Germany). PCR amplification of the mitochondrial 12S rRNA gene was performed using the following primers: forward, 5′- tggctttaacatatctgaacaca-3′, and reverse, 5′-ctcctaagtgtaagttgggtgct-3′. For the identification of the A1555G mutation, the PCR products were analyzed using PCR-RFLP with BsmAI (New England Biolabs, Ipswich, MA, USA) [28]. To confirm the A1555G mutation, the PCR products were purified with the Exo-SAP enzyme (USB, Cleveland, OH, USA) and analyzed through direct sequencing on an ABI 3130 Genetic Analyzer (Applied Biosystems Corps., Foster City, CA, USA) using the Big-Dye Terminator Cycle Sequencing Kit (Applied Biosystems Corps., Foster City, CA, USA).
The complete mtDNA sequences of the subjects with the A1555G mutation were amplified (Table S1), purified with the Exo-SAP enzyme (USB, Cleveland, OH, USA), and analyzed through direct sequencing. All of the mtDNA sequences were compared with the updated consensus Cambridge Sequence (GenBank accession number: NC_012920).
Mutation Analysis of the GJB2 and TRMU Genes
PCR amplification of the exon 2 of the GJB2 gene was performed using the following primers: forward, 5′-gcattcgtcttttccagagc-3′, and reverse, 5′-cctcatccctctcatgctgt-3′. The PCR products were purified with the Exo-SAP enzyme (USB, Cleveland, OH, USA) and analyzed through direct sequencing. The results were compared with the sequence of the wild-type GJB2 gene (GenBank accession number: NM_004004) to identify mutations. For the identification of the TRMU gene mutation G28T (A10S), the PCR amplification of exon 1 of the TRMU gene was performed using a previously reported primer, and the PCR products were analyzed using PCR-RFLP with Bsp1286I (New England Biolabs, Ipswich, MA, USA) [22]. The digested products were analyzed on a 2% agarose gel.
Results
Mutational Screening of the 12S rRNA Gene in Korean Subjects with Non-syndromic Hearing Loss
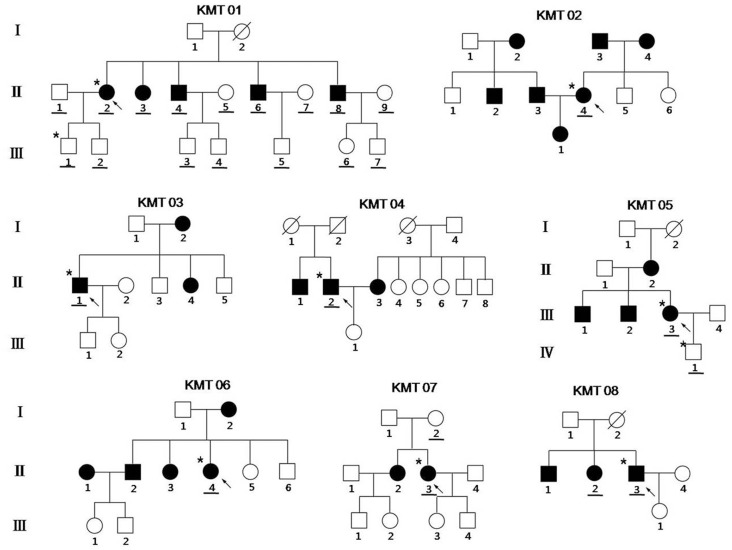
We performed a sequence analysis to identify the A1555G mutation in 281 Korean unrelated subjects with non-syndromic hearing loss, excluding those subjects with complete autosomal recessive inheritance patterns. First, the genomic DNA of each subject was amplified using the appropriate primers, and these products were digested using BsmAI and analyzed on a 2% agarose gel. Nine of the subjects had the A1555G mutation, which was further confirmed using PCR and subsequent DNA sequence analysis. Next, we performed mutational screening for the A1555G mutation in the available matrilineal relatives of those subjects except for KMT09 family who was not available for pedigree data. The A1555G mutation was detected in all matrilineal relatives. The penetrance of hearing loss (affected matrilineal relatives/total matrilineal relatives) of the eight pedigrees ranged from 28.6% to 75%, with an average of 60.8% (Fig. 1) [17], [19], [23], [24].
Figure 1. Eight Korean pedigrees presenting with nonsyndromic hearing loss were carrying the A1555G mtDNA mutation.
Hearing-impaired individuals are indicated by filled symbols. Arrows denote probands. Subjects used for whole mtDNA sequence analysis are indicated by asterisks. Subjects used for the A1555G mutation screening are underlined.
Clinical and Genetic Evaluation of the Nine Korean Pedigrees
We obtained a comprehensive history and performed physical and audiological examinations to identify any syndromic symptoms, the history of aminoglycoside use and genetic factors in all of the available subjects of the nine pedigrees carrying the A1555G mutation (Table 1). The results showed that the probands and members of the nine pedigrees showed no other clinical abnormalities, including diabetes, muscular diseases, visual dysfunction, and neurological disorders.
Table 1. Summary of clinical features and molecular data for nine patients carrying the A1555G mutation.
| Patient no | Gender | Age at test | Age of onset (Pre- or Postlingual) | Audiometric configuration | Exposure to aminoglycosides | PTA | (dB)* | Degree of Hearing loss | mtDNA haplogroup |
| Right ear | Left ear | ||||||||
| KMT 01 | F | 51 | – | Slope | No | 90 | 93 | Severe | D4b1b1a |
| KMT 02 | F | 26 | Congenital (prelingual) | Slope | No | 101 | 94 | Severe | D4a |
| KMT 03 | M | 49 | – | Slope | – | 105 | 96 | Profound | M7a1a |
| KMT 04 | M | 57 | Childhood (postlingual) | – | – | 111 | 112 | Profound | D5a2a |
| KMT 05 | F | 41 | R: Late childhood, L: 38 (postlingual) | Slope | No | 104 | 101 | Profound | D5b1b1 |
| KMT 06 | F | 47 | Late childhood (postlingual) | Slope | No | 97 | 102 | Profound | G1a1a |
| KMT 07 | F | 45 | Childhood (postlingual) | Slope | Yes | 73 | 67 | Moderate | D4a |
| KMT 08 | M | 50 | Congenital (prelingual) | Slope | No | 103 | 117 | Profound | M11b |
| KMT 09 | F | 67 | Childhood (postlingual) | Slope | Yes | 102 | 95 | Profound | D4 |
PTA, pure-tone audiometry; dB, decibels.
The probands of each pedigree exhibited hearing loss ranging from moderate to profound, with a slope-shaped pattern of audiological evaluation (Fig. 2). Only the probands of the KMT 07 and KMT 09 families had a history of exposure to aminoglycosides. For the age at onset, the probands of the KMT 02 and KMT 08 families showed prelingual hearing loss, and those of the KMT 04, KMT 07 and KMT 09 families had postlingual hearing loss (Table 1).
Figure 2. Air audiograms for pure tone audiometry (PTA) of the available subjects with the A1555G mutation.
Arrows indicate no responses; Symbols, (X) left ear (O) right ear; dB HL, decibel hearing level.
The examination of the clinical information of the KMT 01 pedigree (Fig. 1 and 2 and Table 1) revealed that subjects II-3 and II-6 of the proband’s siblings (II-3, II-4, II-6, and II-8) had profound hearing loss with a flat-shaped pattern, and subjects II-4 and II-8 had severe hearing loss with a slope-shaped pattern. However, the proband’s son, subject III-1, had normal hearing, with only high-frequency hearing loss (30 dB in the right ear and 40 dB in the left ear at 8000 Hz). Husband II-3 of proband II-4 in the KMT 02 family had acquired hearing loss, and the proband’s daughter III-1 had congenital hearing loss. The reason for the hearing loss of subject III-1 was not known. Proband III-3 in the KMT 05 family had prelingual hearing loss in the right ear, but the hearing in the left ear became poor at 38 years of age. The onset of hearing loss in her siblings (III-1 and III-2) occurred during childhood and adulthood, respectively. In generations II and III, the penetrance of the A1555G mutation was 100%, but subject IV-1 had normal hearing. The proband’s siblings II-1 and II-2 of the KMT 08 family all had hearing loss, and the hearing loss of proband II-3 and subject II-2 was congenital.
Haplogroup Analysis of the mtDNA Genome
To determine whether mtDNA variants or haplogroups modulated the variable phenotype of hearing loss in patients carrying the A1555G mutation, we performed a complete mtDNA sequence analysis of the probands and matrilineal members of the nine pedigrees. As shown in Table 2, the members of each pedigree had distinct mtDNA polymorphisms. Of the known nucleotide variations in the complete mtDNA sequence, we found thirty-five in the D-loop, six in the tRNA gene, nine in the 12S rRNA gene, and seven in the 16S rRNA gene. There were 83 variants in the protein-coding genes, including 56 silent variants and 27 missense variants. One novel variant of the 27 amino acid substitution variants was G3496A in the ND1 gene, which resulted in the substitution of the amino acid alanine with threonine (Table 2).
Table 2. mtDNA variants in nine Korean families with hearing loss.
| Conservation | KMT 01 | KMT 01 | KMT 02 | KMT 03 | KMT 04 | KMT 05 | KMT 05 | KMT 06 | KMT 07 | KMT 07 | KMT 08 | KMT 09 | ||||
| Gene | Position | Replacement | (H/B/M/X)a | II-2 | III-1 | II-4 | II-1 | II-2 | III-3 | IV-1 | II-4 | II-3 | I-2 | II-3 | Previously reportedb | |
| D-loop | 44 | C>CC | CC | Yes | ||||||||||||
| 73 | A>G | G | G | G | G | G | G | G | G | G | G | G | G | Yes | ||
| 146 | T>C | C | Yes | |||||||||||||
| 150 | C>T | T | T | T | T | Yes | ||||||||||
| 152 | T>C | C | C | C | C | Yes | ||||||||||
| 183 | A>G | G | G | Yes | ||||||||||||
| 215 | A>G | G | Yes | |||||||||||||
| 228 | G>A | A | A | Yes | ||||||||||||
| 263 | A>G | G | G | G | G | G | G | G | G | G | G | G | G | Yes | ||
| 310 | T>CTC | CTC | CTC | CCTC | CTC | CTC | CCTC | CCTC | TC | CTC | CTC | CTC | CTC | Yes | ||
| 318 | T>C | C | Yes | |||||||||||||
| 326 | A>G | G | Yes | |||||||||||||
| 431 | C>T | T | T | Yes | ||||||||||||
| 456 | C>T | T | T | Yes | ||||||||||||
| 489 | T>C | C | C | C | C | C | C | C | C | C | C | C | C | Yes | ||
| 515 | Del AC | Del AC | Del AC | Del AC | Del AC | Yes | ||||||||||
| 681 | T>C | C | C | Yes | ||||||||||||
| 16092 | T>C | C | Yes | |||||||||||||
| 16129 | G>A | A | A | A | Yes | |||||||||||
| 16164 | A>G | G | Yes | |||||||||||||
| 16182 | A>C | C | C | C | Yes | |||||||||||
| 16183 | A>C | C | C | C | Yes | |||||||||||
| 16189 | T>C | C | C | C | Yes | |||||||||||
| 16209 | T>C | C | Yes | |||||||||||||
| 16223 | C>T | T | T | T | T | T | T | T | T | T | T | T | T | Yes | ||
| 16266 | C>T | T | Yes | |||||||||||||
| 16287 | C>T | T | T | Yes | ||||||||||||
| 16319 | G>A | A | A | Yes | ||||||||||||
| 16324 | T>C | C | Yes | |||||||||||||
| 16325 | T>C | C | Yes | |||||||||||||
| 16357 | T>C | C | C | Yes | ||||||||||||
| 16362 | T>C | C | C | C | C | C | C | C | C | C | C | Yes | ||||
| 16399 | A>G | G | G | Yes | ||||||||||||
| 16497 | A>G | G | Yes | |||||||||||||
| 16519 | T>C | C | C | C | C | C | Yes | |||||||||
| 12s rRNA | 709 | G>A | G/G/A/− | A | Yes | |||||||||||
| 750 | A>G | A/G/G/− | G | G | G | G | G | G | G | G | G | G | G | G | Yes | |
| 752 | C>T | T | Yes | |||||||||||||
| 1048 | C>T | T | T | Yes | ||||||||||||
| 1095 | T>C | C | Yes | |||||||||||||
| 1107 | T>C | T/CT/T | C | C | C | Yes | ||||||||||
| 1310 | C>T | T | Yes | |||||||||||||
| 1438 | A>G | A/A/A/G | G | G | G | G | G | G | G | G | G | G | G | Yes | ||
| 1462 | G>A | A | Yes | |||||||||||||
| 1555 | A>G | A/A/A/A | G | G | G | G | G | G | G | G | G | G | G | G | Yes | |
| 16s rRNA | 1811 | A>G | G | Yes | ||||||||||||
| 2626 | T>C | C | Yes | |||||||||||||
| 2706 | A>G | A/G/A/A | G | G | G | G | G | G | G | G | G | G | G | G | Yes | |
| 2772 | C>T | T | Yes | |||||||||||||
| 3010 | G>A | A | A | A | A | A | A | Yes | ||||||||
| 3107 | Del C | Del C | Del C | Del C | Del C | Del C | Del C | Del C | Del C | Del C | Del C | Del C | Del C | Yes | ||
| 3206 | C>T | T | T | T | Yes | |||||||||||
| ND1 | 3316 | G>A (Ala to Thr) | A | Yes | ||||||||||||
| 3496 | G>A (Ala to Thr) | A/A/L/S | A | A | No | |||||||||||
| 3759 | A>G | G | G | Yes | ||||||||||||
| TQ | 4386 | T>C | C | Yes | ||||||||||||
| ND2 | 4769 | A>G | G | G | G | G | G | G | G | G | G | G | G | G | Yes | |
| 4793 | A>G | G | Yes | |||||||||||||
| 4833 | A>G (Thr to Ala) | G | Yes | |||||||||||||
| 4859 | T>C | C | C | Yes | ||||||||||||
| 4883 | C>T | T | T | T | T | T | T | T | T | T | Yes | |||||
| 4958 | A>G | G | Yes | |||||||||||||
| 5108 | T>C | C | Yes | |||||||||||||
| 5147 | G>A | A | A | Yes | ||||||||||||
| 5153 | A>G | G | G | Yes | ||||||||||||
| 5178 | C>A (Leu to Met) | L/T/T/T | A | A | A | A | A | A | A | A | A | Yes | ||||
| 5276 | A>G | G | G | Yes | ||||||||||||
| 5301 | A>G (Ile to Val) | I/I/M/L | G | G | G | Yes | ||||||||||
| TC | 5802 | T>C | T/T/T/C | C | C | Yes | ||||||||||
| NC5 | 5895 | C>CC | CC | CC | Yes | |||||||||||
| CO1 | 6253 | T>C (Met to Thr) | C | C | Yes | |||||||||||
| 6410 | C>T | T | T | Yes | ||||||||||||
| 6455 | C>T | T | Yes | |||||||||||||
| 6531 | C>T | T | Yes | |||||||||||||
| 6551 | C>T | N/−/N/− | T | T | Yes | |||||||||||
| 6689 | C>T | T | T | Yes | ||||||||||||
| 7028 | C>T | T | T | T | T | T | T | T | T | T | T | T | T | Yes | ||
| 7403 | A>G | G | G | Yes | ||||||||||||
| 7444 | G>A (Ter to Lys) | A | Yes | |||||||||||||
| CO2 | 7642 | G>A | A | Yes | ||||||||||||
| 7867 | C>T | T | Yes | |||||||||||||
| 8020 | G>A | A | A | Yes | ||||||||||||
| 8071 | A>G | G | Yes | |||||||||||||
| 8108 | A>G (Ile to Val) | I/I/I/I | G | Yes | ||||||||||||
| 8176 | T>C | C | Yes | |||||||||||||
| 8200 | T>C | C | Yes | |||||||||||||
| TK | 8308 | A>G | G | Yes | ||||||||||||
| ATP8 | 8414 | C>T (Leu to Phe) | L/F/M/W | T | T | T | T | T | T | Yes | ||||||
| 8473 | T>C | C | C | C | Yes | |||||||||||
| ATP6 | 8701 | A>G (Thr to Ala) | T/S/L/Q | G | G | G | G | G | G | G | G | G | G | G | G | Yes |
| 8860 | A>G (Thr to Ala) | T/A/A/T | G | G | G | G | G | G | G | G | G | G | G | G | Yes | |
| 9180 | A>G | G | G | G | Yes | |||||||||||
| CO3 | 9254 | A>G | G | Yes | ||||||||||||
| 9531 | A>G (Thr to Ala) | G | Yes | |||||||||||||
| 9540 | T>C | C | C | C | C | C | C | C | C | C | C | C | C | Yes | ||
| 9824 | T>C | C | Yes | |||||||||||||
| 9948 | G>A (Val to Ile) | A | A | Yes | ||||||||||||
| 9950 | T>C | C | Yes | |||||||||||||
| ND3 | 10084 | T>C (Ile to Thr) | C | C | Yes | |||||||||||
| 10181 | C>T | T | T | Yes | ||||||||||||
| 10397 | A>G | G | G | G | Yes | |||||||||||
| 10398 | A>G (Thr to Ala) | T/T/T/A | G | G | G | G | G | G | G | G | G | G | G | G | Yes | |
| 10400 | C>T | T | T | T | T | T | T | T | T | T | T | T | T | Yes | ||
| TR | 10438 | A>G | A/A/A/G | G | Yes | |||||||||||
| ND4L | 10685 | G>A | A | Yes | ||||||||||||
| ND4 | 10867 | C>T | I/F/L/L | T | T | Yes | ||||||||||
| 10873 | T>C | C | C | C | C | C | C | C | C | C | C | C | C | Yes | ||
| 11017 | T>C | C | Yes | |||||||||||||
| 11084 | A>G (Thr to Ala) | G | Yes | |||||||||||||
| 11719 | G>A | A | A | A | A | A | A | A | A | A | A | A | A | Yes | ||
| 11914 | G>A | A | Yes | |||||||||||||
| 11944 | T>C | C | Yes | |||||||||||||
| 11969 | G>A (Ala to Thr) | A | Yes | |||||||||||||
| 12026 | A>G (Ile to Val) | G | Yes | |||||||||||||
| 12100 | A>G | L/L/L/L | G | Yes | ||||||||||||
| TH | 12172 | A>G | G | G | Yes | |||||||||||
| ND5 | 12705 | C>T | T | T | T | T | T | T | T | T | T | T | T | T | Yes | |
| 12771 | G>A | A | Yes | |||||||||||||
| 13074 | A>G | G | Yes | |||||||||||||
| 13278 | A>G | G | Yes | |||||||||||||
| 13528 | A>G (Thr to Ala) | G | G | Yes | ||||||||||||
| 13890 | C>T | T | Yes | |||||||||||||
| 13928 | G>T (Ser to Ile) | S/T/S/T | T | Yes | ||||||||||||
| ND6 | 14364 | G>A | A | Yes | ||||||||||||
| 14569 | G>A | A | Yes | |||||||||||||
| 14668 | C>T | T | T | T | T | T | T | Yes | ||||||||
| CYB | 14766 | C>T (Thr to Ile) | T/S/I/S | T | T | T | T | T | T | T | T | T | T | T | T | Yes |
| 14783 | T>C | C | C | C | C | C | C | C | C | C | C | C | C | Yes | ||
| 14790 | A>G (Asn to Ser) | G | Yes | |||||||||||||
| 14979 | T>C (Ile to Thr) | I/I/L/L | C | C | C | Yes | ||||||||||
| 15043 | G>A | A | A | A | A | A | A | A | A | A | A | A | A | Yes | ||
| 15265 | C>T | T | Yes | |||||||||||||
| 15301 | G>A | A | A | A | A | A | A | A | A | A | A | A | A | Yes | ||
| 15323 | G>A (Ala to Thr) | A | Yes | |||||||||||||
| 15326 | A>G (Thr to Ala) | T/M/I/I | G | G | G | G | G | G | G | G | G | G | G | G | Yes | |
| 15440 | T>C | C | C | Yes | ||||||||||||
| 15497 | G>A (Gly to Ser) | A | Yes | |||||||||||||
| 15724 | A>G | G | G | Yes | ||||||||||||
| 15748 | T>C | C | C | Yes | ||||||||||||
| 15860 | A>G (Ile to Val) | G | Yes | |||||||||||||
| TT | 15951 | A>G | G | G | Yes |
Conservation of amino acids for polypeptides or nucleotides for RNAs in human (H), bovine (B), mouse (M), and Xenopus laevis (X).
See the online mitochondrial genome database http://www.mitomap.org.
Three major haplogroups, D, M and G, were detected in the mtDNA haplogroup analysis of the nine pedigrees. Haplogroups D4, D4b1b1a, D5a2a, D5b1b1, G1a1a, M7a1a and M11b were each found in seven pedigrees, and haplogroup D4a was found in two pedigrees. Haplogroup D was found in six pedigrees and was the most prevalent haplogroup in the nine pedigrees (Table 1).
Mutational Analysis of the GJB2 and TRMU Genes
To assess the role of the GJB2 gene in the variable phenotype or existence of the mutation in the patients carrying the A1555G mutation, we performed a sequence analysis of the GJB2 gene in all of the subjects with the A1555G mutation. None of the subjects had mutations in the GJB2 gene. Additionally, the A10S mutation of the TRMU gene has been reported to be a modifier gene in hearing loss with the A1555G mutation. We also analyzed the DNA of the subjects using PCR-RFLP, and the A10S mutation was not detected in any of the subjects (data not shown).
Discussion
The present study was performed in Korean subjects with non-syndromic clinically variable hearing loss carrying the A1555G mutation of the 12S rRNA gene in the mtDNA. To explain these variable phenotypes, we searched for mtDNA variants that acted as modifying factors of the variable phenotypes using complete mtDNA sequence analysis. First, the homoplasmic A1555G mutation of the 12S rRNA gene was detected in nine of the 281 unrelated subjects with non-syndromic hearing loss. Their pedigrees were characterized for clinical, genetic and molecular characteristics. The Korean pedigrees with hearing loss presented with wide penetrance and expressivity. The penetrance of the eight pedigrees (excluding the pedigree for KMT 09) ranged from 28.6% to 75%, with an average of 60.8%. These results were higher than the 29.5% penetrance observed in the previously reported Chinese population [26] but similar to the 65.4% and 54.1% penetrance of a large Arab-Israeli pedigree and nineteen Spanish pedigrees, respectively [9], [29]. This result suggested that the penetrance of hearing loss with the A1555G mutation was variable even within the same eastern population and appeared to differ among ethnic groups.
Mitochondrial haplogroups have been reported to be associated with diseases, including blindness [30], ageing [31], male infertility[32], Alzheimer’s [33], and diabetes [34]. In addition, mtDNA haplogroups have been shown to alter the phenotypic expression of syndromic and non-syndromic hearing loss. Lu et al. (2010) identified ten haplogroups in 69 pedigrees with hearing loss carrying the A1555G mutation: A, B, C, D, F, G, M, N, R and Y. Haplogroup D was found at a higher frequency in the hearing loss pedigrees than in 93 controls. In contrast, haplogroups A and M were found at lower frequencies in the hearing loss pedigrees than in the controls [26]. The mtDNA haplogroup analysis of the Spanish pedigrees revealed the following haplogroups: H, I, J, K, T, U, V and L [27], [35]. These haplogroups did not overlap with the haplogroups of the eastern Asian population. In the study of the Spanish pedigrees, 45.1% of individuals in the control group and 76% of the individuals in the hearing loss group were of haplogroup H, revealing a significantly higher percentage of this haplogroup in the hearing loss group [27]. In the present study, three major haplogroups, D, M and G, were detected in the nine pedigrees. Haplogroups D4, D4b1b1a, D5a2a, D5b1b1, G1a1a, M7a1a and M11b were each present in seven pedigrees, and haplogroup D4a was present in two pedigrees. A study analyzing the mtDNA haplogroups of 593 Koreans showed the following haplogroups: 4.9% haplogroup D4 and D4a, 2% D4b1, 2.2% D5a2, 2.7% G1a1, 1.3% M7a1 and 0.8% M11 [36]. Mitochondrial DNA haplogroups are restricted among ethnic populations. Haplogroup D in the eastern Asian population and haplogroup H in the Europe-Caucasian population are associated with hearing loss with the A1555G mutation [26], [27]. In the present study, haplogroup D was the most represented, similar to that found for the Chinese pedigrees. However, more pedigrees may be needed to estimate the association between an mtDNA haplogroup and hearing loss due to the mtDNA mutation.
Nuclear modifier genes have been reported to influence the variable phenotype of hearing loss with the A1555G mutation [22]. The mutant allele of the MTO2 gene that encode mitochondrial proteins in yeast S. cerevisiae manifests a respiratory-deficient phenotype only when coupled with the paromomycin-resistance mitochondrial 15S rRNA 1409 C to T mutation [37]. This mutation corresponds to the human 12S rRNA 1494 C to T mutation. The MTO2 gene is evolutionarily conserved and display sequence similarity to the human TRMU gene. Indeed, the missense mutation c.G28T (p.A10S) of the TRMU gene has been reported in hearing loss patients with the A1555G mutation in some ethnic populations [22]. However, the p.A10S mutation of the TRMU gene was not detected in the nine Korean pedigrees with hearing loss in this study.
Mitochondrial DNA variations have also been reported to influence the variable phenotype of mitochondrial disease, including the variable phenotype of hearing loss patients carrying the A1555G mutation. For example, the following mitochondrial tRNA variants may contribute to the phenotype: tRNAThr G15927A [18], tRNACys T5802C [18], tRNAArg T10454C [24], tRNASer(AGY) C12224T [26], tRNACys G5821A [19], tRNAGlu A14693G [24], tRNAThr T15908C [24], T12338C [18] of ND5, G7444A [23] of tRNASer(UCN)/CO1 and G11696A [26] of the ND4 gene. These mtDNA variants have been suggested to have significant effects on the penetrance and expressivity of hearing loss with the A1555G mutation. In this study, one novel mtDNA variant, G3496A of theND1 gene, was identified. Additionally, two variants of known modifier factors, tRNACys T5802C and G7444A of the tRNASer(UCN)/CO1 gene, were identified. The G3496A variant, which causes a substitution of alanine to threonine at position 64 (p.A64T) of the ND1 gene, was analyzed for protein biochemical changes using the PolyPhen 2 (http://genetics.bwh.harvard.edu/pph2/), SNPs&GO (http://snps-and-go.biocomp.unibo.it/snps-and-go/) and Panther (http://www.pantherdb.org/tools/csnpScoreForm.jsp) programs. This substitution appears to have no association with the variable phenotype because all of the in silico tools predicted that this substitution is a benign polymorphism. The tRNACys T5802C variant, however, has been reported to alter the structure of tRNAs and lead to a defect in tRNA metabolism [18]. Another variant, G7444A of the tRNASer(UCN)/CO1 gene, was not sufficient to produce a clinical phenotype [38]. Therefore, additional modifier factors, including nuclear backgrounds, environmental factors, and mitochondrial haplogroups, must alter the phenotypic manifestation. The variant G7444A of the tRNASer(UCN)/CO1 gene has been detected in several haplogroups, including C4a, B4 and D4a [26], [38], and was found in haplogroup D4a in this study. This result indicates that this variant was sporadic, similarly to the A1555G mutation. Additional studies are necessary to determine whether this variant affects the variable phenotype or is a simple polymorphism.
This study is the first to perform complete mtDNA sequencing to identify mtDNA haplogroups or variants in Korean pedigrees with non-syndromic hearing loss carrying the A1555G mutation. The haplogroups in the Korean population of the eastern Asian population are similar to those of the Chinese population but differ from the haplogroups of the Spanish populations of the Europe-Caucasian population. The mtDNA variants as modifier factors were also found to be similar to those of the Chinese population. The mtDNA haplogroups and variants are similar to the eastern Asian population but appear to have different phenotypes, although some subjects had the same variants [39], [40]. These results suggest that both the ethnic population and environmental factors lead to the variable phenotype of the A1555G mutation. However, this observation requires further pedigree and clinical evaluations to fully elucidate the mechanisms of the phenotypic manifestation of the A1555G mutation.
Supporting Information
Primer sequences used for whole mtDNA genome analysis. Bold sequences denote primers using PCR. Sequences of the rest are used for internal sequence primers.
(DOC)
Funding Statement
This work was supported by Biomedical Research Institute Grant, Kyungpook National University Hospital (2011) (K.Y. Lee). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Morton NE, Shields DC, Collins A (1991) Genetic epidemiology of complex phenotypes. Ann Hum Genet 55: 301–314. [DOI] [PubMed] [Google Scholar]
- 2. Prezant TR, Agapian JV, Bohlman MC, Bu X, Oztas S, et al. (1993) Mitochondrial ribosomal RNA mutation associated with both antibiotic-induced and non-syndromic deafness. Nat Genet 4: 289–294. [DOI] [PubMed] [Google Scholar]
- 3. Moraes CT, DiMauro S, Zeviani M, Lombes A, Shanske S, et al. (1989) Mitochondrial DNA deletions in progressive external ophthalmoplegia and Kearns-Sayre syndrome. N Engl J Med 320: 1293–1299. [DOI] [PubMed] [Google Scholar]
- 4. Shoffner JM, Lott MT, Lezza AM, Seibel P, Ballinger SW, et al. (1990) Myoclonic epilepsy and ragged-red fiber disease (MERRF) is associated with a mitochondrial DNA tRNA(Lys) mutation. Cell 61: 931–937. [DOI] [PubMed] [Google Scholar]
- 5. Goto Y, Nonaka I, Horai S (1990) A mutation in the tRNA(Leu)(UUR) gene associated with the MELAS subgroup of mitochondrial encephalomyopathies. Nature 348: 651–653. [DOI] [PubMed] [Google Scholar]
- 6. Ballinger SW, Shoffner JM, Hedaya EV, Trounce I, Polak MA, et al. (1992) Maternally transmitted diabetes and deafness associated with a 10.4 kb mitochondrial DNA deletion. Nat Genet 1: 11–15. [DOI] [PubMed] [Google Scholar]
- 7. Guan MX (2004) Molecular pathogenetic mechanism of maternally inherited deafness. Ann N Y Acad Sci 1011: 259–271. [DOI] [PubMed] [Google Scholar]
- 8. Van Camp G, Smith RJ (2000) Maternally inherited hearing impairment. Clin Genet 57: 409–414. [DOI] [PubMed] [Google Scholar]
- 9. Estivill X, Govea N, Barcelo E, Badenas C, Romero E, et al. (1998) Familial progressive sensorineural deafness is mainly due to the mtDNA A1555G mutation and is enhanced by treatment of aminoglycosides. Am J Hum Genet 62: 27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li Z, Li R, Chen J, Liao Z, Zhu Y, et al. (2005) Mutational analysis of the mitochondrial 12S rRNA gene in Chinese pediatric subjects with aminoglycoside-induced and non-syndromic hearing loss. Hum Genet 117: 9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Usami S, Abe S, Akita J, Namba A, Shinkawa H, et al. (2000) Prevalence of mitochondrial gene mutations among hearing impaired patients. J Med Genet 37: 38–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fischel-Ghodsian N, Prezant TR, Chaltraw WE, Wendt KA, Nelson RA, et al. (1997) Mitochondrial gene mutation is a significant predisposing factor in aminoglycoside ototoxicity. Am J Otolaryngol 18: 173–178. [DOI] [PubMed] [Google Scholar]
- 13. Guan MX, Fischel-Ghodsian N, Attardi G (2000) A biochemical basis for the inherited susceptibility to aminoglycoside ototoxicity. Hum Mol Genet 9: 1787–1793. [DOI] [PubMed] [Google Scholar]
- 14. Hamasaki K, Rando RR (1997) Specific binding of aminoglycosides to a human rRNA construct based on a DNA polymorphism which causes aminoglycoside-induced deafness. Biochemistry 36: 12323–12328. [DOI] [PubMed] [Google Scholar]
- 15. Casano RA, Johnson DF, Bykhovskaya Y, Torricelli F, Bigozzi M, et al. (1999) Inherited susceptibility to aminoglycoside ototoxicity: genetic heterogeneity and clinical implications. Am J Otolaryngol 20: 151–156. [DOI] [PubMed] [Google Scholar]
- 16. Matthijs G, Claes S, Longo-Mbenza B, Cassiman JJ (1996) Non-syndromic deafness associated with a mutation and a polymorphism in the mitochondrial 12S ribosomal RNA gene in a large Zairean pedigree. Eur J Hum Genet 4: 46–51. [DOI] [PubMed] [Google Scholar]
- 17. Tang X, Yang L, Zhu Y, Liao Z, Wang J, et al. (2007) Very low penetrance of hearing loss in seven Han Chinese pedigrees carrying the deafness-associated 12S rRNA A1555G mutation. Gene 393: 11–19. [DOI] [PubMed] [Google Scholar]
- 18. Chen B, Sun D, Yang L, Zhang C, Yang A, et al. (2008) Mitochondrial ND5 T12338C, tRNA(Cys) T5802C, and tRNA(Thr) G15927A variants may have a modifying role in the phenotypic manifestation of deafness-associated 12S rRNA A1555G mutation in three Han Chinese pedigrees. Am J Med Genet A 146A: 1248–1258. [DOI] [PubMed] [Google Scholar]
- 19. Zhao L, Wang Q, Qian Y, Li R, Cao J, et al. (2005) Clinical evaluation and mitochondrial DNA sequence analysis in two Chinese families with aminoglycoside-induced and non-syndromic hearing loss. Biochem Biophys Res Commun 336: 967–973. [DOI] [PubMed] [Google Scholar]
- 20. Guan MX, Fischel-Ghodsian N, Attardi G (1996) Biochemical evidence for nuclear gene involvement in phenotype of non-syndromic deafness associated with mitochondrial 12S rRNA mutation. Hum Mol Genet 5: 963–971. [DOI] [PubMed] [Google Scholar]
- 21. Wang X, Lu J, Zhu Y, Yang A, Yang L, et al. (2008) Mitochondrial tRNAThr G15927A mutation may modulate the phenotypic manifestation of ototoxic 12S rRNA A1555G mutation in four Chinese families. Pharmacogenet Genomics 18: 1059–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Guan MX, Yan Q, Li X, Bykhovskaya Y, Gallo-Teran J, et al. (2006) Mutation in TRMU related to transfer RNA modification modulates the phenotypic expression of the deafness-associated mitochondrial 12S ribosomal RNA mutations. Am J Hum Genet 79: 291–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yuan H, Qian Y, Xu Y, Cao J, Bai L, et al. (2005) Cosegregation of the G7444A mutation in the mitochondrial COI/tRNA(Ser(UCN)) genes with the 12S rRNA A1555G mutation in a Chinese family with aminoglycoside-induced and nonsyndromic hearing loss. Am J Med Genet A 138A: 133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Young WY, Zhao L, Qian Y, Li R, Chen J, et al. (2006) Variants in mitochondrial tRNAGlu, tRNAArg, and tRNAThr may influence the phenotypic manifestation of deafness-associated 12S rRNA A1555G mutation in three Han Chinese families with hearing loss. Am J Med Genet A 140: 2188–2197. [DOI] [PubMed] [Google Scholar]
- 25. Ding Y, Li Y, You J, Yang L, Chen B, et al. (2009) Mitochondrial tRNA(Glu) A14693G variant may modulate the phenotypic manifestation of deafness-associated 12S rRNA A1555G mutation in a Han Chinese family. J Genet Genomics 36: 241–250. [DOI] [PubMed] [Google Scholar]
- 26. Lu J, Qian Y, Li Z, Yang A, Zhu Y, et al. (2010) Mitochondrial haplotypes may modulate the phenotypic manifestation of the deafness-associated 12S rRNA 1555A>G mutation. Mitochondrion 10: 69–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Torroni A, Cruciani F, Rengo C, Sellitto D, Lopez-Bigas N, et al. (1999) The A1555G mutation in the 12S rRNA gene of human mtDNA: recurrent origins and founder events in families affected by sensorineural deafness. Am J Hum Genet 65: 1349–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li R, Greinwald JH Jr, Yang L, Choo DI, Wenstrup RJ, et al. (2004) Molecular analysis of the mitochondrial 12S rRNA and tRNASer(UCN) genes in paediatric subjects with non-syndromic hearing loss. J Med Genet 41: 615–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bykhovskaya Y, Shohat M, Ehrenman K, Johnson D, Hamon M, et al. (1998) Evidence for complex nuclear inheritance in a pedigree with nonsyndromic deafness due to a homoplasmic mitochondrial mutation. Am J Med Genet 77: 421–426. [DOI] [PubMed] [Google Scholar]
- 30. Torroni A, Petrozzi M, D'Urbano L, Sellitto D, Zeviani M, et al. (1997) Haplotype and phylogenetic analyses suggest that one European-specific mtDNA background plays a role in the expression of Leber hereditary optic neuropathy by increasing the penetrance of the primary mutations 11778 and 14484. Am J Hum Genet 60: 1107–1121. [PMC free article] [PubMed] [Google Scholar]
- 31. Coskun PE, Ruiz-Pesini E, Wallace DC (2003) Control region mtDNA variants: longevity, climatic adaptation, and a forensic conundrum. Proc Natl Acad Sci U S A 100: 2174–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ruiz-Pesini E, Lapena AC, Diez-Sanchez C, Perez-Martos A, Montoya J, et al. (2000) Human mtDNA haplogroups associated with high or reduced spermatozoa motility. Am J Hum Genet 67: 682–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. van der Walt JM, Dementieva YA, Martin ER, Scott WK, Nicodemus KK, et al. (2004) Analysis of European mitochondrial haplogroups with Alzheimer disease risk. Neurosci Lett 365: 28–32. [DOI] [PubMed] [Google Scholar]
- 34. Fuku N, Park KS, Yamada Y, Nishigaki Y, Cho YM, et al. (2007) Mitochondrial haplogroup N9a confers resistance against type 2 diabetes in Asians. Am J Hum Genet 80: 407–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. del Castillo FJ, Rodriguez-Ballesteros M, Martin Y, Arellano B, Gallo-Teran J, et al. (2003) Heteroplasmy for the 1555A>G mutation in the mitochondrial 12S rRNA gene in six Spanish families with non-syndromic hearing loss. J Med Genet 40: 632–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lee HY, Yoo JE, Park MJ, Chung U, Shin KJ (2006) Mitochondrial DNA control region sequences in Koreans: identification of useful variable sites and phylogenetic analysis for mtDNA data quality control. Int J Legal Med 120: 5–14. [DOI] [PubMed] [Google Scholar]
- 37. Yan Q, Li X, Faye G, Guan MX (2005) Mutations in MTO2 related to tRNA modification impair mitochondrial gene expression and protein synthesis in the presence of a paromomycin resistance mutation in mitochondrial 15 S rRNA. J Biol Chem 280: 29151–29157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhu Y, Qian Y, Tang X, Wang J, Yang L, et al. (2006) Aminoglycoside-induced and non-syndromic hearing loss is associated with the G7444A mutation in the mitochondrial COI/tRNASer(UCN) genes in two Chinese families. Biochem Biophys Res Commun 342: 843–850. [DOI] [PubMed] [Google Scholar]
- 39. Dai P, Liu X, Han D, Qian Y, Huang D, et al. (2006) Extremely low penetrance of deafness associated with the mitochondrial 12S rRNA mutation in 16 Chinese families: implication for early detection and prevention of deafness. Biochem Biophys Res Commun 340: 194–199. [DOI] [PubMed] [Google Scholar]
- 40. Young WY, Zhao L, Qian Y, Wang Q, Li N, et al. (2005) Extremely low penetrance of hearing loss in four Chinese families with the mitochondrial 12S rRNA A1555G mutation. Biochem Biophys Res Commun 328: 1244–1251. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primer sequences used for whole mtDNA genome analysis. Bold sequences denote primers using PCR. Sequences of the rest are used for internal sequence primers.
(DOC)