Abstract
Appropriately-placed hydrogen bond surrogates have been demonstrated to efficiently nucleate helical conformations. Herein we describe an efficient method for the synthesis of thioether-based hydrogen bond surrogate (teHBS) helices. A teHBS helix is shown to adopt a stable conformation and target its cognate protein receptor with high affinity.
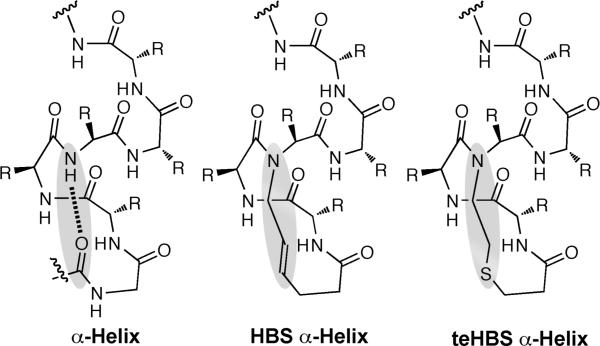
Stabilized α-helices and helix mimetics have emerged as powerful antagonists of model protein-protein interactions.1-10 Our laboratory has developed a hydrogen bond surrogate (HBS) approach that reproduces the conformation of proteinaceous α-helices in short peptide sequences.11 HBS α-helices feature a hydrocarbon linkage in place of an N-terminal i→ i+4 hydrogen bond (Fig. 1), which nucleates the desired helical conformation in the appended peptide chain.12, 13 One of the key advantages of the HBS approach is that all amino acid side-chains remain available for molecular recognition. HBS helices have been shown to bind chosen protein targets in cell free and cell-based assays.2, 3, 14, 15
Fig. 1.
Comparison of a canonical α-helix featuring an i→i+4 hydrogen bond with the hydrocarbon linkage of an HBS α-helix and the thioether linkage of a teHBS α-helix.
The hydrocarbon linkage of an HBS peptide is installed using a ring closing olefin metathesis reaction between an N-terminal 4-pentenoic acid residue, formally occupying the ith position on the helix, and an i+4 N-allyl group.16-18 The optimized metathesis conditions require high reaction temperatures and catalyst loadings, which can result in product mixtures that are difficult to purify. Purification difficulties have restricted the use of HBS helices; specifically, we have been unable to synthesize diverse helix libraries to optimize specificity and affinity for target proteins. We decided to investigate the use of a thioether linkage (teHBS in Fig. 1) as an alternative to the all hydrocarbon linkage of a traditional HBS.
Several peptide cyclization strategies have exploited thioether formation using nucleophilic substitutions of primary alkyl halides.19-22 We envisaged that a substitution reaction or a conjugate addition reaction would provide ready access to teHBS helices. The conditions required to affect these reactions are mild and the resulting thioether linkages have been shown to be stable in biological systems.23 Herein, we describe the efficient synthesis of a teHBS α-helix that mimics the p53 activation domain. We examined the solution conformation of the teHBS p53 helix in aqueous buffers by circular dichroism and 2D NMR spectroscopies, and investigated its potential to target Mdm2 by a fluorescence polarization competition assay. Our results suggest that the thioether linkage nucleates the helical conformation and targets protein receptors as well as the hydrocarbon system.
We designed teHBS 1, an analog of a previously reported HBS helix 2, to compare the helicities and protein binding capabilities of the two systems (Table 1). HBS helix 2, derived from screening several HBS peptides, mimics the p53 activation domain and has been shown to target Mdm2 with high affinity and selectivity.14 Interaction of p53 with Mdm2 is intimately involved in regulating the crucial process of programmed cell death.24 This complex has been targeted with several different types of synthetic inhibitors,25-31 making it a model protein-protein interaction for inhibitor design.
Table 1.
Summary of biophysical data for HBS and teHBS p53 helices.
| Compound | Sequencea | % Helicityb | Kd for Mdm2 (nM)c |
|---|---|---|---|
| teHBS α-helix 1 |
|
54 | 224 ± 20 |
| HBS α-helix 2 |
|
48 | 232 ± 34 |
X denotes pentenoic acid and thiopropionic acid residues in the HBS and teHBS macrocycles, respectively.
Values obtained from circular dichroism spectroscopy studies.
From fluorescence polarization competition assay.
We evaluated two different approaches for solid-phase synthesis of teHBS α-helices consisting of a Michael reaction (Method A in Figure 2) and the nucleophilic substitution method (Method B). The precursor peptides 3 and 4 were synthesized as described in the Supplementary Information. We tested various bases and solvents with peptides 3 and 4, and model tetrapeptide sequences, to establish the optimal cyclization conditions. For example, peptide 3 or 4 was treated with 5 equivalents of triethylamine, N,N-diisopropylethylamine, n-butylamine or DBU, in DMF in separate reaction vessels, and each reaction was monitored periodically using a qualitative on resin Ellman test.32, 33 After 12 h only the DBU catalyzed reaction indicated complete thiol consumption for 3; however, HPLC traces of the crude reaction revealed a complex mixture of products (Fig. 2). For Method B and peptide 4, DBU was again observed to be the most effective base. In this instance, HPLC and mass spectrometry analysis indicated a significant improvement in the yield of the desired product. Identity of the peptide was confirmed by MS/MS sequencing (Supplementary Information).
Fig. 2.
Synthesis of teHBS α-helices through conjugate addition (Method A) and nucleophilic substitution (Method B) reactions.
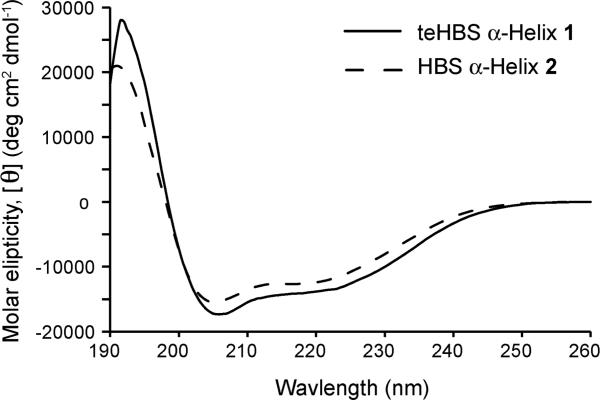
After identifying an efficient synthetic method, we utilized NMR and circular dichroism spectroscopies to examine the conformation of teHBS α-helix 1. Circular dichroism studies were performed in 10% trifluoroethanol in phosphate buffered saline (PBS). As expected for a canonical α-helix, double minima were observed near 208 nm and 222 nm and a maximum near 190 nm (Fig. 3). The percent helicity of 1 was estimated by the mean residue ellipticity at 222 nm to be 54%, although such assessments typically underestimate helical contents of short peptides.34-36 Significantly, the CD spectrum of 1 indicates that it has similar conformational stability to HBS 2 (Fig. 3). Helical content for 2 was calculated to be 48%. teHBS 1 is roughly 40% helical in PBS alone, while the unconstrained derivative provides a spectrum typical of unstructured peptides (Supplementary Information).
Fig. 3.
CD spectra of HBS, teHBS and unconstrained peptide in 10% trifluoroethanol in phosphate buffered saline.
We next utilized NMR spectroscopy to obtain a detailed analysis of the peptide conformation at the atomic level. An initial 1D 1H NMR spectrum was acquired in d3-ACN with 5% d6-DMSO to enable solubility. We observed two sets of NMR peaks in this solution (Supplementary Information). When the spectrum was acquired in d6-DMSO alone, a single set of peaks was observed, indicating the presence of either two slowly equilibrating conformers in d3-ACN/d6-DMSO or peptide aggregation. In 20% trifluoroethanol (TFE) in 1 mM PBS (pH 3.5), two sets of signals were again observed with the major conformer present in a 10:1 ratio. Analysis of the NMR spectra obtained in this solution focused on the major conformer.
2D TOCSY and NOESY spectra of teHBS 1 enabled full assignment of the fingerprint region. Sequential NN (i and i + 1) NOESY cross-peaks, a signature of helical structure, were observed for 1 as shown in the NOESY correlation chart (Fig. 4), although spectral overlap prevented assignment of some key cross-peaks. The NOESY spectrum further reveals several nonsequential medium range NOEs, for example, dαN(i, i + 3) and dαN(i, i + 4), that provide strong evidence of a helical structure (fig. 4a-b).37 The 3JNHCHα coupling constant provides a measure of the ϕ angle and affords intimate details about the local conformation in peptides and proteins.37 The 3JNHCHα values typically range between 4 and 6 Hz (-70 < ϕ < -30) for α-helices, and a series of three or more coupling constants in this range are indicative of the α-helical structure.37 With the exception of Q1, and S12 JNHCHα coupling constants and calculated ϕ angles are consistent with values expected for an α-helix (Table 2). The value for Q1 is not unexpected because it is situated within the macrocycle. The ϕ angle for S12 suggests greater flexibility near the C-terminus.
Fig. 4.
(a) Short-range and (b) medium-range NOE's observed for 1. (c) The NOESY correlation chart for 1. The glycine-3 residue is N-alkylated. Filled rectangles indicate relative intensity of the NOE cross-peaks. Empty rectangles indicate NOE that could not be unambiguously assigned because of overlapping signals. Spectra were acquired in 20% trifluoroethanol-d3 and PBS at 25 °C.
Table 2.
3JNHCHα coupling constants and calculated ϕangles.
| Q1 | E2 | G3 | F4 | S5c | D6c | L7 | W8 | K9 | L10 | L11 | S12 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3 J NHαCH a | 9.75 | 5.15 | N/A | 6.95 | 6.45 | 4.85 | 3.70 | 3.20 | 4.35 | 4.00 | 5.15 | 7.20 |
| ϕ (deg)b | -120 | -68 | N/A | -81 | -78 | -65 | -55 | -56 | -61 | -58 | -68 | -84 |
J values are in Hz
Calculated using the Karplus equation
Coupling constants were derived from 1D 1H NMR spectra acquired at 313 K due to overlapping resonances at 298 K.
The CD and NMR data suggest that the thioether hydrogen bond surrogate can efficiently nucleate a helical conformation in the attached peptide sequence. To ascertain that the thioether linkage does not interfere with the ability of these artificial helices to target their cognate protein receptors, we compared the ability of teHBS 1, and HBS 2, to bind Mdm2 in a fluorescence polarization competition assay.14 We find that both artificial helices target Mdm2 with similar affinities; the calculated Kd's for 1 and 2 are 224 ± 20 and 232 ± 34 nM, respectively.
We have presented a facile and efficient synthesis of thioether-linked hydrogen-bond surrogate α-helices. The traditional hydrocarbon-linked HBS helices have proven to be an exciting class of protein domain mimetics; however, their difficult synthesis has limited their usage. Facile synthesis of the thioether linkage allows us to bypass the ring-closing metathesis reaction – one of the key difficult steps. We find that teHBS compares favourably to HBS α-helices in conformational stability and protein targeting potential. Studies are currently under way to use the method developed here for the synthesis of focused libraries of stabilized helices.
Supplementary Material
Fig. 5.
Determination of teHBS 1 and HBS 2 binding affinity for Mdm2 by a fluorescence-polarization assay.
Acknowledgments
We thank Drs. Susan and Neal Zondlo (University of Delaware) for the His-Mdm2 construct. This work was supported by the National Institutes of Health (GM073943). Funding from the National Science Foundation (CHE-0958457) in the form of an instrumentation grant is gratefully acknowledged.
Footnotes
† Electronic Supplementary Information (ESI) available: Synthetic procedures; characterization including 2D NMR spectra. See DOI: 10.1039/b000000x/
References
- 1.Edwards TA, Wilson AJ. Amino Acids. 2011:1–12. doi: 10.1007/s00726-011-0880-8. [DOI] [PubMed] [Google Scholar]
- 2.Patgiri A, Yadav K, Arora PS, Bar-Sagi D. Nature Chem. Biol. 2011;7:585–587. doi: 10.1038/nchembio.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Henchey LK, Kushal S, Dubey R, Chapman RN, Olenyuk BZ, Arora PS. J. Am. Chem. Soc. 2010;132:941–943. doi: 10.1021/ja9082864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moellering RE, Cornejo M, Davis TN, Del Bianco C, Aster JC, Blacklow SC, Kung AL, Gilliland DG, Verdine GL, Bradner JE. Nature. 2009;462:182–188. doi: 10.1038/nature08543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walensky LD, Kung AL, Escher I, Malia TJ, Barbuto S, Wright RD, Wagner G, Verdine GL, Korsmeyer SJ. Science. 2004;305:1466–1470. doi: 10.1126/science.1099191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harrison RS, Shepherd NE, Hoang HN, Ruiz-Gomez G, Hill TA, Driver RW, Desai VS, Young PR, Abbenante G, Fairlie DP. Proc. Natl. Acad. Sci. U. S. A. 2010;107:11686–11691. doi: 10.1073/pnas.1002498107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horne WS, Johnson LM, Ketas TJ, Klasse PJ, Lu M, Moore JP, Gellman SH. Proc. Natl. Acad. Sci. U. S. A. 2009;106:14751–14756. doi: 10.1073/pnas.0902663106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horne WS, Gellman SH. Acc. Chem. Res. 2008;41:1399–1408. doi: 10.1021/ar800009n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seebach D, Gardiner J. Acc. Chem. Res. 2008;41:1366–1375. doi: 10.1021/ar700263g. [DOI] [PubMed] [Google Scholar]
- 10.Cummings CG, Hamilton AD. Curr. Opin. Chem. Biol. 2010;14:341–346. doi: 10.1016/j.cbpa.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 11.Patgiri A, Jochim AL, Arora PS. Acc. Chem. Res. 2008;41:1289–1300. doi: 10.1021/ar700264k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chapman R, Kulp JL, 3rd, Patgiri A, Kallenbach NR, Bracken C, Arora PS. Biochemistry. 2008;47:4189–4195. doi: 10.1021/bi800136m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang D, Chen K, Dimartino G, Arora PS. Org. Biomolec. Chem. 2006;4:4074–4081. doi: 10.1039/b612891b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henchey LK, Porter JR, Ghosh I, Arora PS. ChemBiochem. 2010;11:2104–2107. doi: 10.1002/cbic.201000378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang D, Lu M, Arora PS. Angew. Chem. Int. Ed. 2008;47:1879–1882. doi: 10.1002/anie.200704227. [DOI] [PubMed] [Google Scholar]
- 16.Patgiri A, Witten MR, Arora PS. Org. Biomol. Chem. 2010;8:1773–1776. doi: 10.1039/c000905a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chapman RN, Arora PS. Org. Lett. 2006;8:5825–5828. doi: 10.1021/ol062443z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dimartino G, Wang D, Chapman RN, Arora PS. Org. Lett. 2005;7:2389–2392. doi: 10.1021/ol0506516. [DOI] [PubMed] [Google Scholar]
- 19.Roberts KD, Lambert JN, Ede NJ, Bray AB. Tet. Lett. 1998;39:8357–8360. [Google Scholar]
- 20.Lung FT, King CR, Roller PP. Lett. Pept. Sci. 1999;6:45–49. [Google Scholar]
- 21.Roberts KD, Ede NJ. J. Pept. Sci. 2007;13:811–821. doi: 10.1002/psc.904. [DOI] [PubMed] [Google Scholar]
- 22.Brunel FM, Dawson PE. Chem. Comm. 2005;20:2552–2554. doi: 10.1039/b419015g. [DOI] [PubMed] [Google Scholar]
- 23.Tugyi R, Mezö G, Fellinger E, Andreu D, Hudecz F. J. Peptide Sci. 2005;11:642–649. doi: 10.1002/psc.669. [DOI] [PubMed] [Google Scholar]
- 24.Joerger AC, Fersht AR. Annu. Rev. Biochem. 2008;77:557–582. doi: 10.1146/annurev.biochem.77.060806.091238. [DOI] [PubMed] [Google Scholar]
- 25.Murray JK, Gellman SH. Biopolymers. 2007;88:657–686. doi: 10.1002/bip.20741. [DOI] [PubMed] [Google Scholar]
- 26.Gemperli AC, Rutledge SE, Maranda A, Schepartz A. J. Am. Chem. Soc. 2005;127:1596–1597. doi: 10.1021/ja0441211. [DOI] [PubMed] [Google Scholar]
- 27.Bernal F, Tyler AF, Korsmeyer SJ, Walensky LD, Verdine GL. J. Am. Chem. Soc. 2007;129:2456–2457. doi: 10.1021/ja0693587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee JH, Zhang Q, Jo S, Chai SC, Oh M, Im W, Lu H, Lim HS. J. Am. Chem. Soc. 2011;133:676–679. doi: 10.1021/ja108230s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shangary S, Wang S. Clin. Cancer Res. 2008;14:5318–5324. doi: 10.1158/1078-0432.CCR-07-5136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Campbell F, Plante JP, Edwards TA, Warriner SL, Wilson AJ. Org. Biomol. Chem. 2010;8:2344–2351. doi: 10.1039/c001164a. [DOI] [PubMed] [Google Scholar]
- 31.Yin H, Lee GI, Park HS, Payne GA, Rodriguez JM, Sebti SM, Hamilton AD. Angew. Chem. Int. Ed. 2005;44:2704–2707. doi: 10.1002/anie.200462316. [DOI] [PubMed] [Google Scholar]
- 32.Ellman GL. Archives Biochem. Biophys. 1959;82:70–77. doi: 10.1016/0003-9861(59)90090-6. [DOI] [PubMed] [Google Scholar]
- 33.Badyal JP, Cameron AM, Cameron NR, Coe DM, Cox R, Davis DG, Oates LJ, Oye G, Steel PG. Tet. Lett. 2001;42:8531–8533. [Google Scholar]
- 34.Wang D, Chen A, Kulp JL, III, Arora PS. J. Am. Chem. Soc. 2006;128:9248–9256. doi: 10.1021/ja062710w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shepherd NE, Hoang HN, Abbenante G, Fairlie DP. J. Am. Chem. Soc. 2005;127:2974–2983. doi: 10.1021/ja0456003. [DOI] [PubMed] [Google Scholar]
- 36.Chin DH, Woody RW, Rohl CA, Baldwin RL. Proc. Natl. Acad. Sci. U.S.A. 2002;99:15416–15421. doi: 10.1073/pnas.232591399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wuthrich K. NMR of Proteins and Nucleic Acids. Wiley; New York: 1986. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.