Abstract
Intraflagellar transport (IFT) proteins are essential for the assembly and maintenance of cilia, which play important roles in development and homeostasis. IFT80 is a newly defined IFT protein. Partial mutation of IFT80 in humans causes diseases such as Jeune asphyxiating thoracic dystrophy (JATD) and short rib polydactyly (SRP) type III with abnormal skeletal development. However, the role and mechanism of IFT80 in osteogenesis is unknown. Here, we first detected IFT80 expression pattern and found that IFT80 was highly expressed in mouse long bone, skull, and during osteoblast differentiation. By using lentivirus-mediated RNA interference (RNAi) technology to silence IFT80 in murine mesenchymal progenitor cell line-C3H10T1/2 and bone marrow derived stromal cells, we found that silencing IFT80 led to either shortening or loss of cilia and the decrease of Arl13b expression - a small GTPase that is localized in cilia. Additionally, silencing IFT80 blocked the expression of osteoblast markers and significantly inhibited ALP activity and cell mineralization. We further found that IFT80 silencing inhibited the expression of Gli2, a critical transcriptional factor in the hedgehog signaling pathway. Overexpression of Gli2 rescued the deficiency of osteoblast differentiation from IFT80-silenced cells, and dramatically promoted osteoblast differentiation. Moreover, introduction of Smo agonist (SAG) promotes osteoblast differentiation, which was partially inhibited by IFT80 silencing. Thus, these results suggested that IFT80 plays an important role in osteogenesis through regulating Hedgehog/Gli signal pathways.
Keywords: Intraflagellar transport, Cilia, osteoblast differentiation, skeleton development, Hedgehog signaling
1. Introduction
Primary cilia are microtubule-based organelles localized on the surface of almost all vertebrate cells including osteoblasts and osteocytes. These organelles are extended and maintained by the transport of particles along the axoneme mediated by intraflagellar transport (IFT) bidirectional machinery. IFT proteins are organized into two complexes. Complex A containing IFT144, 140, 139, and 122 proteins mediates retrograde transport of cargoes from the tip to the base of the cilia, while complex B containing IFT172, 88, 81, 80, 74/72, 57/55, 52, 46, 27, and 20 mediates anterograde transport of specific cargoes from the base to the tip. The movement of IFT proteins is carried out by two different microtubule-based motors: the anterograde (towards the cilia tip) motor is kinesin-II, which is composed of Kif3a and Kif3b motor subunits; the retrograde (towards the cell body) motor is cytoplasmic dynein-Ib. IFT complexes carry axonemal subunits to the site of assembly at the tip of the axoneme and are necessary for axonemal growth [1]. In recent years, the importance of IFT proteins for the development and function of the skeleton has been demonstrated due to the findings of skeletal abnormalities in human cilia-associated disorders [2–6] and in IFT-related mouse knockout studies [7–13]. Increasing studies have shown that primary cilia/IFT regulate embryonic bone development [8, 14–18] and mechanically regulate bone formation in adults[19, 20]. However, the function and mechanism of IFT/cilia proteins in osteoblast differentiation and function are still largely undefined.
Most recently, Xiao et al [21] reported that targeted deletion of PKD1 (polycystin-1) in osteoblasts results in osteopenia phenotype, and impaired osteoblastic differentiation. While Kif3a deficiency reverses the skeletal abnormalities in Pkd1 deficient mice by restoring the balance between osteogenesis and adipogenesis[18]. Qiu et al [17] demonstrated that osteoblast specific deletion of Kif3a causes increased cell proliferation, impaired osteoblastic differentiation, and enhanced adipogenesis in vitro. They further found that conditionally deleted Kif3a in osteoblasts results in the reduction or shorten of primary cilia and develops osteopenia in vivo and suggested that Kif3a regulates osteoblastic differentiation and function through multiple pathways including hedgehog, intracellular calcium and Wnt signaling. These finding highlighted important roles of IFT and cilia related proteins in osteoblast differentiation and bone development.
A number of studies have shown that the skeletal phenotypes observed in a variety of IFT and ciliary component knockout lines can be attributed to abnormal hedgehog signaling (Hh) [8, 12, 22]. Hh signaling is one of the major signaling pathways that regulate osteogenesis and embryonic bone development and post-embryonic bone homeostasis [23, 24]. In vertebrates, the Hh family consists of three members: Sonic Hh (Shh), Indian Hh (Ihh), and Desert Hh (Dhh)[24]. Hh protein binding to the transporter-like receptor Patched (Ptch) releases Ptch inhibition of Smoothened (Smo) allowing the transduction of the Hh signal to the primary cilium. This in turn activates Gli transcription factors that mediate the transcription of Hh target genes in cells [25–27]. Without a cilium, hedgehog signaling is abrogated, leading to a variety of skeletal malformations as well as embryonic lethality. For example, deletion of IFT88 in limb mesenchyme resulted in shortening of the bone in the limbs due to alterations in Ihh signaling and endochondral bone formation [8]. Conditional deletion of IFT88 or Kif3α in chondrocyte lineage by using Col2α1-cre lead to abnormal hedgehog signaling topography and apparent growth plate dysfunction [22, 28], which are similar to conditional deletion of Ihh in postnatal cartilage (Ihhflox/flox, Col2a-CreER)[29].
IFT80 is a newly identified IFT protein, which encodes a 777-residue protein that contains seven WD40 domains and is a component of the IFT complex B [30]. WD40 domains are short motifs of approximately 40 amino acids that form circular beta propeller structures. During intraflagellar transport, this complex helps carry materials from the base to the tip of cilia. Partial mutations of IFT80 in humans cause Jeune asphyxiating thoracic dystrophy (JATD) and short rib polydactyly type III (SRPIII). Both diseases have severe bone abnormalities including shortening of the long bones and constriction of the thoracic cage [31–33]. SRP type III is a more severe disorder with a range of extra skeletal malformations, including cleft lip or palate, cystic renal disease, gastrointestinal, urogenital, brain and/or cardiac malformations. These two diseases often lead to death prenatally or in infancy due to respiratory insufficiency. However, currently, it is still unclear if the abnormal bone phenotype result from the effect of IFT80 mutation on osteogenesis or indirect effect of mutation of IFT80 in human tissues. Therefore, in this study, to identify the role and mechanism of IFT80 in osteoblast differentiation, we first identified the gene expression pattern of this newly discovered protein in various mouse tissues, including skull and bone among others, and confirmed IFT80 is predominantly expressed in bone as well as during osteoblast differentiation. We further determined the effect of IFT80 on osteoblast differentiation and activation and on the Hh/Gli signaling transduction pathway. Our results demonstrated that the IFT80 gene plays an essential role in osteoblast differentiation and likely is involved in Hh/Gli signal pathway.
2. Materials and Methods
2.1. Cell lines and cell culture
HEK293T human embryonic kidney cell line, C3H10T1/2 murine mesenchymal progenitor cell line and RAW264.7 murine monocyte/macrophage cell line were obtained from American Type Culture Collection (ATCC). For preparation of mouse BMMs and BMSCs, animal procedures were conducted in accordance with the protocol approved by the Institutional Animal Care and Use Committee (IACUC) of University at Buffalo (UB). Both femora and tibias were removed and soft tissues were detached from the bone of 6-week-old C57BL/J wild type mice. The metaphysis from both ends was resected and BMMs were collected by flushing the diaphysis with PBS. BMSCs were expanded as described previously [34]. For osteoblast differentiation, C3H10T1/2 cells and BMSCs were respectively seeded at a density of 5×104 cells/cm2 and maintained in complete growth media for 24 hrs (α-modified Eagle’s medium with 10% fetal bovine serum (FBS, Gibco), 100U/ml penicillin and 100mg/ml streptomycin) in a humidified atmosphere of 5% CO2 at 37°C, then the cells were induced with osteogenic medium (OS media), i.e. the growth medium with 10−8M dexamethasone, 50 μg/ml ascorbic acid and 10 mM β-glycerol-phosphate [35] for the indicated times based on different experiments. For osteoclast differentiation, RAW264.7 cells and BMMs were respectively induced with 10ng/ml RANKL and 20ng/ml M-CSF for 0, 24 and 96 hours [36].
2.2. IFT80 shRNA lentivirus packaging, tittering and cell infection
To identify the function of IFT80 gene in osteoblast differentiation and osteoblastic signaling pathways, we used lentivirus-mediated mouse pGIPZ-IFT80 shRNA plasmids and control pGIPZ-scrambled shRNA (Open Biosystems, RMM4532-NM 026641) to package IFT80 recombinant lentivirus according to the manufacturer’s instructions. Briefly, five individual vector pGIPZ-IFT80 shRNAs were respectively co-transfected with the packaging plasmids, pCMV-Dr8.2 and pCMV-VSVG (Addgene) [37] into HEK293T cells (ATCC) using calcium phosphate co-precipitation method. The medium was replaced with fresh complete growth media after co-transfection for 8 hours. After 48–72 hrs, the lentiviral supernatant was harvested and the titer was determined by infecting HEK293T cells with serial dilutions of concentrated lentivirus in the presence of 4 μg/ml polybrene (Sigma). For infection of target cells, the viral supernatant was added to C3H10T1/2 cells or BMSCs. Following incubation for 24 hrs, the virus-containing media was removed and replaced with fresh growth medium. At 48 hrs following the transfection, the cells were analyzed by RT-PCR, western blot and immunostaining to test silent efficiency of the IFT80 gene. For osteoblast differentiation, after the infection of the cells for 24 hrs, the virus-containing media was replaced with OS media for the indicated times based on the different experiments.
2.3. Production of recombinant retrovirus
Retroviral vector pBMN-Gli2 was constructed by inserting a full-length 6.18kb Gli2 cDNA (access no. NM_027770) into the EcoRI and Not I site of pBMN- I-GFP vector (pBMN) (Addgene), and packaging was performed as the protocol from Dr. Garry Nolan Laboratory, Stanford University. Briefly, retrovirus vectors pBMN-I-GFP (control vector) and pBMN-Gli2 were separately transfected into the phoenix-ecotropic packaging cell line by CaCl2 precipitation method [38]. Following the transfection, the cells were placed in a 32°C humidified incubator for 48 hrs. The media containing infectious virus was harvested and filtered through a 0.45 mm filter for tittering assay. The retrovirus carrying Gli2 cDNA was then used to infect 70–80% subconfluent proliferating C3H10T1/2 cells with the presence of 8μg/ml Polybrene to enhance the efficiency of infection. The Gli2 protein expression were confirmed by performing immunostaining and western blot [36].
2.4. RT-PCR and Quantitative real time RT-PCR analyses
All animal procedures were approved by IACUC of UB. Total RNA was extracted from 14-day-old C57BL/6J mouse tissues including the lung, spleen, kidney, muscle, liver, heart, brain and calvaria (free of periosteum), OS media-BMSCs and C3H10T1/2 cells, or RANKL/M-CSF- induced BMMs and RAW264.7 cells using TRIzol reagent (Invitrogen). cDNA was synthesized from 1 μg total RNA using the SuperScript III reverse transcriptase kit (Invitrogen) in a final volume of 20 μl. Primers were designed with the IDT SCI primer design tool (Integrated DNA Technologies, San Diego, California). RT-PCR experiments were performed with a Bio-Rad C1000 thermal cycler (Bio-Rad, Hercules, CA), and real-time PCR experiments were performed with an ABI prism 7500 (Applied Biosystems, Grand Island, NY) in triplicate. Sequence and product lengths for each primer pair were: BMP2, forward primer 5′ GTTTGG CCTGAAGCAGAGAC 3′, reverse primer 5′ TCTAAATGGGCCACTTCCAC 3′; ALP, forward primer 5′ GGGCATTGTGACTACCACTC 3′, reverse primer 5′ AGTCAGGTTGTT CCGATTCA 3′; RUNX2, forward primer 5′ CACTGCCACCTCTGACTTCT 3′, reverse primer 5′ CACCATCATTCTGGTTAGGC 3′; IFT80, forward primer 5′ AAGGAACCAAAGCAT CAAGAATTAG 3′, reverse primer 5′ AG ATGTCATCAGGCAGCTTGAC 3′; GAPDH forward primer 5′-ACCACAGTCCATGCCATCAC-3′; reverse primer 5′-TCCACCACCC TGTTGCTGTA-3′. Samples were analyzed in triplicate and the raw data consisted of PCR cycle numbers required to reach a fluorescence threshold level. The relative expression level of target genes was normalized with geNorm software (Primer Design Ltd) using GADPH gene as reference to determine the normalization factor [39]. The data were expressed as the mean ± SE.
2.5. Alkaline phosphatase (ALP) activity
ALP activity was determined using the ALP assay kit (Sigma) according to the manufacturer’s instructions. Briefly, cells were lysed with 0.5% Triton X-100. ALP assay was performed in alkaline buffer solution (1.5 M, pH 10.3) containing 10 mM p-nitrophenyl phosphate as a substrate. Following the addition of the stop solution (3 M NaOH solution), the optical density was determined using a microplate reader set at 405 nm. ALP activity was normalized with the value of DNA content and expressed as nmol of p-nitrophenol produced per minute per mg of total DNA. DNA concentration was measured according to the method of Schneider [40].
2.6. Alizarin red staining
To measure extracellular matrix Ca deposits for bone nodule formation, cellular matrix was stained using Alizarin red dye, which combines with Ca in the matrix as previously described [41, 42]. At 14 days following the induction with OS media, cells were washed with PBS twice and then fixed with 2.0% formaldehyde. The cells were stained with 40 mmol/L of Alizarin red solution (pH 4.4) for 40 mins at room temperature and rinsed with deionized water twice. The images of stained cells were captured using a phase contrast microscope with a digital camera (IM50, Leica, Germany). The concentration of Alizarin Red staining in the samples was determined by comparing the absorbance values with those obtained from Alizarin Red standards. Briefly, the cells were destained for 15 mins with equal amount of 10% (w/v) cetylpyridinium chloride (sigma) in 10 mM sodium phosphate (pH 7.0). The equal volume of the destained solution was then transferred to a 96-well plate, and measured at the absorbance of 562 nm [43].
2.7. Von Kossa staining
Because Ca coprecipitates with the phosphate ion in the matrix, von kossa staining (which stains phosphate ions) was also used for determining mineralization in the cultures [44, 45]. Cells were washed with PBS and fixed with 2.0% formaldehyde for 15 mins. After washing with deionized water 3 times, cells were incubated with 5% silver nitrate at room temperature under ultraviolet light for 1 hour. After washing with deionized water, the images of the stained cells were photographed using phase-contrast microscope.
2.8. Immunocytochemistry staining
Mouse eyes, calvarial bone and kidneys tissue were dissected from wild type C57BL/J mice in accordance with the protocol approved by IACUC of UB. Immunofluorescence staining analysis of IFT80 expression in mouse tissues were performed as described previously [38]. The cells were seeded at 1×105 cells/well on 24-well plates and incubated overnight, followed by infection with lentivirus IFT80 shRNA and/or retrovirus pBMN-Gli2 for 24 hours. The cells were sequentially induced with OS media with 10% FBS for 3 days. After 48 hours of serum starvation (without FBS), the cells were fixed with 100% ice-cold methanol for 10 min and washed 3 times with PBS. Fixed cells were blocked with 5% BSA in PBS for 60 min. The cells were then incubated with primary antibody diluted in PBS containing 1% BSA for 1 hour. The primary antibodies used were as follows: anti-IFT80 antibody (Abnova, Walnut, CA. 1:500), anti-γ tubulin (Sigma, T3320, AK-151:1000) and mouse monoclonal anti-acetylated tubulin antibody (Sigma, T7451, 6-11B-1, 1:1,000). After washing 3 times in PBS, Alexa Fluor 488-, Alexa Fluor 568- or Alexa Fluor 647 (Invitrogen, Grand Island, NY)-conjugated anti-rabbit or anti-mouse IgG was added in PBS with 1% BSA for 1 hour. In the final washes, 6-diamidino-2-phenylindole (DAPI) (Sigma) was added and used as a counterstain for nuclei. Fluorescence images were acquired using a Zeiss Axioimager microscope.
2.9. Western blot analysis
Cells were lysed with NP40 buffer (1% NP-40, 0.15 M NaCl, 50mM Tris, pH 8.0) containing protease inhibitors (Sigma). Protein quantitation was performed with BCA protein assay reagent (Pierce, Rockford, IL, USA). Equal amounts of protein from the different groups were denatured in SDS sample buffer and separated on 8–10% polyacrylamide-SDS gel based on the protein molecular weight. Proteins were transferred to polyvinylidene difluoride membrane. The antibodies to osteocalcin (abcam 13418), collagen I (abcam 34710), Gli2 (abcam 7195), IFT80 (Abnova, PAB15842), GAPDH (cell signaling technology, 14C10) and β-catenin (Cell signaling technology, 9562) were used to detect the target proteins. The antigen-antibody complexes were detected using the appropriate secondary antibodies (Sigma) and the enhanced chemiluminescence detection system, as described in [38, 46]. The blots were visualized and quantified using Fluor-S Multi-Imager and Multi-Analyst software (Bio-Rad).
2.10. Statistical analyses
Statistical analysis was performed using software SPSS-17.0. Data were analyzed using one-way analysis of variance and Tukey HSD test was applied as a post hoc test if significance was determined. Statistical significance for two groups was assessed using Student’s t-test. All experiments were at least triplicated. All values were expressed as means ± SD. The probability level (P) at which differences were considered significant was at P<0.05.
3. Results
3.1. Ift80 is highly expressed in osteoblast progenitor cells, osteoblasts, and bone
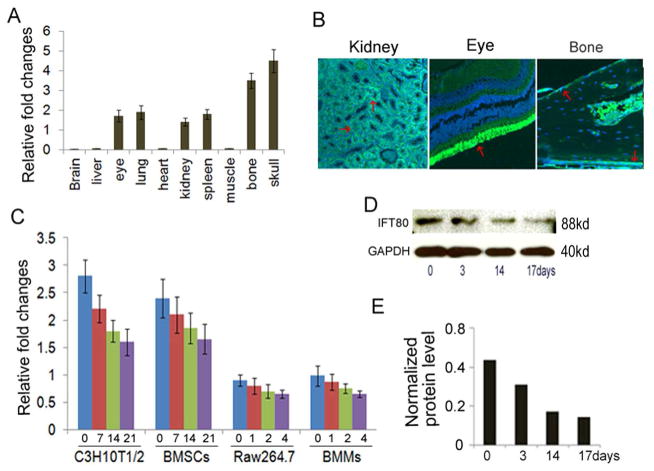
To identify the expression pattern of IFT80, we first analyzed IFT80 mRNA expression on 10 kinds of mouse tissues by performing real time RT-PCR. The result showed that IFT80 was highly expressed in the long bone and skull. It is abundant in eye, lung, spleen and kidney tissues. IFT80 was also expressed in muscle, heart, liver, and brain tissues, but to a far lesser extent (Fig. 1A). This result was further confirmed by immunostaining in mouse long bone, eye and kidney tissues (Fig 1B). To further characterize the expression pattern of IFT80 during osteoblast and osteoclast differentiation, real time RT-PCR analysis were performed for the time course of IFT80 expression in C3H10T1/2 cells and primary mouse bone marrow derived stromal cells (BMSCs) for osteoblast differentiation and in primary mouse bone marrow-derived monocytes (BMMs) and RAW264.7 cells for osteoclast differentiation. As show in Fig. 1C, IFT80 was expressed in C3H10T1/2 cells, RAW264.7 cells, and primary BMSCs and BMMs. The expression of IFT80 was slightly decreased about 5% – 19% during osteoblast and osteoclast differentiation. The levels of IFT80 during osteoblast differentiation are much higher than those during osteoclast differentiation. To further confirm these results in protein level, C3H10T1/2 cells were induced with OS media for 0, 3, 14 and 17 days, the protein level of IFT80 was detected by western blot. Similar to the results from Real time PCR, IFT80 was expressed in preosteoblast and mature osteoblasts (Fig. 1D–E). These results indicate that IFT80 protein is predominantly expressed in early stages of osteoblast, and it continues to be expressed throughout the entirety of differentiation.
Fig. 1.
IFT80 is prominently expressed in bone tissue and during osteoblast differentiation. (A) Real time RT-PCR analysis of IFT80 expression in mouse tissues. Total RNA was extracted from mouse tissues: brain, liver, eye, lung, heart, kidney, spleen, muscle, long bone and skull. (B) Immunostaining for IFT80 expression. Bright green is positive for expression of IFT80 (red arrows). (C) Real time RT-PCR analysis for IFT80 at transcriptional level during osteoblast and osteoclast differentiation. C3H10T1/2 cells and BMSCs were respectively induced with OS media for 0, 7 14 and 21 days respectively. Raw264.7 cells and BMMs were respectively induced with M-CSF/RANKL (20ng/ml) for 0, 1, 2 and 4 days. (D) Western Blot analysis of IFT80 protein expression during osteoblast differentiation. C3H10T1/2 cells were induced with OS media for 0, 3, 7, 14 and 17 days. (E) Quantification of protein levels from immunblots as in D. The protein levels of IFT80 were normalized to GAPDH.
3.2. Silencing IFT80 blocks IFT80 expression and impairs cilia formation
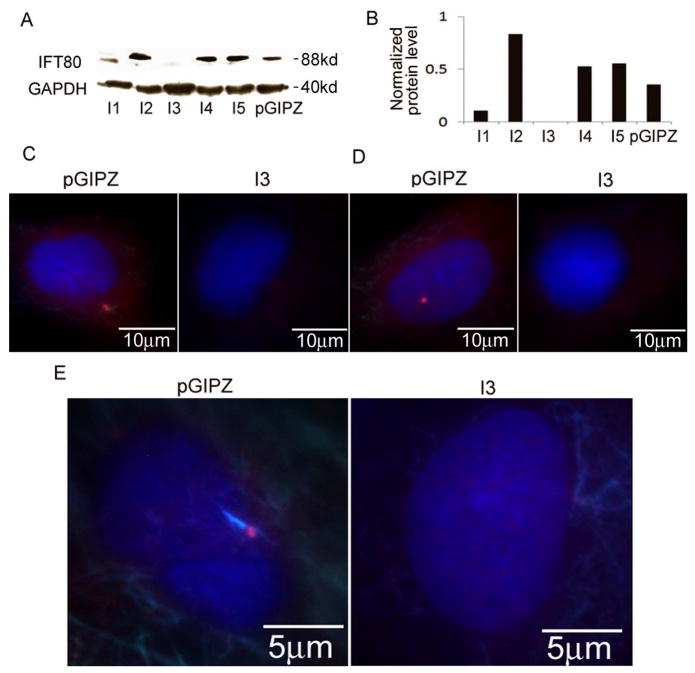
The primary cilium is an organelle present in almost all eukaryotic cells, including osteoblasts (33). IFT proteins are required for primary cilia biogenesis [47–49]. To determine the effects of silencing IFT80 on cilia formation, we used lentivirus-based RNAi gene silencing technology to knockdown IFT80 gene expression in C3H10T1/2 cells and mouse primary BMSCs. After transfection of five unique pGIPZ-IFT80 shRNA constructs respectively, the recombinant lentiviruses (I1, I2, I3, I4 and I5) were used to assess silencing efficiency. As shown in Fig. 2A–B, IFT80 expression was diminished and/or undetectable in the I1 and I3 constructs compared to cells infected with the control pGIPZ-scrambled shRNA (pGIPZ) lentivirus as well as the other constructs (I2, I4, I5). Fluorescent immunostaining analysis showed that the expression of IFT80, and Arl13b, a small GTPase that is localized in cilia and required for cilia formation [50], was blocked in the IFT80-silenced cells compared with that of the control cells (Fig. 2C–D). To further assess the IFT80 silencing effects on cilia formation in mouse BMSCs, the cells were co-stained with the antibodies to gamma tubulin (red), acetylated α-tubulin (bright blue), and DAPI (Blue) for detecting cilia. As shown in Fig. 2E, cilia, as visualized by localization of gamma tubulin (red) and acetylated α-tubulin (bright blue), about 75–80% cells showed apparent cilia formation on the control cells (pGIPZ). However, in IFT80-silenced cells, only about 15–20% cells have cilia formation, and among those cells, about 5–10% cells showed shorten cilia compared to the control. These results indicated that silencing IFT80 impairs cilia formation.
Fig. 2.
Silencing IFT80 inhibits the expression of Arl13b, IFT80, gamma tubulin and acetylated α-tubulin, and impairs cilia formation. (A) Western blot analysis of IFT80 expression. C3H10T1/2 cells were infected with pGIPZ-IFT80 shRNA (I1–I5) or pGIPZ-scrambled shRNA (PGIPZ) lentiviruses for 48 hrs, and then the protein was extracted to detect IFT80 expression. IFT80 expression was silenced in I1 and I3. (B) Quantitative analysis of protein levels from immunoblots as in A. The protein levels of IFT80 were normalized to GAPDH. (C) Immunofluorescence staining for IFT80 location and expression. IFT80 was expressed in control group (pGIPZ, red), but blocked in IFT80-silenced cells. Nuclear were stained with DAPI (blue). (D) Immunofluorescence staining for Arl13b location and expression. Arl13b was expressed in control group (pGIPZ, red)), but not in IFT80-silenced cells. Nuclear were stained with DAPI (blue). (E) Immunofluorescence staining for cilia by detecting the location and expression of gamma tubulin (red) and acetylated α-tubulin (bright blue). Nuclear were stained with DAPI (dark blue). Cilia existed in control cells, but not in IFT80-silenced cells.
3.3. Silencing IFT80 inhibits osteoblastic differentiation and mineralization
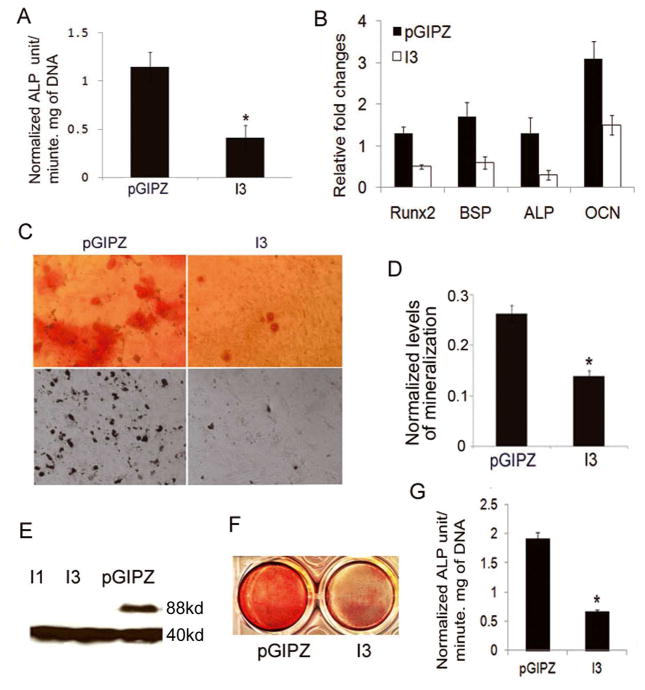
ALP activity and matrix mineralization are important factors for bone formation. Alkaline phosphatase hydrolyzes pyrophosphate and provides inorganic phosphate to promote mineralization in osteoblast [51]. To determine whether silencing IFT80 affects ALP activity and osteoblast marker gene expression, C3H10T1/2 cells were infected with pGIPZ-IFT80 shRNA (I3) or pGIPZ-scrambled shRNA (pGIPZ) lentiviruses and induced with OS media for 7 days. ALP activity was then examined as described in the Materials and Methods section. As shown in Fig. 3A, silencing IFT80 significantly inhibited ALP activity. ALP activity in the control group (pGIPZ) was 2.9 fold of that in IFT80-silenced group (I3) (n=6, p<0.01). Meanwhile, real time RT-PCR showed that silencing IFT80 significantly suppressed the expression of osteoblast marker genes - RUNX2, OCN, BSP and ALP as shown in Fig. 3B. The level of RUNX2, BSP, ALP and OCN in control cells are respectively 3.5, 3, 5 and 2 folds of that in IFT80 silenced cells, indicating IFT80 regulates osteoblast marker gene expression. To test whether IFT80 affects cell mineralization, C3H10T1/2 cells were infected with I3 or pGIPZ, and then induced with OS media for 14 days. Von Kossa and Alizarin Red staining assays detected the phosphate and calcium deposits on the cells, respectively. As shown in Fig. 3C–D, silencing IFT80 led to the apparent reduction of calcium and phosphate deposits (Fig. 3C). Quantitative mineralization levels based on Alizarin Red staining in IFT80-silenced cells was significantly lower than that in control cells (n=6, p<0.05) (Fig. 3D). This indicates that IFT80 plays a critical role in osteoblast differentiation, activation, and cell mineralization. To confirm our findings on primary BMSCs, BMSCs were infected with I3 and pGIPZ, and then induced with OS media for 2 days for western blotting, 7 days for ALP assays and 14 days for alizarin red staining. Similar to the results from osteoblast progenitor cell line, the expression of IFT80 was undetectable in IFT80-silenced cells (Fig. 3E), and silencing IFT80 significantly inhibited cell mineralization (Fig. 3F). ALP activity in pGIPZ group was 2.7 fold higher than that in the IFT80-silenced group (Fig. 3G); indicating IFT80 plays an important role in osteogenesis and cell mineralization.
Fig. 3.
Silencing IFT80 inhibits osteoblastic ALP activity, osteoblast marker gene expression, and cell mineralization. (A) ALP activity assay. C3H10T1/2 cells were infected with pGIPZ-IFT80 shRNA (I3) or pGIPZ-scrambled shRNA (pGIPZ) lentiviruses, induced with OS media for 7 days, and then ALP activity was examined. ALP activity in control group was 2.9 fold of that in IFT80-silenced group (n=6, *p<0.01). (B) Real time RT-PCR for analyzing the expression of RUNX2, OCN, BSP and ALP. C3H10T1/2 cells were infected with pGIPZ-IFT80 shRNA (I3) or pGIPZ-scrambled shRNA (pGIPZ) lentiviruses and induced with OS media for 7 days. (C) Von Kossa and Alizarin Red staining. C3H10T1/2 cells were infected with I3 or pGIPZ as described in Material and Methods, and then induced with OS media for 14 days. (D) Quantitative mineralization level based on Alizarin Red staining. The mineralized level in IFT80-silenced cells was significantly lower than that in control cells (n=6, *p<0.05). (E–G) Silence of IFT80 in mouse BMSCs. Primary BMSCs, derived from mouse bone marrow, were infected with I3 or pGIPZ and then induced with OS media for: E, 2 days for western blotting; F, 14 days for Alizarin Red staining; G, 7 days for ALP assays (n=6, *p<0.05).
3.4. Ectopic expression of Gli2 rescues the impaired osteoblast differentiation and mineralization resulting from silencing IFT80
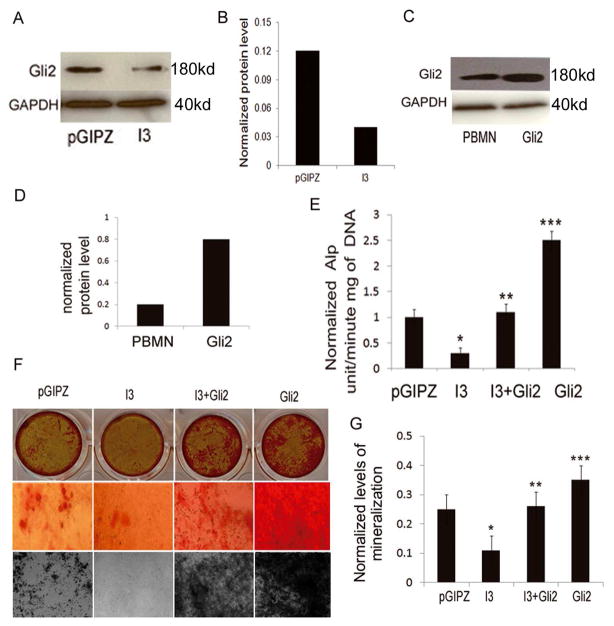
It has been reported that intraflagellar transport is essential for efficient hedgehog signaling (Hh) [52–54]. Gli transcriptional factors play essential roles in Hh-mediated pathway, bone development and osteoblast differentiation [55–57]. To investigate whether IFT80 controls osteoblast differentiation through the Hh-Gli pathway, we first analyzed whether silencing IFT80 affects Gli2 gene expression in C3H10T1/2 cells by western blot. As shown in Fig. 4A–B, silencing IFT80 markedly inhibited Gli2 gene expression. Next we looked at whether ectopic expression of Gli2 can rescue the deficiency of osteogenesis, resulting from silencing IFT80, and restore osteoblast differentiation. A recombinant retrovirus of Gli2 gene (pBMN-Gli2) was generated by using the pBMN-I-GFP retrovirus system. Then pBMN-I-GFP (the empty vector as a control) or pBMN-Gli2 were separately transfected into Phoenix-ecotropic cells and packaged into recombinant retroviruses. C3H10T1/2 cells were infected with the Gli2-expressing virus (Gli2) or with the control virus (pBMN) as described [38]. To confirm the ectopic expression of Gli2, we detected Gli2 protein by Western blot analysis. As shown in Fig. 4C–D, the protein level of Gli2 was significantly increased in Gli2-overexpresed cells compared with that in the control cells. Notably, the expression of Gli2 in IFT80-silenced cells could rescue the impaired ALP activity and cell mineralization as shown in Fig. 4E–G. ALP activity in Gli2 group was 10, 2.5, and 2.2 folds greater than that of the I3, pGIPZ, and I3+Gli2 groups, respectively (n=6, p<0.01). Moreover, overexpression of Gli2 significantly promoted ALP activity and cell mineralization of normal osteoblastic progenitor cells. These results indicate that IFT80 regulates osteoblast differentiation and mineralization through the Gli2-mediated pathway.
Fig. 4.
Ectopic expression of Gli2 rescues the impaired osteoblast differentiation and mineralization resulting from IFT80 silencing. (A) Western blot analysis of Gli2 expression. C3H10T1/2 cells were infected with pGIPZ-shRNA IFT80 (I3) or pGIPZ-scrambled shRNA (pGIPZ) lentiviruses for 48 hrs, at which time the protein was extracted for detecting Gli2 expression. (B) Quantitative analysis of protein levels from immunoblots as in a. The protein levels of Gli2 were normalized to GAPDH. (C) Western blot analysis for ectopic expression of Gli2. C3H10T1/2 cells were infected with the Gli2-expressing retrovirus (Gli2) or with a control retrovirus (pBMN). Gli2 protein level was significantly increased in Gli2-overexpresed cells compared with the control cells. (D) Quantitative analysis of protein levels from immunoblots as in c. The protein levels of Gli2 were normalized to GAPDH. (E–F) ALP activity assay. C3H10T1/2cells were first infected with either I3 or pGIPZ for 48hrs, followed by infection with pBMN or Gli2 for an additional 48 hours. The cells were then induced with OS media for 7 days before detection of the ALP activity assay. Each bar is expressed as the mean ± SD (unit/min*mg DNA) of six determinations: Gli2 vs. I3, pGIPZ, or I3+Gli2, ***p30.001; I3 vs. I3+Gli2 or pGIPZ, *p30.001; I3+Gli2 vs. pGIPZ, **p>0.05. (f) Von Kossa and Alizarin Red staining. After the infection protocol outlined above in e, cells were induced in OS media for 14 days prior to Von Kossa and Alizarin Red staining. (G) Quantitative mineralization level based on Alizarin Red staining. Each bar is expressed as the mean ± SD (unit/min*mg DNA) of three determinations. Gli2 vs. I3, pGIPZ, or I3+Gli2, ***p30.001; I3 vs. I3+Gli2 or pGIPZ, *p30.001; I3+Gli2 vs. pGIPZ, **p>0.05.
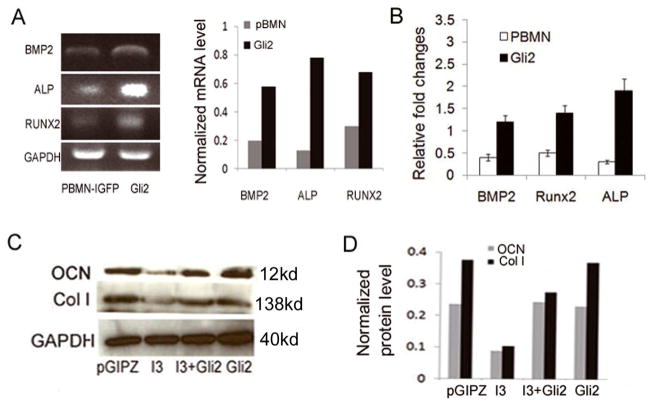
3.5. IFT80 and Gli2 regulate osteoblast marker gene expression
The osteoblast-associated molecules such as type I collagen (Col I), Runx2 (early differentiation marker), and osteocalcin (OCN) (late differentiation marker) are considered osteoblast markers [39, 58, 59]. To investigate the effect of IFT80 and Gli2 on the expression of osteoblast marker genes, C3H10T1/2 cells were infected with the IFT80-silenced (I3) or control (pGIPZ) lentivirus for 48 hrs followed by infection with Gli2-overexpressed (Gli2) or control (pBMN) retrovirus for 48 hrs, and then induced with OS media for 7 days. The expression of bone morphogenetic protein 2 (BMP2), ALP, and RUNX2 was detected by RT-PCR and quantitative Real Time RT-PCR (Fig 5A–B). We found that overexpression of Gli2 significantly increased BMP2, ALP and RUNX2 expression compared with that in the control cells, indicating that IFT80 affects osteoblast differentiation and mineralization through regulating osteoblast marker gene expression. Consistent with the above result, Western Blot analysis revealed that the expression of Col I and OCN was attenuated in IFT80-silenced cells, but could be increased in Gli2-overexpressed cells after being induced with OS media for 7 days (Fig. 5C–D). This demonstrates that IFT80 facilitates osteoblast differentiation and mineralization through regulating the expression of Gli2 and osteoblastic marker genes BMP2, Runx2, Col I, and OCN.
Fig. 5.
IFT80/Gli2 regulates osteoblast marker gene expression. C3H10T1/2 cells were first infected with either I3 or pGIPZ for 48hrs, followed by infection with pBMN or Gli2 for an additional 48 hours, and then induced with OS media for 7 days. (A) RT-PCR. Overexpression of Gli2 significantly increased BMP2, ALP and RUNX2 expression compared with that in the control cells. (B) Real time RT-PCR. Overexpression of Gli2 increased by 2.7, 2.8 and 3.9 fold the expression of BMP2, ALP and RUNX2, respectively. (C) Western Blot. The expression of Col I and OCN was attenuated in IFT80-silenced cells, but rescued and increased in Gli2- overexpressed cells. (D) Quantitative analysis of protein levels from immunoblots as in C. The protein levels were normalized to GAPDH.
3.6. IFT80 acts downstream of smoothened in Hh signaling pathway
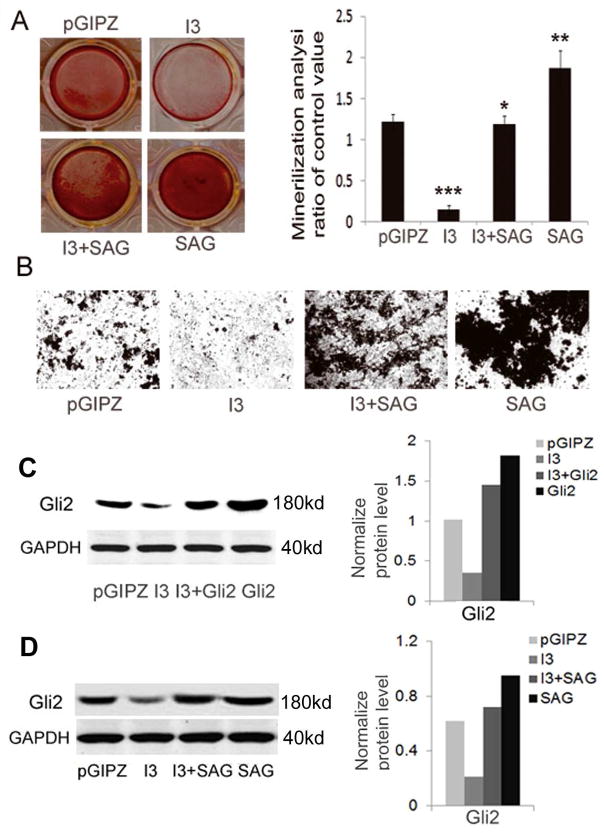
Hh signaling is characterized by Smoothened (Smo)-dependent activation of Gli1 and Gli2, which transcriptionally regulate target genes. Smo is activated upon Hh stimulation or treatment with the agonist of Smo such as SAG, [27, 60–62]. Knowing that silence of IFT80 down-regulates Gli2 expression, and ectopic expression of Gli2 is able to rescue the deficiency of osteoblast differentiation in IFT80-silenced cells, we further explored whether the activation of Hh and Smo signaling can promote osteoblast differentiation and whether IFT80-silencing can block the function of Smo. We found that treatment with SAG (10nM) could significantly promote osteoblast differentiation; however, when silencing IFT80, this promotion was partially inhibited (Fig 6A–B). The normalized level of mineralization in the I3+SAG group is 0.64 folds that of the SAG group. By performing western blot, we further show that overexpression of Gli2, or treatment with SAG, up-regulated the expression of Gli2 in IFT80-silenced cells and control cells as shown in Fig 6C–D. These results provide the first evidence that IFT80 regulates osteogenesis through Hh/Gli pathway, and IFT80 acts downstream of Hh-Smo and upstream of Glis in this pathway.
Fig. 6.
IFT80 silencing inhibits the SAG induced osteoblast differentiation and mineralization. (A–B) C3H10T1/2 cells were infected with pGIPZ-IFT80 shRNA (I3) or pGIPZ-scrambled shRNA (pGIPZ) lentiviruses, and treated with OS media with 10nM SAG for 24 days to study effects on cell mineralization. (A) Alizarin Red staining. Quantitative mineralization level based on Alizarin Red staining reported as the mean ± SD from tests of samples in triplicate. There are significant differences between the SAG group vs. other groups (**p<0.01), and between I3 vs. pGIPZ or I3+SAG (***P<0.001). There is no different between pGIPZ vs. I3+SAG (* P>0.05), indicating that SAG completely rescued the deficiency of osteoblast mineralization resulting from silencing IFT80. (B) Von kossa staining. (C–D) Western blots. C3H10T1/2 cells were infected with I3 or pGIPZ, followed by co-infection with Gli2-expressing retrovirus (Gli2) or with a control retrovirus (pBMN) for 24 hrs. The infected cells were treated with OS media with or without 10nM SAG for 7 days for analysis of Gli2 expression. Overexpression of Gli2, or treatment with SAG, up-regulates the expression of Gli2 in IFT80-silenced cells as well as control cells.
4. Discussion
It has been reported that partially mutation of IFT80 in human causes diseases such as Jeune asphyxiating thoracic dystrophy (JATD) and short rib polydactyly (SRP) type III with abnormal skeletal development [31, 33, 63]. However, based on these mutation observations, it is still unclear if the abnormal skeletal phenotype were resulted from indirect effect of mutation of IFT80 in human tissues or due to the effect of IFT80 mutation on osteogenesis. Therefore, in this study, we have addressed this question by using lentivirus mediated gene silencing method to knockdown of IFT80 in osteoblast progenitor cells. We demonstrated that IFT80 expression was predominant in bone tissues, and that IFT80 silencing impaired osteoblast differentiation and cell mineralization and decreased the expression of the osteoblast-specific genes. Also IFT80 silencing inhibited cilia formation. Gain-of-function experiments via overexpression of Gli2 showed the enhancement of osteoblast differentiation and mineralization in control cells and could rescue the deficiency of osteogenesis from silencing IFT80. Moreover, treatment of SAG could promote osteoblast differentiation which was partially inhibited by silencing IFT80. Those finding suggest that IFT80 regulates osteoblast differentiation and mineralization through Hh/Gli signal pathway and IFT80 functions downstream of Smo and upstream of Glis. This report is the first to demonstrate that IFT80 is directly involved in the regulation of osteoblast differentiation through Hh signaling pathway.
Our results showed that IFT80 was highly expressed in adult long bone and skull. It is abundant in eye, lung, spleen and kidney tissues. IFT80 was also expressed in muscle, heart, liver, and brain tissues, but to a far lesser extent. We further found that IFT80 expressed in all stages of osteoblast differentiation, but the expression level of IFT80 gradually decreased during the later stages of osteoblast differentiation. These results suggest diversity in the function of IFT80. In agreement with this, cilia with IFT particles are expressed on the surface of almost all vertebrate cells including osteoblasts and osteocytes[18]. In addition, the loss of the IFT proteins in mice resulting in a complex series of pathologies involving not only the skeleton, but also kidney, liver, brain, testis, eye, pancreas, etc [17–19, 22, 32, 33, 64–66], indicating that IFT proteins/cilia may have different functions in specific tissues and cell types or during distinct stages of cell differentiation. In this study, by silencing IFT80 in osteoblastic precursor cell stage, we have investigated the effect of IFT80 silencing on osteoblast differentiation and mineralization. However, we cannot rule out the possibility that IFT80 might also play an important role in mature osteoblast mechanotransduction, because cilia are not only involved in cell differentiation and function, but also acts as a mechanical sensor [19, 20].
Intraflagellar transport proteins are microtubule-based transport machinery, which are essential for the assembly and maintenance of cilia [47, 67]. Accumulated evidence has shown that defective IFT could result in impaired cilia [48, 54] with a class of human genetic disorders. Conditional deletion of IFT88 in the developing limb showed the disrupted cilia on different cell populations of developing limb and abnormal endochondral bone formation [8]. Most recent studies have shown that disruption of Kif3a significantly inhibited primary cilia formation in osteoblasts and reduced osteoblast differentiation in vitro and bone formation in vivo [17, 18]. Knockdown of IFT80 in zebrafish, and Tetrahymena thermophila, which produced shortened or absent cilia [31, 68] with symptoms associated with JATD such as abnormal skeletal development resulting in a long, narrow thorax, short ribs, shortened long bones, and occasional polydactyly [69]. Consistent with these previous studies, our results show that silencing IFT80 from osteoblast progenitor cells cause loss of cilia in 80–85% cells and the shorten cilia length in 5–10% cells. Moreover, we found that silencing IFT80 inhibits the expression of cilia marker protein- Arl13b, which plays a direct role in the assembly or maintenance of the axoneme [70]. Thus, we provide the first evidence that IFT80 plays an essential role in cilia formation in osteoblast progenitor cells.
Much evidence revealed that there are connections between the Hedgehog (Hh) signal pathway and the primary cilium as well as IFT [52, 54, 71]. Kif3a-deficient mouse has a phenotype similar to that of an Indian Hh (Ihh)-deficient mouse [22], suggesting the relationship between Ihh and primary cilia and their role in skeletogenesis. Gli transcription factors including Gli1, Gli2 and Gli3 are key effectors of the Hh signaling system [72]. Mutant studies revealed that Gli2 function primarily as the main transcriptional activator whereas Gli3 as the transcriptional repressor of the mammalian Hh pathway [73–75]. In contrast, Gli1 function is transcriptional activator and dispensable for development, as Gli1 null mice are viable and appear normal [76]. It has been reported that loss of Kif3a and IFT88 also led to a significant decrease of Gli2 expression, reduced cilia number and length[8, 22]. Consistent with those findings, our results showed that silencing IFT80 disrupted cilia formation and suppressed the expression of Gli2. In addition, our results show that overexpression of Gli2 could promote osteoblast differentiation and rescued the deficiency of osteogenesis from IFT80 silencing. These results indicate that IFT80 acts upstream of Glis and regulates Gli2 expression in Hh signaling pathway, and Gli2 directly regulates osteogenesis. In agreement with this view, Andrea Kesper et al [57] found the expression of Gli2 in preosteoblasts is sufficient for the induction of osteoblast differentiation in absence of Gli3. Shimoyama et al [77] reported that overexpression of Gli2- but not Gli3-induced osteoblast differentiation; whereas dominant-negative Gli2 markedly inhibited Ihh-dependent osteoblast differentiation. Most recently, Hojo et al [78] demonstrated SAG induces osteoblast differentiation and osteoblast marker gene expression, and SAG induction is inhibited in Gli1−/− and Gli2−/− cells relative to that in wild type cells. This report agree with our finding that treatment with SAG in control cells promoted osteoblast differentiation and silencing IFT80 could decrease Gli2 expression and partially inhibit this SAG enhancement. Currently, we have no evidence for the precise mechanism. It is most likely because 15–20% of IFT80 silenced cells still have cilia existed which can response to ectopic higher level of Hh and Smo signaling to compensate the reduced Hh signaling transduction in IFT80 silencing cells. It is also possible that similar to the suppressor of fused (Sufu) [79], which regulates Gli protein levels in cilium-independent manner, SAG activated, high level of smo regulates Gli2 protein level through both cilia independent and dependent pathways in osteoblast differentiation.
It has been shown that there is a definite lineage-specific differentiation of mesenchymal stem cells into osteoblasts upon Hh protein stimulation [80, 81]. Furthermore, the expression of molecules that stimulate osteoblast differentiation, including BMP-2, -4, and -7 is regulated by Hh signaling [82, 83]. Shimoyama et al. [77] reported that overexpression of Gli2 and treatment with Ihh promotes osteoblast differentiation by regulating RUNX2 expression and function. Zhao, et al [84] reported that Gli2 plays a critical role in osteoblast differentiation by up-regulating BMP-2 expression in osteoblast precursor cells. Our results showed that silencing IFT80 significantly inhibited osteoblast differentiation, calcium deposition in extracellular matrix and the expression of osteoblast marker gene expression – RUNX2, BSP, BMP2, Col I and OCN. The level of RUNX2, BSP, ALP and OCN in control cells are respectively 3.5, 3, 5 and 2 folds of that in IFT80 silenced cells. Overexpression of Gli2 could rescue these deficiencies, indicating that Gli2 plays critical role in IFT80-mediated osteoblast differentiation. Thus, our finding supports a model that IFT80 regulates the expression of Gli proteins, allowing them to acquire full transcriptional activity and regulate osteoblast differentiation and osteoblast marker gene expression.
In summary, our results demonstrate that IFT80 plays a critical role in osteoblast differentiation through regulating Hh/Gli pathway and osteoblast marker gene expression. The study presented here adds to the increasing body of evidence suggesting that IFT80 acts on osteoblastic progenitor cells to promote osteoblast differentiation. Most recently, Rix et al. group [66] generated a murine IFT80 gene-trap line, and Hypomorphic levels of IFT80 result in more than 97% embryonic lethality. This early embryonic lethality precluded direct analysis of the roles of IFT80 in bone developmental processes in those mutants. Generating IFT80 conditional knockout model would facilitate to elucidate the other possible pathways such as wnt signaling, calcium signaling pathways coordinated by primary cilia and IFT80 for osteogenesis and mature osteoblast function such as acting as a mechanosensor, like PKD1 or PKD2 genes [21, 85]. The findings on those studies would provide novel gene therapeutic approaches and drug targets for treatment of JATD, SRPIII and other bone diseases.
Highlights.
IFT80 was highly expressed in bone tissue and during osteoblast differentiation.
Silencing IFT80 led to either shortening or loss of cilia and blocked osteogenesis
Overexpression of Gli2 rescued osteogenic deficiency of IFT80-silenced cells
Introduction of SAG promoted osteogenesis, which was inhibited by silencing IFT80
IFT80 stimulates osteogenesis through regulating Hedgehog/Gli signal pathway
Acknowledgments
We thank Drs. Rosemary Dziak, Douglas Olson and Mr. David Hadbawnik for critical reading of the manuscript, and Dr. Wade J. Sigurdson, the director of the Confocal Microscope Facility in the School of Medicine and Biomedical Sciences, University at Buffalo for the assistance with fluorescence microscopy. This work was supported by a National Institute of Health grant AR055678 (S. Yang) and AR061052 (S. Yang).
Abbreviations
- IFT
intraflagellar transport
- BBS
Bardet-Biedl syndrome
- JATD
Jeune asphyxiating thoracic dystrophy
- SRPIII
short rib polydactyly type III
- ALP
alkaline phosphatase
- BMP
bone morphogenetic protein
- RUNX2
Runt-related transcription factor 2
- OCN
osteocalcin
- BSP
Bone sialoprotein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Serra R. Intraflagellar Transport In Skeletal Development. J Musculoskelet Neuronal Interact. 2007;7:302–3. [PubMed] [Google Scholar]
- 2.Bisgrove Bw, Yost Hj. The Roles Of Cilia In Developmental Disorders And Disease. Development. 2006;133:4131–43. doi: 10.1242/dev.02595. [DOI] [PubMed] [Google Scholar]
- 3.Davenport Jr, Yoder Bk. An Incredible Decade For The Primary Cilium: A Look At A Once-Forgotten Organelle. Am J Physiol Renal Physiol. 2005;289:F1159–69. doi: 10.1152/ajprenal.00118.2005. [DOI] [PubMed] [Google Scholar]
- 4.Pan J, Wang Q, Snell Wj. Cilium-Generated Signaling And Cilia-Related Disorders. Lab Invest. 2005;85:452–63. doi: 10.1038/labinvest.3700253. [DOI] [PubMed] [Google Scholar]
- 5.Mykytyn K, Mullins Rf, Andrews M, Chiang Ap, Swiderski Re, Yang B, Braun T, Casavant T, Stone Em, Sheffield Vc. Bardet-Biedl Syndrome Type 4 (Bbs4)-Null Mice Implicate Bbs4 In Flagella Formation But Not Global Cilia Assembly. Proc Natl Acad Sci U S A. 2004;101:8664–9. doi: 10.1073/pnas.0402354101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mykytyn K, Sheffield Vc. Establishing A Connection Between Cilia And Bardet-Biedl Syndrome. Trends Mol Med. 2004;10:106–9. doi: 10.1016/j.molmed.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 7.Haycraft Cj, Banizs B, Aydin-Son Y, Zhang Q, Michaud Ej, Yoder Bk. Gli2 And Gli3 Localize To Cilia And Require The Intraflagellar Transport Protein Polaris For Processing And Function. Plos Genet. 2005;1:E53. doi: 10.1371/journal.pgen.0010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haycraft Cj, Zhang Q, Song B, Jackson Ws, Detloff Pj, Serra R, Yoder Bk. Intraflagellar Transport Is Essential For Endochondral Bone Formation. Development. 2007;134:307–16. doi: 10.1242/dev.02732. [DOI] [PubMed] [Google Scholar]
- 9.Kolpakova-Hart E, Jinnin M, Hou B, Fukai N, Olsen Br. Kinesin-2 Controls Development And Patterning Of The Vertebrate Skeleton By Hedgehog- And Gli3-Dependent Mechanisms. Dev Biol. 2007;309:273–84. doi: 10.1016/j.ydbio.2007.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu A, Wang B, Niswander La. Mouse Intraflagellar Transport Proteins Regulate Both The Activator And Repressor Functions Of Gli Transcription Factors. Development. 2005;132:3103–11. doi: 10.1242/dev.01894. [DOI] [PubMed] [Google Scholar]
- 11.Mcglashan Sr, Haycraft Cj, Jensen Cg, Yoder Bk, Poole Ca. Articular Cartilage And Growth Plate Defects Are Associated With Chondrocyte Cytoskeletal Abnormalities In Tg737orpk Mice Lacking The Primary Cilia Protein Polaris. Matrix Biol. 2007;26:234–46. doi: 10.1016/j.matbio.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 12.Ruiz-Perez Vl, Blair Hj, Rodriguez-Andres Me, Blanco Mj, Wilson A, Liu Yn, Miles C, Peters H, Goodship Ja. Evc Is A Positive Mediator Of Ihh-Regulated Bone Growth That Localises At The Base Of Chondrocyte Cilia. Development. 2007;134:2903–12. doi: 10.1242/dev.007542. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Q, Murcia Ns, Chittenden Lr, Richards Wg, Michaud Ej, Woychik Rp, Yoder Bk. Loss Of The Tg737 Protein Results In Skeletal Patterning Defects. Dev Dyn. 2003;227:78–90. doi: 10.1002/dvdy.10289. [DOI] [PubMed] [Google Scholar]
- 14.Haycraft Cj, Serra R. Cilia Involvement In Patterning And Maintenance Of The Skeleton. Curr Top Dev Biol. 2008;85:303–32. doi: 10.1016/S0070-2153(08)00811-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang Cf, Ramaswamy G, Serra R. Depletion Of Primary Cilia In Articular Chondrocytes Results In Reduced Gli3 Repressor To Activator Ratio, Increased Hedgehog Signaling, And Symptoms Of Early Osteoarthritis. Osteoarthritis Cartilage. 2012;20:152–61. doi: 10.1016/j.joca.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Serra R. Role Of Intraflagellar Transport And Primary Cilia In Skeletal Development. Anat Rec (Hoboken) 2008;291:1049–61. doi: 10.1002/ar.20634. [DOI] [PubMed] [Google Scholar]
- 17.Martins Aa, Paiva A, Morgado Jm, Gomes A, Pais Ml. Quantification And Immunophenotypic Characterization Of Bone Marrow And Umbilical Cord Blood Mesenchymal Stem Cells By Multicolor Flow Cytometry. Transplant Proc. 2009;41:943–6. doi: 10.1016/j.transproceed.2009.01.059. [DOI] [PubMed] [Google Scholar]
- 18.Berbari Nf, O’connor Ak, Haycraft Cj, Yoder Bk. The Primary Cilium As A Complex Signaling Center. Curr Biol. 2009;19:R526–35. doi: 10.1016/j.cub.2009.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiao Z, Dallas M, Qiu N, Nicolella D, Cao L, Johnson M, Bonewald L, Quarles Ld. Conditional Deletion Of Pkd1 In Osteocytes Disrupts Skeletal Mechanosensing In Mice. Faseb J. 2011;25:2418–32. doi: 10.1096/fj.10-180299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kalogeropoulos M, Varanasi Ss, Olstad Ok, Sanderson P, Gautvik Vt, Reppe S, Francis Rm, Gautvik Km, Birch Ma, Datta Hk. Zic1 Transcription Factor In Bone: Neural Developmental Protein Regulates Mechanotransduction In Osteocytes. Faseb J. 2010;24:2893–903. doi: 10.1096/fj.09-148908. [DOI] [PubMed] [Google Scholar]
- 21.Xiao Z, Zhang S, Cao L, Qiu N, David V, Quarles Ld. Conditional Disruption Of Pkd1 In Osteoblasts Results In Osteopenia Due To Direct Impairment Of Bone Formation. J Biol Chem. 2010;285:1177–87. doi: 10.1074/jbc.M109.050906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koyama E, Young B, Nagayama M, Shibukawa Y, Enomoto-Iwamoto M, Iwamoto M, Maeda Y, Lanske B, Song B, Serra R, Pacifici M. Conditional Kif3a Ablation Causes Abnormal Hedgehog Signaling Topography, Growth Plate Dysfunction, And Excessive Bone And Cartilage Formation During Mouse Skeletogenesis. Development. 2007;134:2159–69. doi: 10.1242/dev.001586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kimura H, Ng Jm, Curran T. Transient Inhibition Of The Hedgehog Pathway In Young Mice Causes Permanent Defects In Bone Structure. Cancer Cell. 2008;13:249–60. doi: 10.1016/j.ccr.2008.01.027. [DOI] [PubMed] [Google Scholar]
- 24.Mcmahon Ap, Ingham Pw, Tabin Cj. Developmental Roles And Clinical Significance Of Hedgehog Signaling. Curr Top Dev Biol. 2003;53:1–114. doi: 10.1016/s0070-2153(03)53002-2. [DOI] [PubMed] [Google Scholar]
- 25.Wang B, Fallon Jf, Beachy Pa. Hedgehog-Regulated Processing Of Gli3 Produces An Anterior/Posterior Repressor Gradient In The Developing Vertebrate Limb. Cell. 2000;100:423–34. doi: 10.1016/s0092-8674(00)80678-9. [DOI] [PubMed] [Google Scholar]
- 26.Marigo V, Johnson Rl, Vortkamp A, Tabin Cj. Sonic Hedgehog Differentially Regulates Expression Of Gli And Gli3 During Limb Development. Dev Biol. 1996;180:273–83. doi: 10.1006/dbio.1996.0300. [DOI] [PubMed] [Google Scholar]
- 27.Corbit Kc, Aanstad P, Singla V, Norman Ar, Stainier Dy, Reiter Jf. Vertebrate Smoothened Functions At The Primary Cilium. Nature. 2005;437:1018–21. doi: 10.1038/nature04117. [DOI] [PubMed] [Google Scholar]
- 28.Song B, Haycraft Cj, Seo Hs, Yoder Bk, Serra R. Development Of The Post-Natal Growth Plate Requires Intraflagellar Transport Proteins. Dev Biol. 2007;305:202–16. doi: 10.1016/j.ydbio.2007.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shahangian A, Chow Ek, Tian X, Kang Jr, Ghaffari A, Liu Sy, Belperio Ja, Cheng G, Deng Jc. Type I Ifns Mediate Development Of Postinfluenza Bacterial Pneumonia In Mice. J Clin Invest. 2009;119:1910–20. doi: 10.1172/JCI35412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang W, Kane Jk, Li Md. Identification And Characterization Of A Long Isoform Of Human Ift80, Ift80-L. Biochem Biophys Res Commun. 2008;373:653–8. doi: 10.1016/j.bbrc.2008.06.085. [DOI] [PubMed] [Google Scholar]
- 31.Beales Pl, Bland E, Tobin Jl, Bacchelli C, Tuysuz B, Hill J, Rix S, Pearson Cg, Kai M, Hartley J, Johnson C, Irving M, Elcioglu N, Winey M, Tada M, Scambler Pj. Ift80, Which Encodes A Conserved Intraflagellar Transport Protein, Is Mutated In Jeune Asphyxiating Thoracic Dystrophy. Nat Genet. 2007;39:727–9. doi: 10.1038/ng2038. [DOI] [PubMed] [Google Scholar]
- 32.Cavalcanti Dp, Huber C, Le Quan Sang Kh, Baujat G, Collins F, Delezoide Al, Dagoneau N, Le Merrer M, Martinovic J, Mello Mf, Vekemans M, Munnich A, Cormier-Daire V. Mutation In Ift80 Gene In A Foetus With A Phenotype Of Verma-Naumoff Provides Molecular Evidence For The Jeune-Verma-Naumoff Dysplasia Spectrum. J Med Genet. 2009 doi: 10.1136/jmg.2009.069468. [DOI] [PubMed] [Google Scholar]
- 33.Dagoneau N, Goulet M, Genevieve D, Sznajer Y, Martinovic J, Smithson S, Huber C, Baujat G, Flori E, Tecco L, Cavalcanti D, Delezoide Al, Serre V, Le Merrer M, Munnich A, Cormier-Daire V. Dync2h1 Mutations Cause Asphyxiating Thoracic Dystrophy And Short Rib-Polydactyly Syndrome, Type Iii. Am J Hum Genet. 2009;84:706–11. doi: 10.1016/j.ajhg.2009.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pittenger Mf, Mackay Am, Beck Sc, Jaiswal Rk, Douglas R, Mosca Jd, Moorman Ma, Simonetti Dw, Craig S, Marshak Dr. Multilineage Potential Of Adult Human Mesenchymal Stem Cells. Science. 1999;284:143–7. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 35.Yang Sy, Wei Dy, Wang D, Phimphilai M, Krebsbach Ph, Franceschi Rt. In Vitro And In Vivo Synergistic Interactions Between The Runx2/Cbfa1 Transcription Factor And Bone Morphogenetic Protein-2 In Stimulating Osteoblast Differentiation. Journal Of Bone And Mineral Research. 2003;18:705–715. doi: 10.1359/jbmr.2003.18.4.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang S, Li Yp. Rgs12 Is Essential For Rankl-Evoked Signaling For Terminal Differentiation Of Osteoclasts In Vitro. J Bone Miner Res. 2007;22:45–54. doi: 10.1359/jbmr.061007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stewart Sa, Dykxhoorn Dm, Palliser D, Mizuno H, Yu Ey, An Ds, Sabatini Dm, Chen Is, Hahn Wc, Sharp Pa, Weinberg Ra, Novina Cd. Lentivirus-Delivered Stable Gene Silencing By Rnai In Primary Cells. Rna. 2003;9:493–501. doi: 10.1261/rna.2192803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang S, Li Yp. Rgs10-Null Mutation Impairs Osteoclast Differentiation Resulting From The Loss Of [Ca2+]I Oscillation Regulation. Genes Dev. 2007;21:1803–16. doi: 10.1101/gad.1544107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vandesompele J, De Paepe A, Speleman F. Elimination Of Primer-Dimer Artifacts And Genomic Coamplification Using A Two-Step Sybr Green I Real-Time Rt-Pcr. Anal Biochem. 2002;303:95–8. doi: 10.1006/abio.2001.5564. [DOI] [PubMed] [Google Scholar]
- 40.Schneider Gb, Whitson Sw, Cooper Lf. Restricted And Coordinated Expression Of Beta3-Integrin And Bone Sialoprotein During Cultured Osteoblast Differentiation. Bone. 1999;24:321–7. doi: 10.1016/s8756-3282(99)00007-1. [DOI] [PubMed] [Google Scholar]
- 41.Ovchinnikov D. Alcian Blue/Alizarin Red Staining Of Cartilage And Bone In Mouse. Cold Spring Harb Protoc 2009. 2009:Pdb Prot5170. doi: 10.1101/pdb.prot5170. [DOI] [PubMed] [Google Scholar]
- 42.Yamakawa K, Iwasaki H, Masuda I, Ohjimi Y, Honda I, Saeki K, Zhang J, Shono E, Naito M, Kikuchi M. The Utility Of Alizarin Red S Staining In Calcium Pyrophosphate Dihydrate Crystal Deposition Disease. J Rheumatol. 2003;30:1032–5. [PubMed] [Google Scholar]
- 43.Reinholz Gg, Getz B, Pederson L, Sanders Es, Subramaniam M, Ingle Jn, Spelsberg Tc. Bisphosphonates Directly Regulate Cell Proliferation, Differentiation, And Gene Expression In Human Osteoblasts. Cancer Res. 2000;60:6001–7. [PubMed] [Google Scholar]
- 44.Rungby J, Kassem M, Eriksen Ef, Danscher G. The Von Kossa Reaction For Calcium Deposits: Silver Lactate Staining Increases Sensitivity And Reduces Background. Histochem J. 1993;25:446–51. doi: 10.1007/BF00157809. [DOI] [PubMed] [Google Scholar]
- 45.Mccauley Lk, Koh Aj, Beecher Ca, Cui Y, Decker Jd, Franceschi Rt. Effects Of Differentiation And Transforming Growth Factor Beta 1 On Pth/Pthrp Receptor Mrna Levels In Mc3t3-E1 Cells. J Bone Miner Res. 1995;10:1243–55. doi: 10.1002/jbmr.5650100815. [DOI] [PubMed] [Google Scholar]
- 46.Yang S, Wei D, Wang D, Phimphilai M, Krebsbach Ph, Franceschi Rt. In Vitro And In Vivo Synergistic Interactions Between The Runx2/Cbfa1 Transcription Factor And Bone Morphogenetic Protein-2 In Stimulating Osteoblast Differentiation. J Bone Miner Res. 2003;18:705–15. doi: 10.1359/jbmr.2003.18.4.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rosenbaum Jl, Witman Gb. Intraflagellar Transport. Nat Rev Mol Cell Biol. 2002;3:813–25. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- 48.Pedersen Lb, Rosenbaum Jl. Intraflagellar Transport (Ift) Role In Ciliary Assembly, Resorption And Signalling. Curr Top Dev Biol. 2008;85:23–61. doi: 10.1016/S0070-2153(08)00802-8. [DOI] [PubMed] [Google Scholar]
- 49.Mukhopadhyay S, Lu Y, Qin H, Lanjuin A, Shaham S, Sengupta P. Distinct Ift Mechanisms Contribute To The Generation Of Ciliary Structural Diversity In C. Elegans Embo J. 2007;26:2966–80. doi: 10.1038/sj.emboj.7601717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gu Y, Liang X, Wu W, Liu M, Song S, Cheng L, Bo L, Xiong C, Wang X, Liu X, Peng L, Yao K. Multicenter Contraceptive Efficacy Trial Of Injectable Testosterone Undecanoate In Chinese Men. J Clin Endocrinol Metab. 2009;94:1910–5. doi: 10.1210/jc.2008-1846. [DOI] [PubMed] [Google Scholar]
- 51.Orimo H. The Mechanism Of Mineralization And The Role Of Alkaline Phosphatase In Health And Disease. J Nihon Med Sch. 2010;77:4–12. doi: 10.1272/jnms.77.4. [DOI] [PubMed] [Google Scholar]
- 52.Huangfu D, Liu A, Rakeman As, Murcia Ns, Niswander L, Anderson Kv. Hedgehog Signalling In The Mouse Requires Intraflagellar Transport Proteins. Nature. 2003;426:83–7. doi: 10.1038/nature02061. [DOI] [PubMed] [Google Scholar]
- 53.Huangfu D, Anderson Kv. Cilia And Hedgehog Responsiveness In The Mouse. Proc Natl Acad Sci U S A. 2005;102:11325–30. doi: 10.1073/pnas.0505328102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eggenschwiler Jt, Anderson Kv. Cilia And Developmental Signaling. Annu Rev Cell Dev Biol. 2007;23:345–73. doi: 10.1146/annurev.cellbio.23.090506.123249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jemtland R, Divieti P, Lee K, Segre Gv. Hedgehog Promotes Primary Osteoblast Differentiation And Increases Pthrp Mrna Expression And Ipthrp Secretion. Bone. 2003;32:611–20. doi: 10.1016/s8756-3282(03)00092-9. [DOI] [PubMed] [Google Scholar]
- 56.Joeng Ks, Long F. The Gli2 Transcriptional Activator Is A Crucial Effector For Ihh Signaling In Osteoblast Development And Cartilage Vascularization. Development. 2009;136:4177–85. doi: 10.1242/dev.041624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kesper Da, Didt-Koziel L, Vortkamp A. Gli2 Activator Function In Preosteoblasts Is Sufficient To Mediate Ihh-Dependent Osteoblast Differentiation, Whereas The Repressor Function Of Gli2 Is Dispensable For Endochondral Ossification. Dev Dyn. 2010;239:1818–26. doi: 10.1002/dvdy.22301. [DOI] [PubMed] [Google Scholar]
- 58.Kinsella Tm, Nolan Gp. Episomal Vectors Rapidly And Stably Produce High-Titer Recombinant Retrovirus. Hum Gene Ther. 1996;7:1405–13. doi: 10.1089/hum.1996.7.12-1405. [DOI] [PubMed] [Google Scholar]
- 59.Yang S, Delgado R, King Sr, Woffendin C, Barker Cs, Yang Zy, Xu L, Nolan Gp, Nabel Gj. Generation Of Retroviral Vector For Clinical Studies Using Transient Transfection. Hum Gene Ther. 1999;10:123–32. doi: 10.1089/10430349950019255. [DOI] [PubMed] [Google Scholar]
- 60.Rohatgi R, Milenkovic L, Scott Mp. Patched1 Regulates Hedgehog Signaling At The Primary Cilium. Science. 2007;317:372–6. doi: 10.1126/science.1139740. [DOI] [PubMed] [Google Scholar]
- 61.Wang Y, Zhou Z, Walsh Ct, Mcmahon Ap. Selective Translocation Of Intracellular Smoothened To The Primary Cilium In Response To Hedgehog Pathway Modulation. Proc Natl Acad Sci U S A. 2009;106:2623–8. doi: 10.1073/pnas.0812110106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rohatgi R, Milenkovic L, Corcoran Rb, Scott Mp. Hedgehog Signal Transduction By Smoothened: Pharmacologic Evidence For A 2-Step Activation Process. Proc Natl Acad Sci U S A. 2009;106:3196–201. doi: 10.1073/pnas.0813373106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cavalcanti Dp, Huber C, Sang Kh, Baujat G, Collins F, Delezoide Al, Dagoneau N, Le Merrer M, Martinovic J, Mello Mf, Vekemans M, Munnich A, Cormier-Daire V. Mutation In Ift80 In A Fetus With The Phenotype Of Verma-Naumoff Provides Molecular Evidence For Jeune-Verma-Naumoff Dysplasia Spectrum. J Med Genet. 2011;48:88–92. doi: 10.1136/jmg.2009.069468. [DOI] [PubMed] [Google Scholar]
- 64.Pazour Gj, Baker Sa, Deane Ja, Cole Dg, Dickert Bl, Rosenbaum Jl, Witman Gb, Besharse Jc. The Intraflagellar Transport Protein, Ift88, Is Essential For Vertebrate Photoreceptor Assembly And Maintenance. J Cell Biol. 2002;157:103–13. doi: 10.1083/jcb.200107108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pazour Gj, Dickert Bl, Vucica Y, Seeley Es, Rosenbaum Jl, Witman Gb, Cole Dg. Chlamydomonas Ift88 And Its Mouse Homologue, Polycystic Kidney Disease Gene Tg737, Are Required For Assembly Of Cilia And Flagella. J Cell Biol. 2000;151:709–18. doi: 10.1083/jcb.151.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rix S, Calmont A, Scambler Pj, Beales Pl. An Ift80 Mouse Model Of Short Rib Polydactyly Syndromes Shows Defects In Hedgehog Signalling Without Loss Or Malformation Of Cilia. Hum Mol Genet. 2011;20:1306–14. doi: 10.1093/hmg/ddr013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Scholey Jm. Intraflagellar Transport. Annu Rev Cell Dev Biol. 2003;19:423–43. doi: 10.1146/annurev.cellbio.19.111401.091318. [DOI] [PubMed] [Google Scholar]
- 68.Hudak Lm, Lunt S, Chang Ch, Winkler E, Flammer H, Lindsey M, Perkins Bd. The Intraflagellar Transport Protein Ift80 Is Essential For Photoreceptor Survival In A Zebrafish Model Of Jeune Asphyxiating Thoracic Dystrophy. Invest Ophthalmol Vis Sci. 2010;51:3792–9. doi: 10.1167/iovs.09-4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Oberklaid F, Danks Dm, Mayne V, Campbell P. Asphyxiating Thoracic Dysplasia. Clinical, Radiological, And Pathological Information On 10 Patients. Arch Dis Child. 1977;52:758–65. doi: 10.1136/adc.52.10.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Caspary T, Larkins Ce, Anderson Kv. The Graded Response To Sonic Hedgehog Depends On Cilia Architecture. Dev Cell. 2007;12:767–78. doi: 10.1016/j.devcel.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 71.Ocbina Pj, Anderson Kv. Intraflagellar Transport, Cilia, And Mammalian Hedgehog Signaling: Analysis In Mouse Embryonic Fibroblasts. Dev Dyn. 2008;237:2030–8. doi: 10.1002/dvdy.21551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hui Cc, Angers S. Gli Proteins In Development And Disease. Annu Rev Cell Dev Biol. 2011;27:513–37. doi: 10.1146/annurev-cellbio-092910-154048. [DOI] [PubMed] [Google Scholar]
- 73.Persson M, Stamataki D, Te Welscher P, Andersson E, Bose J, Ruther U, Ericson J, Briscoe J. Dorsal-Ventral Patterning Of The Spinal Cord Requires Gli3 Transcriptional Repressor Activity. Genes Dev. 2002;16:2865–78. doi: 10.1101/gad.243402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Litingtung Y, Chiang C. Specification Of Ventral Neuron Types Is Mediated By An Antagonistic Interaction Between Shh And Gli3. Nat Neurosci. 2000;3:979–85. doi: 10.1038/79916. [DOI] [PubMed] [Google Scholar]
- 75.Bai Cb, Joyner Al. Gli1 Can Rescue The In Vivo Function Of Gli2. Development. 2001;128:5161–72. doi: 10.1242/dev.128.24.5161. [DOI] [PubMed] [Google Scholar]
- 76.Park Hl, Bai C, Platt Ka, Matise Mp, Beeghly A, Hui Cc, Nakashima M, Joyner Al. Mouse Gli1 Mutants Are Viable But Have Defects In Shh Signaling In Combination With A Gli2 Mutation. Development. 2000;127:1593–605. doi: 10.1242/dev.127.8.1593. [DOI] [PubMed] [Google Scholar]
- 77.Shimoyama A, Wada M, Ikeda F, Hata K, Matsubara T, Nifuji A, Noda M, Amano K, Yamaguchi A, Nishimura R, Yoneda T. Ihh/Gli2 Signaling Promotes Osteoblast Differentiation By Regulating Runx2 Expression And Function. Mol Biol Cell. 2007;18:2411–8. doi: 10.1091/mbc.E06-08-0743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hojo H, Ohba S, Yano F, Saito T, Ikeda T, Nakajima K, Komiyama Y, Nakagata N, Suzuki K, Takato T, Kawaguchi H, Chung Ui. Gli1 Protein Participates In Hedgehog-Mediated Specification Of Osteoblast Lineage During Endochondral Ossification. J Biol Chem. 2012;287:17860–9. doi: 10.1074/jbc.M112.347716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen Mh, Wilson Cw, Li Yj, Law Kk, Lu Cs, Gacayan R, Zhang X, Hui Cc, Chuang Pt. Cilium-Independent Regulation Of Gli Protein Function By Sufu In Hedgehog Signaling Is Evolutionarily Conserved. Genes Dev. 2009;23:1910–28. doi: 10.1101/gad.1794109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Spinella-Jaegle S, Rawadi G, Kawai S, Gallea S, Faucheu C, Mollat P, Courtois B, Bergaud B, Ramez V, Blanchet Am, Adelmant G, Baron R, Roman-Roman S. Sonic Hedgehog Increases The Commitment Of Pluripotent Mesenchymal Cells Into The Osteoblastic Lineage And Abolishes Adipocytic Differentiation. J Cell Sci. 2001;114:2085–94. doi: 10.1242/jcs.114.11.2085. [DOI] [PubMed] [Google Scholar]
- 81.Van Der Horst G, Farih-Sips H, Lowik Cw, Karperien M. Hedgehog Stimulates Only Osteoblastic Differentiation Of Undifferentiated Ks483 Cells. Bone. 2003;33:899–910. doi: 10.1016/j.bone.2003.07.004. [DOI] [PubMed] [Google Scholar]
- 82.Kawai S, Sugiura T. Characterization Of Human Bone Morphogenetic Protein (Bmp)-4 And -7 Gene Promoters: Activation Of Bmp Promoters By Gli, A Sonic Hedgehog Mediator. Bone. 2001;29:54–61. doi: 10.1016/s8756-3282(01)00470-7. [DOI] [PubMed] [Google Scholar]
- 83.Krishnan V, Ma Y, Moseley J, Geiser A, Friant S, Frolik C. Bone Anabolic Effects Of Sonic/Indian Hedgehog Are Mediated By Bmp-2/4-Dependent Pathways In The Neonatal Rat Metatarsal Model. Endocrinology. 2001;142:940–7. doi: 10.1210/endo.142.2.7922. [DOI] [PubMed] [Google Scholar]
- 84.Zhao M, Qiao M, Harris Se, Chen D, Oyajobi Bo, Mundy Gr. The Zinc Finger Transcription Factor Gli2 Mediates Bone Morphogenetic Protein 2 Expression In Osteoblasts In Response To Hedgehog Signaling. Mol Cell Biol. 2006;26:6197–208. doi: 10.1128/MCB.02214-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xiao Zs, Quarles Ld. Role Of The Polycytin-Primary Cilia Complex In Bone Development And Mechanosensing. Ann N Y Acad Sci. 2010;1192:410–21. doi: 10.1111/j.1749-6632.2009.05239.x. [DOI] [PMC free article] [PubMed] [Google Scholar]