Abstract
Point mutations targeting muscle thin filament proteins are the cause of a number of cardiomyopathies. In many cases, biological effects of the mutations are well-documented, whereas their structural and mechanical impact on filament assembly and regulatory function is lacking. In order to elucidate molecular defects leading to cardiac dysfunction, we have examined the structural mechanics of two tropomyosin mutants, E180G and D175N, which are associated with hypertrophic cardiomyopathy (HCM). Tropomyosin is an α–helical coiled-coil dimer which polymerizes end-to-end to create an elongated superhelix that wraps around F-actin filaments of muscle and non-muscle cells, thus modulating the binding of other actin-binding proteins. Here, we study how flexibility changes in the E180G and D175N mutants might affect tropomyosin binding and regulatory motion on F-actin. Electron microscopy and Molecular Dynamics simulations show that E180G and D175N mutations cause an increase in bending flexibility of tropomyosin both locally and globally. This excess flexibility is likely to increase accessibility of the myosin-binding sites on F-actin, thus destabilizing the low-Ca2+ relaxed-state of cardiac muscle. The resulting imbalance in the on-off switching mechanism of the mutants will shift the regulatory equilibrium towards Ca2+-activation of cardiac muscle, as is observed in affected muscle, accompanied by enhanced systolic activity, diastolic dysfunction, and cardiac compensations associated with HCM and heart failure.
Keywords: actin, cardiomyopathy, electron microscopy, Molecular Dynamics, tropomyosin
1. Introduction
The elongated protein tropomyosin, present along the surface of actin-based thin filaments in virtually all eukaryotic cells, is composed of two α-helical chains that dimerize as a parallel coiled-coil. Pseudo-repeating domains (seven in muscle tropomyosin) bind to successive actin monomers along thin filaments. Tropomyosin polymerizes end-to-end to form a continuous superhelical strand extending over the length of the filament (reviewed in [1,2]). This arrangement strengthens the thin filament and increases filament flexural rigidity, but perhaps more importantly allows tropomyosin to function as a “gatekeeper” controlling access of other actin-binding proteins onto thin filaments [1,2]. Tropomyosin is best known for its action in muscle, where under the influence of troponin and Ca2+, it reversibly gates myosin interaction on thin filaments, and consequently regulates contraction. The size of the thin filament “regulatory unit”, namely the length of the thin filament over which a single troponin constrains the tropomyosin strand to cooperatively block myosin-binding sites on actin in resting muscle (or conversely Ca2+-saturated troponin or myosin molecules influence tropomyosin in activated muscle), is thought to be 12 to 15 actin monomers long [3,4]. This distance represents about 1.5 to 2 times the span of a single tropomyosin molecule on F-actin. Thus, end-to-end tropomyosin linkage appears to carry information between neighboring molecules, suggesting that tropomyosin operates as a stiff or semi-rigid cable [2,5].
We have proposed [2] that isolated tropomyosin is “preshaped” to fit to the helical contours of F-actin, thus allowing it to bind to actin filaments effectively while being sufficiently rigid to allow cooperative gating. This hypothesis has been validated by examination of EM images of purified cardiac and smooth muscle tropomyosin, which showed that these ~40 nm long molecules are appropriately curved, and by Molecular Dynamics simulations, which showed that tropomyosin is only moderately flexible with an average structure to match the F-actin helix perfectly [5,6]. The high observed stiffness confirms that tropomyosin would move on F-actin as a cooperative unit in response to the effects of troponin- or myosin-binding.
Given that tropomyosin plays a central role in controlling the mechanical and functional properties of the muscle thin filament, it is not surprising that mutations in tropomyosin are the cause of a variety of devastating muscle disorders [reviewed in 7]. Indeed, mutations in many contractile proteins, including those in α-tropomyosin, can lead to cardiomyopathy (HCM and DCM) and heart failure. In the current study, we have compared the bending flexibility of wild-type tropomyosin to that of two mutant tropomyosins, Asp175Asn and Glu180Gly, known to be associated with HCM [7], reasoning that defects in the material properties of the mutant proteins may correlate with thin filament dysfunction. We chose these mutations for study since they are located in a critical area of the tropomyosin molecule that is known to interact with actin [1,8,9] and the core-domain of the troponin complex [10], where troponin-I, troponin-C and troponin-T converge.
2. Materials and methods
Wild-type [11] and mutant α(cardiac)-tropomyosins [12] were prepared as previously. Electron microscopy and Molecular Dynamics were carried out as described recently [5,6].
3. Results and Discussion
3.1. Electron microscopy of isolated tropomyosin molecules
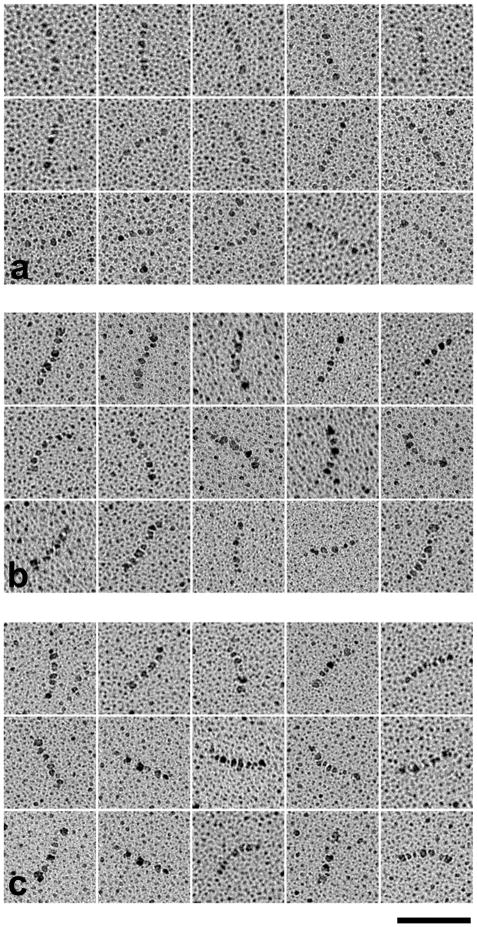
Electron microscopy was carried out on isolated tropomyosin molecules following adsorption and rotary-shadowing of the tropomyosins on untreated, freshly cleaved mica [6]. Images of both wild-type and mutant tropomyosin molecules showed gentle curvature and no obvious kinking as illustrated in Fig. 1. Some molecules appeared to be fairly straight whereas others displayed a degree of end-to-end curvature ranging up to about 55°. The molecules were skeletonized by cubic interpolation using the MatLab PCHIP program as described by Li et al. [5]. End-to-end curvature measurements (θ, see Supplementary Material, Fig. S1) indicated that the mutant molecules are on average slightly more curved than are those of wild-type tropomyosin (38.1°, D175N; 37.7°, E180G; 33.4°, wild-type).
Fig. 1.
EM images of representative tropomyosin molecules used for apparent persistence length measurement. (a) wild-type α-tropomyosin, (b) D175N tropomyosin, (c) E180G tropomyosin. Bar = 50 nm.
3.2. Tropomyosin persistence length
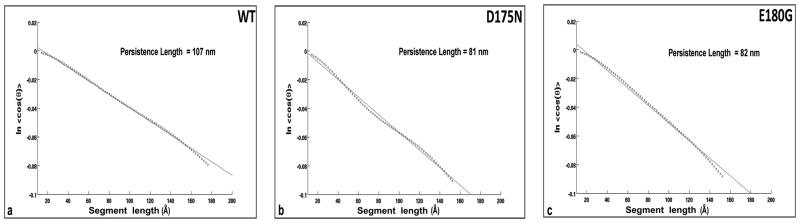
Persistence length values are often determined to quantify the magnitude of bending fluctuations of a rod-like material. However, the “apparent” persistence length (PLa) normally reported reflects the rod’s compounded deviation (angle θ in Supplementary Material, Fig. S1) from a hypothetical straight structure, which can be due to either flexible bending and/or because the structure is curved on average. Note that PLa is not to be confused with the dynamical persistence length, PLd, which quantifies the true flexibility by measuring the actual bending fluctuations (δ in Fig. S1) about the average structure, be it straight OR curved [5,13]. PLd is defined as the length along the rod over which its direction (viz. its longitudinal tangent) becomes highly uncorrelated (i.e. where the bending angle δ reaches on average the large value of 68.4° [13]). Using the relationship cos(θ)= e−40nm/2PL [13,14], 2D-images of tropomyosin (whose length is 40 nm) yield only PLa, not PLd, because the distinction between flexibly bent molecules and the projection of permanently curved molecule is not possible a priori. Simple visual inspection of the bending of tropomyosin molecules shown in Fig. 1 indicates that the apparent persistence length of tropomyosin molecules (wild-type or mutant) is between 80 and 120 nm, consistent with values of end-to-end curvature noted above.
To determine the apparent persistence length rigorously, over 200 skeletonized molecules from each data set (wild-type and mutants) were randomly chosen and persistence length quantified by the tangent correlation method, as previously described [13,14] (see Fig. 2). The resulting PLa of 107 nm for wild-type molecules agrees with previous determinations [5,6]. For the two mutants, we find PLa of ~80 nm, suggesting that the mutants are more curved and/or more flexible than the wild-type, in agreement with the trend observed in preliminary reports by others [15].
Fig. 2.
Quantifying apparent persistence length of isolated tropomyosin. The tangent correlation method [5,6,13,14] was applied to >200 skeletonized tropomyosin molecules from two preparations to obtain PL. (a) wild-type α-tropomyosin, (b) D175N tropomyosin, (c) E180G tropomyosin.
3.3. Molecular Dynamics of tropomyosin
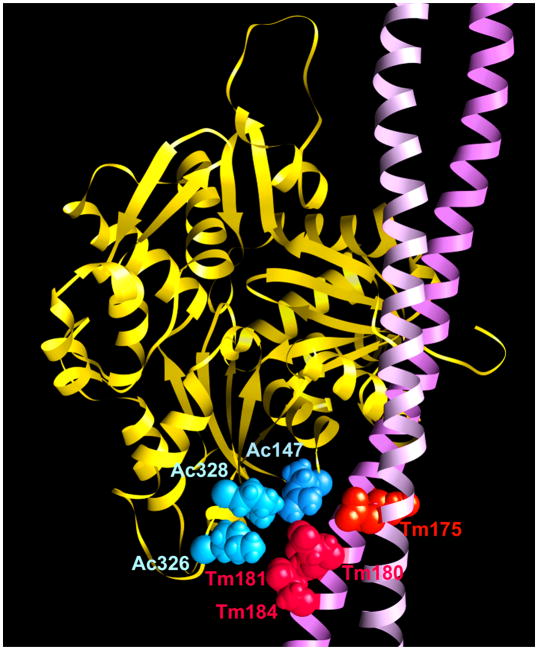
Given the 3 to 5 nm resolution limit achieved by rotary shadowing of single molecules, local flexibility at the mutation sites themselves could not be inferred using this procedure beyond the fact that no obvious kinks along tropomyosin could be observed. In contrast, MD simulation performed on atomic models of tropomyosin molecules is ideally suited to determine the effects of local perturbation of tropomyosin structure and flexibility. The MD show (Table 1) that both mutations lead to slightly greater flexibility in the surrounds of residue 180, which is part of a cluster of acidic residues that binds to lysine and arginine residues on actin [1,8,9]. This increased local flexibility is also observed when MD is carried out on tropomyosin bound to F-actin. However, neither mutation affected local flexibility at residue Tm175 (Table 1), which is not known to contact actin, but may interact with troponin-T [10] (Fig. 3).
Table 1.
Local and global flexibility of tropomyosin. Flexibility determined from MD simulations of either isolated tropomyosin molecules or tropomyosin when bound to F-actin [5,8,13]. (D175N was not studied on F-actin, since MD runs of actin-tropomyosin are extremely expensive.) δ measures true flexibility as the mean deviation angle (see Fig. S1) from the time-averaged position between the two ends of a given segment of the rod [5,8,13].
| Tropomyosin examined
|
δ - F-actin-free tropomyosin (°)
|
δ - Tropomyosin bound to F-actin (°)
|
|---|---|---|
| Wild-type | ||
| Local δ - at residue 175 | 4.45 ± 0.05 | 3.59 ± 0.19 |
| Local δ - at residue 180 | 4.21 ± 0.05 | 3.60 ± 0.21 |
| δ over entire tropomyosin | 22.0 ± 1.9 | 9.89 ± 0.11 |
| D175N | ||
| Local δ - at residue 175 | 4.39 ± 0.02 | -- |
| Local δ - at residue 180 | 5.86 ± 0.08* | -- |
| δ over entire tropomyosin | 30.5 ± 0.19* | -- |
| E180G | ||
| Local δ - at residue 175 | 4.47 ± 0.09 | 3.85 ± 0.01 |
| Local δ - at residue 180 | 5.30 ± 0.08* | 4.94 ± 0.30* |
| δ over entire tropomyosin | 27.6 ± 1.3* | 10.9 ± 0.50* |
Local δ: segment is taken over 9 residues centered at either residue 175 or 180. δ over entire tropomyosin: segment is the whole tropomyosin.
p<0.01 compared to wild-type; error of corresponding half-data sets reported.
Fig. 3.
Location of residues Tm180 and Tm175 on an atomic model of the F-actin – tropomyosin [8]. Tm180 lies in a cluster of acidic amino acids including Glu181 and Glu184 (red) that likely interact with Lys326, Lys328 and Arg147 (blue) on actin. Tm175 (orange-red), lying close to this cluster, does not itself interact with actin yet may contact troponin-T [10].
Quantitation of the end-to-end deflection from the mean structure (δ in Fig. S1) shows a greater overall flexibility of the mutants (<δ> is 30.5° and 27.6° for D175N and E180G respectively, Table 1) relative to that of the wild-type tropomyosin (<δ>=22.0°). However, when the MD simulations are performed with tropomyosin bound to F-actin (in explicit water), the end-to-end deflections are significantly dampened (<δ> is 9.9° and 10.9° for wild-type and E180G respectively, Table 1). This reduction can be accounted for by the electrostatic interactions between tropomyosin and actin (there are approximately 30 amino acids on the F-actin filament that interact electrostatically with each tropomyosin molecule [8]). Thus, measurements of the dynamic persistence length of isolated tropomyosin, while giving an indication of the upper boundary of the size of the thin filament regulatory unit, cannot be directly equated to the length of this regulatory unit, as some have previously inferred [4]. Moreover, local conformational changes associated with the HCM mutations must be considered, as charge neutralization at the site of the mutations (Fig. 3) is likely to interfere with normal electrostatic interactions between tropomyosin, troponin-T and actin on the thin filament.
3.4. Functional implications of tropomyosin mutation
We have shown that each of tropomyosin mutation studied increases global flexibility, as well as local flexibility, in the surrounds of tropomyosin residue 180 (the latter local effect persists even when tropomyosin binds to F-actin). The excess flexibility is likely to increase accessibility to the myosin binding-sites on F-actin, thus destabilizing the low-Ca2+ relaxed-state of cardiac muscle [16]. This can result in a shift of the regulatory equilibrium towards Ca2+-activation in intact cardiac muscle. Moreover, the mutations affect the average overall tropomyosin curvature, which may interfere with proper tropomyosin assembly onto thin filaments, as the mutant tropomyosins will less likely be pre-shaped appropriately to the contours of the F-actin helix. Such a modification would lower the affinity of the mutant molecules for F-actin, as is observed [12]. We expect that local charge neutralization affects the distribution of side-chains of neighboring residues, and, in turn, this influences the binding of residue E181 to F-actin, which is critical for proper actin-tropomyosin interaction on thin filaments. Perturbing this association may reduce the steric hindrance of myosin-binding, again causing an increase in Ca2+-sensitivity of muscle activation noted experimentally [4]. In fact, an increase in myosin-binding to actin may influence troponin-actin interactions that indirectly enhance Ca2+-binding by troponin-C, hence leading to increased Ca2+-sensitivity [17]. Moreover, D175N mutations may directly affect tropomyosin interaction with TnT on the thin filament [10], further destabilizing Ca2+-regulation. No doubt, imbalances in the normal on-off switching mechanism needed to regulate cardiac contractility is accompanied by chronic complications (prolonged systolic activation, diastolic dysfunction [4]) and cardiac compensations associated with HCM and heart failure.
Supplementary Material
Research Highlights.
Well-known tropomyosin mutants, D175N and E180G are linked to cardiomyopathies.
The structural mechanics of D175N and E180G tropomyosins have been investigated.
D175N and E180G mutations increase both local and global tropomyosin flexibility.
In muscle, this increased flexibility will enhance myosin interactions on actin.
Extra myosin interaction can alter cardiac Ca2+-switching, leading to dysfunction.
Acknowledgments
This work was supported by grants from the NIH (HL36153 and HL86655 to W.L.), the Welcome Trust Program (085309 to M.A.G.) and the European Union “BIG-HEART,” Seventh Framework Program (241577 to S.B.M).
Abbreviations
- DCM
dilated cardiomyopathy
- HCM
hypertrophic cardiomyopathy
- EM
electron microscopy
- MD
Molecular Dynamics
- PL
persistence length
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Brown JH, Cohen C. Regulation of muscle contraction by tropomyosin and troponin: how structure illuminates function. Adv Protein Chem. 2005;71:121–159. doi: 10.1016/S0065-3233(04)71004-9. [DOI] [PubMed] [Google Scholar]
- 2.Holmes KC, Lehman W. Gestalt-binding of tropomyosin to actin filaments. J Muscle Res Cell Motil. 2008;29:213–219. doi: 10.1007/s10974-008-9157-6. [DOI] [PubMed] [Google Scholar]
- 3.Lehrer SS, Geeves MA. Dynamics of the muscle thin filament regulatory switch: the size of the cooperative unit. Biophys J. 1994;67:273–282. doi: 10.1016/S0006-3495(94)80478-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Loong CK, Badr MA, Chase PB. Tropomyosin flexural rigidity and single Ca(2+) regulatory unit dynamics: implications for cooperative regulation of cardiac muscle contraction and cardiomyocyte hypertrophy. Front Physiol. 2012;3:1–10. doi: 10.3389/fphys.2012.00080. #80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li XE, Holmes KC, Lehman W, Jung H, Fischer S. The shape and flexibility of tropomyosin coiled coils: implications for actin filament assembly and regulation. J Mol Biol. 2010;395:327–339. doi: 10.1016/j.jmb.2009.10.060. [DOI] [PubMed] [Google Scholar]
- 6.Sousa D, Cammarato A, Jang K, Graceffa P, Tobacman LS, Li XE, Lehman W. Electron microscopy and persistence length analysis of semi-rigid smooth muscle tropomyosin strands. Biophys J. 2010;99:862–868. doi: 10.1016/j.bpj.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marston SB, Ingwall JS, Glueck SB. Calcium, contractions, and tropomyosin Focus on “divergent abnormal muscle relaxation by hypertrophic cardiomyopathy and nemaline myopathy mutant tropomyosins”. Physiol Genomics. 2002;9:57–58. doi: 10.1152/physiolgenomics.00038.2002. [DOI] [PubMed] [Google Scholar]
- 8.Li XE, Tobacman LS, Mun JY, Craig R, Fischer S, Lehman W. Tropomyosin position on F-actin revealed by EM reconstruction and computational chemistry. Biophys J. 2011;100:1005–1013. doi: 10.1016/j.bpj.2010.12.3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barua B, Pamula MC, Hitchcock-DeGregori SE. Evolutionarily conserved surface residues constitute actin-binding sites of tropomyosin. Proc Natl Acad Sci USA. 2011;108:10150–10155. doi: 10.1073/pnas.1101221108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mudalige AW, Lehrer SS. What region of tropomyosin interacts with the N-terminal half of troponin T? Biophys J. 2010;98:351a. [Google Scholar]
- 11.Tobacman LS, Adelstein RS. Mechanism of regulation of cardiac actin-myosin subfragment 1 by troponin-tropomyosin. Biochemistry. 1986;25:798–802. doi: 10.1021/bi00352a010. [DOI] [PubMed] [Google Scholar]
- 12.Kremneva E, Boussouf S, Nikolaeva O, Maytum R, Geeves MA, Levitsky DI. Effects of two familial hypertrophic cardiomyopathy mutations in alpha-tropomyosin, Asp175Asn and Glu180Gly, on the thermal unfolding of actin-bound tropomyosin. Biophys J. 2004;87:3922–3933. doi: 10.1529/biophysj.104.048793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li XE, Lehman W, Fischer S. The relationship between curvature, flexibility and persistence length in the tropomyosin coiled-coil. J Struct Biol. 2010;170:313–318. doi: 10.1016/j.jsb.2010.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frontali C, Dore E, Ferrauto A, Gratton E, Bettini A, Pozzan MR, Valdevit E. An absolute method for the determination of the persistence length of native DNA from electron micrographs. Biopolymers. 1979;18:1353–1373. doi: 10.1002/bip.1979.360180604. [DOI] [PubMed] [Google Scholar]
- 15.Loong C, Zhou HX, Chase PB. Persistence length of human cardiac a-tropomyosin implies near-neighbor cooperative activation of cardiac thin filaments. Biophys J. 2012;102:230a. [Google Scholar]
- 16.Bing W, Knott A, Redwood C, Esposito G, Purcell I, Watkins H, Marston S. Effect of hypertrophic cardiomyopathy mutations in human cardiac muscle alpha -tropomyosin (Asp175Asn and Glu180Gly) on the regulatory properties of human cardiac troponin determined by in vitro motility assay. J Mol Cell Cardiol. 2000;32:1489–1498. doi: 10.1006/jmcc.2000.1182. [DOI] [PubMed] [Google Scholar]
- 17.Lehrer SS. The 3-state model of muscle regulation revisited: is a fourth state involved? J Muscle Res Cell Motil. 2011;32:203–208. doi: 10.1007/s10974-011-9263-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.