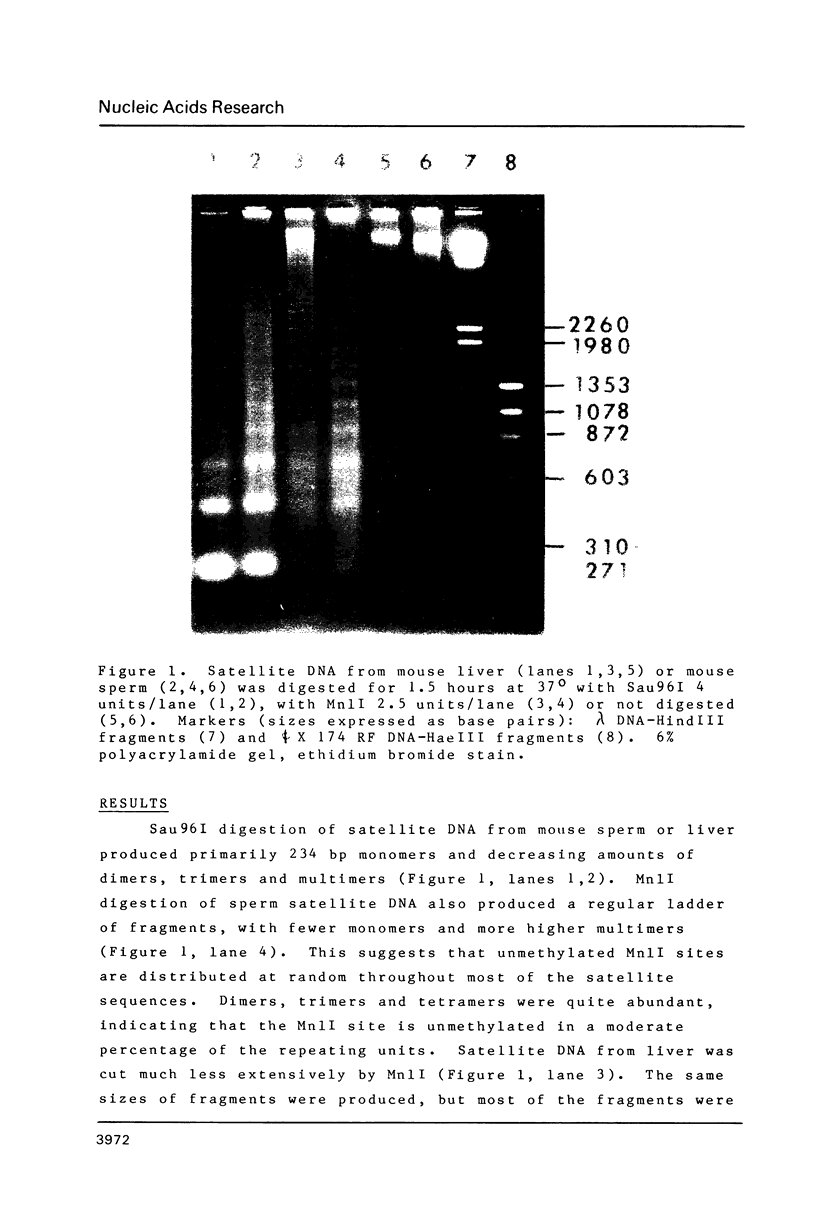
Abstract
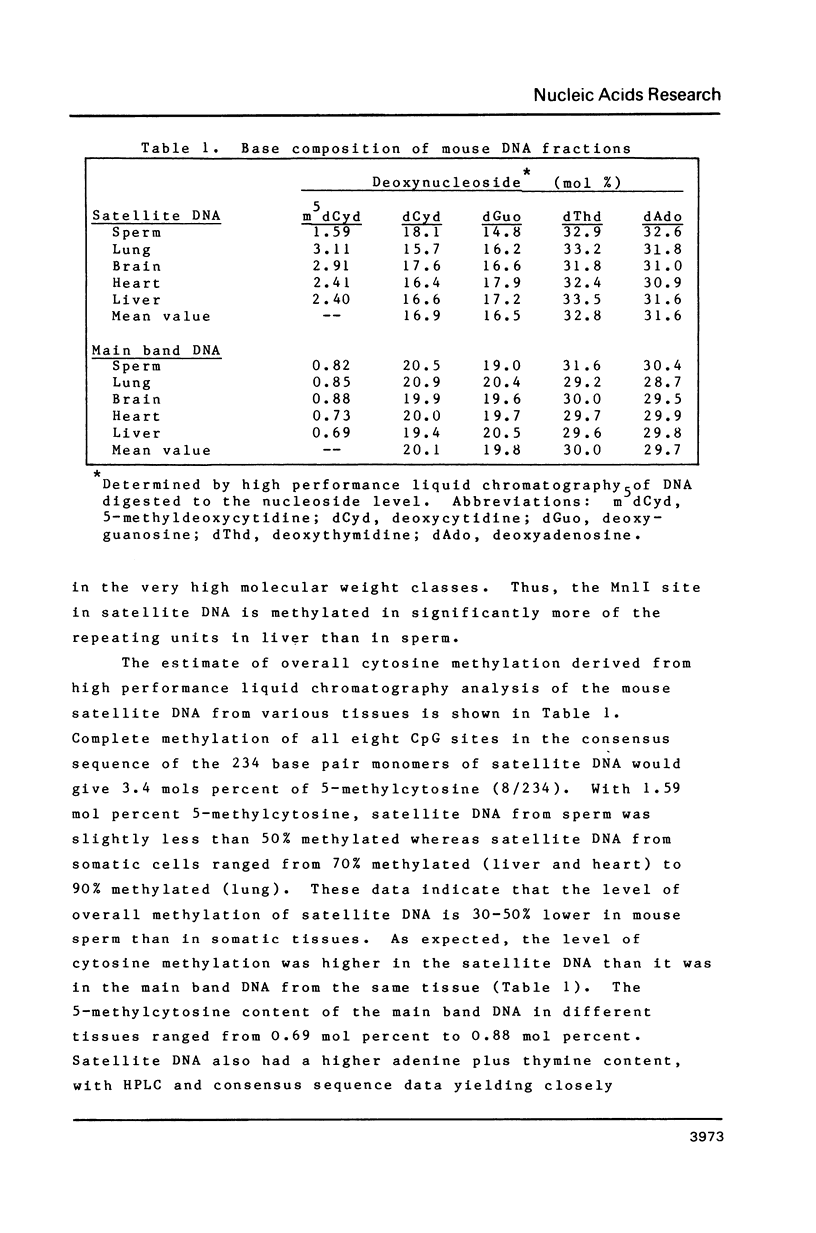
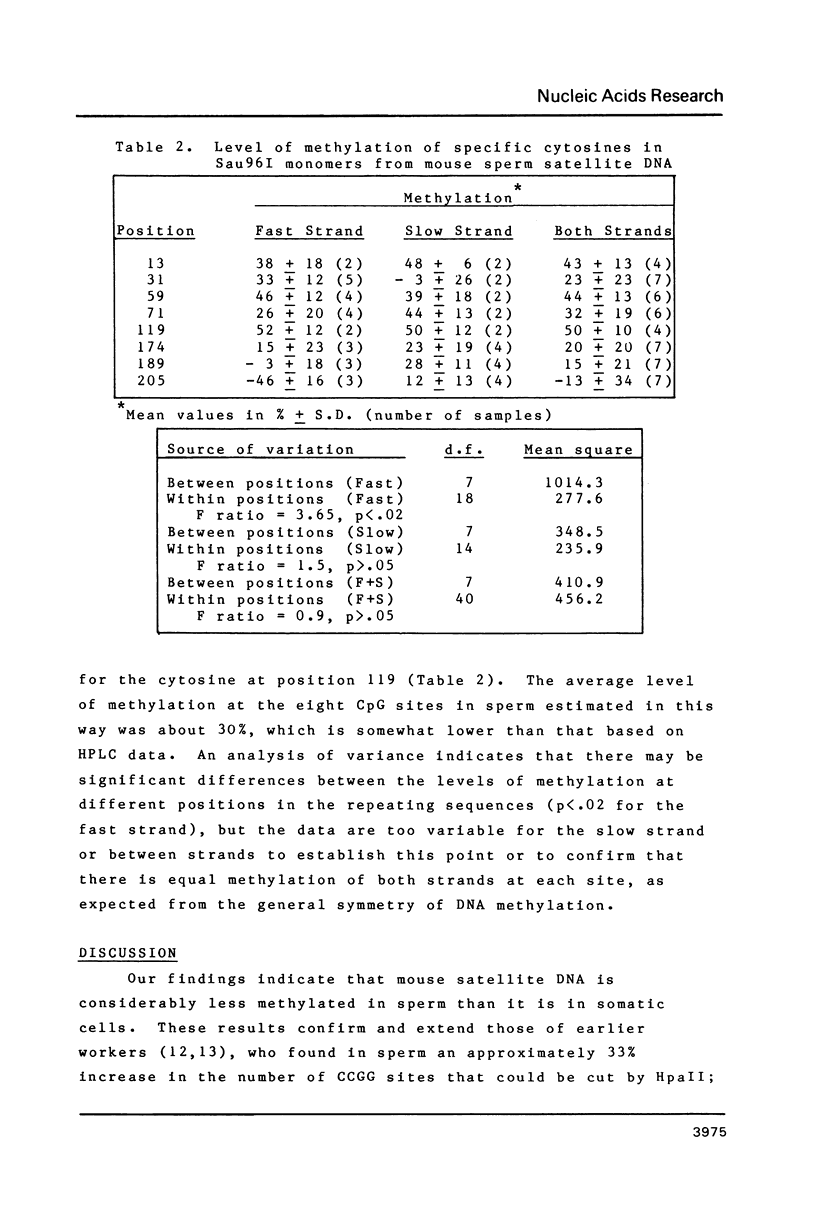
Enzymatic hydrolysis and base analysis by high performance liquid chromatography showed that mouse satellite DNA had 30-50% less 5-methylcytosine in sperm than in somatic tissue (1.59 mols % vs 2.40-3.11 mols %). Maxam-Gilbert sequencing and analysis of the intensity of the cytosine bands indicated that the level of methylation of the eight CpGs of the consensus sequence in sperm satellite DNA ranged from 0 to about 50%, considerably lower than the levels reported in somatic tissues. The Mn1I site containing one of these CpGs was cut much more extensively in satellite DNA from sperm than from liver, confirming the undermethylation of this site in sperm DNA.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chapman V., Forrester L., Sanford J., Hastie N., Rossant J. Cell lineage-specific undermethylation of mouse repetitive DNA. Nature. 1984 Jan 19;307(5948):284–286. doi: 10.1038/307284a0. [DOI] [PubMed] [Google Scholar]
- Cooke H. J., Schmidtke J., Gosden J. R. Characterisation of a human Y chromosome repeated sequence and related sequences in higher primates. Chromosoma. 1982;87(5):491–502. doi: 10.1007/BF00333470. [DOI] [PubMed] [Google Scholar]
- Ehrlich M., Ehrlich K., Mayo J. A. Unusual properties of the DNA from Xanthomonas phage XP-12 in which 5-methylcytosine completely replaces cytosine. Biochim Biophys Acta. 1975 Jun 16;395(2):109–119. doi: 10.1016/0005-2787(75)90149-5. [DOI] [PubMed] [Google Scholar]
- Ehrlich M., Gama-Sosa M. A., Huang L. H., Midgett R. M., Kuo K. C., McCune R. A., Gehrke C. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues of cells. Nucleic Acids Res. 1982 Apr 24;10(8):2709–2721. doi: 10.1093/nar/10.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gama-Sosa M. A., Midgett R. M., Slagel V. A., Githens S., Kuo K. C., Gehrke C. W., Ehrlich M. Tissue-specific differences in DNA methylation in various mammals. Biochim Biophys Acta. 1983 Jun 24;740(2):212–219. doi: 10.1016/0167-4781(83)90079-9. [DOI] [PubMed] [Google Scholar]
- Gama-Sosa M. A., Wang R. Y., Kuo K. C., Gehrke C. W., Ehrlich M. The 5-methylcytosine content of highly repeated sequences in human DNA. Nucleic Acids Res. 1983 May 25;11(10):3087–3095. doi: 10.1093/nar/11.10.3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrke C. W., McCune R. A., Gama-Sosa M. A., Ehrlich M., Kuo K. C. Quantitative reversed-phase high-performance liquid chromatography of major and modified nucleosides in DNA. J Chromatogr. 1984 Sep 28;301(1):199–219. doi: 10.1016/s0021-9673(01)89189-5. [DOI] [PubMed] [Google Scholar]
- Gruenbaum Y., Stein R., Cedar H., Razin A. Methylation of CpG sequences in eukaryotic DNA. FEBS Lett. 1981 Feb 9;124(1):67–71. doi: 10.1016/0014-5793(81)80055-5. [DOI] [PubMed] [Google Scholar]
- Hörz W., Altenburger W. Nucleotide sequence of mouse satellite DNA. Nucleic Acids Res. 1981 Feb 11;9(3):683–696. doi: 10.1093/nar/9.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahana Z. E., Miller O. J., Erlanger B. F. Immunochemical studies on the 5-methylcytosine content of African green monkey satellite DNA. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):397–400. doi: 10.1101/sqb.1978.042.01.041. [DOI] [PubMed] [Google Scholar]
- Kuo K. C., McCune R. A., Gehrke C. W., Midgett R., Ehrlich M. Quantitative reversed-phase high performance liquid chromatographic determination of major and modified deoxyribonucleosides in DNA. Nucleic Acids Res. 1980 Oct 24;8(20):4763–4776. doi: 10.1093/nar/8.20.4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manuelidis L. A simplified method for preparation of mouse satellite DNA. Anal Biochem. 1977 Apr;78(2):561–568. doi: 10.1016/0003-2697(77)90118-x. [DOI] [PubMed] [Google Scholar]
- Manuelidis L. Complex and simple sequences in human repeated DNAs. Chromosoma. 1978 Mar 22;66(1):1–21. doi: 10.1007/BF00285812. [DOI] [PubMed] [Google Scholar]
- Manuelidis L. Consensus sequence of mouse satellite DNA indicates it is derived from tandem 116 basepair repeats. FEBS Lett. 1981 Jun 29;129(1):25–28. doi: 10.1016/0014-5793(81)80746-6. [DOI] [PubMed] [Google Scholar]
- Miller O. J., Schnedl W., Allen J., Erlanger B. F. 5-Methylcytosine localised in mammalian constitutive heterochromatin. Nature. 1974 Oct 18;251(5476):636–637. doi: 10.1038/251636a0. [DOI] [PubMed] [Google Scholar]
- Pietras D. F., Bennett K. L., Siracusa L. D., Woodworth-Gutai M., Chapman V. M., Gross K. W., Kane-Haas C., Hastie N. D. Construction of a small Mus musculus repetitive DNA library: identification of a new satellite sequence in Mus musculus. Nucleic Acids Res. 1983 Oct 25;11(20):6965–6983. doi: 10.1093/nar/11.20.6965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponzetto-Zimmerman C., Wolgemuth D. J. Methylation of satellite sequences in mouse spermatogenic and somatic DNAs. Nucleic Acids Res. 1984 Mar 26;12(6):2807–2822. doi: 10.1093/nar/12.6.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly J. G., Thomas C. A., Jr, Sen A. DNA methylation in mouse cells in culture as measured by restriction enzymes. Biochim Biophys Acta. 1982 Apr 26;697(1):53–59. doi: 10.1016/0167-4781(82)90044-6. [DOI] [PubMed] [Google Scholar]
- Salomon R., Kaye A. M., Herzberg M. Mouse nuclear satellite DNA: 5-methylcytosine content, pyrimidine isoplith distribution and electron microscopic appearance. J Mol Biol. 1969 Aug 14;43(3):581–592. doi: 10.1016/0022-2836(69)90360-x. [DOI] [PubMed] [Google Scholar]
- Sanford J., Forrester L., Chapman V., Chandley A., Hastie N. Methylation patterns of repetitive DNA sequences in germ cells of Mus musculus. Nucleic Acids Res. 1984 Mar 26;12(6):2823–2836. doi: 10.1093/nar/12.6.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano H., Sager R. Tissue specificity and clustering of methylated cystosines in bovine satellite I DNA. Proc Natl Acad Sci U S A. 1982 Jun;79(11):3584–3588. doi: 10.1073/pnas.79.11.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnedl W., Dev V. G., Tantravahi R., Miller D. A., Erlanger B. F., Miller O. J. 5-Methylcytosine in heterochromatic regions of chromosomes: chimpanzee and gorilla compared to the human. Chromosoma. 1975 Sep 15;52(1):59–66. doi: 10.1007/BF00285789. [DOI] [PubMed] [Google Scholar]
- Schnedl W., Erlanger B. F., Miller O. J. 5-methylcytosine in heterochromatic regions of chromosomes in Bovidae. Hum Genet. 1976 Jan 28;31(1):21–26. doi: 10.1007/BF00270395. [DOI] [PubMed] [Google Scholar]
- Shiurba R., Nandi S. Isolation and characterization of germ line DNA from mouse sperm. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3947–3951. doi: 10.1073/pnas.76.8.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm K. S., Taylor J. H. Distribution of 5-methylcytosine in the DNA of somatic and germline cells from bovine tissues. Nucleic Acids Res. 1981 Sep 25;9(18):4537–4546. doi: 10.1093/nar/9.18.4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venolia L., Gartler S. M. Transformation of Hprt gene with sperm DNA. Somatic Cell Genet. 1983 Sep;9(5):617–627. doi: 10.1007/BF01574262. [DOI] [PubMed] [Google Scholar]
- Wigler M., Sweet R., Sim G. K., Wold B., Pellicer A., Lacy E., Maniatis T., Silverstein S., Axel R. Transformation of mammalian cells with genes from procaryotes and eucaryotes. Cell. 1979 Apr;16(4):777–785. doi: 10.1016/0092-8674(79)90093-x. [DOI] [PubMed] [Google Scholar]