Summary
Medulloblastoma is a malignant childhood brain tumour comprising four discrete subgroups. To identify mutations that drive medulloblastoma we sequenced the entire genomes of 37 tumours and matched normal blood. One hundred and thirty-six genes harbouring somatic mutations in this discovery set were sequenced in an additional 56 medulloblastomas. Recurrent mutations were detected in 41 genes not yet implicated in medulloblastoma: several target distinct components of the epigenetic machinery in different disease subgroups, e.g., regulators of H3K27 and H3K4 trimethylation in subgroup-3 and 4 (e.g., KDM6A and ZMYM3), and CTNNB1-associated chromatin remodellers in WNT-subgroup tumours (e.g., SMARCA4 and CREBBP). Modelling of mutations in mouse lower rhombic lip progenitors that generate WNT-subgroup tumours, identified genes that maintain this cell lineage (DDX3X) as well as mutated genes that initiate (CDH1) or cooperate (PIK3CA) in tumourigenesis. These data provide important new insights into the pathogenesis of medulloblastoma subgroups and highlight targets for therapeutic development.
Medulloblastoma is the most common malignant childhood brain tumor1. The disease includes four subgroups (Sonic Hedgehog (SHH)-subgroup, WNT-subgroup, subgroup-3 and subgroup-4) defined primarily by gene expression profiling that display differences in karyotype, histology and prognosis2. Studies of genetically engineered mice show these tumours arise from different cell types: SHH-subgroup medulloblastomas develop from committed cerebellar granule neuron progenitors (GNPs) in Ptch1+/− mice3,4; WNT-subgroup tumours are generated by lower rhombic lip progenitors (LRLPs) in Blbp-Cre ; Ctnnb1+/lox(Ex3) ; Tp53flx/flx mice5; while subgroup-3 medulloblastomas likely arise from an undefined class of cerebellar progenitors6. The identification of medulloblastoma subgroups has not changed clinical practice. All patients currently receive the same combination of surgery, radiation and chemotherapy. This aggressive treatment fails to cure two thirds of patients with subgroup-3 disease, and probably over-treats children with WNT-subgroup medulloblastoma who invariably survive with long term cognitive and endocrine side effects2,7. Drugs targeting the genetic alterations that drive each medulloblastoma subgroup could prove more effective and less toxic, but the identity of these alterations remains largely unknown.
The genomic landscape of medulloblastoma
To identify genetic alterations that drive medulloblastoma, we performed whole genome sequencing (WGS) of DNA from 37 tumours and matched normal blood (discovery cohort). Tumours were subgrouped by gene expression (WNT-subgroup, n=5; SHH-subgroup, n=5; subgroup-3, n=6; subgroup-4, n=19; ‘unclassified’ [profiles not available], n=2. Figure 1; Supplementary Figures 1-3 and Supplementary Table 1). Validation of all putative somatic alterations including single nucleotide variations (SNVs), insertion/deletions (indels) and structural variations (SVs) identified by CREST8, was conducted for 12 tumours using custom capture arrays and Illumina-based DNA sequencing (Supplementary Table 2). Putative coding alterations and SVs were validated in the remaining 25 ‘discovery cohort’ cases by polymerase chain reaction and Sanger-based sequencing. Mutation frequency was determined in a separate ‘validation cohort’ of 56 medulloblastomas (WNT-subgroup, n=6; SHH-subgroup, n=8; subgroup-3, n=11; subgroup-4, n=19; unclassified, n=12; Figure 1, Supplementary Table 1).
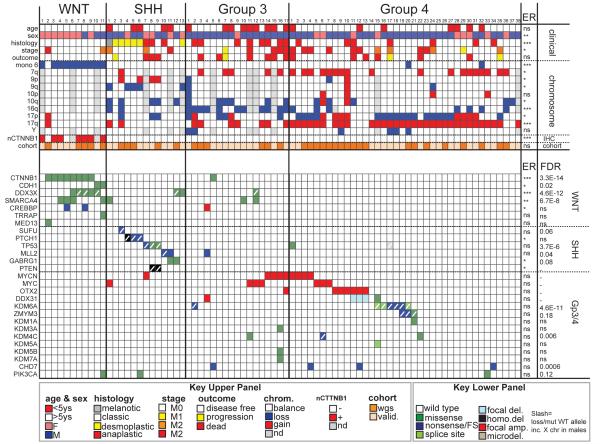
Figure 1. The genomic landscape of medulloblastoma.
Top: clinical, histologic, gross chromosomal, nuclear CTNNB1 (nCTNNB1), and cohort (discovery or validation) details of 79 medulloblastomas by subgroup. Below: genetic alterations detected in 27 genes of particular interest. Color key at bottom. ANOVA (continuous) or Fisher’s exact (categorical) p-value is shown right. False discovery estimates (FDR) of each mutation are shown right. ***=P<0.0005; **=P<0.005; *=P<0.05; ns=not significant.
WGS of the ‘discovery cohort’ detected 22,887 validated or high-quality somatic sequence mutations (SNVs and indels), 536 validated or curated SVs, and 5,802 copy number variations (CNVs, 92% concordant with 6.0 SNP mapping arrays; Supplementary Tables 3-6, Supplementary Figures 4-7). In all but five tumours with the highest mutation rates, >50% of SNVs were C>T/G>A transitions (Supplementary Figure 8). The mean missense:silent mutation ratio was 3.6:1 and 40% of all missense mutations were predicted to be deleterious, suggesting a selective pressure for SNVs that impact protein coding (Supplementary Table 5). Global patterns of total SNVs and amplifications varied significantly among medulloblastoma subgroups, even when corrected for age and sex, supporting the notion that these tumours are distinct pathological entities (Figure 1, Supplementary Figure 6). Custom capture-based analysis of the allele frequency of all somatic mutations in 12 medulloblastomas allowed us to predict the ancestry of certain genetic alterations, suggesting that aneuploidy precedes widespread sequence mutation in medulloblastomas with highly mutated genomes (Supplementary Figures 9-11).
Novel copy number variations, structural alterations and heritable mutations are rare in medulloblastoma
The repertoire of focally amplified or deleted genes appears to be very limited in medulloblastoma. We detected expected2 gains of MYC, MYCN and OTX2 in subgroup-3 and 4, but no novel recurrent amplifications (Figure 1, Supplementary Figure 12, Supplementary Table 7). In keeping with recent reports9, high-level amplification of MYCN in subgroup-3 sample #16 (sample numbering as Figure 1) was generated by chromothripsis; although chromothripsis was observed infrequently (n=2/37 of ‘discovery cohort’; Supplementary Figure 13).
Focal homo- or heterozygous deletion of genes previously implicated in medulloblastoma were also detected (e.g., PTCH1, PTEN, Figure 1)10,11 but novel recurrent focal deletions were rare. Three subgroup-4 tumours (#11-13) and one unclassified tumour, deleted DDX31, AK8 and TSC1 at 9q34.14 in concert with OTX2 amplification, suggesting these alterations are cooperative (P<0.0005, Fisher’s exact). The breakpoint in this deletion occurs in DDX31 and two samples contained a missense mutation (subgroup-4 #15) and complex rearrangement (unidentified case SJMB026) in this gene, suggesting DDX31 is the target of these alterations (Supplementary Figure 14).
Over 50% of SVs detected by WGS broke the coding region of at least one gene, but less than 2% (n=6/314, excluding two tumours with excessive SVs) encode potential in-frame fusion proteins (Supplementary Figure 15); none affect the same gene or signal pathway. Therefore, fusion proteins are likely to be an uncommon transforming mechanism in medulloblastoma.
Although germline mutations in TP53, PTCH1, APC, and CREBBP, predispose to medulloblastoma11-14, only 23 mutations previously associated with cancer were detected in ‘discovery cohort’ germlines: only one of these - in a known case of Turcot’s syndrome - was accompanied with a somatic mutation (germline APC Y935*/somatic deletion: WNT-subgroup #11, Supplementary Table 8). Thus, inherited forms of medulloblastoma appear to be rare in our cohort.
Novel recurrent sequence mutations target distinct medulloblastoma subgroups
Since SVs and CNVs are unlikely to drive most medulloblastomas, we looked to see if recurrent (>2 samples), somatic SNVs and/or indels might target discrete genes and pathways. This analysis identified 49 genes, across all 93 tumors, that were targeted by non-silent, recurrent, somatic mutations: 84% (n=41/49) are not yet implicated in medulloblastoma (Supplementary Tables 9 and 10). Several of these congregated in disease subgroups and converged on specific cell pathways (Figure 1; Supplementary Figure 8 and Table 11).
Writing, reading and erasing of histone methylation is deregulated in subgroup-3 and 4 medulloblastomas
The H3K27 trimethyl mark (H3K27me3) represses lineage specific genes in stem cells (Supplementary Figure 8)15. H3K27me3 is written by the polycomb repressive complex 2 (PRC2) that includes the methylase EZH216,17, and erased during differentiation by the demethylase KDM6A18. As H3K27me3 is erased, chromatin remodelers recruited to H3K4me3 promote differentiation e.g., CHD719,20. This process is tightly controlled during development and deregulated in cancers: EZH2 is mutated in lymphomas21, and upregulated in breast22 and prostate23 cancer; while biallelic inactivation of KDM6A (Xp11.2) or KDM6A and its paralog UTY (Yq11), occurs in adult female and male cancers, respectively24.
Hypergeometric distribution analyses revealed selective mutation of histone modifiers in subgroup-3 and 4 medulloblastomas (Supplementary Table 11). Six subgroup-4, one subgroup-3, and one unclassified medulloblastoma contained novel inactivating mutations in KDM6A (Figures 1 and 2; Supplementary Figures 8 and 16). The single female with a KDM6A splice site mutation deleted the second allele that escapes X inactivation25 (subgroup-4 #15), and 57% (n=4/7) of KDM6A-mutant male medulloblastomas deleted chromosome Y, compared with only 6% (n=3/51) of male, KDM6A wild-type tumours (P<0.005, Fisher’s exact; Figure 1). Thus, a two-hit model of KDM6A–UTY tumour suppression appears to operate in subgroup-4 medulloblastomas. Notably, mutations in six other KDM family members (KDM1A, KDM3A, KDM4C, KDM5A, KDM5B and KDM7A) were detected exclusively in subgroup-3 and 4 tumours, implicating broad disruption of lysine demethylation in these medulloblastomas (Figure 1, Supplementary Table 11; Supplementary Figure 16).
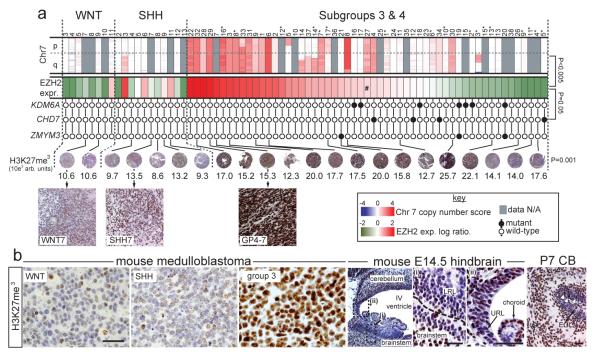
Figure 2. Deregulation of H3K27me3 in subgroup-3 and 4 human and mouse medulloblastoma.
(a) Top row, SNP profiles of chromosome 7 copy number in medulloblastomas (samples as Figure 1; *=subgroup-3 cases). Second row, expression of EZH2. Subgroup-3 and 4 tumours are ordered left to right by expression level, #=median expression point (Bonferroni corrected p-value of EZH2 expression vs. chromosome 7 gain). Third row, mutation status of KDM6A, CHD7 and ZMYM3 (p-value, Fisher’s exact mutations vs. EZH2 expression). Fourth row, H3K27me3 immunohistochemistry (numbers=colorimetry, p-value ANOVA). (b) H3K27me3 expression (right) in mouse Blbp-Cre ; Ctnnb1+/lox(Ex3) ; Tp53flx/flx (WNT), Ptch1+/−; Tp53−/− (SHH) and Myc ; Ink4c−/− (group 3) medulloblastomas and (left) developing hindbrain. high power views of E14.5 (i) LRL and (ii) upper rhombic lip (URL). IGL=internal granule layer, EGL=external germinal layer. Scale bar=50μm. White arrows in P7 cerebellum pinpoint H3K27me3 cells in the EGL.
Subgroup-3 and 4 medulloblastomas also gained and overexpressed EZH2 (7q35-34) that writes H3K27me3, and contained novel inactivating mutations in effectors and regulators of the H3K4me3 mark26 (Figure 2a; Supplementary Figure 8). Gain of 7q was significantly enriched among subgroup-3 and 4 medulloblastomas (P<0.005 Fisher’s exact) and correlated directly with EZH2 expression. Indeed, EZH2 was the 8th most significantly overexpressed gene on chromosome 7 among subgroup-3 and 4 medulloblastomas that gained chromosome 7q relative to those with diploid chromosome 7 (P<0.005, Bonferroni correction). Nonsense and frameshift mutations were detected in CHD7 in four subgroup-3 and 4 tumours. ZMYM3 (Xq13.1) that participates in a protein complex with KDM1A to regulate gene expression at the H3K4me3 mark27 was targeted by novel frameshift, nonsense and missense mutations in three male subgroup-4 medulloblastomas. All three tumours with mutations in ZMYM3 also mutated KDM6A (subgroup-4 #19, 20) or KDM1A (subgroup-4 #21) suggesting these alterations are cooperative. Remarkably, KDM6A, CHD7 and ZMYM3 mutations were confined to subgroup-3 and 4, and clustered in samples with sub-median EZH2 expression levels (Figure 2a; P<0.05, Fisher’s exact). These data suggest that subgroup-3 and 4 medulloblastomas retain a stem-like epigenetic state by aberrantly writing (EZH2 upregulation) or preserving (KDM6A-UTY inactivation) H3K27me3, or disrupting H3K4me3 associated transcription (CHD7 and ZMYM3 inactivation). Indeed, human and mouse subgroup-3 and 4 medulloblastomas contained significantly more H3K27me3 than did WNT or SHH-subgroup tumours (Figure 2b). Thus, gain of EZH2 and loss of KDM6A likely maintains H3K27me3 in subgroup-3 and 4 medulloblastomas.
Finally, we looked to see if the differential expression of H3K27me3 among medulloblastoma subgroups reflects ancestral chromatin marking in the progenitors that generate these tumours (Figure 2b). Relatively low levels of H3K27me3 were detected in LRLPs and committed GNPs that generate WNT and SHH-subgroup medulloblastomas respectively3-5, potentially explaining why mutations that preserve this epigenetic mark are absent from these tumours. We recently showed that subgroup-3 medulloblastomas arise from a rare fraction of cerebellar progenitors6. We are currently investigating if these progenitors are found among the H3K27me3 positive cells seen in the external germinal layer (Figure 2b).
Novel mutations in WNT-subgroup medulloblastomas target CTNNB1-associated chromatin remodelers and regulators of LRLPs
WNT-subgroup medulloblastomas contained mutations in epigenetic regulators that are different to those seen in subgroup-3 and 4 disease. CTNNB1, the principal effector of the WNT pathway, forms a transcription factor with the T cell factor/lymphoid enhancer factor (TCF/LEF)28. The c-terminus of CTNNB1 then recruits a series of protein complexes that remodel chromatin and promote transcription at WNT-responsive genes (Supplementary Figure 8). These include: histone acetyltransferases (e.g., CREBBP and TRRAP-TIP60 complexes)28,29; ATPases of the SWI/SNF family (e.g., SMARCA4)30; and the Mediator complex that coordinates RNA polymerase II placement (e.g., MED13)31. As expected, >70% (n=8/11) of WNT-medulloblastomas contained mutations that stabilise CTNNB1 (Figure 1 and Supplementary Figure 8; P<0.0001, Fishers exact)32,33. A single subgroup-3 case (#5) also mutated CTNNB1, but this mutation has not been reported in cancer, did not upregulate nuclear CTNNB1 (Figure 1) and is of unclear significance. Remarkably, six WNT-subgroup medulloblastomas mutated chromatin modifiers that are recruited to TCF/LEF WNT-responsive genes by CTNNB1 (Figure 1, Supplementary Figure 8): four WNT-subgroup tumours contained heterozygous missense mutations in the helicase domain of SMARCA4 (P<0.002, Fisher’s exact); two samples, including one with a SMARCA4 mutation (#5), contained nonsense mutations in CREBBP (WNT-subgroup enrichment P<0.02, Fisher’s exact); and missense mutations in TRRAP and MED13 were detected in a single WNT-subgroup medulloblastoma each. Thus, in addition to stabilization of CTNNB1, the development of WNT-subgroup medulloblastoma may require disruption of chromatin remodeling at WNT-responsive genes.
A small number of WNT-subgroup medulloblastomas lack mutations in CTNNB1 or APC, suggesting alternative mechanisms drive aberrant WNT-signals in these tumours. Three WNT-subgroup medulloblastomas in our series contained wild-type CTNNB1 (#1, 10 and 11, Figure 1). Sample #11 inactivated APC as the sole case of Turcot’s Syndrome in our study, but this tumour and sample #10 also contained novel missense mutations in CDH1 (R63G, V329F; Figure 1 WNT-subgroup enrichment P<0.05, Fisher’s exact). CDH1 sequesters CTNNB1 at the cell membrane34, and mutations that disrupt this interaction promote WNT signaling in adult cancers35,36. The functional consequences of CDH1R63G and CDH1V329F remain to be determined, but their restriction to WNT-subgroup tumours; mutual exclusivity with CTNNB1 mutations; and adjacency to residues mutated in breast cancer (http://www.sanger.ac.uk/genetics/CGP/cosmic/), suggest these might promote aberrant WNT signals in medulloblastoma.
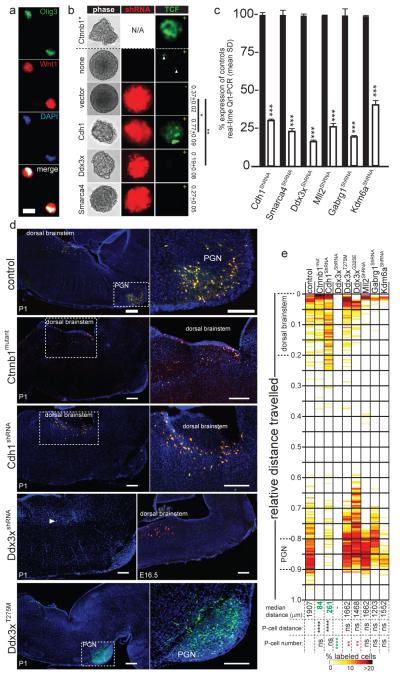
We showed previously that mutant Ctnnb1 initiates WNT-subgroup medulloblastoma by arresting the migration of LRLPs from the embryonic dorsal brainstem to the pontine grey nucleus (PGN)5. Therefore, to test if disruption of CDH1 might substitute for mutant CTNNB1 in medulloblastoma, we used shRNAs to knock down Cdh1 in embryonic day (E) 14.5 mouse LRLPs (Figure 3a to c). Deletion of Cdh1 expression upregulated Tcf/Lef mediated gene transcription in LRLPs and more than doubled their self-renewal capacity (Figure 3b). Furthermore, in utero electroporation of LRLPs with Cdh1 shRNAs impeded their migration from the dorsal brainstem to the PGN with an efficiency similar to that of mutant Ctnnb1 (Figure 3d,e; see Supplementary Methods). These data support the hypothesis that CDH1 suppresses the formation of WNT-subgroup medulloblastoma by regulating WNT-signals in LRLPs.
Figure 3. Genes mutated in WNT-subgroup medulloblastomas regulate LRLPs.
(a) Isolated Olig3+/Wnt1+ LRLPs were transduced in (b) with mutant Ctnnb1 (above hashed line) or the indicated shRNA-Red Fluorescence Protein construct (below hashed line). LRLPs were also transduced (+) or not (−) with a Tcf-Lef-enhanced green fluorescence (Tcf) reporter. Numbers right report clonal % 2′ to 3′ passage neurosphere formation (+SD). (c) Knock-down of genes targeted by shRNA relative to control transduced cells. (d) Immunofluorescence of P1 mouse hindbrains electroporated in utero at E14.5 with GFP (to control for equivalence of electroporation between embryos control) and the indicated construct. High-power views of indicated areas are shown right. Cells targeted by Ddx3x-shRNA are present 48 hours post electroporation but ablated by P1. Scale=200μm. (e) Heatmap reporting the distribution of GFP+/RFP+ cells in eletroporated mice at P1. Median distance migrated by cells, and p-values of migration distance and cell number relative to controls is shown (****, p<0.00005; ***, p<0.0005; **, p<0.005; *, p<0.05. Red and green text reports significant increase or decrease, respectively relative to control).
WNT-subgroup medulloblastomas were also enriched for novel, recurrent somatic missense mutations in the DEAD-Box RNA helicase DDX3X at Xp11.3 (P<0.0001, Fisher’s exact; Figure 1). DDX3X regulates several critical cell processes including chromosome segregation37, cell cycle progression38, gene transcription and translation39. Previously reported cancer associated mutations in DDX3X disrupt the ATPase activity of the protein, but seven of eight mutations identified in our series clustered in the DEAD-box domain (Supplementary Information; Supplementary Figure 8). Structural modeling predicts that these mutations interfere with nucleic acid binding, possibly altering specificity and/or affinity for RNA substrates, rather than inactivating DDX3X (Supplementary Figures 17-22). Indeed, the wild-type allele of DDX3X that escapes X inactivation25 was retained by two of three DDX3X-mutant female medulloblastomas, and knock-down of Ddx3x halved the self-renewal rate of mouse LRLPs, suggesting this protein is important for the proliferation and/or maintenance of the LRLP lineage (Figure 3b).
To better understand the role of DDX3X in WNT-subgroup medulloblastoma, we employed our in utero migration assay to assess the impact of Ddx3x shRNAs, mutant-Ddx3xT275M (identified in WNT-sample #9) or mutant-Ddx3xG325E (WNT-sample #8) on LRLPs. Remarkably, while Ddx3x shRNAs were expressed abundantly in E14.5 brainstem cells within 48 hours of electroporation, <0.5% of Ddx3x- shRNA-positive cells were present by postnatal day 1, confirming the critical importance of this gene to maintain the LRLP lineage (Figure 3d,e). In stark contrast, mice electroporated with either mutant-Ddx3xT275M or Ddx3xG325E consistently contained ~50% more labeled cells at postnatal day (P) 1 than did controls, although these cells migrated normally (Figure 3d,e and data not shown). Thus, mutations in DDX3X may contribute to WNT-subgroup medulloblastoma by increasing LRLP proliferation rather than perturbing the migration of their daughters. Notably, comparable knock-down in utero of Mll2, Gabrg1, and Kdm6a that were selectively mutated in non-WNT medulloblastomas had no apparent impact on LRLPs; supporting the value of our assay for assessing WNT-subgroup specific mutations and underscoring the importance of cell context for functional studies of genes mutated in cancer subgroups.
PIK3CA mutations promote but don’t initiate WNT-subgroup medulloblastoma
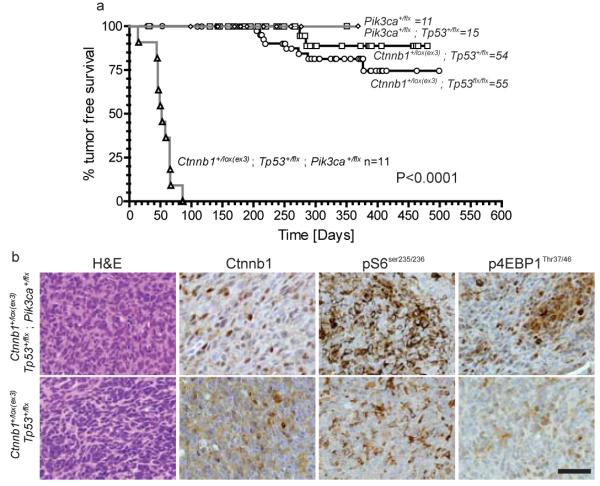
Cancer-associated, activating mutations in PIK3CA were detected in a single case each of WNT (PIK3CAQ546K), SHH (PIK3CAH1047R) and subgroup-4 (PIK3CAN345K) medulloblastoma (Figure 1; Supplementary Figure 23). Although PIK3CA mutations are common in adult cancers40 and reported in medulloblastoma41, their role in tumourigenesis remains controversial. In particular it is not known if these mutations initiate or progress cancer. To test this, we generated mice that express a conditional allele of the Pik3caE545K mutation. Mice harboring Pik3caE545K or Pik3caE545K and Tp53flx/flx were bred with Blbp-Cre that drives efficient recombination in LRLPs5. Blbp-Cre ; Pik3caE545K mice, with or without Tp53flx/flx, survived tumour free for a median of 212 days with no evidence of aberrant LRLP migration (Figure 4a and data not shown). In stark contrast, 100% (n=11/11) of Blbp-Cre ; Ctnnb1+/lox(Ex3) ; Tp53+/flx ; Pik3caE545K mice developed WNT-subgroup medulloblastomas by 3 months of age: only 4% (n=2/54) of Blbp-Cre ; Ctnnb1+/lox(Ex3) ; Tp53+/flx mice develop WNT-medulloblastoma by 11 months (Figure 4a,b). Pik3ca wild-type and mutant mouse medulloblastomas displayed similar ‘classic’ histologies and nuclear Ctnnb1+, but Pik3caE545K mutant tumors contained greater AKT pathway activity as measured by pS6 and p4EBP1 immunostaining. Thus mutations in PIK3CA likely activate the AKT pathway to progress, rather than initiate, WNT-medulloblastoma.
Figure 4. Pik3caE545K accelerates but does not initiate WNT-subgroup medulloblastoma.
Tumour free survival of mice of the indicated genotype. All mice carry the Blbp-cre allele. Log Rank P<0.0001. (b) Hematoxylin and eosin and immunohistochemical stains of indicated tumors. Scale=50 μm.
SHH-subgroup medulloblastomas
Four of 13 SHH-subgroup medulloblastomas contained expected biallelic inactivating alterations in SUFU or PTCH1. What drives aberrant SHH-signals in the remaining cases remains unclear. These tumours contained mutations in MLL2, TP53, and PTEN that have been reported previously in medulloblastoma42; but these mutations occur in other subgroups and are not known to activate SHH signals. Two SHH-subgroup tumours (#11 and 12) contained identical novel T48M mutations in gamma-aminobutyric acid (GABA) A receptor, gamma 1 that is predicted to be deleterious (Figure 1, Supplementary Table 9). Disruption of GABAA receptors can enhance neural stem cell proliferation43, suggesting these mutations might deregulate the proliferation of GNPs that generate SHH-subgroup medulloblastomas.
Discussion
We have identified several, new, recurrent, somatic mutations in specific subgroups of medulloblastoma. Alterations affecting EZH2, KDM6A, CHD7 and ZMYM3 appear to disrupt chromatin marking of genes in subgroup-3 and 4 tumours. Further epigenetic studies will be required to uncover the identity of these genes, but evidence suggests these may include OTX2, MYC and MYCN44,45. Since amplification of these genes was detected almost exclusively in subgroup-3 and 4 tumours that lacked mutations in KDM6A, CHD7 or ZMYM3, it is tempting to speculate that these genetic alterations target common transforming pathways. A recent study detected recurrent mutations in three other chromatin remodelers in medulloblastoma42: SMARCA4, MLL2 and MLL3, but this study did not include details of tumour subgroup. Here, we show that mutations in SMARCA4, CREBBP, TRRAP and MED13 are enriched in WNT-subgroup medulloblastomas; thereby uncovering potential cooperative mutations in chromatin remodelers and their binding-partner oncogene, CTNNB1. Thus, disruptions in the epigenetic machinery of medulloblastoma are likely to be subgroup specific and may cooperate with other oncogenic mutations. The low incidence of MLL2 mutations detected in our study relative to Parsons et al.,42 likely reflects differences the our study populations (see Supplementary Results).
Although medulloblastoma is more prevalent in males, especially subgroup-3 and 4 disease46, the reason for this sex bias is unknown. One potential explanation is the location of medulloblastoma oncogenes or tumour suppressor genes on chromosome X47. Three of the most recurrently mutated genes detected in our study are located on chromosome X, of which two (ZMYM3 and KDM6A) were observed almost exclusively in males. Mutation of these genes might explain some of the male sex-bias in medulloblastoma. The third mutated X chromosome gene, DDX3X, is more likely to be a WNT-medulloblastoma oncogene. Three of four female medulloblastomas carried heterozygous mutations in DDX3X that escapes X inactivation25 and our functional data indicate that mutations in this gene provide a proliferative advantage to LRLPs that generate these tumours.
Our findings also have important implications for drug development. Inhibitors of the epigenetic machinery, especially those that maintain H3K27me3 e.g., EZH2 methylases, may be useful treatments of subgroup-3 and 4 disease. These tumours include the most aggressive forms of medulloblastoma for which treatment options are limited. Mutations that activate PIK3CA and DDX3X in WNT-subgroup tumors might also be targeted with novel therapeutic strategies48,49. Future clinical trials of drugs that target these mutant proteins must recruit the appropriate patient populations, since we show mutations display subgroup-specificity in medulloblastoma. Our accurate mouse models of WNT, SHH and subgroup-3 medulloblastoma should help considerably with future studies of the biological and therapeutic significance of the novel genetic alterations described in this study.
Methods summary
Human tumour and matched blood samples were obtained with informed consent through an institutional review board approved protocol at St Jude Children’s Research Hospital. Whole genome sequencing (WGS) and analysis of WGS data were performed as previously described50. Details of sequence coverage, custom capture and other validation procedures are provided in Supplementary Information (Supplementary Tables 12-15). Sequence and SNP array data were deposited in dbGaP (dbGaP accession number: phs000409, SRA accession number: SRP008292). Immunohistochemistry and immunofluorescence of human and mouse tissues were performed using routine techniques and primary antibodies of the appropriate tissues as described (Supplementary Methods). Medulloblastoma mRNA and DNA profiles were generated using Affymetrix U133v2 and SNP 6.0 arrays, respectively (Supplementary Methods). Reverse transcriptase Real Time-PCR analysis of genes targeted in mouse LRLPs by shRNAs were performed as described previously32. LRLPs were isolated and transduced with indicated lentiviruses in stem cell cultures or targeted in utero with shRNAs or mutant cDNA sequences by electroporation as described (Supplementary Information)5. Mice harbouring a cre-inducible Pik3caE545K allele were generated using homologous recombination: A lox-puro-STOP-lox cassette was introduced immediately upstream of the exon containing the initiation codon, exon 9 was replaced with an exon containing the E545K mutation. Pik3caE545K mice were bred with Blbp-Cre, Ctnnb1lox(ex3)/lox(ex3) and Tp53flx/flx mice to generate progeny of the appropriate genotype and subjected to clinical surveillance.
Supplementary Material
Acknowledgements
This research was supported as part of the St. Jude Children’s Research Hospital, Washington University Pediatric Cancer Genome Project. This work was supported by grants from the National Institutes of Health (R01CA129541, P01CA96832 and P30CA021765, R.J.G), the Collaborative Ependymoma Research Network (CERN), Musicians against Childhood Cancer (MACC), The Noyes Brain Tumour Foundation, and by the American Lebanese Syrian Associated Charities (ALSAC). We are grateful to Sally Temple for the generous gift of reagents and the staff of the Hartwell Center for Bioinformatics and Biotechnology and ARC at St Jude Children’s Research Hospital for technical assistance.
Footnotes
Author contributions. G.R., M.P., T.A.K, C.L., X.C., L.D., T.N.P., E.H., W.L., X.Z., N.C., R.H., N.C., R.T., J.W., G.W., M.R., X.H., J.B., P.G., J.M., J.E., B.V., S.P., D.Z., D.K., D.F., contributed to the design and conduct of experiments and to the writing S.J.B., R.K., M.F.R., R.S.F., L.L.F., D.J.D., K.O., E.R.M., contributed to experimental design and to the writing A.G., D.W.E., C.C.L., E.B., T.H., S.G., R.C., provided clinical expertise. R.K.W., J.R.D., J.Z., and R.J.G., conceived the research and contributed to the design, direction and reporting of the study. No competing financial interests.
Competing interests. None.
Supplementary Information. Supplementary Information is available online.
References
- 1.CBTRUS . Statistical Report: Primary Brain Tumors in the United States, 1995-1999. Central Brain Tumor Registry of the United States; Hinsdale, IL: 2006. 2002. [Google Scholar]
- 2.Taylor MD, et al. Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol. 2012;123:465–472. doi: 10.1007/s00401-011-0922-z. Epub 2011 Dec 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schuller U, et al. Acquisition of granule neuron precursor identity is a critical determinant of progenitor cell competence to form Shh-induced medulloblastoma. Cancer Cell. 2008;14:123–134. doi: 10.1016/j.ccr.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang ZJ, et al. Medulloblastoma can be initiated by deletion of Patched in lineage-restricted progenitors or stem cells. Cancer Cell. 2008;14:135–145. doi: 10.1016/j.ccr.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gibson P, et al. Subtypes of medulloblastoma have distinct developmental origins. Nature. 2010;468:1095–1099. doi: 10.1038/nature09587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kawauchi D, et al. A mouse model of the most aggressive subgroup of human medulloblastoma. Cancer Cell. 2012;21:168–180. doi: 10.1016/j.ccr.2011.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mulhern RK, et al. Neurocognitive Consequences of Risk-Adapted Therapy for Childhood Medulloblastoma. Journal of Clinical Oncology. 2005;23:5511–5519. doi: 10.1200/JCO.2005.00.703. [DOI] [PubMed] [Google Scholar]
- 8.Wang J, et al. CREST maps somatic structural variation in cancer genomes with base-pair resolution. Nat Meth. 2011;8:652–654. doi: 10.1038/nmeth.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rausch T, et al. Genome Sequencing of Pediatric Medulloblastoma Links Catastrophic DNA Rearrangements with TP53 Mutations. Cell. 2012;148:59–71. doi: 10.1016/j.cell.2011.12.013. doi:10.1016/j.cell.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Castellino RC, et al. Heterozygosity for Pten Promotes Tumorigenesis in a Mouse Model of Medulloblastoma. PLoS One. 2010;5:e10849. doi: 10.1371/journal.pone.0010849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hahn H, et al. Mutations of the human homolog of Drosophila patched in the nevoid basal cell carcinoma syndrome. Cell. 1996;85:841–851. doi: 10.1016/s0092-8674(00)81268-4. [DOI] [PubMed] [Google Scholar]
- 12.Malkin D, et al. Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science. 1990;250:1233–1238. doi: 10.1126/science.1978757. [DOI] [PubMed] [Google Scholar]
- 13.Hamilton SR, et al. The molecular basis of Turcot’s syndrome. N Engl J Med. 1995;332:839–847. doi: 10.1056/NEJM199503303321302. [DOI] [PubMed] [Google Scholar]
- 14.Taylor MD, et al. Medulloblastoma in a child with Rubenstein-Taybi Syndrome: case report and review of the literature. Pediatr Neurosurg. 2001;35:235–238. doi: 10.1159/000050428. [DOI] [PubMed] [Google Scholar]
- 15.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cao R, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. Epub 2002 Sep 1026. [DOI] [PubMed] [Google Scholar]
- 17.Czermin B, et al. Drosophila Enhancer of Zeste/ESC Complexes Have a Histone H3 Methyltransferase Activity that Marks Chromosomal Polycomb Sites. Cell. 2002;111:185–196. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- 18.Agger K, et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 19.Schnetz MP, et al. Genomic distribution of CHD7 on chromatin tracks H3K4 methylation patterns. Genome Research. 2009;19:590–601. doi: 10.1101/gr.086983.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sauvageau M, Sauvageau G. Polycomb Group Proteins: Multi-Faceted Regulators of Somatic Stem Cells and Cancer. Cell Stem Cell. 2010;7:299–313. doi: 10.1016/j.stem.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morin RD, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet. 2010;42:181–185. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kleer CG, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proceedings of the National Academy of Sciences. 2003;100:11606–11611. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Varambally S, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 24.van Haaften G, et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat Genet. 2009;41:521–523. doi: 10.1038/ng.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang F, Babak T, Shendure J, Disteche CM. Global survey of escape from X inactivation by RNA-sequencing in mouse. Genome Research. 2010;20:614–622. doi: 10.1101/gr.103200.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Christensen J, et al. RBP2 Belongs to a Family of Demethylases, Specific for Tri-and Dimethylated Lysine 4 on Histone 3. Cell. 2007;128:1063–1076. doi: 10.1016/j.cell.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 27.Lee MG, Wynder C, Cooch N, Shiekhattar R. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature. 2005;437:432–435. doi: 10.1038/nature04021. [DOI] [PubMed] [Google Scholar]
- 28.Mosimann C, Hausmann G, Basler K. [beta]-Catenin hits chromatin: regulation of Wnt target gene activation. Nat Rev Mol Cell Biol. 2009;10:276–286. doi: 10.1038/nrm2654. [DOI] [PubMed] [Google Scholar]
- 29.Hecht A, Vleminckx K, Stemmler MP, van Roy F, Kemler R. The p300/CBP acetyltransferases function as transcriptional coactivators of [beta]-catenin in vertebrates. EMBO J. 2000;19:1839–1850. doi: 10.1093/emboj/19.8.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barker N, et al. The chromatin remodelling factor Brg-1 interacts with [beta]-catenin to promote target gene activation. EMBO J. 2001;20:4935–4943. doi: 10.1093/emboj/20.17.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carrera I, Janody F, Leeds N, Duveau F, Treisman JE. Pygopus activates Wingless target gene transcription through the mediator complex subunits Med12 and Med13. Proceedings of the National Academy of Sciences. 2008;105:6644–6649. doi: 10.1073/pnas.0709749105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson MC, et al. Genomics identifies medulloblastoma subgroups that are enriched for specific genetic alterations. J Clin Oncol. 2006;24:1924–1931. doi: 10.1200/JCO.2005.04.4974. Epub 2006 Mar 1927. [DOI] [PubMed] [Google Scholar]
- 33.Kool M, et al. Integrated genomics identifies five medulloblastoma subtypes with distinct genetic profiles, pathway signatures and clinicopathological features. PLoS One. 2008;3:e3088. doi: 10.1371/journal.pone.0003088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Orsulic S, Huber O, Aberle H, Arnold S, Kemler R. E-cadherin binding prevents beta-catenin nuclear localization and beta-catenin/LEF-1-mediated transactivation. Journal of Cell Science. 1999;112:1237–1245. doi: 10.1242/jcs.112.8.1237. [DOI] [PubMed] [Google Scholar]
- 35.Risinger JI, Berchuck A, Kohler MF, Boyd J. Mutations of the E-cadherin gene in human gynecologic cancers. Nat Genet. 1994;7:98–102. doi: 10.1038/ng0594-98. [DOI] [PubMed] [Google Scholar]
- 36.Becker K-F, et al. E-Cadherin Gene Mutations Provide Clues to Diffuse Type Gastric Carcinomas. Cancer Research. 1994;54:3845–3852. [PubMed] [Google Scholar]
- 37.Pek JW, Kai T. DEAD-box RNA helicase Belle/DDX3 and the RNA interference pathway promote mitotic chromosome segregation. Proc Natl Acad Sci U S A. 2011;108:12007–12012. doi: 10.1073/pnas.1106245108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lai MC, Chang WC, Shieh SY, Tarn WY. DDX3 regulates cell growth through translational control of cyclin E1. Mol Cell Biol. 2010;30:5444–5453. doi: 10.1128/MCB.00560-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schroder M. Human DEAD-box protein 3 has multiple functions in gene regulation and cell cycle control and is a prime target for viral manipulation. Biochem Pharmacol. 2010;79:297–306. doi: 10.1016/j.bcp.2009.08.032. [DOI] [PubMed] [Google Scholar]
- 40.Samuels Y, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 41.Broderick DK, et al. Mutations of PIK3CA in anaplastic oligodendrogliomas, high-grade astrocytomas, and medulloblastomas. Cancer Res. 2004;64:5048–5050. doi: 10.1158/0008-5472.CAN-04-1170. [DOI] [PubMed] [Google Scholar]
- 42.Parsons DW, et al. The genetic landscape of the childhood cancer medulloblastoma. Science. 2011;331:435–439. doi: 10.1126/science.1198056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Andang M, et al. Histone H2AX-dependent GABAA receptor regulation of stem cell proliferation. Nature. 2008;451:460–464. doi: 10.1038/nature06488. [DOI] [PubMed] [Google Scholar]
- 44.Pasini D, et al. Coordinated regulation of transcriptional repression by the RBP2 H3K4 demethylase and Polycomb-Repressive Complex 2. Genes & Development. 2008;22:1345–1355. doi: 10.1101/gad.470008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khan A, Shover W, Goodliffe JM. Su(z)2 Antagonizes Auto-Repression of Myc in Drosophila, Increasing Myc Levels and Subsequent Trans-Activation. PLoS One. 2009;4:e5076. doi: 10.1371/journal.pone.0005076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Northcott PA, et al. Medulloblastoma comprises four distinct molecular variants. J Clin Oncol. 2011;29:1408–1414. doi: 10.1200/JCO.2009.27.4324. Epub 2010 Sep 1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spatz A, Borg C, Feunteun J. X-Chromosome Genetics and Human Cancer. Nat Rev Cancer. 2004;4:617–629. doi: 10.1038/nrc1413. [DOI] [PubMed] [Google Scholar]
- 48.Lindqvist L, et al. Selective Pharmacological Targeting of a DEAD Box RNA Helicase. PLoS One. 2008;3:e1583. doi: 10.1371/journal.pone.0001583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Engelman JA. Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat Rev Cancer. 2009;9:550–562. doi: 10.1038/nrc2664. [DOI] [PubMed] [Google Scholar]
- 50.Zhang J, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–163. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.