Abstract
The homeodomain transcription factor Pdx1 is essential for pancreas formation and functions in pancreatic islets cells to regulate genes involved in maintenance of glucose homeostasis. In order to investigate a role for Pdx1 in intestinal cells, we analyzed the functions and networks associated with genes differentially expressed by Pdx1 overexpression in human Caco-2 cells. In agreement with previous results for intestine isolated from mice with Pdx1 inactivation, functional analysis of genes differentially expressed with Pdx1 overexpression revealed functions significantly associated with nutrient metabolism. Similarly, network analysis examining the interactions among the differentially expressed genes revealed gene networks involved in lipid metabolism. Consistent with defects in maternal nutrient metabolism, mouse pups born to dams with intestine-specific Pdx1 inactivation are underweight and fail to thrive in the neonatal period compared to pups born to control dams. We conclude that Pdx1 mediates lipid metabolism gene networks in intestinal cells and that maternal expression is essential for perinatal growth in mice.
Keywords: transcription factors, gene regulation, expression profile
INTRODUCTION
The homeodomain-containing transcription factor Pdx1 is essential for pancreas formation during embryogenesis and subsequent maintenance of islet function for normal glucose homeostasis. Pdx1 regulates expression of genes in the pancreas necessary for maintaining pancreatic identity and function including insulin, glucose transporter 2, glucokinase, islet amyloid polypeptide and somatostatin.1–8 Mutations in the human Pdx1 gene are linked to maturity-onset diabetes of the young, type 4 (MODY4) and type 2 diabetes mellitus.9–12 With respect to nutritional and hormonal regulation controlling pancreatic Pdx1 expression, glucose, GLP-1, insulin, T3, HB-EGF, and TNF-α all positively regulate the Pdx1 gene promoter in pancreatic β- cells.13 The role of Pdx1 expressed in the intestine, however, is not well defined.
Pdx1 is expressed in the anterior duodenal region of the small intestine and decreases in expression distally.14 Mice homozygous for a Pdx1 null mutation (Pdx1−/−) fail to form a pancreas and die in the neonatal period within a week of birth.15, 16 Therefore, in order to investigate roles for Pdx1 expressed in the intestine, mice with Pdx1 inactivation restricted to the intestinal epithelium (Pdx1flox/flox;VilCre) have been generated.17 Pdx1flox/flox;VilCre mice survive through adulthood and have pancreata and small intestines with gross morphologies that are indistinguishable from those of controls. Expression profiling identified genes differentially expressed in duodenal segments isolated from mature Pdx1flox/flox;VilCre and control mice.18 Pathway analysis of the differentially expressed genes revealed functions that are significantly associated with metabolism of nutrients such as lipids, carbohydrates, amino acids, vitamins and minerals. In addition, network analysis examining the interactions among the differentially expressed genes revealed gene networks involved in metabolism of lipids and minerals.18
In order to identify additional novel Pdx1-regulated genes, human intestinal epithelial Caco-2 cells have been engineered to overexpress Pdx1 and gene expression profiles relative to control cells were assessed.19 Fatty acid binding protein 1, liver, FABP1, a gene with known intestinal cell expression, was identified as a candidate Pdx1 target through such analysis. In the present study, we sought to analyze the functions and networks associated with genes differentially expressed by Pdx1 overexpression in human intestinal Caco-2 cells. Upon identifying associations between Pdx1 overexpression and gene networks associated with nutrient and lipid metabolism, we proceeded to investigate the role of intestinal Pdx1 expression in supporting maternal-fetal nutrition in mice.
MATERIALS AND METHODS
Animals
Mice with intestinal epithelium-specific Pdx1 inactivation (Pdx1flox/flox;VilCre) were generated by intercross mating between VilCre and Pdx1flox/flox mouse strains as previously described.17 To investigate the extent of defects in nutrient metabolism in mothers with Pdx1 inactivation, a Pdx1flox/flox;VilCre virgin female was mated with a Pdx1flox/flox;VilCre stud male. A littermate control Pdx1flox/flox virgin female was mated with the same Pdx1flox/flox;VilCre stud in the same cage. Identical normal diet feed was accessible to all mice. The protocol for animal use was reviewed and approved by the Stanford University Institutional Animal Care and Use Committee (IACUC).
Functional analysis of genes differentially expressed in Caco-2 cells with Pdx1 overexpression
Microarray data, previously generated for human intestinal epithelial Caco-2 cells stably transfected with a vector driving mouse Pdx1 cDNA expression or with empty vector alone19, was analyzed with the web-based software and database, Ingenuity Pathways Analysis (IPA version 8.8, Ingenuity Systems, www.ingenuity.com). Specifically, functions and interactions of genes exhibiting significant differential expression >4-fold were analyzed.
IPA Functional analysis was performed to find significant associations of the differentially expressed genes to molecular and cellular functions. Under the primary categories, subcategories were classified, consisting of specific, basic level functions populated with a group of genes or chemicals, based on the findings stored in the Ingenuity Knowledge Base. Statistically significant, non-random associations of the differentially expressed genes with the specific functions and subcategories were indicated by a p value less than 0.05 following right-tailed Fisher’s exact test.
Network analysis of differentially expressed genes
IPA Network analysis was performed to examine and visualize interactions among genes exhibiting significant changes in expression with Pdx1 overexpression in Caco-2 cells by generating statistically significant, non random networks. The differentially expressed genes served as “seeds” and connected to other genes or chemicals in the Ingenuity Knowledge Base via direct or indirect interactions. Networks were limited to 35 genes or chemicals to maximize specificity of the connections. Network analysis complements functional analysis, because functional analysis considers the differentially expressed genes alone. The statistical significance, or scores, of generated networks were calculated with right-tailed Fisher’s Exact Test. The higher the score, the lower the probability of finding the observed number of differentially expressed genes in a given network by random chance
RESULTS
Pdx1 overexpression differentially regulates genes with functions associated with nutrient metabolism
In order to identify functions for genes differentially expressed in intestinal cells with Pdx1 overexpression, microarray data was analyzed for human intestinal epithelial Caco-2 cells engineered to overexpress Pdx1 relative to control cells. The microarray data was previously generated from experiments using RNA isolated from Caco-2 cells stably transfected with a Pdx1 cDNA expression or with empty vector alone at 9 days post confluency.19
Microarray data was analyzed by a computer software, Ingenuity Pathway Analysis (IPA), for significant association (p<0.05) with biological functions represented by the genes differentially expressed > 4-fold by Pdx1 overexpression in post-confluent Caco-2 cells. The association was examined by analysis for molecular and cellular functional annotations (Table 1). In post-confluent Caco-2 cells, metabolism of drugs and nutrients such as lipids, carbohydrates, amino acids, nucleic acids, vitamins and minerals was likely affected by Pdx1 overexpression (Table 1).
Table 1.
Pdx1 overexpression in Caco-2 cells alters expression of genes with functions significantly associated with metabolism of nutrients and drugs (p<0.05).
| Category | P Value |
|---|---|
| Lipid Metabolism | 1.30E-04 – 1.31E-02 |
| Carbohydrate Metabolism | 3.98E-04 – 1.10E-02 |
| Nucleic Acid Metabolism | 1.59E-03 – 5.22E-03 |
| Drug Metabolism | 1.90E-03 – 6.65E-03 |
| Vitamin and Mineral Metabolism | 6.65E-03 – 6.65E-03 |
| Amino Acid Metabolism | 1.90E-03 – 1.28E-02 |
The findings are in agreement with the previous gene profiling study using mice with intestinal epithelium-specific Pdx1 inactivation, showing that functions of the genes differentially expressed in mature duodenum are also significantly associated with nutrient and drug metabolism.18 In post-confluent Caco-2 cells overexpressing Pdx1, products of the differentially expressed genes involved in lipid metabolism include biosynthetic enzymes, transport proteins, kinases, ligand-dependent nuclear receptors and cytokines (Table 2).
Table 2.
Genes with functions in lipid metabolism increased or decreased in expression >4-fold by Pdx1 overexpression in Caco-2 cells (p<0.05)
| Genes with Functions in Lipid Metabolism | Gene Symbol | Fold Change |
|---|---|---|
| acyl-CoA synthetase long-chain family member 1 | ACSL1 | 4.04 |
| angiotensinogen (serpin peptidase inhibitor, clade A, member 8) | AGT | −6.24 |
| apolipoprotein M | APOM | −4.32 |
| cytochrome P450, family 27, subfamily A, polypeptide 1 | CYP27A1 | −4.95 |
| estrogen receptor 1 | ESR1 | −4.18 |
| fatty acid binding protein 1, liver | FABP1 | −4.07 |
| fibronectin 1 | FN1 | 11.17 |
| kininogen 1 | KNG1 (includes EG:3827) | −14.58 |
| mitogen-activated protein kinase 8 | MAPK8 | −5.62 |
| sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A | SEMA3A | 4.20 |
| tumor necrosis factor (ligand) superfamily, member 10 | TNFSF10 | −5.17 |
| UDP glucuronosyltransferase 2 family, polypeptide B4 | UGT2B4 | −10.54 |
| UDP glucuronosyltransferase 2 family, polypeptide B7 | UGT2B7 | −7.69 |
| UDP glucuronosyltransferase 2 family, polypeptide B15 | UGT2B15 | −11.06 |
Network analysis of differentially expressed genes indicates that Pdx1 overexpression may impact lipid metabolism
To complement the functional analysis described above, networks were generated to analyze the relational interactions among differentially expressed genes. Gene networks visualize the relationships among genes differentially expressed > 4-fold in response to Pdx1 overexpression in post-confluent Caco-2 cells. The relationships examined include direct and indirect interactions between the genes of interest. Direct interactions refer to physical binding relationships such as protein-DNA binding, while examples for indirect interactions include activation, transcription, phosphorylation, or localization. The differentially expressed genes were used as “seeds” and connected as many of them into a network. Other molecules (genes or chemicals) in the Ingenuity Knowledge Base were also included to connect multiple smaller gene networks into a larger network, thus providing insights into possible functional roles for Pdx1 in the intestinal cell culture.
The representative network shown in Figure 1 has a high significance score of 30 and contains a high number (16) of genes differentially expressed > 4-fold in response to Pdx1 overexpression in post-confluent Caco-2 cells. The score of 30 indicates that the chance is 1 in 1030 to form a network of 35 molecules by randomly selecting from the Ingenuity database and including at least 16 differentially expressed genes by Pdx1 overexpression. Network analysis examining the interactions among the differentially expressed genes revealed genes involved in lipid metabolism, including acyl-CoA synthetase long-chain family member 1 (ACSL1), fatty acid binding protein 1, liver (FABP1) and UDP glucuronosyltransferase 2 family, polypeptide B15 (UGT2B15). This network suggests a functional role of Pdx1 in modulating lipid metabolism.
Figure 1. Network analysis suggesting a role for Pdx1 in modulating lipid metabolism in Caco-2 cells.
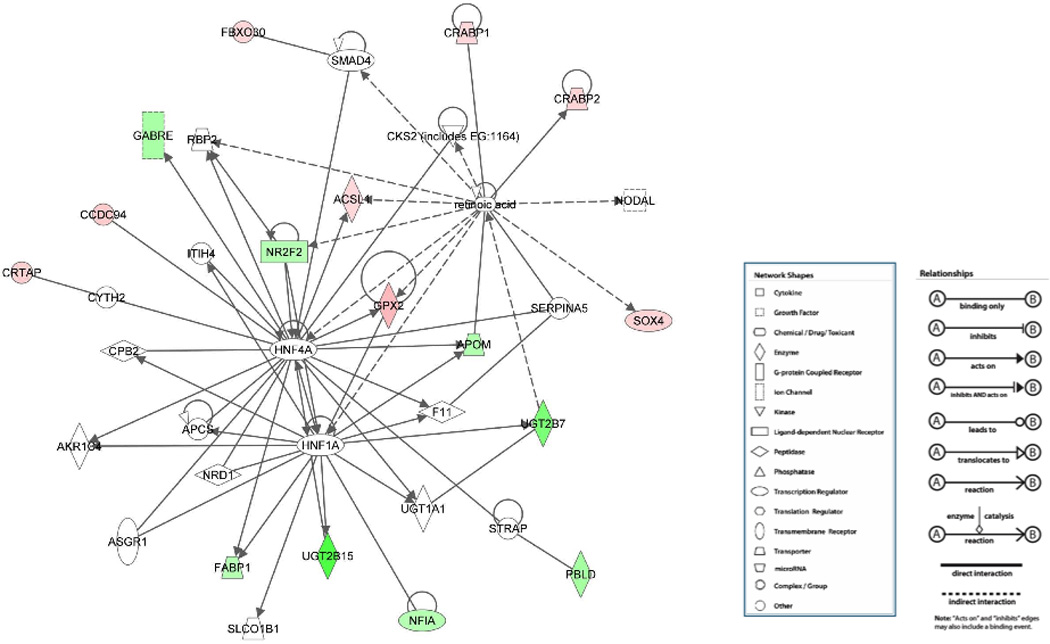
Genes significantly upregulated in expression by >4-fold are in red and genes significantly downregulated are in green. This network scored high statistical significance with right-tailed Fisher’s Exact Test and contained a high fraction of differentially expressed genes. Genes or chemicals in white were not included in the 129 genes with >4-fold changes in expression19 or on the microarray chips screened.
Offspring of Pdx1flox/flox;VilCre dam were underweight and failed to thrive postnatally
To investigate the extent of defects in nutrient metabolism in mice with Pdx1 inactivation, a Pdx1flox/flox;VilCre virgin female was mated with a Pdx1flox/flox;VilCre stud male, along with a littermate control Pdx1flox/flox virgin female. Identical normal diet feed was accessible to all mice. The weight and condition of the offspring from both dams were tracked upon birth through weaning and compared (Figure 2). The offspring from both dams were born on the same day and the size of the litters was similar (7 pups to Pdx1flox/flox;VilCre dam and 10 to the littermate control dam). However, the size of the litter to Pdx1flox/flox;VilCre dam continued to decrease; with 5 pups remaining 3 days after birth, 3 pups 10 days after birth, and 1 surviving until weaning at 23 days that died shortly thereafter, compared with 10 pups born to the control dam that thrived throughout the postnatal period.
Figure 2. Maternal Pdx1 inactivation effects perinatal growth.
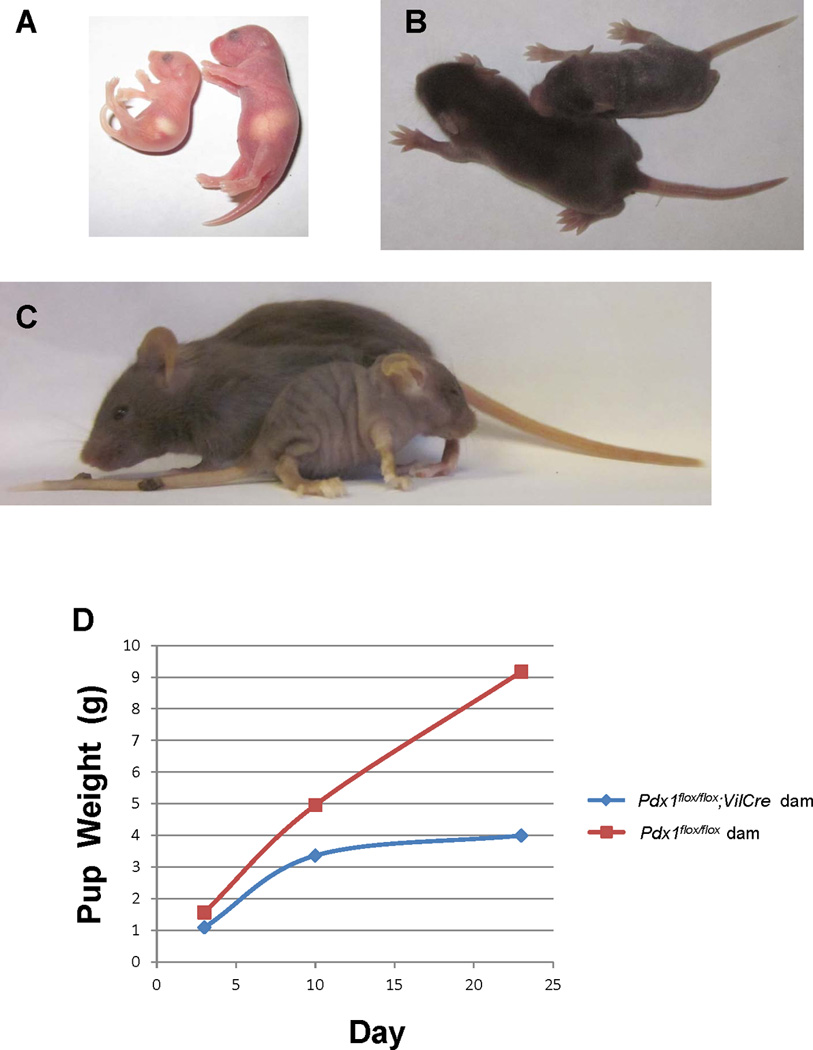
Pups born to Pdx1flox/flox;VilCre dams are underweight (A) and fail to thrive (B,C) compared to those born to littermate control Pdx1flox/flox dams. D). Weights of of pups born to a Pdx1flox/flox;VilCre dam (blue line) compared to pups born to a littermate control Pdx1flox/flox dam (red line).
At postnatal day 3, the offspring of Pdx1flox/flox;VilCre dam appeared smaller in size, pale and lethargic compared to those of the control dam (Figure 2A). Although milk was present in the stomach of pups from Pdx1flox/flox;VilCre dam, indicating that they were fed, the average weight of the pups (1.1g) was less than that of control pups (1.6g) (Figure 2D). Weight gain was delayed at postnatal day 10 for pups from the Pdx1flox/flox;VilCre dam, with an average weight of 3.4g in contrast to that of 5g for control pups (Fig. 2D). In comparison to control pups at postnatal day 10, the pups born to the Pdx1flox/flox;VilCre dam remained smaller overall in size, appeared pale in extremities, and lacked fur coat coverage from the ear down (Figure 2B). At postnatal day 23, pups were weaned. The remaining pup from the Pdx1flox/flox;VilCre dam showed further growth retardation, weighing less than half (4g) of the average weight (9.2g) of the control pups (Figure 2D). The appearance of the remaining pup was also consistent with growth retardation showing small physique, fur coat abnormality, pale limbs, ears and tail (Figure 2C). Pups born to the littermate control dam had no apparent developmental abnormalities.
DISCUSSION
In order to investigate a role for Pdx1 in intestinal cells, the functions and networks associated with genes differentially expressed by Pdx1 overexpression in human intestinal Caco-2 cells were analyzed. The functions of genes differentially expressed by Pdx1 overexpression are significantly associated with nutrient and lipid metabolism. Analysis of the relationships among the genes also supports a role for Pdx1 in mediating networks associated with lipid metabolism. These findings are in agreement with our previous gene profiling study of mice with intestinal epithelium-specific Pdx1 inactivation, which showed that functions of the genes differentially expressed in mature duodenum are also significantly associated with nutrient and lipid metabolism.18
Having identified associations between both Pdx1 overexpression and Pdx1 inactivation and gene networks associated with nutrient and lipid metabolism, we investigated the role of intestinal Pdx1 expression in supporting maternal-fetal nutrition in mice. Consistent with defects in maternal nutrient metabolism, Pdx1flox/flox;VilCre mice born to dams with intestine-specific Pdx1 inactivation are underweight and fail to thrive in the neonatal period compared to pups born to control dams. These findings are also consistent with our previous report demonstrating that mature Pdx1flox/flox;VilCre mice with intestinal epithelium-specific Pdx1 inactivation have altered metabolism for nutrients such as lipid and iron, even when a normal diet was fed.18 Failure to gain weight and the growth abnormalities observed in the offspring from Pdx1flox/flox;VilCre dam are most likely due to malnutrition resulting from the defects in maternal nutrient metabolism following Pdx1 inactivation in the dam’s intestinal epithelium, regardless of the offspring’s own genotypes, because the pups born to the littermate control dam show no developmental abnormalities. We conclude that Pdx1 mediates nutrient metabolism gene networks in intestinal cells and that maternal expression is essential for perinatal growth in mice.
Highlights.
Pdx1 overexpression regulates genes with functions associated with nutrient metabolism.
Network analysis indicates that Pdx1 overexpression may impact lipid metabolism.
Offspring of mothers with intestine-specific Pdx1 inactivation fail to thrive.
ACKNOWLEDGEMENTS
This research was supported by National Institute of Diabetes and Digestive and Kidney Diseases Grants DK72416 and DK60715 (to E. Sibley) and DK56339 (to the Stanford Digestive Diseases Center).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DISCLOSURES
No conflicts of interest are declared by the author(s).
REFERENCES
- 1.Bretherton-Watt D, Gore N, Boam DS. Insulin upstream factor 1 and a novel ubiquitous factor bind to the human islet amyloid polypeptide/amylin gene promoter. Biochem J. 1996;313(Pt 2):495–502. doi: 10.1042/bj3130495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carty MD, Lillquist JS, Peshavaria M, Stein R, Soeller WC. Identification of cis- and trans-active factors regulating human islet amyloid polypeptide gene expression in pancreatic beta-cells. J Biol Chem. 1997;272:11986–11993. doi: 10.1074/jbc.272.18.11986. [DOI] [PubMed] [Google Scholar]
- 3.Leonard J, Peers B, Johnson T, Ferreri K, Lee S, Montminy MR. Characterization of somatostatin transactivating factor-1, a novel homeobox factor that stimulates somatostatin expression in pancreatic islet cells. Mol Endocrinol. 1993;7:1275–1283. doi: 10.1210/mend.7.10.7505393. [DOI] [PubMed] [Google Scholar]
- 4.Miller CP, McGehee RE, Jr, Habener JF. IDX-1: a new homeodomain transcription factor expressed in rat pancreatic islets and duodenum that transactivates the somatostatin gene. Embo J. 1994;13:1145–1156. doi: 10.1002/j.1460-2075.1994.tb06363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ohlsson H, Karlsson K, Edlund T. IPF1, a homeodomain-containing transactivator of the insulin gene. Embo J. 1993;12:4251–4259. doi: 10.1002/j.1460-2075.1993.tb06109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Serup P, Jensen J, Andersen FG, et al. Induction of insulin and islet amyloid polypeptide production in pancreatic islet glucagonoma cells by insulin promoter factor 1. Proc Natl Acad Sci U S A. 1996;93:9015–9020. doi: 10.1073/pnas.93.17.9015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Waeber G, Thompson N, Nicod P, Bonny C. Transcriptional activation of the GLUT2 gene by the IPF-1/STF-1/IDX-1 homeobox factor. Mol Endocrinol. 1996;10:1327–1334. doi: 10.1210/mend.10.11.8923459. [DOI] [PubMed] [Google Scholar]
- 8.Watada H, Kajimoto Y, Umayahara Y, et al. The human glucokinase gene beta-celltype promoter: an essential role of insulin promoter factor 1/PDX-1 in its activation in HITT15 cells. Diabetes. 1996;45:1478–1488. doi: 10.2337/diab.45.11.1478. [DOI] [PubMed] [Google Scholar]
- 9.Cockburn BN, Bermano G, Boodram LL, et al. Insulin promoter factor-1 mutations and diabetes in Trinidad: identification of a novel diabetes-associated mutation (E224K) in an Indo-Trinidadian family. J Clin Endocrinol Metab. 2004;89:971–978. doi: 10.1210/jc.2003-031282. [DOI] [PubMed] [Google Scholar]
- 10.Stoffers DA, Stanojevic V, Habener JF. Insulin promoter factor-1 gene mutation linked to early-onset type 2 diabetes mellitus directs expression of a dominant negative isoprotein. J Clin Invest. 1998;102:232–241. doi: 10.1172/JCI2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hani EH, Stoffers DA, Chevre JC, et al. Defective mutations in the insulin promoter factor-1 (IPF-1) gene in late-onset type 2 diabetes mellitus. J Clin Invest. 1999;104:R41–R48. doi: 10.1172/JCI7469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Macfarlane WM, Frayling TM, Ellard S, et al. Missense mutations in the insulin promoter factor-1 gene predispose to type 2 diabetes. J Clin Invest. 1999;104:R33–R39. doi: 10.1172/JCI7449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Campbell SC, Macfarlane WM. Regulation of the pdx1 gene promoter in pancreatic beta-cells. Biochem Biophys Res Commun. 2002;299:277–284. doi: 10.1016/s0006-291x(02)02633-5. [DOI] [PubMed] [Google Scholar]
- 14.Guz Y, Montminy MR, Stein R, et al. Expression of murine STF-1, a putative insulin gene transcription factor, in beta cells of pancreas, duodenal epithelium and pancreatic exocrine and endocrine progenitors during ontogeny. Development. 1995;121:11–18. doi: 10.1242/dev.121.1.11. [DOI] [PubMed] [Google Scholar]
- 15.Jonsson J, Carlsson L, Edlund T, Edlund H. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature. 1994;371:606–609. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- 16.Offield MF, Jetton TL, Labosky PA, et al. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development. 1996;122:983–995. doi: 10.1242/dev.122.3.983. [DOI] [PubMed] [Google Scholar]
- 17.Chen C, Fang R, Davis C, Maravelias C, Sibley E. Pdx1 inactivation restricted to the intestinal epithelium in mice alters duodenal gene expression in enterocytes and enteroendocrine cells. Am J Physiol Gastrointest Liver Physiol. 2009;297:G1126–G1137. doi: 10.1152/ajpgi.90586.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen C, Sibley E. Expression profiling identifies novel gene targets and functions for Pdx1 in the duodenum of mature mice. Am J Physiol Gastrointest Liver Physiol. 2012;302:G407–G419. doi: 10.1152/ajpgi.00314.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen C, Fang R, Chou LC, Lowe AW, Sibley E. PDX1 regulation of FABP1 and novel target genes in human intestinal epithelial Caco-2 cells. Biochem Biophys Res Commun. 2012;423:183–187. doi: 10.1016/j.bbrc.2012.05.113. [DOI] [PMC free article] [PubMed] [Google Scholar]


