Abstract
Understanding gene regulation is a major objective in molecular biology research. Frequently, transcription is driven by transcription factors (TFs) that bind to specific DNA sequences. These motifs are usually short and degenerate, rendering the likelihood of multiple copies occurring throughout the genome due to random chance as high. Despite this, TFs only bind to a small subset of sites, thus prompting our investigation into the differences between motifs that are bound by TFs and those that remain unbound. Here we constructed vectors representing various chromatin- and sequence-based features for a published set of bound and unbound motifs representing nine TFs in the budding yeast Saccharomyces cerevisiae. Using a machine learning approach, we identified a set of features that can be used to discriminate between bound and unbound motifs. We also discovered that some TFs bind most or all of their strong motifs in intergenic regions. Our data demonstrate that local sequence context can be strikingly different around motifs that are bound compared to motifs that are unbound. We concluded that there are multiple combinations of genomic features that characterize bound or unbound motifs.
Keywords: Gene regulation, yeast, transcription factors, genomic features, machine learning
1 Introduction
Control of gene expression is fundamental to all forms of life. Transcription initiation is controlled primarily by transcription factor (TF) binding to key DNA sequence motifs. In many cases, the sequence motifs recognized by DNA binding proteins are short and degenerate, thus rendering it highly likely that they may appear multiple times in the genome due to random chance. This is especially true for large eukaryotic genomes. Using sequence motifs alone to predict TF binding leads to an unacceptable level of false positives (Fickett, 1996; D’Haeseleer, 2006). Given this, many transcription factor binding site prediction methods incorporate other sources of information in addition to sequence similarity. For example, previous studies have incorporated information about the sequence conservation between species (Blanchette and Tompa, 2003; Xie et al., 2005) or the fact that different TFs frequently bind DNA in clusters (Frith et al., 2003; Tharakaraman et al., 2008). Combining sequence conservation or clustering of TFBS into a single tool can improve predictive performance; however how a TF selects the appropriate motif out of all its occurrences in vivo across the genome remains unknown. While conservation between species is a useful computational tool for identifying potential regulatory regions, this information is not available in vivo to guide a TF to the correct binding location. Recent advances in high-throughput techniques [e.g., chromatin immunoprecipitation with microarray technology (ChIP-chip) and chromatin immunoprecipitation sequencing (ChIP-seq)] have produced high-quality maps of genome-wide TF binding. In addition, machine-learning techniques have been used successfully to predict TF binding (Holloway et al., 2005; Holloway et al., 2007; Bauer et al., 2010). In this study, we used both these techniques to compare the local genomic environment near established TF binding sites with unbound motifs to identify biological features associated with bound motifs in vivo. Our approach is to use machine-learning techniques not primarily in an attempt to predict TF binding sites but rather to gain insight into local genomic features of bound and unbound motifs.
2 Materials and Methods
2.1 Data sets
Histone modification data was obtained from a previous study (Pokholok et al., 2005). We obtained the raw data from the ArrayExpress Archive (http://www.ebi.ac.uk/arrayexpress/) and performed MA2C normalization (Peng et al., 2007). Nucleosome occupancy data was previously published (Kaplan et al., 2009), and the nucleosome occupancy scores as calculated by the authors were used unchanged. PWMs used in this study were obtained from three different sources. The matrices used to produce the set of bound and unbound motifs were taken from (MacIsaac et al., 2006). This was performed in an attempt to be consistent with the matrices that were used by MacIsaac, et al. to produce a map of TF binding across the yeast genome. We used 143 matrices from Badis, et al. and Zhu, et al. (Badis et al., 2008; Zhu et al., 2009) to identify motifs near bound or unbound motifs. Many of the DNA specificity matrices supplied by the authors were position frequency matrices, which we converted to PWMs as described previously (Wasserman and Sandelin, 2004). Gene coordinates were obtained from the UCSC genome browser (http://genome.ucsc.edu/).
2.2 Construction of bound and unbound motif datasets
We obtained the binding locations within Saccharomyces cerevisiae intergenic regions of 118 TFs that were mapped using ChIP-chip (MacIsaac et al., 2006). MacIsaac, et al. analyzed a previously published ChIP-chip dataset (Harbison et al., 2004). Selected for further study were those TFs with at least 100 experimentally mapped binding sites (n = 12). We used the mapped binding sites generated under the strictest criteria as defined by MacIsaac, et al. Briefly, sites defined as bound required a PWM match of 60% of the maximum possible log-likelihood PWM score, conservation in at least three of four sensu stricto yeast species, and a p-value of less than 0.001 for the probe containing the motif. A recent study using Chip-exo argues the false positive rate for Chip-chip may be as high as 50%(Rhee and Pugh, 2011). By using both motif information and conservation between species MacIsacc, et al was able to identify bound motifs with high confidence.
The bound sites as obtained by MacIsaac et al. are presumably centered at motif occurrences. But the bound sites as listed do not have information we would like to study such as the average strength of the bound motifs. Hence it was necessary to remap motif locations and link motifs with bound sites as identified by MacIsacc, et al. To do so we scanned yeast intergenic regions with the PWMs corresponding to the 12 selected TFs and all occurrences of motifs with a score of 60% or greater maximum log-likelihood score were collected. Motifs that overlapped the experimentally mapped binding sites as defined by MacIsaac et al. were labeled as “bound” motifs. With the restriction of only one bound motif allowed per experimentally mapped site. Given that bound sites as defined by MacIsaac, et al. must include a motif instance, it seems that all experimentally determined bound sites taken from the MacIsaac study should overlap a motif as identified by us. In practice this was largely the case (Supplemental Table 1) with one exception, DIG1. Other experimentally mapped TF binding sites as defined by MacIsaac et al. also do not exhibit complete overlap with a motif as defined by us (Supplemental Table 1 second column). This is likely due to MacIsacc using an older yeast genome build. The 11 TFs retained for further analysis were: REB1, GCN4, MBP1, PHD1, SKN7, STE12, SUT1, SWI4, SWI6, ABF1, and CBF1. Only binding sites as determined by MacIsaac et al. for which we could link a motif were defined as “bound” motifs.
To produce the set of unbound motifs, we obtained ChIP-chip binding data for the 11 TFs used in this study (Harbison et al., 2004). A p-value of binding was assigned to each intergenic region in yeast according to Harbison, et al. A set of unbound motifs was produced by scanning yeast intergenic regions with PWMs corresponding to the 11 TFs; all occurrences of motifs with a 60% or greater maximum log-likelihood score were identified. Motifs found in intergenic regions whose p-value of binding was 0.5 or greater in all experimental conditions studied by Harbison, et al. were labeled as unbound.
To check the robustness of our approach we repeated the analysis shown in Figures 2, 3 and 4 using p-value cutoffs of 0.4 and 0.6 in calling unbound motifs, our results did not change.
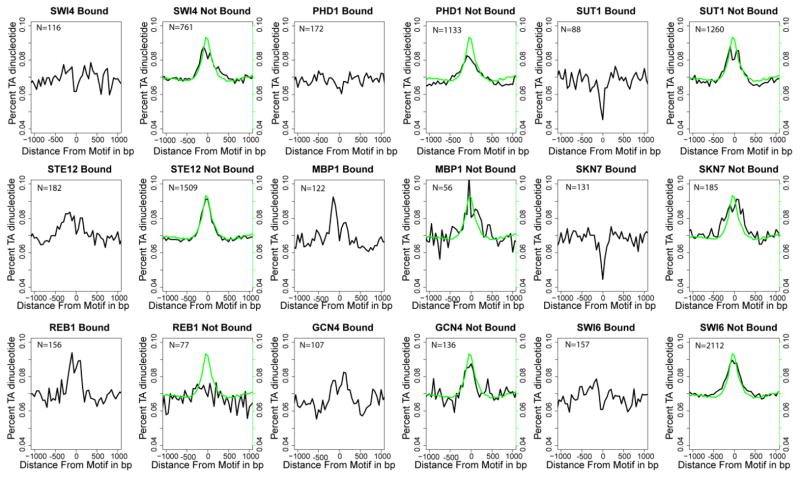
Figure 2. TA dinucleotide content around bound or unbound motifs.
Motifs classified as bound or unbound were aligned. The TA dinucleotide content was binned in 50-bp windows moving upstream and downstream from the motif. Zero on the x-axis represents the center of the aligned motif. Black: The average percentage of TA, which is defined as the fraction of dinucleotides that are TA within each 50 bp window. Green: The background TA content calculated by randomly selecting locations in intergenic regions and repeating the procedure as described.
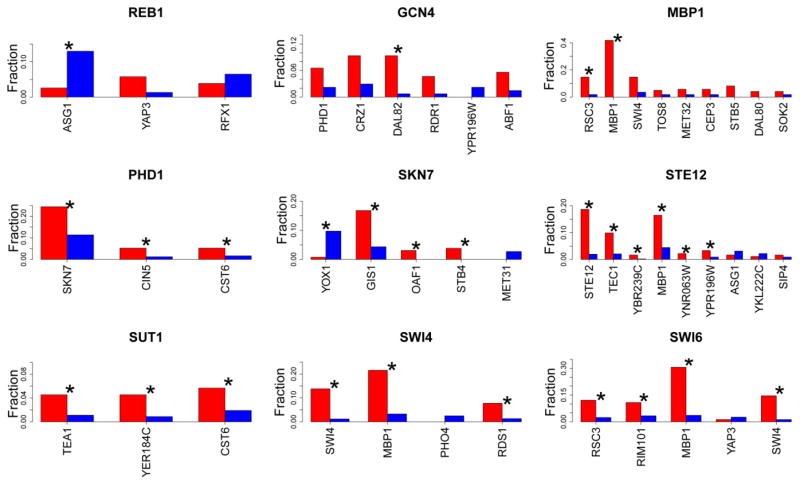
Figure 3. Motifs enriched near bound or unbound motifs.
The fraction of bound (red) or unbound (blue) motifs that exhibit at least one of the labeled motifs within 100 bp is plotted for the nine TFs shown. p-values were calculated using the z-test for two proportions, and corrected for multiple testing using Benjamini, Hochberg, and Yekutieli correction (Benjamini and Yekutieli, 2001). Comparisons with a q-value < 0.05 are marked with an asterisk.
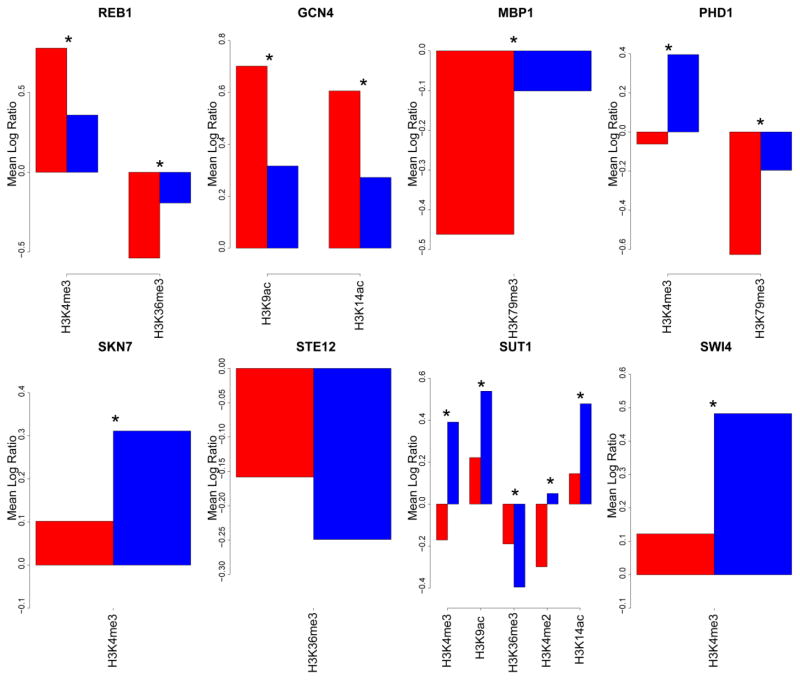
Figure 4. Histone modification-based features.
Histone modification-based features are plotted for the eight TFs for which a histone modification feature was selected as important. Red bars represent the average log ratio of the given histone modification within a 200-bp window centered at bound sites. Blue bars represent the average value of the given nucleosome-based feature within a 200- bp window centered at unbound sites. P-values were calculated using the Wilcox rank sum test, and corrected for multiple testing using the Benjamini, Hochberg, and Yekutieli correction (q-values) (Benjamini and Yekutieli, 2001). Comparisons with a q-value < 0.05 are marked with a asterisk.
2.3 Generation of feature vectors
Generation of nucleosome-based features
Pokholok, et al. (Pokholok et al., 2005) used tiling arrays to map histone modifications in S. cerevisiae. We used this data to calculate the level of histone modification around bound and unbound motifs. For each 200-bp window centered on a motif, we obtained the degree of enrichment by averaging the normalized log ratio values of the probes within that region. For example, the feature “H3K14ac” represents the average degree of acetylation of lysine 14 in histone H3 for the given window. A similar approach was used for each histone modification mark.
To calculate the degree of nucleosome occupancy, we used a dataset produced by Kaplan, et al. (Kaplan et al., 2009). For most positions in the genome, Kaplan and co-authors calculated a nucleosome occupancy score. The average nucleosome occupancy was normalized to zero. A value greater than zero represented nucleosome enrichment relative to the genome-wide average, while a value less than zero signified nucleosome depletion. For each window centered at a motif, nucleosome occupancy was calculated by averaging the nucleosome occupancy scores for that window. Eight features were chromatin-based: “Nucleosome occupancy,” “H3K14ac,” “H3K36me3,” “H3K4me1,” “H3K4me2,” “H3K4me3,” “H3K79me3,” and “H3K9ac.”
Generation of motif-based features
We scripted our own program in Perl to scan yeast intergenic regions with a library of 143 PWMs obtained as described above. Motif matches of 70% or better of the maximum possible log-likelihood score for the given PWM were retained. For each bound and unbound motif for the set of nine TFs analyzed by feature selection, the number of motif matches within 100 bp of every motif represented in our PWM library was calculated. We did not consider any motif match that was within 10 bp of the bound or unbound motifs. 143 out of the 171 features were generated in this fashion. An additional motif-based feature was motif strength, which was simply the log-likelihood PWM score at bound or unbound motifs. Also included in the set of motif-based features was the distance in base pairs to the closest TSS. Finally, a motif-based feature was constructed by calculating the average number of nearby motifs in a 200-bp window centered at every bound or unbound motif; this analysis resulted in a total of 146 motif-based features.
Generation of sequence-based features
Of the 17 sequence-based features, 16 represented the normalized frequency of dinucleotides within a 200-bp window centered at bound or unbound motifs. For example, the feature measuring TA content would be calculated as the number of times the “TA” 2-mer was found within the 200-bp window divided by the number of k-mers of size 2 found in the window. Hence, this feature represents the enrichment of TA relative to all 2-mers. Also included was a sequence-based feature reflecting the overall content. Removing the reverse complement of a given dinucleotide (e.g., CG is the same as GC in the complementary strand) could further reduce the sequence features. Whether the reverse complement is redundant is based on whether strand-specific processes act at bound or unbound motifs. Since TF binding can be strand-specific, reverse complements were retained in the final set of features.
Feature selection
Feature selection can be described as finding the subset of features from the set of all possible combinations of features that can best distinguish classes of interest. In our case, the two classes of interest are bound and unbound motifs. Because the search space of all possible combinations of features grows exponentially with the number of features, it is rarely feasible to perform an exhaustive search. Instead, various heuristic search methods can be used to identify meaningful feature subsets. Here, we used three different feature selection algorithms to identify those features that are consistently selected by the different methods. We used two algorithms implemented in the open source software package ‘weka’ (Hall et al., 2009) and ‘galgo’, an R package (Trevino and Falciani, 2006).
Feature selection consists of two parts: the first are methods to score how well a feature subset predicts the correct class; the second are methods to search the space of all possible feature subsets and achieve convergence to an optimal feature subset.
The first ‘weka’-based feature selection algorithm used was a correlation subset scoring approach paired with a best first search algorithm. Correlation subset scoring is based on the idea that a good subset of features contains those that are highly correlated with the class and yet do not correlate with each other (Hall, 1999). Feature subsets that have this characteristic are scored highly. This scoring function was paired with a best first search method, which searches the space of feature subsets using a greedy hill climbing approach augmented with backtracking.
The second ‘weka’-based feature selection algorithm used was a consistency subset scoring approach paired with a linear forward selection search algorithm. Consistency subset scoring is based on the concept that the best features are those that are most consistent with a class. Thus, good features are consistently similar within a class but very different between classes (Liu and Setiono, 2000). This scoring function was paired with a linear forward search algorithm. Briefly, the search algorithm initially ranks all features individually using the consistency subset scoring method. Then the algorithm starts with an empty set of features and adds them one at a time based on ranking until performance can no longer be improved. The ‘weka’ version does allow for some backtracking and restarting of the search to help prevent quick convergence to small local optima. For more information, see Gutlein, et al. (Gutlein et al., 2009).
‘Galgo’ is a genetic algorithm-based feature selection approach. To score a feature subset, a nearest shrunken centroid classifier is built using only the feature subset. The score assigned to a feature subset represents how well the nearest shrunken centroid classifier performs in classifying held out test datasets of bound or unbound motifs (Trevino and Falciani, 2006).
All of the feature selection approaches used require a training dataset. Unfortunately, training datasets were frequently imbalanced, with far more examples of unbound motifs than bound motifs or vice versa. Many traditional machine learning-based approaches have lower accuracy when trained on imbalanced datasets (Japkowicz and Stephen, 2002). We performed repetitive random under-sampling to deal with the imbalanced dataset problem due to its simplicity and ability to improve performance (Van Hulse et al., 2007; Van Hulse et al., 2009). Randomly removing examples from the majority class until balanced datasets are achieved is a common solution to the data imbalance problem. One drawback to this method is the possibility of discarding potentially useful information. Repetitive random under-sampling attempts to address this issue by combining the results from several rounds of random sampling. Studies have demonstrated that this method can potentially improve performance over simple under-sampling or not performing any sampling (Van Hulse et al., 2009; Xu-Ying et al., 2009).
We will describe our overall approach using the PHD1 dataset as an example. The PHD1 dataset is a highly imbalanced dataset consisting of 172 bound motifs and 1,133 unbound motifs giving a total of 1,305 motifs. For each motif, a vector of length 171 was constructed to produce a matrix with 1,305 rows and 171 columns. Two-thirds of the rows representing bound and unbound motifs were randomly selected and set aside as a training dataset. This resulted in a training dataset with 114 rows representing bound motifs and 755 rows representing unbound motifs for a total dataset matrix of 869 rows with 171 columns. The remaining data was used as the test set. The training dataset then underwent random sampling without replacement selecting rows representing unbound motifs until a balanced dataset was achieved with 114 examples of bound motifs and 114 examples of randomly selected unbound motifs. This matrix was used as an input into the three feature selection methods, and the resulting feature subsets were stored. The process of randomly sampling from the training dataset to create a balanced dataset was repeated 10,000 times. The resulting 10,000 feature subsets were combined by counting the number of times each feature was observed. Features were then ranked based on the number of times each feature was selected. For example, if the “PWM_score” feature was included in 9,000 out of the 10,000 feature subsets, it would rank higher than a feature selected in 1,000 out of the 10,000 feature subsets. Each feature selection method produced a ranked list of features in this manner. Selecting features that were ranked in the top 10% by at least two of the three feature selection methods produced the final subset of features presented in Supplemental Table 1 and Supplemental Table 2. The above approach was used for each of the nine TFs that were analyzed by feature selection.
The features were globally ranked by pooling how often each feature was selected by each of the feature selection algorithms. Those features selected most often across all feature selection algorithms were presumed to be more important than features selected less often. Features listed in order of rank are provided in Supplemental Table 2.
Accuracy, sensitivity, and specificity were calculated by first producing a dataset using only those features selected using the feature selection approach. For example, the training dataset for PHD1 would consist of 869 rows and 16 columns with each column representing one of the 16 features listed in Supplemental Table 1. A balanced dataset was produced by random sampling and the resulting matrix was given as an input training dataset to build a random forest classifier (Breiman, 2001). The resulting classifier then works to correctly predict the class of motifs (bound or unbound) in the testing dataset. This procedure was repeated 10 times. The mean, accuracy, sensitivity, and specificity are presented in Supplemental Table 1. Hence, how well the features selected can discriminate between bound and unbound motifs was assessed on a testing dataset that was not used in feature selection.
3 Results
We obtained experimentally mapped binding sites in intergenic regions for 118 TFs (MacIsaac et al., 2006) in the yeast Saccharomyces cerevisiae genome. We selected twelve TFs for further study since they have at least 100 experimentally mapped binding sites. A cutoff of 100 was used to ensure enough bound sites existed so useful statistics could be performed. For each of these, a set of motifs bound by the TF and a set of motifs likely unbound by the TF were obtained (see Materials and Methods). DIG1’s motif as described by MacIsaac et al. was present in only a minority of the experimentally-proven DIG1 binding sites (n = 41), suggesting the possibility of an error in the position weight matrix (PWM) used. As a result, DIG1 was not further analyzed in this study. For each of the motifs in the datasets, a 200-bp window centered on the motif and a vector containing 171 elements was calculated. Each element of the vector represented a measurement of a biological feature for that window. For example, vector element one is a score indicating the degree of nucleosome occupancy averaged over the 200-bp window. Feature selection was then applied to identify the subset of vector elements (hereby referred to as features) that were most informative in correctly predicting whether a motif is actually bound by the respective TF [for a review of the use of feature selection in bioinformatics see (Saeys et al., 2007)]. We applied three different feature selection techniques (see Materials and Methods), two of which were implemented in ‘weka’ (Hall et al., 2009) while the third is ‘galgo’, (Trevino and Falciani, 2006). Features selected by at least two of the three methods were examined in more detail (Supplemental Table 1). While feature selection can identify a set of promising candidates that differ between bound and unbound motifs, further analyses of the selected features are necessary to verify biologically significant differences. In general, there was good agreement between features selected by all three methods (Supplemental Figure 1).
3.1 Correlation between motif strength and binding
In order to determine which biological features are associated with motifs bound by their TFs, it is necessary to compare the local environment of motifs bound by protein with motifs unbound by protein. Therefore our analysis required obtaining a dataset of motifs that are unlikely to be bound by a TF. We created this set for the 11 TFs examined from a published Chip-chip dataset (Harbison et al., 2004), in which a p-value was calculated representing the degree of evidence regarding binding to each intergenic region in the yeast genome (in general the lower the p-value the stronger the ChIP-chip evidence the given TF binds somewhere in the intergenic region). Our unbound motif dataset consisted of motifs that occurred in intergenic regions with a p-value greater than 0.5 in all experimental conditions studied by Harbison et al. Using these criteria, we discovered that two out of the 11 TFs, CBF1 and ABF1, exhibited too few non-bound motif matches (8 and 38, respectively); therefore, these TFs were eliminated from further feature selection analysis.
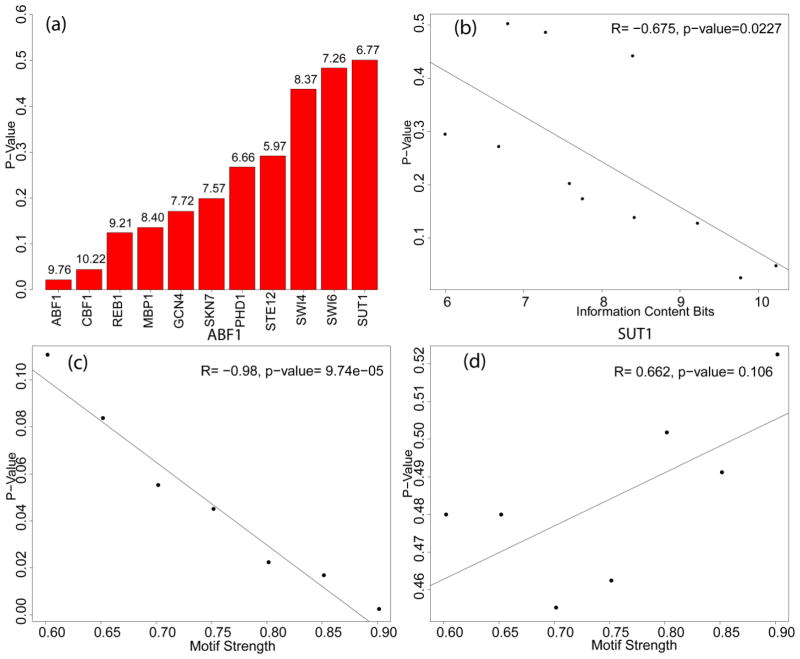
To further explore the relationship between the presence of a motif match and the p-value for binding as measured by Harbison, et al., we plotted the average p-values for intergenic regions that contain strong motif matches (i.e., matches > 80% of the maximum possible PWM log likelihood score; Figure 1, panel a). Although there was low information content for the majority of the motifs, the presence of a strong motif was a surprisingly good predictor of binding for many of the TFs (Figure 1, panel a). ABF1 and CBF1 had the two lowest average p-values for binding 0.022 and 0.044, respectively.
Figure 1. Correlation between motif strength and p-value of binding.
(a) Plotted is the mean p-value of binding for intergenic regions whose average motif strength was >80% of the maximum possible log-likelihood score. The p-value of binding was obtained from (Harbison et al., 2004). The number above each bar is the information content for the given motif in bits. The smaller the information content, the more likely that motif is to occur by random chance in a sequence. (b) For every motif, the average p-value of binding in intergenic regions containing high scoring motifs was calculated as described above (y-axis). The x-axis is the information content of the motifs in bits. (c and d) Plots of the p-value of binding versus motif strength for (c) ABF1 and (d) SUT1. The x-axis denotes the motif strength of a given TF as a percentage of the maximum possible PWM log-likelihood score. Higher motif strength correlates with closer proximity to the consensus sequence. The average p-value of binding for the collected intergenic regions that met the given motif strength threshold was calculated (y-axis). ABF1 and SUT1 were plotted because they represent the two extremes.
It is reasonable to expect a connection between motifs with high information content and a higher probability of binding to a strong motif. Indeed, a positive correlation between information content and p-value for binding to strong motifs (r = −0.67, p-value = 0.02) was observed (Figure 1, panel b). Next, we plotted the average p-value for binding at different motif strengths for ABF1 and SUT1 the two extreme cases (Figure 1, panels c and d). ABF1 showed an almost perfect correlation between motif strength and p-value (r = −0.98, p-value = 0.00009). In contrast, a positive correlation was found for SUT1 (r = 0.66, p-value = 0.1055); however, this correlation was not statistically significant (alpha = 0.05).
3.2 Comparison of sequence-based features surrounding bound and unbound motifs
Some of the features assessed by the feature selection algorithms were sequence-based (e.g., dinucleotide content, see Supplemental Table 1). To explore this further, we plotted the percentage of dinucleotides surrounding bound and unbound motifs (Figures 2 and Supplemental Figure 3). In this analysis, we masked the actual motif and 15 bp flanking both sides. The percentage of the dinucleotide TA present near bound and unbound motifs was calculated as the ratio of TAs present in a given sequence (Figure 2). Out of the 16 dinucleotides examined, TA was selected by our feature selection approach for all nine TFs as an important feature that discriminates between bound and unbound motifs (Supplemental Table 1, Supplemental Table 2). The peak observed in the control dataset is due to the fact that intergenic regions are TA-rich compared to coding regions (Figure 2, green line).
In general, the sequence surrounding bound motifs was depleted of TA dinucleotides compared to unbound motifs (Figure 2) with six (SWI4, PHD1, SKN7, SUT1, STE12, and SWI6) out of the nine TFs clearly showing this pattern. Two TFs, MBP1 and GCN4, did not exhibit strong differences in the percentage of TA between bound and unbound motifs. REB1 showed slightly higher levels of the TA dinucleotide around bound sites compared to unbound sites. SWI6 may not bind DNA directly but instead be recruited to the genes it regulates by other TFs. It is known that SWI4 and MBP1 can bind to DNA as a complex with SWI6 (Andrews and Moore, 1992; Leem et al., 1998). Given the indirect binding of SWI6 to DNA, it is likely the motif identified for SWI6 is a combination of the motifs recognized by the proteins that recruit SWI6 to DNA. Indeed, the core SWI6 motif CGCG is found in both the SWI4 motif and the MBP1 motif (Supplemental Table 3). Furthermore, the local TA dinucleotide content surrounding the bound SWI6 motif closely resembles that of bound SWI4 motifs (Figure 2). Many genes have a tendency to be regulated by multiple TFs. Hence it is possible that the differences in sequence composition when comparing bound to unbound motifs is due to bound motifs having multiple other motifs nearby which may affect local sequence composition. To control for this we obtained all motifs with some evidence of being bound by protein (MacIsaac et al., 2006) and masked these motifs.
Additionally, a number of TFs show the same general trend with regards to differences in sequence composition when comparing bound to unbound motifs. The example given above is the six TFs that all show the same pattern of depleted TA dinucleotides around bound motifs compared to unbound. Since it is unlikely these six TFs all share the same regulatory partners, it is unlikely that the same pattern of depleted TA dinucleotides is due to the potential confounding effect of having multiple other bound TF motifs nearby The same motifs will likely not be present around all six TFs since they do not share the same regulatory partners (see Figure 3).
Several studies have shown TFs in yeast exhibiting distinct positional preferences relative to the transcription start site (TSS) (Harbison et al., 2004; Hansen et al., 2010; Lin et al., 2010). Thus, it is possible that the differences in dinucleotide content are due to bound motifs predominantly occurring −100 to −500 bp upstream of the TSS (Harbison et al., 2004). To investigate this, we extracted the noncoding sequence −100 to −500 bp upstream of all yeast TSSs and calculated the TA dinucleotide content for these regions. The percentage of TA was slightly lower in sequences −100 to −500 bp upstream of the TSS than in intergenic regions as a whole (0.091 compared to 0.099). However, this phenomenon was insufficient to explain the pronounced depletion of TA around bound motifs found for many of the TFs (Figure 2). For example, the average percentage of TA within a 200-bp window centered at bound SUT1 motifs was 0.054. Hence, reduced TA content is not universal throughout promoter regions, but is instead generally found in sequences surrounding motifs bound by TFs. Additionally TFs, in general share, similar location binding preferences, but do not always share the same pattern of dinucleotide frequency around bound motifs. For example PHD1 prefers to bind on average ~340 bp upstream of the TSS while SWI4 prefers to bind on average ~380 bps upstream of a TSS. PHD1’s bound motifs in general are not embedded in GG rich sequence, while SWI4s motifs are (Supplemental Figure 2).
While for TA the trend is for bound motifs to be embedded in TA depleted sequence compared to unbound motifs, this is not the case for other dinucleotides. For example, the GG dinucleotide shows a tendency to be enriched around bound motifs relative to unbound motifs (Supplemental Figure 2). In general, the dinucleotide content of unbound motifs was similar to the overall background intergenic content, while the dinucleotide content around bound motifs was either enriched or depleted relative to background (with the exception of REB1). Given the apparent strong dependence of ABF1 on its motif, we examined the dinucleotide content surrounding its bound motifs relative to background (Supplemental Figure 2). Contrary to the trend observed for many of the other nine TFs, the dinucleotide content surrounding bound ABF1-specific motifs does not show strong deviations from the background dinucleotide content.
3.3 Comparison of motif-based features surrounding bound and unbound motifs
For all nine TFs, the feature selection algorithms selected the distance from the motif to the nearest TSS as a significant discriminator between bound and unbound motifs. Many of the TFs exhibited a striking difference between bound and unbound motifs. For instance, the median distance to the nearest TSS for PHD1 was −430 and −155 for bound and unbound motifs, respectively. This result was expected given the strong positional preference relative to the TSS seen for many yeast TFs (Harbison et al., 2004; Tharakaraman et al., 2005; Kim et al., 2008; Hansen et al., 2010; Lin et al., 2010).
Included in the set of motif-based features were a number of characteristics designed to take advantage of the fact that TFs have a tendency to bind in clusters (Ptashne, 1988; Frith et al., 2003). To construct these features, we obtained a library of PWMs representing 143 TFs (Badis et al., 2008; Zhu et al., 2009). For each bound and unbound motif, we counted the number of motif matches within 100 bp for each of the TFs in our PWM library. By comparing the number of motif matches near bound and unbound sites, we identified motifs that are commonly found near bound and unbound motifs (Figure 3). Interestingly, three TFs (MBP1, STE12, and SWI4) bound motifs had a tendency to have repeated copies of their motifs surrounding their binding sites. Such homotypic clusters of the same motif have been observed in other organisms including vertebrates and invertebrates (Lifanov et al., 2003; Gotea et al., 2010). As our results and others (Harbison et al., 2004) have shown, homotypic clustering is also present in yeast, suggesting an evolutionarily conserved regulatory mechanism.
Regulation of transcription initiation is facilitated by the binding of multiple TFs to the promoter region of a gene. Indeed many of the TFs whose motifs are enriched at bound sites relative to unbound sites showed signs of cooperative binding. For example, in budding yeast numerous genes are induced early in the cell cycle with SWI4 and MBP1 as the predominant regulators of these genes (Koch et al., 1993; Sidorova and Breeden, 1993). In some cases these genes cooperate in regulating the same gene (Bean et al., 2005). Unsurprisingly we observed enrichment of MBP1 motifs surrounding SW4-bound motifs compared to unbound motifs. SWI4 motifs were also enriched around MBP1-bound motifs compared to unbound motifs; however, this enrichment (q-value = 0.07) did not meet our q-value cutoff of 0.05 (Figure 3). In addition, enrichment of the TEC1 motif near bound STE12 motifs exhibited a similar pattern. STE12 is necessary for the proper regulation of mating, haploid invasion, and pseudohyphal development (Herskowitz, 1995). STE12 binds with TEC1 cooperatively to achieve developmental specificity (Madhani and Fink, 1997), which is consistent with our observation that the TEC1 motif is enriched around STE12-bound motifs compared to unbound ones.
Since TFs have a tendency to bind cooperatively, a greater number of motifs can be found enriched around bound motifs compared to unbound ones. Therefore, the feature measuring the average number of nearby motifs was selected for five out of the nine TFs (MBP1, SKN7, STE12, SWI4, and SWI6), further indicating the tendency for the enrichment of multiple motifs surrounding TF binding sites. However, there were also instances of enrichment of motifs around unbound motifs compared to bound motifs. For example, four of the nine TFs (GCN4, PHD1, SKN7, and SWI4) exhibited statistically significant enrichment of the PHO2 motif around unbound motifs compared to bound ones. This unique enrichment may occur because unbound motifs are generally located within TA- rich regions of the genome (Figure 2). Because the PHO2 motif is TA/AT-rich and information poor, it is not surprising that this motif is widespread across yeast intergenic regions (~32,000 PHO2 motifs in intergenic regions N = 32,562. This widespread occurrence may explain why the PHO2 motif, as well as those of SIG1 and GLN3, is enriched near unbound motifs. To control for the tendency of information poor motifs to be strongly influenced by local sequence context we filtered motifs on the basis of information content. And only counted motifs near bound and not bound motifs with at least 8 bits of information. This filtering removed the PHO2, SIG1 and GLN3 motifs from consideration (Figure 3).
However, the above explanation does not account for all cases of motif enrichment around unbound motifs. For instance, despite lacking in AT/TA dinucleotides, the ASG1 motif is enriched around unbound REB1 and STE12 motifs. Given the enrichment of the ASG1 motif around unbound motifs it is possible that ASG1 may be acting as a repressor.
The average motif strength for bound and unbound motifs (i.e., feature “PWM_score,” Supplemental Table 1 and Supplemental Table 2) was selected for all TFs but SKN7 and SUT1. On average, bound motifs were stronger than unbound motifs.
3.4 Comparison of nucleosome-based features surrounding bound and unbound motifs
REB1 possesses nucleosome-modifying properties and functions to create regions of open chromatin (Chasman et al., 1990). Hence, nucleosome occupancy was selected as an important feature to discriminate between REB1-bound and unbound motifs, with bound motifs located in nucleosome-depleted regions (data not shown). Interestingly, although nucleosome occupancy was selected as an important feature for SKN7, bound sites had a higher nucleosome occupancy score than unbound sites (data not shown). This result is contrary to the overall trend of bound sites occurring predominantly in nucleosome-depleted regions (Kaplan et al., 2009). Nevertheless, a recent study mapping nucleosome occupancy suggested that the presence of SKN7 leads to higher nucleosome occupancy at its binding site (Kaplan et al., 2009).
Several histone post-translational modifications have been associated with either bound or unbound motifs. Often, the level of histone modification is associated closely with gene activity (Pokholok et al., 2005). Hence a correlation between active binding sites and histone modification levels is expected. Surprisingly, we also observed an enrichment of active histone marks reported to be associated with active genes, namely H3K4me3, H3K9ac, and H3K14ac (Pokholok et al., 2005), around unbound motifs. Our data demonstrate that these marks are enriched around unbound motifs for a number of TFs (PHD1, SKN7, SUT1, and SWI4) (Figure 4).
These histone marks are present at the highest levels near the TSS (Pokholok et al., 2005). Consistent with this, higher levels of histone modification can be found around motifs that are closest to the TSS. TFs have a tendency to avoid binding 0 to approximately 100 bp upstream of the TSS (Harbison et al., 2004; Lin et al., 2010). Thus, enrichment of active histone marks around unbound motifs may occur because a larger fraction of these motifs is located within 0 to 100 bp of the TSS. Indeed, our data supports this hypothesis. The percentage of bound sites within 100 bp of the TSS for PHD1, SKN7, SUT1, and SWI4 was 5.24%, 4.58%, 1.14%, and 1.72%, respectively. Meanwhile, the percentage of unbound motifs within 100 bps upstream of the TSS for PHD1, SKN7, SUT1, and SWI4 was 21.27%, 13.51%, 12.78%, and 19.06%, respectively. Because the region 0 to approximately 140 bp upstream of the TSS is free of nucleosomes (Lee et al., 2007; Shivaswamy et al., 2008; Kaplan et al., 2009), motifs located within this region are most likely in an open chromatin configuration and accessible for TF binding, which raises the question what mechanism is repressing binding at these motifs.
4 Discussion
Given the prominent role TFs play in gene regulation throughout the genome, mapping TF binding is very important to gaining a thorough understanding of transcription. In recent years, ChIP-chip and ChIP-seq have become widely used experimental tools in identifying binding locations for TFs. Unfortunately, even with these techniques, mapping the binding sites for large numbers of TFs is still a substantial undertaking. Computational prediction of TF binding sites has the potential to provide high-quality predictions of TF binding with precision and low cost. Indeed this has been an active area of research for computational biologists (Elemento and Tavazoie, 2005; Xie et al., 2009; Ernst et al., 2010; Pique-Regi et al., 2011). Our primary goal in this analysis is not prediction of TF binding site but identifying differences in local genomic context comparing bound motifs to unbound motifs.
We examined a subset of yeast TFs so our results cannot be generalized across all TFs. While it is true the total number of TFs studied was low in comparison to the total number of TFs in yeast. There was high diversity in the DNA binding domains; the nine different TFs represent six different DNA binding domain families (Supplemental Table 3). There were several broad trends that were universal or mostly universal across all TFs studied. For example, every TF examined in this study showed differences in local sequence composition around bound motifs compared to unbound motifs. With the sequence composition surrounding unbound motifs in general corresponding to the background sequence composition.
Experimental data suggests local sequence content may be important in transcription factor binding site functioning (Starr et al., 1995; Meierhans et al., 1997; Ponomarenko et al., 1999). Our results are consistent with these findings. However it is not apparent what role sequence context plays in transcription factor binding. In some cases it is clear that sequence context plays a direct role in stabilizing binding (Starr et al., 1995; Meierhans et al., 1997). It is also possible that sequence context is important indirectly though mediating nucleosome binding. Indeed nucleosome occupancy around TF binding sites is depleted of nucleosomes in vitro strongly suggesting sequence context plays a role in excluding nucleosomes (Kaplan et al., 2009). Interestingly, sequences containing 9–11 bp periodic TA dinucleotides have recently been shown in vitro to have strong nucleosome forming potential (Takasuka and Stein, 2010).
The TA dinucleotide was identified by our approach as being important for all 9 TFs in distinguishing between bound and unbound motifs. In general sequences around bound motifs were depleted of the TA dinucleotide compared to unbound motifs (Figure 2); this effect is stronger for some TFs than others. An exception to this general trend is the REB1 motif which showed increased TA frequency around bound motifs compared to unbound motifs. The REB1 protein has chromatin modifying properties with the ability to form nucleosome free regions (Chasman et al., 1990). Indeed if TA dinucleotides in certain sequence contexts increase nucleosome formation, the depletion of TA dinucleotides around bound motifs we observe would presumably discourage nucleosome formation.
It is however unlikely that the differences in sequence context comparing bound to unbound motifs are entirely explained by nucleosome sequence preferences. The TFs examined do not always exhibit consistent sequence preferences. For example SWI4, SUT1 and SKN7 all have enriched GG dinucleotide content around bound motifs while STE12 and PHD1 do not (Supplemental figure 3). Nucleosome sequence preferences should theoretically be consistent within a cell, hence if the differences in sequence composition observed is entirely due to nucleosome sequence preference this preference would be expected to be consistent for all TFs.
Another possible explanation for the differences in sequence composition comparing bound to unbound motifs is direct stabilization of binding. The recognition of binding locations by DNA binding proteins is dependent on two difference approaches: first nucleotide sequence specific formation of hydrogen bonds and second non base pair specific interactions between the protein body and DNA (Rohs et al., 2009). It has recently been shown that the binding of arginine residues to narrow minor groves is a common mechanism assisting in protein-DNA recognition (Rohs et al., 2009). Differences in sequence composition around an embedded motif could either enhance or inhibit such interactions by affecting the DNA shape or width of the major/minor grove. It would be of interest to measure the binding affinity of the same motif embedded in different sequence contexts.
It is also possible that differences in sequence context between bound and unbound motifs are reflective of differences in sequence composition between regions of regulatory sequence and non-regulatory sequence. While this is a possibility we observed there is little difference in TA dinucleotide composition in promoter sequence compared to background yeast intergenic regions. This is not surprising since in, yeast, intergenic regions are compact with promoter sequence being a large fraction of intergenic sequence. This suggests that the differences in dinucleotide frequency comparing bound to unbound motifs are not due to a general trend observed in regulatory sequence.
Understanding gene regulation is a fundamental question in molecular biology. Many genes are regulated by TFs recognizing and binding to short DNA sequence motifs. In most cases, only subsets of the genomic regions that match TF binding sites are actually bound by the TF in vivo. Thus, it is critical to understand the difference between motifs that are bound and unbound by a given TF. Here, we begin to investigate this question by performing a systematic genome-wide comparison of motifs that are bound in vivo compared to motifs that are unbound. To our knowledge, this is the first such study. Further work could extend the set of biological features being examined. Our analysis cannot answer whether any of the differences we identify are a causal component of TF binding specificity.
For ABF1 and CBF1, our results suggest that the presence of a strong motif is a good predictor of binding in intergenic regions. Both proteins have chromatin-modifying properties (Yarragudi et al., 2004) (Kent et al., 2004). In agreement with our results, genome localization studies indicate that the majority of CBF1 motifs in intergenic regions are most likely bound by the TF (Lee et al., 2002; Kent et al., 2004).
Our results suggest a range of strategies is employed in determining DNA binding specificity. For some TFs (e.g. ABF1 and CBF1 (Figure 1)) the information contained in their motif is apparently sufficient to mostly determine specificity. Little additional information from genomic context is needed. Every place a strong copy of their motif is found it may be likely the protein will bind. We reported previously that the ABF1 motif is strongly biased to occur predominantly in potential regulatory regions. We also showed that the ABF1 motif exhibits a strong positional preference relative to the TSS (Hansen et al., 2010).
For other TFs whose motifs are information poor and found in high abundance throughout the genome the information contained in the motif is not sufficient to determine specificity and input from the local genomic environment may play a dominant role in determining specificity. The majority of TFs may fall somewhere between these two scenarios depending to a greater or lesser extent on genomic context to determine specificity.
It is also likely there is interplay between these two approaches. ABF1 is an abundant general regulatory factor essential to cell growth (Halfter et al., 1989). This factor acts in part by creating a bubble of open chromatin (Yarragudi et al., 2004). In many cases, ABF1 alone is insufficient to activate robust transcription and requires the cooperation of other regulatory factors (Goncalves et al., 1995). A recent study indicates that ABF1 may play an important role in determining chromatin structure throughout the genome, with weaker motifs showing evidence of ABF1 binding and chromatin remodeling (Ganapathi et al., 2011). Genome-wide interaction studies have identified ABF1 to be a network “hub,” suggesting that it plays a central role in gene regulation (Zhang et al., 2006).
Given these results, ABF1 may act in part as an important pioneer TF that binds chromatin and acts to create regions of open chromatin that allows other factors to bind similar to pioneer factors in higher organisms (Zaret et al., 2008). ABF1 could be acting to create a local genomic environment conducive for other TFs to bind, while ABF1’s binding specificity is dependent mostly on the presence of its motif.
Supplementary Material
Highlights.
The bound and unbound motifs of 11 transcription factors (TF) in yeast were studied
Two of the TFs appear to depend mostly on their motifs for specificity
Differences in chromatin structure including histone modifications were found
For every TF, local sequence composition varied comparing bound motifs to unbound
Acknowledgments
This research was supported by the Intramural Research Program of the National Institutes of Health, National Library of Medicine and National Center for Biotechnology Information.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andrews BJ, Moore LA. Interaction of the yeast Swi4 and Swi6 cell cycle regulatory proteins in vitro. Proceedings of the National Academy of Sciences, USA. 1992;89:11852–6. doi: 10.1073/pnas.89.24.11852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badis G, Chan ET, van Bakel H, Pena-Castillo L, Tillo D, Tsui K, Carlson CD, Gossett AJ, Hasinoff MJ, Warren CL, Gebbia M, Talukder S, Yang A, Mnaimneh S, Terterov D, Coburn D, Li Yeo A, Yeo ZX, Clarke ND, Lieb JD, Ansari AZ, Nislow C, Hughes TR. A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Molecular Cell. 2008;32:878–87. doi: 10.1016/j.molcel.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer AL, Hlavacek WS, Unkefer PJ, Mu F. Using sequence-specific chemical and structural properties of DNA to predict transcription factor binding sites. PLoS Computational Biology. 2010;6:e1001007. doi: 10.1371/journal.pcbi.1001007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bean JM, Siggia ED, Cross FR. High functional overlap between MluI cell-cycle box binding factor and Swi4/6 cell-cycle box binding factor in the G1/S transcriptional program in Saccharomyces cerevisiae. Genetics. 2005;171:49–61. doi: 10.1534/genetics.105.044560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Yekutieli D. The Control of the False Discovery Rate in Multiple Testing under Dependency. The Annals of Statistics. 2001;29:1165–1188. [Google Scholar]
- Blanchette M, Tompa M. FootPrinter: A program designed for phylogenetic footprinting. Nucleic Acids Research. 2003;31:3840–2. doi: 10.1093/nar/gkg606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman L. Random Forests. Mach Learn. 2001;45:5–32. [Google Scholar]
- Chasman DI, Lue NF, Buchman AR, LaPointe JW, Lorch Y, Kornberg RD. A yeast protein that influences the chromatin structure of UASG and functions as a powerful auxiliary gene activator. Genes & Development. 1990;4:503–14. doi: 10.1101/gad.4.4.503. [DOI] [PubMed] [Google Scholar]
- Cherry JM, Hong EL, Amundsen C, Balakrishnan R, Binkley G, Chan ET, Christie KR, Costanzo MC, Dwight SS, Engel SR, Fisk DG, Hirschman JE, Hitz BC, Karra K, Krieger CJ, Miyasato SR, Nash RS, Park J, Skrzypek MS, Simison M, Weng S, Wong ED. Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Research. 2012;40:D700–5. doi: 10.1093/nar/gkr1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Haeseleer P. What are DNA sequence motifs? Nature Biotechnology. 2006;24:423–5. doi: 10.1038/nbt0406-423. [DOI] [PubMed] [Google Scholar]
- Elemento O, Tavazoie S. Fast and systematic genome-wide discovery of conserved regulatory elements using a non-alignment based approach. Genome Biology. 2005;6:R18. doi: 10.1186/gb-2005-6-2-r18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Plasterer HL, Simon I, Bar-Joseph Z. Integrating multiple evidence sources to predict transcription factor binding in the human genome. Genome Research. 2010;20:526–36. doi: 10.1101/gr.096305.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fickett JW. Quantitative discrimination of MEF2 sites. Molecular and Cellular Biology. 1996;16:437–41. doi: 10.1128/mcb.16.1.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frith MC, Li MC, Weng Z. Cluster-Buster: Finding dense clusters of motifs in DNA sequences. Nucleic Acids Research. 2003;31:3666–8. doi: 10.1093/nar/gkg540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganapathi M, Palumbo MJ, Ansari SA, He Q, Tsui K, Nislow C, Morse RH. Extensive role of the general regulatory factors, Abf1 and Rap1, in determining genome-wide chromatin structure in budding yeast. Nucleic Acids Research. 2011;39:2032–44. doi: 10.1093/nar/gkq1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncalves PM, Griffioen G, Minnee R, Bosma M, Kraakman LS, Mager WH, Planta RJ. Transcription activation of yeast ribosomal protein genes requires additional elements apart from binding sites for Abf1p or Rap1p. Nucleic Acids Research. 1995;23:1475–80. doi: 10.1093/nar/23.9.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotea V, Visel A, Westlund JM, Nobrega MA, Pennacchio LA, Ovcharenko I. Homotypic clusters of transcription factor binding sites are a key component of human promoters and enhancers. Genome Research. 2010;20:565–77. doi: 10.1101/gr.104471.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutlein M, Frank E, Hall M, Karwath A. Large-scale attribute selection using wrappers, Computational Intelligence and Data Mining, 2009. CIDM ‘09 IEEE Symposium on. 2009:332–339. [Google Scholar]
- Halfter H, Kavety B, Vandekerckhove J, Kiefer F, Gallwitz D. Sequence, expression and mutational analysis of BAF1, a transcriptional activator and ARS1-binding protein of the yeast Saccharomyces cerevisiae. EMBO Journal. 1989;8:4265–72. doi: 10.1002/j.1460-2075.1989.tb08612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall M. Correlation-based Feature Subset Selection for Machine Learning. Department of Computer Science, University of Waikato; 1999. [Google Scholar]
- Hall M, Frank E, Holmes G, Pfahringer B, Reutemann P, Witten IH. The WEKA data mining software: an update. SIGKDD Explor Newsl. 2009;11:10–18. [Google Scholar]
- Hansen L, Marino-Ramirez L, Landsman D. Many sequence-specific chromatin modifying protein-binding motifs show strong positional preferences for potential regulatory regions in the Saccharomyces cerevisiae genome. Nucleic Acids Research. 2010;38:1772–9. doi: 10.1093/nar/gkp1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J, Jennings EG, Zeitlinger J, Pokholok DK, Kellis M, Rolfe PA, Takusagawa KT, Lander ES, Gifford DK, Fraenkel E, Young RA. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskowitz I. MAP kinase pathways in yeast: for mating and more. Cell. 1995;80:187–97. doi: 10.1016/0092-8674(95)90402-6. [DOI] [PubMed] [Google Scholar]
- Holloway DT, Kon M, DeLisi C. Integrating genomic data to predict transcription factor binding. Genome Inform. 2005;16:83–94. [PubMed] [Google Scholar]
- Holloway DT, Kon M, Delisi C. Machine learning for regulatory analysis and transcription factor target prediction in yeast. Systems and synthetic biology. 2007;1:25–46. doi: 10.1007/s11693-006-9003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Japkowicz N, Stephen S. The Class Imbalance Problem: A Systematic Study. Intelligent Data Analysis. 2002;6:429–449. [Google Scholar]
- Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, Segal E. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–6. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent NA, Eibert SM, Mellor J. Cbf1p is required for chromatin remodeling at promoter-proximal CACGTG motifs in yeast. Journal of Biological Chemistry. 2004;279:27116–23. doi: 10.1074/jbc.M403818200. [DOI] [PubMed] [Google Scholar]
- Kim NK, Tharakaraman K, Marino-Ramirez L, Spouge JL. Finding sequence motifs with Bayesian models incorporating positional information: an application to transcription factor binding sites. BMC Bioinformatics. 2008;9:262. doi: 10.1186/1471-2105-9-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch C, Moll T, Neuberg M, Ahorn H, Nasmyth K. A role for the transcription factors Mbp1 and Swi4 in progression from G1 to S phase. Science. 1993;261:1551–7. doi: 10.1126/science.8372350. [DOI] [PubMed] [Google Scholar]
- Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, Hannett NM, Harbison CT, Thompson CM, Simon I, Zeitlinger J, Jennings EG, Murray HL, Gordon DB, Ren B, Wyrick JJ, Tagne JB, Volkert TL, Fraenkel E, Gifford DK, Young RA. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- Lee W, Tillo D, Bray N, Morse RH, Davis RW, Hughes TR, Nislow C. A high-resolution atlas of nucleosome occupancy in yeast. Nature Genetics. 2007;39:1235–44. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- Leem SH, Chung CN, Sunwoo Y, Araki H. Meiotic role of SWI6 in Saccharomyces cerevisiae. Nucleic Acids Research. 1998;26:3154–8. doi: 10.1093/nar/26.13.3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifanov AP, Makeev VJ, Nazina AG, Papatsenko DA. Homotypic regulatory clusters in Drosophila. Genome Research. 2003;13:579–88. doi: 10.1101/gr.668403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Wu WS, Liang H, Woo Y, Li WH. The spatial distribution of cis regulatory elements in yeast promoters and its implications for transcriptional regulation. BMC Genomics. 2010;11:581. doi: 10.1186/1471-2164-11-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Setiono R. A Probabilistic Approach to Feature Selection - A Filter Solution. 2000:319–327. [Google Scholar]
- MacIsaac KD, Wang T, Gordon DB, Gifford DK, Stormo GD, Fraenkel E. An improved map of conserved regulatory sites for Saccharomyces cerevisiae. BMC Bioinformatics. 2006;7:113. doi: 10.1186/1471-2105-7-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhani HD, Fink GR. Combinatorial control required for the specificity of yeast MAPK signaling. Science. 1997;275:1314–7. doi: 10.1126/science.275.5304.1314. [DOI] [PubMed] [Google Scholar]
- Meierhans D, Sieber M, Allemann RK. High affinity binding of MEF-2C correlates with DNA bending. Nucleic Acids Research. 1997;25:4537–44. doi: 10.1093/nar/25.22.4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng S, Alekseyenko AA, Larschan E, Kuroda MI, Park PJ. Normalization and experimental design for ChIP-chip data. BMC Bioinformatics. 2007;8:219. doi: 10.1186/1471-2105-8-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pique-Regi R, Degner JF, Pai AA, Gaffney DJ, Gilad Y, Pritchard JK. Accurate inference of transcription factor binding from DNA sequence and chromatin accessibility data. Genome Research. 2011;21:447–55. doi: 10.1101/gr.112623.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, Zeitlinger J, Lewitter F, Gifford DK, Young RA. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–27. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- Ponomarenko JV, Ponomarenko MP, Frolov AS, Vorobyev DG, Overton GC, Kolchanov NA. Conformational and physicochemical DNA features specific for transcription factor binding sites. Bioinformatics. 1999;15:654–68. doi: 10.1093/bioinformatics/15.7.654. [DOI] [PubMed] [Google Scholar]
- Ptashne M. How eukaryotic transcriptional activators work. Nature. 1988;335:683–9. doi: 10.1038/335683a0. [DOI] [PubMed] [Google Scholar]
- Rhee HS, Pugh BF. Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell. 2011;147:1408–19. doi: 10.1016/j.cell.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohs R, West SM, Sosinsky A, Liu P, Mann RS, Honig B. The role of DNA shape in protein-DNA recognition. Nature. 2009;461:1248–53. doi: 10.1038/nature08473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeys Y, Inza I, Larranaga P. A review of feature selection techniques in bioinformatics. Bioinformatics. 2007;23:2507–17. doi: 10.1093/bioinformatics/btm344. [DOI] [PubMed] [Google Scholar]
- Shivaswamy S, Bhinge A, Zhao Y, Jones S, Hirst M, Iyer VR. Dynamic remodeling of individual nucleosomes across a eukaryotic genome in response to transcriptional perturbation. PLoS Biology. 2008;6:e65. doi: 10.1371/journal.pbio.0060065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidorova J, Breeden L. Analysis of the SWI4/SWI6 protein complex, which directs G1/S-specific transcription in Saccharomyces cerevisiae. Molecular and Cellular Biology. 1993;13:1069–77. doi: 10.1128/mcb.13.2.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr DB, Hoopes BC, Hawley DK. DNA bending is an important component of site-specific recognition by the TATA binding protein. Journal of Molecular Biology. 1995;250:434–46. doi: 10.1006/jmbi.1995.0388. [DOI] [PubMed] [Google Scholar]
- Takasuka TE, Stein A. Direct measurements of the nucleosome-forming preferences of periodic DNA motifs challenge established models. Nucleic Acids Research. 2010;38:5672–80. doi: 10.1093/nar/gkq279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharakaraman K, Bodenreider O, Landsman D, Spouge JL, Marino-Ramirez L. The biological function of some human transcription factor binding motifs varies with position relative to the transcription start site. Nucleic Acids Research. 2008;36:2777–86. doi: 10.1093/nar/gkn137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharakaraman K, Marino-Ramirez L, Sheetlin S, Landsman D, Spouge JL. Alignments anchored on genomic landmarks can aid in the identification of regulatory elements. Bioinformatics. 2005;21(Suppl 1):i440–8. doi: 10.1093/bioinformatics/bti1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevino V, Falciani F. GALGO: an R package for multivariate variable selection using genetic algorithms. Bioinformatics. 2006;22:1154–6. doi: 10.1093/bioinformatics/btl074. [DOI] [PubMed] [Google Scholar]
- Van Hulse J, Khoshgoftaar T, Napolitano A. ICML ‘07: Proceedings of the 24th international conference on Machine learning. ACM, Corvalis; Oregon: 2007. Experimental perspectives on learning from imbalanced data; pp. 935–942. [Google Scholar]
- Van Hulse J, Khoshgoftaar TM, Napolitano A. An empirical comparison of repetitive undersampling techniques, Information Reuse & Integration, 2009; IRI ‘09 IEEE International Conference on; 2009. pp. 29–34. [Google Scholar]
- Wasserman WW, Sandelin A. Applied bioinformatics for the identification of regulatory elements. Nature Reviews: Genetics. 2004;5:276–87. doi: 10.1038/nrg1315. [DOI] [PubMed] [Google Scholar]
- Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–45. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Rigor P, Baldi P. MotifMap: a human genome-wide map of candidate regulatory motif sites. Bioinformatics. 2009;25:167–74. doi: 10.1093/bioinformatics/btn605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu-Ying L, Jianxin W, Zhi-Hua Z. Exploratory Undersampling for Class-Imbalance Learning. Systems, Man, and Cybernetics, Part B: Cybernetics, IEEE Transactions on. 2009;39:539–550. doi: 10.1109/TSMCB.2008.2007853. [DOI] [PubMed] [Google Scholar]
- Yarragudi A, Miyake T, Li R, Morse RH. Comparison of ABF1 and RAP1 in chromatin opening and transactivator potentiation in the budding yeast Saccharomyces cerevisiae. Molecular and Cellular Biology. 2004;24:9152–64. doi: 10.1128/MCB.24.20.9152-9164.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS, Watts J, Xu J, Wandzioch E, Smale ST, Sekiya T. Pioneer factors, genetic competence, and inductive signaling: programming liver and pancreas progenitors from the endoderm. Cold Spring Harbor Symposia on Quantitative Biology. 2008;73:119–26. doi: 10.1101/sqb.2008.73.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Liu C, Skogerbo G, Zhu X, Lu H, Chen L, Shi B, Zhang Y, Wang J, Wu T, Chen R. Dynamic changes in subgraph preference profiles of crucial transcription factors. PLoS Computational Biology. 2006;2:e47. doi: 10.1371/journal.pcbi.0020047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Byers KJ, McCord RP, Shi Z, Berger MF, Newburger DE, Saulrieta K, Smith Z, Shah MV, Radhakrishnan M, Philippakis AA, Hu Y, De Masi F, Pacek M, Rolfs A, Murthy T, Labaer J, Bulyk ML. High-resolution DNA-binding specificity analysis of yeast transcription factors. Genome Research. 2009;19:556–66. doi: 10.1101/gr.090233.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.