Abstract
The WWOX tumor suppressor participates in a diverse array of cellular activities by virtue of its ability to recognize WBP1 and WBP2 signaling adaptors among a wide variety of other ligands. Herein, using a multitude of biophysical techniques, we provide evidence that while the WW1 domain of WWOX binds to PPXY motifs within WBP1 and WBP2 in a physiologically-relevant manner, the WW2 domain exhibits no affinity toward any of these PPXY motifs. Importantly, our data suggest that while R25/W44 residues located within the binding pocket of triple-stranded β-fold of WW1 domain are critical for the recognition of PPXY ligands, they are replaced by the chemically-distinct E66/Y85 duo at structurally-equivalent positions within the WW2 domain, thereby accounting for its failure to bind PPXY ligands. Predictably, introduction of E66R/Y85W double-substitution within the WW2 domain not only results in gain-of-function but the resulting engineered domain, hereinafter referred to as WW2_RW, also appears to be a much stronger binding partner of WBP1 and WBP2 than the wild type WW1 domain. We also show that while the WW1 domain is structurally disordered and folds upon ligand binding, the WW2 domain not only adopts a fully structured conformation but also aids stabilization and ligand binding to WW1 domain. This salient observation implies that the WW2 domain likely serves as a chaperone to augment the physiological function of WW1 domain within WWOX. Collectively, our study lays the groundwork for understanding the molecular basis of a key protein-protein interaction pertinent to human health and disease.
Keywords: WW-ligand thermodynamics, WW domain chaperone, WW domain engineering, WW tandem domains
INTRODUCTION
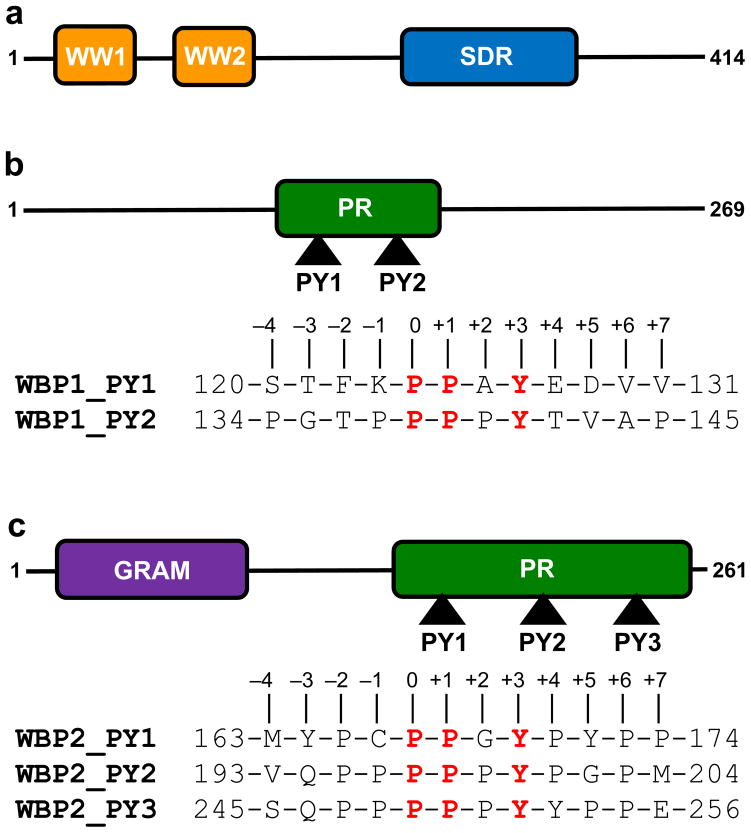
The WWOX tumor suppressor orchestrates a diverse array of cellular activities, including growth, proliferation, apoptosis and tumor suppression (1–4). In particular, the WWOX gene spans the 16q23 fragile chromosomal region involved in cancer. Tellingly, aberrant expression of WWOX tumor suppressor is believed to be linked to the progression of many forms of cancer, including that of breast and prostate (5–11). Moreover, disruption of WWOX gene in mice results in defective metabolism, impaired growth and post-natal lethality, implying that WWOX serves a non-redundant role that cannot be compensated by other cellular proteins (12–14). This argument is further supported by the fact that WWOX is expressed in several distinct isoforms across various tissues, thereby further complicating its role in cellular signaling networks central to health and disease. Importantly, the modular architecture of WWOX tumor suppressor, comprised of a tandem copy of WW domains ( designated WW1 and WW2) located N-terminal to the short-chain dehydrogenase/reductase (SDR) domain (Figure 1a), exquisitely befits its role as an interacting partner of a diverse array of ligands such as p73, RUNX, AP2 and NF-κB transcription factors as well as many other cellular proteins including SIMPLE, ErbB4, Ezrin and WBP1 (1, 15–18). The WWOX-ligand interactions are believed to be largely driven by the binding of proline-rich PPXY motifs found within potential ligands to the WW domains of WWOX in a canonical manner (19–21). Notably, many of these ligands also bind to the YAP transcriptional regulator and, in so doing, mediate the Hippo tumor suppressor pathway involved in regulating the size of organs and in the suppression of tumors through inhibiting cellular proliferation and promoting apoptosis (22–29). For example, both YAP and WWOX bind to PPXY motifs located within the cytoplasmic tails of ErbB4 receptor tyrosine kinase (17, 23). Thus, while YAP-ErbB4 interaction upregulates transactivation function of ErbB4, WWOX-ErbB4 interaction results in transcriptional suppression. Accordingly, the ability or lack thereof of WWOX to compete with YAP for cellular partners may be a defining event in the development of cancer. Despite such physiological significance, the mechanisms underlying the specificity of WWOX-ligand interactions remain largely elusive. In an effort toward this goal, we undertook here detailed biophysical analysis of the binding of WW domains of WWOX to PPXY-peptides derived from WBP1 and WBP2 adaptors (Figures 1b and 1c).
Figure 1.
Modular organization of human WWOX tumor suppressor and human WBP proteins. (a) WWOX is comprised of a tandem copy of WW domains, designated WW1 and WW2, located N-terminal to the short-chain dehydrogenase/reductase (SDR) domain. (b) WBP1 contains a central proline-rich (PR) domain flanked between long stretches of uncharacterized regions. The PR domain of WBP1 contains two PPXY motifs, designated PY1 and PY2. (c) WBP2 contains the GRAM domain located N-terminal to the proline-rich (PR) domain. The PR domain of WBP2 contains three PPXY motifs, designated PY1, PY2 and PY3. Note that the amino acid sequences of peptides containing the PPXY motifs and flanking residues within both WBP1 and WBP2 are provided. The numerals indicate the nomenclature used in this study to distinguish residues within and flanking the motifs relative to the first proline within the PPXY motifs, which is arbitrarily assigned zero.
RESULTS and DISCUSSION
WWOX physically interacts with WBP2 via its WW1 domain
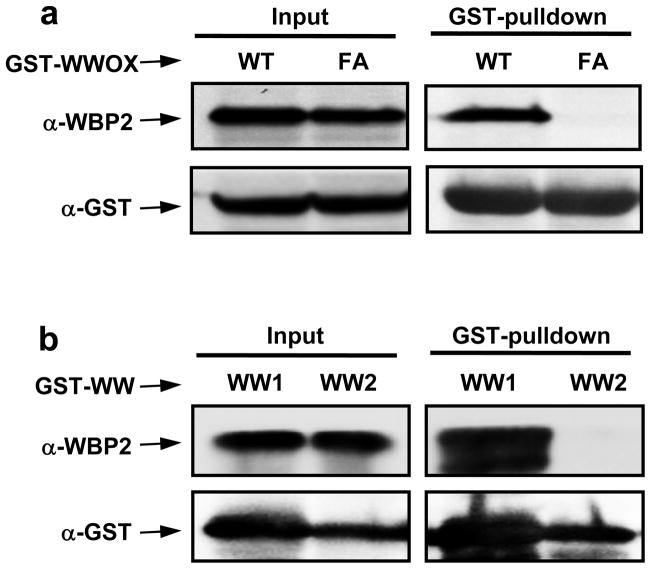
It has been previously reported that WWOX binds to WBP1 via the classical WW-PPXY interaction (18). To examine whether WWOX physically interacts with WBP2 and to what extent this interaction is mediated by the WW1 domain, we conducted GST-pulldown and Western blot analysis on WWOX-WBP2 interaction in mammalian cells (Figure 2a). More specifically, we analyzed and compared the binding of wild type WWOX (WT) and a mutant WWOX containing the W44F/P47A double-substitution (FA). We note that the W44/P47 residues, located within the WW1 domain of WWOX, are essential for its structural integrity and ligand binding, and the W44F/P47A double-mutation is sufficient to abolish interaction of WWOX with its partners as noted previously (16, 30). Importantly, our analysis reveals that while GST-pulldown of WT construct of WWOX co-precipitates WBP2, the FA mutant construct of WWOX fails to do so (Figure 2a). This observation unequivocally demonstrates that WWOX specifically binds to WBP2 and does so via its WW1 domain. To further demonstrate that WWOX binds WBP2 via its WW1 domain but not WW2 domain, we also conducted the above analysis on isolated WW domains of WWOX (Figure 2b). Consistent with our analysis on full-length WWOX, our data suggest that only WW1 domain co-precipitates WBP2 upon GST-pulldown. Interestingly, previous studies have also indicated that WWOX binds to its cellular partners through WW1 domain (1, 15–18), raising the question as to the physiological role of WW2 domain. We hypothesize that the WW2 domain of WWOX is an orphan WW domain that is devoid of ligand binding capability and that it likely serves as a chaperone to augment the physiological function of WW1 domain within WWOX.
Figure 2.
GST-pulldown and Western blot analysis for WWOX-WBP2 interaction in mammalian cells. (a) HEK293 cells were transiently transfected with WBP2 and either wild type GST-tagged WWOX (WT) or GST-tagged WWOX containing the W44F/P47A double-mutation (FA). After harvesting for 24–30h, SDS-PAGE analysis was conducted on either whole cell lysate (Input) or after GST-pulldown and the resolved bands were detected by immunoblotting with anti-WBP2 (α-WBP2) and anti-GST (α-GST) antibodies. (b) HEK293 cells were transiently transfected with WBP2 and either GST-tagged WW1 domain (WW1) or GST-tagged WW2 domain (WW2) of WWOX. After harvesting for 24–30h, SDS-PAGE analysis was conducted on either whole cell lysate (Input) or after GST-pulldown and the resolved bands were detected by immunoblotting with anti-WBP2 (α-WBP2) and anti-GST (α-GST) antibodies.
WW1 domain but not WW2 domain of WWOX binds to PPXY motifs within WBP1 and WBP2
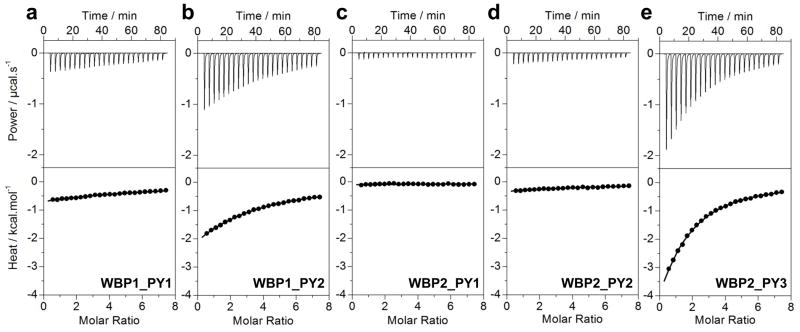
In order to further understand and dissect the role of WW domains in mediating the WWOX-WBP interactions, we analyzed the binding of isolated WW1 and WW2 domains of WWOX to PPXY-peptides derived from potential WWOX-binding sites in WBP1 and WBP2 using ITC (Figure 3 and Table 1). Our data suggest that while the WW1 domain binds to PY2 motif in WBP1 (WBP1_PY2) and PY3 motif in WBP2 (WBP2_PY3) with affinities in the physiologically-relevant micromolar range, the WW2 domain does not appear to interact with any PPXY motifs within either WBP1 or WBP2 within the sensitivity of our measurements. These observations are consistent with our cell-based studies demonstrating that WWOX binds to WBP2 exclusively via its WW1 domain.
Figure 3.
ITC isotherms for the binding of WW1 domain of WWOX to PPXY peptides containing WBP1_PY1 (a), WBP1_PY2 (b), WBP2_PY1 (c), WBP2_PY2 (d) and WBP2_PY3 (e) motifs. The upper panels show the raw ITC data expressed as change in thermal power with respect to time over the period of titration. In the lower panels, change in molar heat is expressed as a function of molar ratio of corresponding peptide to WW1 domain of WWOX. The solid lines in the lower panels show the fit of data to a one-site model, as embodied in Eq [1], using the ORIGIN software.
Table 1.
Thermodynamic parameters obtained from ITC measurements for the binding of WW1 domain of WWOX to various PPXY-peptides derived from WBP1 and WBP2
| Sequence | Kd/μM | ΔH/kcal.mol−1 | TΔS/kcal.mol−1 | ΔG/kcal.mol−1 | |
|---|---|---|---|---|---|
| WBP1_PY1 |
|
NBD | NBD | NBD | NBD |
| WBP1_PY2 |
|
366 ± 23 | −16.14 ± 0.29 | −11.44 ± 0.25 | −4.69 ± 0.04 |
| WBP2_PY1 |
|
NBD | NBD | NBD | NBD |
| WBP2_PY2 |
|
NBD | NBD | NBD | NBD |
| WBP2_PY3 |
|
122 ± 1 | −12.61 ± 0.11 | −7.27 ± 0.12 | −5.34 ± 0.01 |
The PPXY motif is colored blue for clarity. Binding stoichiometries generally agreed to within ±10%. Errors were calculated from at least three independent measurements. All errors are given to one standard deviation. NBD indicates no binding determined for the particular peptides either due to lack of any observable heat change or due to poor accuracy of ITC analysis involving such weak interactions (Kd > 1 mM).
Notably, binding of WW1 domain of WWOX to both WBP1_PY2 and WBP2_PY3 motifs is largely driven by favorable enthalpic forces accompanied by unfavorable entropic changes, implying that the intermolecular interactions such as hydrogen bonding and van der Waals contacts likely account for the specificity of this key protein-protein interaction — a hallmark of molecular recognition underlying WW-ligand interactions in general (19–21). More importantly, the WW1 domain of WWOX binds to WBP2_PY3 motif with an affinity that is three-fold greater than that observed for binding to WBP1_PY2 motif, implying that residues within and flanking the PPXY motifs play an important role in mediating WWOX-WBP interaction. In an attempt to fully understand the role of these residues, we next performed alanine scan on the WBP2_PY3 peptide and measured the binding of each alanine mutant to WW1 domain of WWOX (Table 2). Interestingly, our data reveal that, in addition to consensus residues within the PPXY motif, proline at −2 position and tyrosine at +4 position, according to the nomenclature outlined in Figures 1b and 1c, are required for high-affinity binding, rendering the PXPPXYY motif the optimal consensus sequence for binding to the WW1 domain of WWOX. Consistent with this observation, both WBP1_PY1 and WBP1_PY2 motifs lack proline at −2 position and tyrosine at +4 position, while WBP2_PY1 and WBP2_PY2 motifs lack tyrosine at +4 position. Nonetheless, the lack of either proline at −2 position or tyrosine at +4 position or lack of both of these residues does not necessarily equate to complete lack of binding but rather a reduction in binding affinity as demonstrated in the case of binding of WW1 domain to WBP1_PY2 motif. It is also important to note that although non-consensus residues within and flanking the PXPPXYY motif have little or negligible effect on the binding affinity of WW1 domain, they may be important for stabilizing the polyproline type II (PPII) helical conformation of PPXY peptides as reported previously (31, 32). Thus, for example, both WBP1_PY1 and WBP1_PY2 lack proline at −2 position and tyrosine at +4 position, but while WW1 domain shows apparent binding to the latter motif, no binding is observed to the former. We believe that such differential behavior of WBP1_PY1 and WBP1_PY2 motifs toward WW1 domain most likely resides in the nature of residue at +2 position. In WBP1_PY1 motif, the +2 residue is alanine, while the same position is occupied by proline in WBP1_PY2 motif. Thus, while P+2 may be important for the stabilization of PPII conformation of WBP1_PY2 motif, substitution to alanine as found within the WBP1_PY1 motif is likely to compromise this conformation, thereby compromising its binding to WW1 domain, as noted previously (32).
Table 2.
Thermodynamic parameters obtained from ITC measurements for the binding of WW1 domain of WWOX to single alanine mutants of the WBP2_PY3 peptide
| Sequence | Kd/μM | ΔH/kcal.mol−1 | TΔS/kcal.mol−1 | ΔG/kcal.mol−1 | |
|---|---|---|---|---|---|
| PY3_WT |
|
122 ± 1 | −12.61 ± 0.11 | −7.27 ± 0.12 | −5.34 ± 0.01 |
| PY3_Q-3A |
|
149 ± 1 | −12.43 ± 0.07 | −7.20 ± 0.07 | −5.23 ± 0.01 |
| PY3_P-2A |
|
250 ± 8 | −17.99 ± 0.10 | −13.07 ± 0.08 | −4.92 ± 0.02 |
| PY3_P-1A |
|
126 ± 4 | −8.42 ± 0.24 | −3.09 ± 0.22 | −5.33 ± 0.02 |
| PY3_P0A |
|
NBD | NBD | NBD | NBD |
| PY3_P+1A |
|
636 ± 37 | −3.75 ± 0.27 | +0.61 ± 0.23 | −4.37 ± 0.03 |
| PY3_P+2A |
|
134 ± 1 | −8.52 ± 0.16 | −3.23 ± 0.16 | −5.29 ± 0.01 |
| PY3_Y+3A |
|
554 ± 35 | −4.95 ± 0.10 | −0.50 ± 0.14 | −4.45 ± 0.04 |
| PY3_Y+4A |
|
229 ± 7 | −8.35 ± 0.39 | −3.37 ± 0.37 | −4.97 ± 0.02 |
| PY3_P+5A |
|
98 ± 6 | −14.68 ± 0.31 | −9.21 ± 0.28 | −5.47 ± 0.03 |
| PY3_P+6A |
|
117 ± 2 | −13.01 ± 0.13 | −7.64 ± 0.12 | −5.37 ± 0.01 |
Note that the alanine substitutions within the WBP2_PY3 peptide are colored red and underlined for clarity. Binding stoichiometries generally agreed to within ±10%. Errors were calculated from at least three independent measurements. All errors are given to one standard deviation. NBD indicates no binding determined for the particular peptides either due to lack of any observable heat change or due to poor accuracy of ITC analysis involving such weak interactions (Kd > 1 mM).
Structural models provide physical basis for the binding of WW1 domain but not WW2 domain of WWOX to PPXY motifs within WBP1 and WBP2
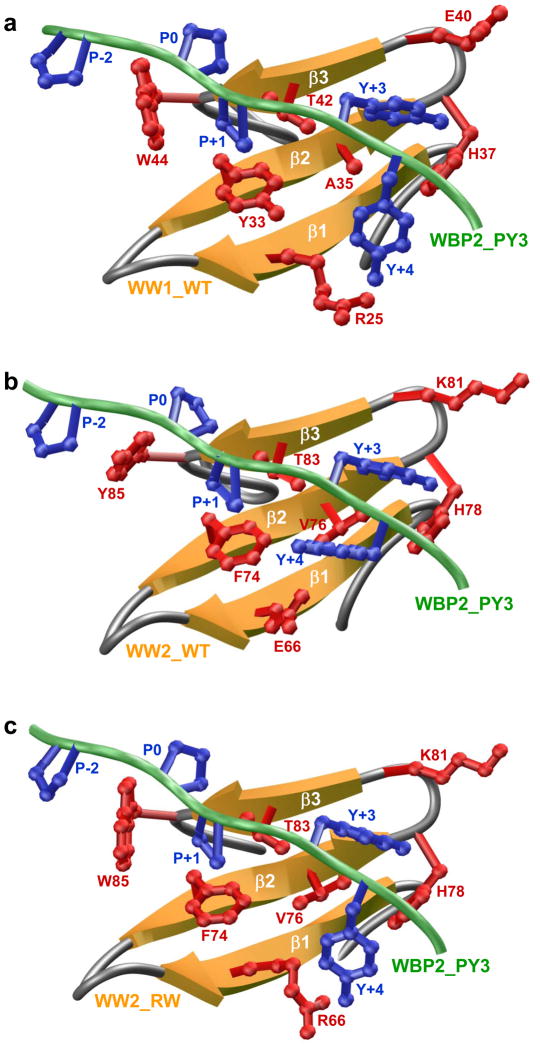
To understand the physical basis of the binding of WW1 domain but not WW2 domain of WWOX to PPXY motifs within WBP1 and WBP2, we built structural models of WW1 and WW2 domains in complex with the WBP2_PY3 peptide (Figures 4a and 4b). Our models show that the PPXY peptide adopts the PPII helical conformation and binds within the hydrophobic groove of the anti-parallel triple-stranded β-sheet fold of the WW1 domain in a canonical manner (31, 33–35). In agreement with our thermodynamic data presented above, the consensus residues within the PXPPXYY motif appear to be engaged in key intermolecular contacts with specific residues lining the hydrophobic groove of the WW1 domain, while non-consensus residues within and flanking the PXPPXYY motif make no discernable contacts (Figure 4a). Notably, the pyrrolidine moieties of P-2 and P0, according to the nomenclature presented in Figures 1b and 1c, stack against and sandwich the indole sidechain of W44 in WW1 domain. Interestingly, W44 is replaced by tyrosine (Y85) at structurally-equivalent position within the WW2 domain (Figure 4b). The chemically-distinguishing features of indole moiety of tryptophan versus phenyl ring of tyrosine imply that the latter cannot substitute in a critical position occupied by tryptophan and doing so would seriously jeopardize van der Waals contacts with the pyrrolidine moities necessary to promote a thermodynamically stable WW-ligand complex.
Figure 4.
Structural models of wild type (WT) and mutant WW domains of WWOX in complex with WBP2_PY3 peptide conatining the PXPPXYY motif. (a) Structural model of WW1_WT domain in complex with WBP2_PY3 peptide. (b) Structural model of WW2_WT domain in complex with WBP2_PY3 peptide. (c) Structural model of WW2_RW mutant domain in complex with WBP2_PY3 peptide. Note that theβ-strands in the WW domains are shown in yellow with loops depicted in gray and the peptide is colored green. The sidechain moities of residues within WW domains engaged in key intermolecular contacts with the peptide are shown in red. The sidechain moieties of residues within the peptide colored blue correspond to the PXPPXYY motif.
The intermolecular association is further stabilized through reverse sandwiching of the pyrrolidine moiety of P+1 within the PXPPXYY motif by the sidechain groups of Y33/T42 in WW1 domain. Remarkably, the Y33/T42 pair of residues lining the hydrophobic groove of WW1 domain appears to be more or less conserved within the WW2 domain (F74/T83). Accordingly, the P+1 residue within the PXPPXYY motif would be expected to be stabilized within the WW2 domain in a manner akin to its stabilization within the WW1 domain. The same argument would also hold true for the Y+3 residue within the PXPPXYY motif. Thus, the phenyl moiety of Y+3 buries deep into the hydrophobic groove and is escorted by sidechain groups of a trio of more or less conserved residues within WW1 (A35/H37/E40) and WW2 (V76/H78/K81) domains. However, major differences are also notable in how WW1 and WW2 domains accommodate the Y+4 residue within the PXPPXYY motif. Thus, while the phenyl moiety of Y+4 appears to be stabilized either via the aliphatic sidechain of R25 and/or via hydrogen bonding between the hydroxyl group of Y+4 and guanidine moiety of R25 within the WW1 domain, substitution of R25 with an acidic residue (E66) at structurally-equivalent position within the WW2 domain likely mitigates such intermolecular association necessary for binding.
Taken collectively, our structural models suggest strongly that the replacement of R25 and W44 within WW1 domain respectively with E66 and Y85 at corresponding structurally-equivalent positions within the WW2 domain most likely accounts for the lack of binding of the latter to PPXY motifs within WBP1 and WBP2 adaptors. Tellingly, our structural model of WW2 domain containing the Y85W/E66R double-substitution, hereinafter referred to as WW2_RW domain, in complex with WBP2_PY3 peptide suggests that binding to PPXY motifs within WBP1 and WBP2 is likely to be recapitulated (Figure 4c).
Engineered WW2_RW Domain of WWOX recognizes PPXY motifs within WBP1 and WBP2 with higher affinity than the wildtype WW1 domain
To test our hypothesis that the lack of binding of WW2 domain of WWOX to PPXY motifs within WBP1 and WBP2 is due to replacement of R25 and W44 within the WW1 domain respectively with E66 and Y85 at structurally-equivalent positions within the WW2 domain, we generated various single and double alanine mutants of WW domains and measured their binding to WBP2_PY3 peptide using ITC (Table 3). First, we generated the R25A (WW1_R25A), R25E (WW1_R25E) and W44Y (WW1_W44Y) single mutants of WW1 domain to test the contributions of R25 and W44 for binding to PPXY motifs. It should be noted that the WW1_R25E and WW1_W44Y mutants mimic the E66 and Y85 residues located at structurally-equivalent positions within the WW2 domain. Interestingly, none of these mutants bound to WBP2_PY3 peptide, implying that R25 and W44 play a critical role in mediating binding to PPXY motifs. Next, we generated E66R (WW2_E66R) and Y85W (WW2_Y85W) single mutants of WW2 domain, so as to mimic their structural equivalents found with the WW1 domain. Neither the WW2_E66R nor the WW2_Y85W mutant bound to WBP2_PY3 peptide (Table 3). Remarkably, the E66R/Y85W double-mutant of WW2 domain (WW2_RW) bound to the WBP2_PY3 peptide with an affinity stronger than that observed for the binding of wildtype WW1 domain (Tables 1 and 3).
Table 3.
Thermodynamic parameters obtained from ITC measurements for the binding of WBP2_PY3 peptide to wild type (WT) and various mutants of WW1 and WW2 domains of WWOX
| Kd/μM | ΔH/kcal.mol−1 | TΔS/kcal.mol−1 | ΔG/kcal.mol−1 | |
|---|---|---|---|---|
| WW1_WT | 122 ± 1 | −12.61 ± 0.11 | −7.27 ± 0.12 | −5.34 ± 0.01 |
| WW1_R25A | NBD | NBD | NBD | NBD |
| WW1_R25E | NBD | NBD | NBD | NBD |
| WW1_W44Y | NBD | NBD | NBD | NBD |
| WW2_WT | NBD | NBD | NBD | NBD |
| WW2_E66R | NBD | NBD | NBD | NBD |
| WW2_Y85W | NBD | NBD | NBD | NBD |
| WW2_RW | 91 ± 2 | −5.20 ± 0.04 | −0.32 ± 0.03 | −5.52 ± 0.01 |
Note that WW2_RW is a double-mutant of WW2 domain containing the E66R and Y85W substitutions. Binding stoichiometries generally agreed to within ±10%. Errors were calculated from at least three independent measurements. All errors are given to one standard deviation. NBD indicates no binding determined for the particular domains either due to lack of any observable heat change or due to poor accuracy of ITC analysis involving such weak interactions (Kd > 1 mM).
To further analyze the ligand binding potential of the engineered WW2_RW Domain, we also measured its binding to all PPXY motifs within WBP1 and WBP2 adaptors. As shown in Table 4, the WW2_RW Domain binds to all but the WBP2_PY1 motif. In summary, structure-based engineering of the WW2 domain with the introduction of E66R/Y85W double-substitution did not only result in gain-of-function but the engineered WW2_RW domain also appears to be a much stronger binding partner of WBP1 and WBP2 than the wild type WW1 domain.
Table 4.
Thermodynamic parameters obtained from ITC measurements for the binding of engineered WW2_RW domain of WWOX to various PPXY-peptides derived from WBP1 and WBP2
| Sequence | Kd/μM | ΔH/kcal.mol−1 | TΔS/kcal.mol−1 | ΔG/kcal.mol−1 | |
|---|---|---|---|---|---|
| WBP1_PY1 |
|
80 ± 2 | −5.59 ± 0.06 | +0.01 ± 0.04 | −5.60 ± 0.02 |
| WBP1_PY2 |
|
123 ± 1 | −12.11 ± 0.20 | −6.77 ± 0.19 | −5.34 ± 0.01 |
| WBP2_PY1 |
|
NBD | NBD | NBD | NBD |
| WBP2_PY2 |
|
69 ± 1 | −8.44 ± 0.06 | −2.75 ± 0.06 | −5.69 ± 0.01 |
| WBP2_PY3 |
|
91 ± 2 | −5.20 ± 0.04 | −0.32 ± 0.03 | −5.52 ± 0.01 |
Note that WW2_RW is a double-mutant of WW2 domain containing the E66R and Y85W substitutions. Binding stoichiometries generally agreed to within ±10%. Errors were calculated from at least three independent measurements. All errors are given to one standard deviation. NBD indicates no binding determined for the particular peptides either due to lack of any observable heat change or due to poor accuracy of ITC analysis involving such weak interactions (Kd > 1 mM).
WW1 domain but not WW2 domain of WWOX appears to be structurally disordered
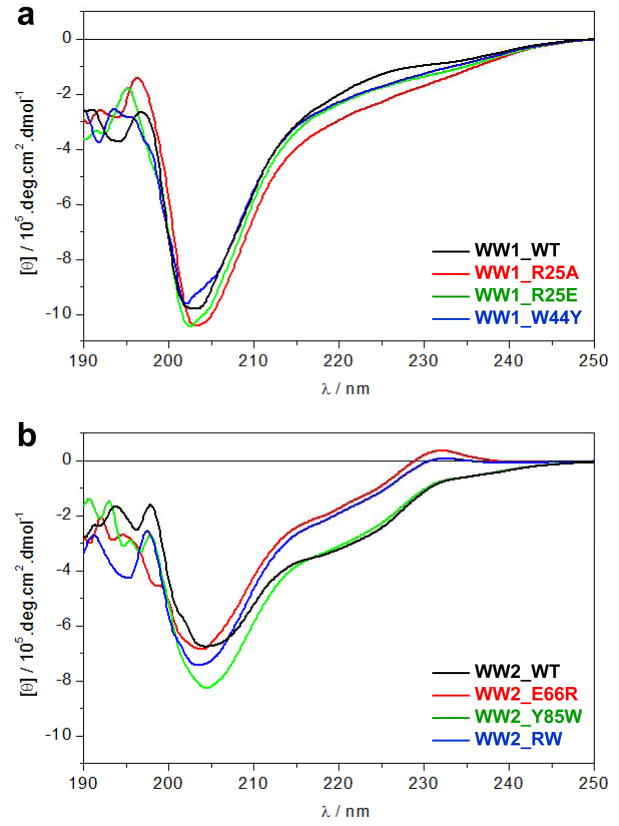
We next wondered whether the lack of binding of wildtype WW2 domain of WWOX to PPXY motifs within WBP1 and WBP2 is in part a manifestaton of its inability to properly fold into a globular conformation best suited for recognizing cognate ligands and to what extent the introduction of E66R/Y85W double-substitution may circumvent this shortcoming. To test our hypothesis, we probed and compared the secondary structures of wildtype WW1 and WW2 domains of WWOX as well as the engineered WW2_RW domain using far-UV CD spectroscopy (Figure 5).
Figure 5.
CD analysis of wild type (WT) and various mutants of WW domains of WWOX. (a) Far-UV spectra of WW1_WT (black), WW1_R25A (red), WW1_R25E (green) and WW1_W44Y (blue). (b) Far-UV spectra of WW2_WT (black), WW2_E66R (red), WW2_Y85W (green) and WW2_RW (blue).
To our surprise, our data suggest that it is the WW1 domain not the WW2 domain that is structurally disordered. Thus, the spectral features of WW1 domain, largely characerized by a negative band centered around 205nm due to random coil contribution, support a relatively unfolded protein (Figure 5a). Furthermore, introduction of R25A, R25E and W44Y substitutions has little or negligible effect on the secondary structure of WW1 domain. In sharp contrast, the WW2 domain also displays a negative band around 205nm due to random coil contribution, but there is also a second negative band around 220nm due to β-sheet contribution, and an additional positive band at around 230nm due to aromatic contribution (Figure 5b). These latter spectral features are characteristic of well-folded WW domains as reported previously (36–38), implying that the wildtype WW2 domain is structurally folded even though it fails to bind to PPXY motifs. Interestingly, while the Y85W substitution appears to destabilize the WW2 domain as marked by an increase in the intensity of 205-nm band, the E66R substitution is accompanied by a decrease in the intensity of 205-nm band coupled with an increase in the intensity of the 220-nm and 230-nm bands, implying that the E66R substitution imparts a noticeable increase in the secondary structural content of WW2 domain. Moreover, the E66R/Y85W double-substitution also results in structural enhancement of WW2 domain in a manner akin to that observed for the E66R substitution alone.
Taken together, our data demonstrate that the lack of structure does not necessarily equate to lack of binding and vice versa in the context of WW-ligand interactions. On the contrary, it is seemingly clear that WW1 domain of WWOX is largely unstructured and undergoes disorder-order transition upon ligand binding to attain a triple-stranded β-sheet fold. Importantly, the presence of a short inter-domain linker between the WW1 and WW2 domains implies that the latter may serve as a structural scaffold or chaperone and aid ligand binding to the WW1 domain in the context of full-length WWOX in agreement with previous studies demonstrating that WWOX binds to its cellular partners through WW1 domain (1, 15–18).
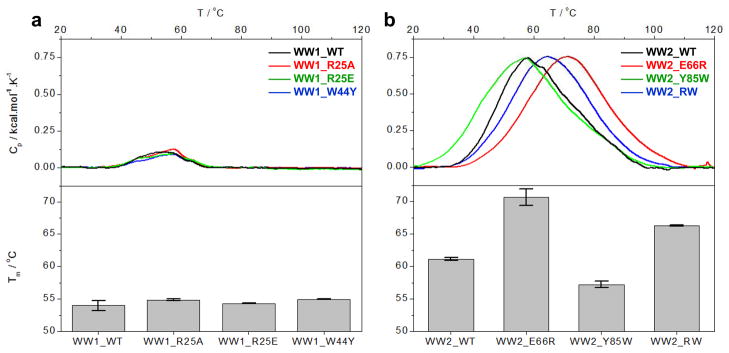
Engineered WW2_RW domain of WWOX displays higher thermal stability than the wildtype WW domains
To further build on our observations that it is the WW2 domain that is structurally folded rather than the WW1 domain and that the E66R/Y85W double-substitution likely results in further folding of the WW2 domain, we next monitored the thermal stability of the wildtype and various mutant WW domains of WWOX using DSC (Figure 6). Our analysis reveals that the unfolding of wildtype WW1 domain is accompanied by a melting tempertaure (Tm) of around 53°C and that the R25A, R25E and W44Y mutations within WW1 domain have little or negligible effect on the Tm value (Figure 6a). In contrast, unfolding of wildtype WW2 domain occurs with a Tm value of around 60°C (Figure 6b), implying that the WW2 domain is thermally more stable than the WW1 domain. Importantly, while the Y85W mutation destabilizes the WW2 domain, the E66R mutation augments thermal stability by about 10°C. Consistent with this observation, the introduction of E66R/Y85W double-substitution also results in the enhancement of thermal stability of WW2 domain but the effect is not as astounding as that observed in the case of E66R mutation alone. Taken together, these data are in an excellent agreement with our far-UV CD analysis and further corroborate the notion that while the WW1 domain is largely devoid of β-sheet content and structurally disordered, the WW2 domain harbors substantial amount of β-sheet content reminiscent of a well-folded WW domain. Notably, the engineered WW2_RW domain is not only structurally more folded than the wildtype WW2 domain but is also thermally more stable.
Figure 6.
DSC analysis of wild type (WT) and various mutants of WW domains of WWOX. (a) DSC analysis of WW1_WT (black), WW1_R25A (red), WW1_R25E (green) and WW1_W44Y (blue). (b) DSC analysis of WW2_WT (black), WW2_E66R (red), WW2_Y85W (green) and WW2_RW (blue). In each case, the top panels show the Cp-T isotherm for each construct and the bottom panels Tm of corresponding construct.
WW2 domain chaperones WW1 domain in the context of WW1-WW2 tandem domains
Our data presenetd above strongly suggest that the WW2 domain may serve as a chaperone and aid ligand binding to the WW1 domain in the context of full-length WWOX in agreement with previous studies demonstrating that WWOX binds to its cellular partners through WW1 domain (1, 15–18). To further test this hypothesis, we next measured and compared the binding of various PPXY-peptides derived from WBP1 and WBP2 to WW1 domain alone and in the context of WW1-WW2 tandem domains using ITC (Tables 1 and 5). Consistent with our hypothesis above, our data suggest that not only does the WW1 domain bind to WBP1_PY2 and WBP2_PY3 peptides in the context of WW1-WW2 tandem domains with a 1:1 stoichiometry in a manner akin to the binding of WW1 domain alone but binding of WW1-WW2 tandem domains is also accompanied by affinities that experience greater than two-fold enhancement. That this is so speaks volumes about the role of WW2 domain in modulating ligand binding to WW1 domain. We also note that while the WW1 domain alone only exhibits binding to WBP1_PY1 and WBP2_PY3 peptides, it additionally also recognizes WBP2_PY2 peptide in the context of WW1-WW2 tandem domains, implying that the WW2 domain not only plays a role in augmenting ligand binding affinity of WW1 domain but also increases its capture radius.
Table 5.
Thermodynamic parameters obtained from ITC measurements for the binding of wildtype WW1-WW2 tandem domains of WWOX to various PPXY-peptides derived from WBP1 and WBP2
| Sequence | n | Kd/μM | ΔH/kcal.mol−1 | TΔS/kcal.mol−1 | ΔG/kcal.mol−1 | |
|---|---|---|---|---|---|---|
| WBP1_PY1 |
|
NBD | NBD | NBD | NBD | NBD |
| WBP1_PY2 |
|
0.99 ± 0.04 | 132 ± 1 | −9.85 ± 0.35 | −4.55 ± 0.35 | −5.30 ± 0.01 |
| WBP2_PY1 |
|
NBD | NBD | NBD | NBD | NBD |
| WBP2_PY2 |
|
1.03 ± 0.02 | 326 ± 8 | −3.36 ± 0.15 | −1.41 ± 0.16 | −4.76 ± 0.01 |
| WBP2_PY3 |
|
1.05 ± 0.01 | 49 ± 1 | −8.46 ± 0.03 | −2.58 ± 0.03 | −5.88 ± 0.01 |
Note that n is the stoichiometry for the binding of various PPXY-peptides to WW1-WW2 tandem domains. Errors were calculated from at least three independent measurements. All errors are given to one standard deviation. NBD indicates no binding determined for the particular peptides either due to lack of any observable heat change or due to poor accuracy of ITC analysis involving such weak interactions (Kd > 1 mM).
To rule out the possibility that ligand binding to WW1 and/or WW2 domain in the context of WW1-WW2 tandem domains is not in any way sterically hindered, we also measured the binding of various PPXY-peptides derived from WBP1 and WBP2 to WW1-WW2rw tandem domains, containing the E66R/Y85W double-substitution within the WW2 domain, using ITC (Table 6). Evidently, our data reveal that the binding of WBP1_PY2 and WBP2_PY3 peptides to WW1-WW2rw tandem domains occurs with a 2:1 stoichiometry, while a 1:1 stoichiometry is observed for the binding of WBP1_PY1 and WBP2_PY2 peptides. These observations are consistent with the fact that while both WW1 and WW2_RW domains alone bind to WBP1_PY2 and WBP2_PY3 peptides, only WW2_RW alone binds to WBP1_PY1 and WBP2_PY2 peptides (Tables 1 and 4). Collectively, these data corroborate the notion that the WW2 domain is a bona fide orphan domain both alone and in the context of WW1-WW2 tandem domains and that ligand binding to WW2 domain can only be restored upon the introduction of E66R/Y85W double-substitution.
Table 6.
Thermodynamic parameters obtained from ITC measurements for the binding of engineered WW1-WW2rw tandem domains to various PPXY-peptides derived from WBP1 and WBP2
| Sequence | n | Kd/μM | ΔH/kcal.mol−1 | TΔS/kcal.mol−1 | ΔG/kcal.mol−1 | |
|---|---|---|---|---|---|---|
| WBP1_PY1 |
|
1.07 ± 0.04 | 103 ± 2 | −8.66 ± 0.13 | −3.21 ± 0.14 | −5.45 ± 0.01 |
| WBP1_PY2 |
|
2.02 ± 0.01 | 101 ± 1 | −10.09 ± 0.25 | −4.64 ± 0.25 | −5.45 ± 0.01 |
| WBP2_PY1 |
|
NBD | NBD | NBD | NBD | NBD |
| WBP2_PY2 |
|
1.02 ± 0.02 | 86 ± 3 | −11.50 ± 0.28 | −5.94 ± 0.30 | −5.55 ± 0.02 |
| WBP2_PY3 |
|
2.00 ± 0.02 | 59 ± 1 | −8.02 ± 0.09 | −2.24 ± 0.10 | −5.78 ± 0.01 |
Note that in the WW1-WW2rw tandem domains, the WW2 domain contains the E66R and Y85W substitutions, while n is the stoichiometry for the binding of various PPXY-peptides to WW1-WW2rw tandem domains. Errors were calculated from at least three independent measurements. All errors are given to one standard deviation. NBD indicates no binding determined for the particular peptides either due to lack of any observable heat change or due to poor accuracy of ITC analysis involving such weak interactions (Kd > 1 mM).
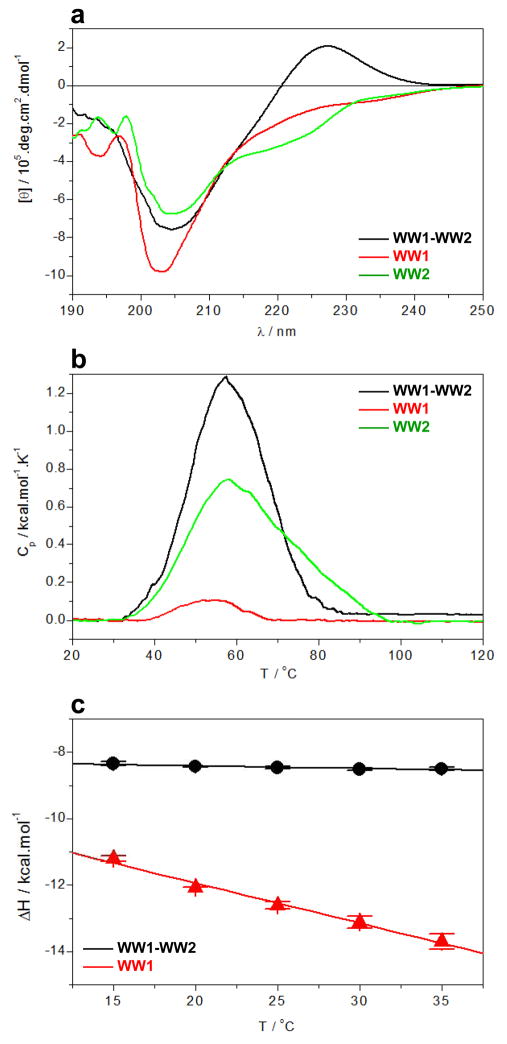
To further establish the role of WW2 domain as a chaperone in the context of WW1-WW2 tandem domains, we next analyzed and compared the secondary structural contents of WW1 and WW2 domains alone and in the context of WW1-WW2 tandem domains using far-UV CD spectroscopy (Figure 7a). As discussed above, our analysis reveals that while WW1 domain alone is structurally unfolded, the spectral features of WW1-WW2 tandem domains suggest a protein with a largely structured β-fold as reported previously (36–38), implying that WW2 domain indeed plays a key scaffolding role in the folding and stabilization of WW1 domain. Consistent with this argument is also the observation that while the thermal stability of WW1-WW2 tandem domains is comparable to that of WW2 domain alone, the enthalpy associated with the unfolding of WW1-WW2 domains is much larger than that of WW1 or WW2 domain alone as probed by DSC (Figure 7b). This implies that while the WW1 domain alone is largely unfolded, it adopts a folded conformation in the context of WW1-WW2 tandem domains. That this is the case is further demonstrated by the markedly differential heat capacity changes (ΔCp) associated with ligand binding to WW1 domain alone and in the context of WW1-WW2 tandem domains as determined from the slopes of corresponding plots of enthalpy change as a function of temperature using ITC (Figure 7c). Thus, while ΔCp associated with the binding of WW1 domain alone to WBP2_PY3 peptide is observed to be around −120 cal/mol/K, this value is less than −10 cal/mol/K for the binding of WW1-WW2 tandem domains. A negative value of ΔCp implies the predominant burial of apolar groups within proteins upon folding and/or ligand binding and, more negative the value of ΔCp the greater the extent of such burial. Thus, an order of magnitude increase in ΔCp associated with ligand binding to WW1 domain alone compared to WW1-WW2 tandem domains is further evidence that while ligand binding to WW1 domain alone induces folding, the WW1 domain is most likely fully folded in the context of WW1-WW2 tandem domains and ligand binding is not associated with any substantial change in structure.
Figure 7.
Comparison of structural and biophysical properties of the wildtype WW1 and WW2 domains alone and in the context of WW1-WW2 tandem domains of WWOX. (a) Far-UV CD analysis of WW1-WW2 tandem domains (black), WW1 domain alone (red) and WW2 domain alone (green). (b) DSC analysis of WW1-WW2 tandem domains (black), WW1 domain alone (red) and WW2 domain alone (green). (c) Dependence of enthalpic change (ΔH) as a function of temperature (T) for the binding of WBP2_PY3 peptide to WW1-WW2 tandem domains (black) and WW1 domain alone (red).
MD simulations provide insights into conformational dynamics of wild type WW1 domain and engineered WW2_RW domain in complex with PPXY peptides
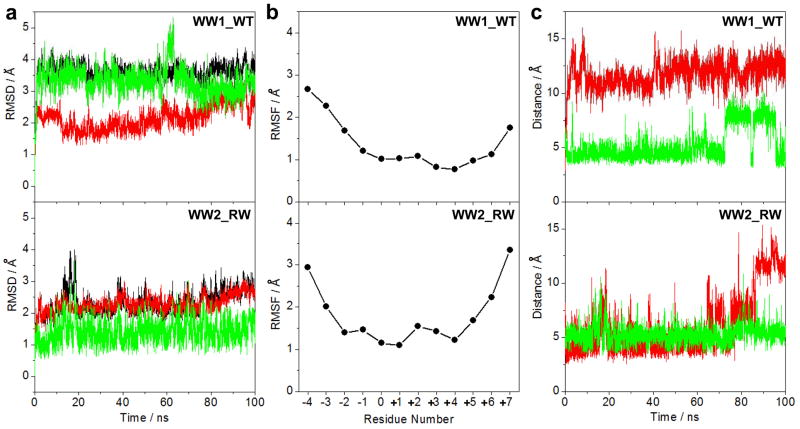
In an effort to test and compare conformational dynamics of wild type WW1 domain versus the engineered WW2_RW domain in complex with PPXY motifs within WBP1 and WBP2, we conducted MD simulations over tens of nanoseconds. As shown in Figure 8a, the MD trajectories reveal that although both the wild type WW1 domain and the engineered WW2_RW domain in complex with WBP2_PY3 peptide reach structural equilibrium after about 20ns, they do so with markedly distinct stabilities. Thus, while the wild type WW1 domain in complex with the WBP2_PY3 peptide equilibrates with a root mean square deviation (RMSD) of ~3.5Å, the engineered WW2_RW domain reaches equilibrium with an RMSD of ~2.5Å. This implies that the engineered WW2_RW domain in complex with WBP2_PY3 peptide is strucutrally more stable than that obtained with the wild type WW1 domain in agreement with our spectroscopic and thermodynamic data. To understand the origin of differential stabilities, we deconvoluted the overall RMSD of these complexes into their constituent components: WW domain alone and peptide alone. Remarkably, it becomes immediately apparent that while both the wild type WW1 domain and the engineered WW2_RW domain more or less undergo conformational dynamics with an RMSD of ~2.5Å, the motional properties of the WBP2_PY3 peptide are quite distinct within each complex. Thus, for example, the WBP2_PY3 peptide in complex with the wild type WW1 domain equilibrates with an RMSD of ~3.5Å, it does so with a much lower RMSD of ~1.5Å in complex with the engineered WW2_RW domain. These observations suggest that lower stability of the wild type WW1 domain in complex with the WBP2_PY3 peptide relative to the engineered WW2_RW domain is most likely due to pronounced conformational dynamics of the peptide ligand.
Figure 8.
MD analysis of wild type (WT) and mutant WW domains of WWOX in complex with WBP2_PY3 peptide conatining the PXPPXYY motif. (a) Root mean square deviation (RMSD) of backbone atoms (N, Cα and C) within each simulated structure relative to the initial modeled structure of the WW1_WT (top panel) and WW2_RW (bottom panel) domains in complex with WBP2_PY3 peptide as a function of simulation time. Note that the overall RMSD for each WW-peptide complex (black) is deconvoluted into the WW domain alone (red) and the peptide alone (green). (b) Root mean square fluctuation (RMSF) of backbone atoms (N, Cα and C) averaged over the entire course of corresponding MD trajectory of the WW1_WT (top panel) and WW2_RW (bottom panel) domains in complex with WBP2_PY3 peptide as a function of peptide residue number (see Figure 1 for nomenclature). Note that RMSF is only displayed for residues within the WBP2_PY3 peptide and residues within the WW domains are omitted for clarity. (c) Distance between the Nη1 guanidine nitorgen of R25 within the WW1_WT domain and the Oη phenolic oxygen of Y+4 within the PXPPXYY motif as a function of simulation time (top panel, red), distance between the Nη1 guanidine nitorgen of R66 within the WW2_RW domain and the Oη phenolic oxygen of Y+4 within the PXPPXYY motif as a function of simulation time (bottom panel, red), distance between the Nε1 indole nitorgen of W44 within the WW1_WT domain and the Cγ pyrrolidine carbon of P-2 within the PXPPXYY motif as a function of simulation time (top panel, green), and distance between the Nε1 indole nitorgen of W85 within the WW2_RW domain and the Cγ pyrrolidine carbon of P-2 within the PXPPXYY motif as a function of simulation time (bottom panel, green).
An alternative means to assess mobility and stability of macromolecular complexes is through an assessment of the root mean square fluctuation (RMSF) of specific atoms over the course of MD simulation. Figure 8b provides such comparative analysis for the backbone atoms of each residue within the WBP2_PY3 peptide in complex with both the wild type WW1 domain and the engineered WW2_RW domain. As expected, the RMSF analysis shows that residues encompassing the N- and C-termini of the WBP2_PY3 peptide within both complexes are conformationally flexible relative to residues located within the core PXPPXYY motif. However, residues within the PXPPXYY motif in complex with the engineered WW2_RW domain appear to exhibit less fluctuation relative to those in complex with the wild type WW1 domain, thereby further providing insights into the differential stabilities of these complexes at atomic level.
Our structural models suggest that the guanidine moieties of R25 within the wild type WW1 domain and R66 within the engineered WW2_RW domain most likely hydrogen bond with the hydroxyl moiety of Y+4 within the PXPPXYY motif of WBP2_PY3 peptide (Figure 4). To test the stability of these potential hydrogen bonds, we plotted the distance between Nη1 guanidine nitrogen atoms of R25/R66 within the WW domains and the Oη phenolic oxygen of Y+4 within the PXPPXYY motif as a function of simulation time (Figure 8c). Our analysis reveals that neither hydrogen bond inferred from our structural models is stable and that R25/R66 within the WW domains as well as Y+4 within the PXPPXYY motif may have a more structural role in stabilizing the conformations of respective interactands in lieu of engaging in intermolecular interactions. Moreover, our structural models also suggest that P-2 residue within the PXPPXYY motif likely stacks against the indole sidechain of W44 within the wild type WW1 domain and W85 within the engineered WW2_RW domain (Figure 4). As shown in Figure 7c, the Nε1 indole nitorgen atoms of W44/W85 within the WW domains and the Cγ pyrrolidine carbon of P-2 within the PXPPXYY motif are spaced apart ~5Å at the start of our MD simulation and this distance remains more or less constant over the entire course of trajectories, albeit with some notable fluctuations after about 70ns in the case of wild type WW1 domain. That this is so suggests strongly that stacking interactions between the sidechain moities of P-2 within the PXPPXYY motif and W44/W85 within the WW domains play an active role in the stabilization of these complexes.
Wildtype WW1 domain and the engineered WW2_RW domain appear to share high sequence similarity with ubiquitin ligase family of WW domains
Given that the E66R/Y85W double-substitution is all that is required to introduce “gain-of-function” within the WW2 domain of WWOX, we wondered the extent to which the combination of an arginine and a tryptophan in structurally-equivalent positions occurred within other WW domains. Of more than several hundred WW domains found within the human proteome, only a handful contain such a combination of an arginine and a tryptophan (Figure 9). Strikingly, all of these WW domains belong to the E3 ubiquitin ligase family of enzymes, such as ITCH and NEDD4, involved in protein turnover and degradation through the proteasome pathway. In light of this consideration, we are tempted to suggest that the WW1 domain of WWOX may compete with cellular partners of ubiquitin ligases and, in so doing, modulate or neutralize their physiological function. Given that tumor suppressors are one of the major targets of ubiquitin ligases, the ability of WW1 domain of WWOX to compete with common cellular partners of ubiquitin ligases may have evolved as a mechanism to prevent rapid turnover and degradation of proteins central to cellular homeostasis. Accordingly, a breakdown in this mechanism, for example due to down-expression of WWOX, may serve as a signal for normal cells to undergo oncogenic transformation. Importantly, the WW domains of ITCH ubiquitin ligase are also believed to play a central role in the Hippo tumor suppressor pathway (39–41). It is thus also conceivable that the WW1 domain of WWOX may have a role to play in regulating the Hippo tumor suppressor pathway.
Figure 9.
Amino acid sequence alignment of human WW1 and WW2_RW domains of WWOX with other WW domains within human proteome, identified through BLAST search, in which an arginine and a tryptophan are conserved at structurally-equivalent positions with respect to E66R and Y85W substitutions within WW2_RW mutant domain. Note that absolutely conserved residues within the WW domains are shown in red, semi-conserved residues in blue, and all other residues are colored black.
CONCLUSIONS
Despite its key role in a diverse array of cellular activities, including growth, proliferation, apoptosis and tumor suppression (1–4), little is known about the molecular origin of how the WWOX tumor suppressor recognizes its cellular partners. In this study, we analyzed the binding of WW1 and WW2 domains of WWOX alone and in the context of WW1-WW2 tandem domains to PPXY motifs located within WBP1 and WBP2 adaptors. Our detailed biophysical analysis reveals that while WW1 domain binds to WBP1_PY2 and WBP1_PY3 motifs in a physiologically-relevant manner, WW2 domain is completely devoid of recognizing any of the PPXY motifs, at least in the context of WBP1 and WBP2 adaptors. Notably, our studies reveal that the high-affinity binding of WW1 domain of WWOX requires the rather extended PXPPXYY motif in lieu of minimal PPXY sequence. Additionally, our data suggest that the WW2 domain of WWOX is likely an orphan WW domain without any physiological ligands and that it may have evolved as a chaperone to impart structural and thermal stability upon the WW1 domain in the context of full-length WWOX. This notion is further corroborated by the fact while WW2 domain appears to exude rather high thermal stability, the WW1 domain is structurally disordered and only undergoes folding upon ligand binding. Additionally, the WW2 domain not only augments ligand binding to WW1 domain but also aids stabilization of WW1 domain in the context of WW1-WW2 tandem domains. In this regard, the WW1-WW2 module of WWOX appears to resemble the WW3-WW4 module of SUDX ubiquitin ligase in that the WW3 domain of the latter is not only unstructured and undergoes folding upon ligand binding but the WW4 domain is also functionally sterile and stabilizes the WW3 domain in a manner akin to the WW2 domain of WWOX (42). It should be noted that the ability of one domain within a module to serves as a structural scaffold to aid ligand binding and stability within another is not unique to WW domains but has also been observed in the case of a tandem pair of PDZ domains (43). Importantly, we have also shown here that the E66R/Y85W double-substitution is all that is required to introduce “gain-of-function” within the WW2 domain of WWOX. This engineered domain that we have referred to herein as WW2_RW appears to bind to PPXY motifs within WBP1 and WBP2 adaptors much stronger than the wild type WW1 domain. In short, our study lends biophysical insights into a key protein-protein interaction pertinent to human health and disease.
MATERIALS and METHODS
GST-pulldown and Western blot analysis
HEK293 cells were transiently transfected with pEBG expression plasmids encoding mammalian WBP2 and either wild type GST-tagged WWOX (WT), or GST-tagged WWOX containing the W44F/P47A double-mutation (FA), or GST-tagged WW1 domain of WWOX (WW1), or GST-tagged WW2 domain of WWOX (WW2), as described earlier (44). After harvesting for 24–30h, cells were lysed in Lysis Buffer (50mM Tris, 150mM NaCl, 10% (v/v) glycerol, pH 7.5) containing 0.5% (v/v) Nonidet P-40 and trace amounts of protease inhibitors. For GST-pulldown, 1mg of whole cell lysate was mixed with glutathione beads and allowed to rock overnight at 4°C. After overnight incubation, the beads were extensively washed with Lysis buffer containing 0.1% (v/v) Nonidet P-40. SDS-PAGE analysis was conducted on either whole cell lysate (Input) or after GST-pulldown and the resolved bands were detected by immunoblotting with anti-WBP2 (α-WBP2) and anti-GST (α-GST) antibodies. Antibodies used for immunoblotting were rabbit polyclonal anti-WBP2 (Santa Cruz Biotechnology, Santa Cruz, CA) and goat polyclonal anti-GST (GE Healthcare, Munich, Germany).
Recombinant protein purification and peptide synthesis
WW1 domain (residues 16–50), WW2 domain (residues 57–91), and WW1-WW2 tandem domains (residues 16–91) of human WWOX were cloned into pET30 bacterial expression vectors with an N-terminal His-tag using Novagen LIC technology. The various mutants of the WW domains of WWOX alone or in the context of WW1-WW2 tandem domains were generated using the PCR primer extension method (45). Briefly, the mutant domains contained the following substitutions: R25A in WW1 domain (WW1_R25A), R25E in WW1 domain (WW1_R25E), W44Y in WW1 domain (WW1_W44Y), E66R in WW2 domain (WW2_E66R), Y85W in WW2 domain (WW2_Y85W), and E66R/Y85W double-substitution in WW2 domain alone (WW2_RW) or in the context of WW1-WW tandem domains (WW1-WW2rw). All recombinant domains were subsequently expressed in Escherichia coli BL21*(DE3) bacterial strain (Invitrogen) and purified on a Ni-NTA affinity column using standard procedures. Briefly, bacterial cells were grown at 20°C in Terrific Broth to an optical density of greater than unity at 600nm prior to induction with 0.5mM isopropyl β-D-1-thiogalactopyranoside (IPTG). The bacterial culture was further grown overnight at 20°C and the cells were subsequently harvested and disrupted using a BeadBeater (Biospec). After separation of cell debris at high-speed centrifugation, the cell lysate was loaded onto a Ni-NTA column and washed extensively with 20mM imidazole to remove non-specific binding of bacterial proteins to the column. The recombinant proteins were subsequently eluted with 200mM imidazole and dialyzed against an appropriate buffer to remove excess imidazole. Further treatment on a Hiload Superdex 200 size-exclusion chromatography (SEC) column coupled in-line with GE Akta FPLC system led to purification of WW domains to apparent homogeneity as judged by SDS-PAGE analysis. Final yield was typically between 50–100mg protein of apparent homogeneity per liter of bacterial culture. Protein concentration was determined by the fluorescence-based Quant-It assay (Invitrogen) and spectrophotometrically on the basis of extinction coefficients calculated for the various wild type and mutant WW domains of WWOX using the online software ProtParam at ExPasy Server (46). 12-mer wild type and mutant peptides spanning various PPXY motifs within human WBP1 and WBP2 proteins were commercially obtained from GenScript Corporation. The sequences of these peptides are shown in Figures 1b and 1c. The peptide concentrations were measured gravimetrically.
Isothermal titration calorimetry
Isothermal titration calorimetry (ITC) experiments were performed on a Microcal VP-ITC instrument and data were acquired and processed using the integrated Microcal ORIGIN software. All measurements were repeated at least three times. Briefly, various WW domain samples alone or in the context of WW1-WW2 tandem domains were prepared in 50mM Sodium phosphate, 100mM NaCl, 1mM EDTA and 5mM β-mercaptoethanol at pH 7.0. The experiments were initiated by injecting 25 × 10μl aliquots of 2–4 mM of each PPXY-peptide from the syringe into the calorimetric cell containing 1.8ml of 100–200 μM of a WW domain solution at 25 °C. The change in thermal power as a function of each injection was automatically recorded using the ORIGIN software and the raw data were further processed to yield binding isotherms of heat release per injection as a function of molar ratio of each peptide to WW domain. The heats of mixing and dilution were subtracted from the heat of binding per injection by carrying out a control experiment in which the same buffer in the calorimetric cell was titrated against each peptide in an identical manner. To extract binding affinity (Kd) and binding enthalpy (ΔH), the ITC isotherms were iteratively fit to the following built-in function by non-linear least squares regression analysis using the integrated ORIGIN software:
| [1] |
where q(i) is the heat release (kcal/mol) for the ith injection, n is the binding stoichiometry, V is the effective volume of WW domain solution in the calorimetric cell (1.46 ml), P is the total concentration of WW domain in the calorimetric cell and L is the total concentration of peptide ligand added for the ith injection. The above equation is derived from the binding of a ligand to a macromolecule using the law of mass action assuming a one-site model (47). The free energy change (ΔG) upon ligand binding was calculated from the relationship:
| [2] |
where R is the universal molar gas constant (1.99 cal/K/mol) and T is the absolute temperature. The entropic contribution (TΔS) to the free energy of binding was calculated from the relationship:
| [3] |
where ΔH and ΔG are as defined above.
Differential scanning calorimetry
Differential scanning calorimetry (DSC) experiments were performed on a TA Nano-DSC instrument and data were acquired and processed using the integrated NanoAnalyze software. All measurements were repeated at least three times. Briefly, various WW domain samples alone or in the context of WW1-WW2 tandem domains were prepared in 50mM Sodium phosphate at pH 7.0. All experiments were conducted on 400–500μM sample of each WW domain in the 20–120°C temperature range at a heating rate (dT/dt) of 1°C/min under an excess pressure of 3atm. The change in thermal power (dQ/dt) as a function of temperature was automatically recorded using the NanoAnalyze software. Control experiments on the buffer alone were also conducted in an identical manner to generate baselines that were subtracted from the raw data to remove the contribution of buffer. The raw data were further processed to yield the melting isotherms of excess heat capacity (Cp) as a function of temperature (T) using the following relationship:
| [4] |
where P is the initial concentration of WW domain loaded into the calorimetric cell and V is the effective volume of calorimetric cell (0.3ml). To determine the melting temperature (Tm) accompanying the unfolding of WW domains, the melting isotherms were iteratively fit to the following built-in function by non-linear least squares regression analysis using the integrated NanoAnalyze software:
| [5] |
where ΔHm is the enthalpy of unfolding, R is the universal molar gas constant (1.99cal/K/mol), T is the absolute temperature, a is the scaling factor that accounts for the difference between the initial protein concentration loaded into the calorimetric cell and the effective protein concentration that is actually available due to the loss to aggregation during the experiment, and K is given by the following relationship:
| [6] |
It should be noted that Eq [5] is derived from the inter-conversion of a macromolecule between a folded and an unfolded state assuming a two-state model (48, 49).
Circular dichroism
Far-UV circular dichroism (CD) measurements were conducted on a Jasco J-815 spectrometer thermostatically controlled at 25°C. Experiments were conducted on a 20μM sample of each WW domain alone or in the context of WW1-WW2 tandem domains in 10mM Sodium phosphate at pH 7.0. Data were collected using a quartz cuvette with a 2-mm pathlength in the 190–250nm wavelength range. Data were normalized against reference spectra to remove the contribution of buffer. Data were recorded with a slit bandwidth of 2nm at a scan rate of 10nm/min. Each data set represents an average of four scans acquired at 0.1nm intervals. Data were converted to molar ellipticity, [θ], as a function of wavelength (λ) of electromagnetic radiation using the equation:
| [16] |
where Δε is the observed ellipticity in mdeg, c is the concentration of WW domain in μM and l is the cuvette pathlength in cm.
Molecular modeling
Molecular modeling (MM) was employed to build structural models of wild type WW1 domain (WW1_WT), wild type WW2 domain (WW2_WT), and the engineered WW2 domain containing the E66R/Y85W double-substitution (WW2_RW) of WWOX in complex with a peptide containing the PY3 motif of WBP2 (WBP2_PY3) using the MODELLER software based on homology modeling (50). In each case, the NMR structure of WW domain of YAP1 bound to a peptide containing the PPXY motif was used as a template (PDB# 1JMQ). Additionally, hydrogen bonding restraints were added between the Nη1/Nη2 nitrogen atoms of guanidine moiety of R25 within WW1_WT domain and Oη oxygen atom of the phenyl moiety of Y+4 within WBP2_PY3 peptide as well as between the Nη1/Nη2 nitrogen atoms of guanidine moiety of R66 within WW2_RW domain and Oη oxygen atom of the phenyl moiety of Y+4 within WBP2_PY3 peptide. Introduction of such hydrogen bonding restraints was necessary to bring the sidechain atoms of respective residues within optimal hydrogen bonding distance in agreement with our thermodynamic data reported here. The atomic distances set for hydrogen bonding restraints between a specific pair of oxygen and nitrogen atoms were 2.8±0.5Å. A total of 100 structural models were calculated and the structure with the lowest energy, as judged by the MODELLER Objective Function, was selected for further analysis. The atomic models were rendered using RIBBONS (51).
Molecular dynamics
Molecular dynamics (MD) simulations were performed with the GROMACS software (52, 53) using the integrated OPLS-AA force field (54, 55). Briefly, the modeled structures of wild type WW1 domain (WW1_WT) and the engineered WW2 domain containing the E66R/Y85W double-substitution (WW2_RW) of WWOX in complex with a peptide containing the PY3 motif of WBP2 (WBP2_PY3) were centered within a cubic box and hydrated using the extended simple point charge (SPC/E) water model (56, 57). The hydrated structures were energy-minimized with the steepest descent algorithm prior to equilibration under the NPT ensemble conditions, wherein the number of atoms (N), pressure (P) and temperature (T) within the system were respectively kept constant at ~17000, 1 bar and 300 K. The Particle-Mesh Ewald (PME) method was employed to compute long-range electrostatic interactions with a 10Å cut-off (58) and the Linear Constraint Solver (LINCS) algorithm to restrain bond lengths (59). All MD simulations were performed under periodic boundary conditions (PBC) using the leap-frog integrator with a time step of 2fs. For the final MD production runs, data were collected every 10ps over a time scale of 100ns.
Acknowledgments
This work was supported by the National Institutes of Health Grants R01-GM083897 (to AF) and R01-DK079217 (to ZN and RIA), the Israeli Science Foundation Grant #1331-08 (to RIA), an award from the Israel Cancer Research Fund (to ZS), funds from the USylvester Braman Family Breast Cancer Institute (to AF and ZN), the Pennsylvania Breast Cancer Coalition Grants #60707 (to MS) and #9200903 (to MS), and by funds from the Geisinger Clinic (to MS). CBM is a recipient of a postdoctoral fellowship from the National Institutes of Health (Award# T32-CA119929).
ABBREVIATIONS
- CD
Circular dichroism
- DSC
Differential scanning calorimetry
- GST
Glutathione S-transferase
- ITC
Isothermal titration calorimetry
- LIC
Ligation-independent cloning
- MD
Molecular dynamics
- MM
Molecular modeling
- PPII
Polyproline type II
- SEC
Size-exclusion chromatography
- WBP1
WW-binding protein 1
- WBP2
WW-binding protein 2
- YAP1
YES-associated protein 1
- YAP2
YES-associated protein 2
- WWOX
WW-containing oxidoreductase
References
- 1.Del Mare S, Salah Z, Aqeilan RI. WWOX: its genomics, partners, and functions. J Cell Biochem. 2009;108:737–745. doi: 10.1002/jcb.22298. [DOI] [PubMed] [Google Scholar]
- 2.Bednarek AK, Laflin KJ, Daniel RL, Liao Q, Hawkins KA, Aldaz CM. WWOX, a novel WW domain-containing protein mapping to human chromosome 16q23.3-24.1, a region frequently affected in breast cancer. Cancer Res. 2000;60:2140–2145. [PubMed] [Google Scholar]
- 3.Bednarek AK, Keck-Waggoner CL, Daniel RL, Laflin KJ, Bergsagel PL, Kiguchi K, Brenner AJ, Aldaz CM. WWOX, the FRA16D gene, behaves as a suppressor of tumor growth. Cancer Res. 2001;61:8068–8073. [PubMed] [Google Scholar]
- 4.Hezova R, Ehrmann J, Kolar Z. WWOX, a new potential tumor suppressor gene. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2007;151:11–15. doi: 10.5507/bp.2007.002. [DOI] [PubMed] [Google Scholar]
- 5.Nunez MI, Ludes-Meyers J, Abba MC, Kil H, Abbey NW, Page RE, Sahin A, Klein-Szanto AJ, Aldaz CM. Frequent loss of WWOX expression in breast cancer: correlation with estrogen receptor status. Breast Cancer Res Treat. 2005;89:99–105. doi: 10.1007/s10549-004-1474-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aqeilan RI, Kuroki T, Pekarsky Y, Albagha O, Trapasso F, Baffa R, Huebner K, Edmonds P, Croce CM. Loss of WWOX expression in gastric carcinoma. Clin Cancer Res. 2004;10:3053–3058. doi: 10.1158/1078-0432.ccr-03-0594. [DOI] [PubMed] [Google Scholar]
- 7.Aqeilan RI, Croce CM. WWOX in biological control and tumorigenesis. J Cell Physiol. 2007;212:307–310. doi: 10.1002/jcp.21099. [DOI] [PubMed] [Google Scholar]
- 8.Aqeilan RI, Hagan JP, Aqeilan HA, Pichiorri F, Fong LY, Croce CM. Inactivation of the Wwox gene accelerates forestomach tumor progression in vivo. Cancer Res. 2007;67:5606–5610. doi: 10.1158/0008-5472.CAN-07-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pluciennik E, Kusinska R, Potemski P, Kubiak R, Kordek R, Bednarek AK. WWOX--the FRA16D cancer gene: expression correlation with breast cancer progression and prognosis. Eur J Surg Oncol. 2006;32:153–157. doi: 10.1016/j.ejso.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 10.Lewandowska U, Zelazowski M, Seta K, Byczewska M, Pluciennik E, Bednarek AK. WWOX, the tumour suppressor gene affected in multiple cancers. J Physiol Pharmacol. 2009;60(Suppl 1):47–56. [PubMed] [Google Scholar]
- 11.Zelazowski MJ, Pluciennik E, Pasz-Walczak G, Potemski P, Kordek R, Bednarek AK. WWOX expression in colorectal cancer--a real-time quantitative RT-PCR study. Tumour Biol. 2011;32:551–560. doi: 10.1007/s13277-010-0150-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aqeilan RI, Trapasso F, Hussain S, Costinean S, Marshall D, Pekarsky Y, Hagan JP, Zanesi N, Kaou M, Stein GS, Lian JB, Croce CM. Targeted deletion of Wwox reveals a tumor suppressor function. Proc Natl Acad Sci U S A. 2007;104:3949–3954. doi: 10.1073/pnas.0609783104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aqeilan RI, Hassan MQ, de Bruin A, Hagan JP, Volinia S, Palumbo T, Hussain S, Lee SH, Gaur T, Stein GS, Lian JB, Croce CM. The WWOX tumor suppressor is essential for postnatal survival and normal bone metabolism. J Biol Chem. 2008;283:21629–21639. doi: 10.1074/jbc.M800855200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aqeilan RI, Hagan JP, de Bruin A, Rawahneh M, Salah Z, Gaudio E, Siddiqui H, Volinia S, Alder H, Lian JB, Stein GS, Croce CM. Targeted ablation of the WW domain-containing oxidoreductase tumor suppressor leads to impaired steroidogenesis. Endocrinology. 2009;150:1530–1535. doi: 10.1210/en.2008-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aqeilan RI, Palamarchuk A, Weigel RJ, Herrero JJ, Pekarsky Y, Croce CM. Physical and functional interactions between the Wwox tumor suppressor protein and the AP-2gamma transcription factor. Cancer Res. 2004;64:8256–8261. doi: 10.1158/0008-5472.CAN-04-2055. [DOI] [PubMed] [Google Scholar]
- 16.Aqeilan RI, Pekarsky Y, Herrero JJ, Palamarchuk A, Letofsky J, Druck T, Trapasso F, Han SY, Melino G, Huebner K, Croce CM. Functional association between Wwox tumor suppressor protein and p73, a p53 homolog. Proc Natl Acad Sci U S A. 2004;101:4401–4406. doi: 10.1073/pnas.0400805101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aqeilan RI, Donati V, Palamarchuk A, Trapasso F, Kaou M, Pekarsky Y, Sudol M, Croce CM. WW domain-containing proteins, WWOX and YAP, compete for interaction with ErbB-4 and modulate its transcriptional function. Cancer Res. 2005;65:6764–6772. doi: 10.1158/0008-5472.CAN-05-1150. [DOI] [PubMed] [Google Scholar]
- 18.Ludes-Meyers JH, Kil H, Bednarek AK, Drake J, Bedford MT, Aldaz CM. WWOX binds the specific proline-rich ligand PPXY: identification of candidate interacting proteins. Oncogene. 2004;23:5049–5055. doi: 10.1038/sj.onc.1207680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Einbond A, Sudol M. Towards prediction of cognate complexes between the WW domain and proline-rich ligands. FEBS Lett. 1996;384:1–8. doi: 10.1016/0014-5793(96)00263-3. [DOI] [PubMed] [Google Scholar]
- 20.Sudol M. Structure and function of the WW domain. Prog Biophys Mol Biol. 1996;65:113–132. doi: 10.1016/s0079-6107(96)00008-9. [DOI] [PubMed] [Google Scholar]
- 21.Sudol M. WW domain. In: Cesareni GGM, Sudol M, Yaffe M, editors. Modular Protein Domains. Wiley VCH, Verlag GmbH & Co; 2004. pp. 59–72. [Google Scholar]
- 22.Yagi R, Chen LF, Shigesada K, Murakami Y, Ito Y. A WW domain-containing yes-associated protein (YAP) is a novel transcriptional co-activator. Embo J. 1999;18:2551–2562. doi: 10.1093/emboj/18.9.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Komuro A, Nagai M, Navin NE, Sudol M. WW domain-containing protein YAP associates with ErbB-4 and acts as a co-transcriptional activator for the carboxyl-terminal fragment of ErbB-4 that translocates to the nucleus. J Biol Chem. 2003;278:33334–33341. doi: 10.1074/jbc.M305597200. [DOI] [PubMed] [Google Scholar]
- 24.Zhao B, Ye X, Yu J, Li L, Li W, Li S, Lin JD, Wang CY, Chinnaiyan AM, Lai ZC, Guan KL. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hao Y, Chun A, Cheung K, Rashidi B, Yang X. Tumor suppressor LATS1 is a negative regulator of oncogene YAP. J Biol Chem. 2008;283:5496–5509. doi: 10.1074/jbc.M709037200. [DOI] [PubMed] [Google Scholar]
- 26.Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, Zheng P, Ye K, Chinnaiyan A, Halder G, Lai ZC, Guan KL. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bertini E, Oka T, Sudol M, Strano S, Blandino G. YAP: at the crossroad between transformation and tumor suppression. Cell Cycle. 2009;8:49–57. doi: 10.4161/cc.8.1.7259. [DOI] [PubMed] [Google Scholar]
- 28.Sudol M. Newcomers to the WW Domain-Mediated Network of the Hippo Tumor Suppressor Pathway. Genes Cancer. 2010;1:1115–1118. doi: 10.1177/1947601911401911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sudol M, Harvey KF. Modularity in the Hippo signaling pathway. Trends Biochem Sci. 2010;35:627–633. doi: 10.1016/j.tibs.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 30.Salah Z, Alian A, Aqeilan RI. WW domain-containing proteins: Retrospectives and the future. Front Biosci. 2012;17:331–348. doi: 10.2741/3930. [DOI] [PubMed] [Google Scholar]
- 31.Pires JR, Taha-Nejad F, Toepert F, Ast T, Hoffmuller U, Schneider-Mergener J, Kuhne R, Macias MJ, Oschkinat H. Solution structures of the YAP65 WW domain and the variant L30 K in complex with the peptides GTPPPPYTVG, N-(n-octyl)-GPPPY and PLPPY and the application of peptide libraries reveal a minimal binding epitope. J Mol Biol. 2001;314:1147–1156. doi: 10.1006/jmbi.2000.5199. [DOI] [PubMed] [Google Scholar]
- 32.McDonald CB, McIntosh SK, Mikles DC, Bhat V, Deegan BJ, Seldeen KL, Saeed AM, Buffa L, Sudol M, Nawaz Z, Farooq A. Biophysical Analysis of Binding of WW Domains of the YAP2 Transcriptional Regulator to PPXY Motifs within WBP1 and WBP2 Adaptors. Biochemistry. 2011;50:9616–9627. doi: 10.1021/bi201286p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Macias MJ, Hyvonen M, Baraldi E, Schultz J, Sudol M, Saraste M, Oschkinat H. Structure of the WW domain of a kinase-associated protein complexed with a proline-rich peptide. Nature. 1996;382:646–649. doi: 10.1038/382646a0. [DOI] [PubMed] [Google Scholar]
- 34.Huang X, Poy F, Zhang R, Joachimiak A, Sudol M, Eck MJ. Structure of a WW domain containing fragment of dystrophin in complex with beta-dystroglycan. Nat Struct Biol. 2000;7:634–638. doi: 10.1038/77923. [DOI] [PubMed] [Google Scholar]
- 35.Kanelis V, Rotin D, Forman-Kay JD. Solution structure of a Nedd4 WW domain-ENaC peptide complex. Nat Struct Biol. 2001;8:407–412. doi: 10.1038/87562. [DOI] [PubMed] [Google Scholar]
- 36.Fernandez-Escamilla AM, Ventura S, Serrano L, Jimenez MA. Design and NMR conformational study of a beta-sheet peptide based on Betanova and WW domains. Protein Sci. 2006;15:2278–2289. doi: 10.1110/ps.062186506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jager M, Dendle M, Kelly JW. Sequence determinants of thermodynamic stability in a WW domain--an all-beta-sheet protein. Protein Sci. 2009;18:1806–1813. doi: 10.1002/pro.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tapia VE, Nicolaescu E, McDonald CB, Musi V, Oka T, Inayoshi Y, Satteson AC, Mazack V, Humbert J, Gaffney CJ, Beullens M, Schwartz CE, Landgraf C, Volkmer R, Pastore A, Farooq A, Bollen M, Sudol M. Y65C missense mutation in the WW domain of the Golabi-Ito-Hall syndrome protein PQBP1 affects its binding activity and deregulates pre-mRNA splicing. J Biol Chem. 2010;285:19391–19401. doi: 10.1074/jbc.M109.084525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salah Z, Melino G, Aqeilan RI. Negative regulation of the Hippo pathway by E3 ubiquitin ligase ITCH is sufficient to promote tumorigenicity. Cancer Res. 2011;71:2010–2020. doi: 10.1158/0008-5472.CAN-10-3516. [DOI] [PubMed] [Google Scholar]
- 40.Salah Z, Aqeilan RI. WW domain interactions regulate the Hippo tumor suppressor pathway. Cell Death Dis. 2011;2:e172. doi: 10.1038/cddis.2011.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ho KC, Zhou Z, She YM, Chun A, Cyr TD, Yang X. Itch E3 ubiquitin ligase regulates large tumor suppressor 1 stability. Proc Natl Acad Sci U S A. 2011;108:4870–4875. doi: 10.1073/pnas.1101273108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fedoroff OY, Townson SA, Golovanov AP, Baron M, Avis JM. The Structure and Dynamics of Tandem WW Domains in a Negative Regulator of Notch Signaling, Suppressor of Deltex. J Biol Chem. 2004;279:34991–35000. doi: 10.1074/jbc.M404987200. [DOI] [PubMed] [Google Scholar]
- 43.Feng W, Shi Y, Li M, Zhang M. Tandem PDZ repeats in glutamate receptor–interacting proteins have a novel mode of PDZ domain–mediated target binding. Nat Struct Biol. 2003;10:972–978. doi: 10.1038/nsb992. [DOI] [PubMed] [Google Scholar]
- 44.Gaudio E, Palamarchuk A, Palumbo T, Trapasso F, Pekarsky Y, Croce CM, Aqeilan RI. Physical association with WWOX suppresses c-Jun transcriptional activity. Cancer Res. 2006;66:11585–11589. doi: 10.1158/0008-5472.CAN-06-3376. [DOI] [PubMed] [Google Scholar]
- 45.Gao X, Yo P, Keith A, Ragan TJ, Harris TK. Thermodynamically balanced inside-out (TBIO) PCR-based gene synthesis: a novel method of primer design for high-fidelity assembly of longer gene sequences. Nucleic Acids Res. 2003;31:e143. doi: 10.1093/nar/gng143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. Protein Identification and Analysis Tools on the ExPASy Server. In: Walker JM, editor. The Proteomics Protocols Handbook. Humana Press; Totowa, New Jersey, USA: 2005. pp. 571–607. [Google Scholar]
- 47.Wiseman T, Williston S, Brandts JF, Lin LN. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal Biochem. 1989;179:131–137. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
- 48.Privalov PL, Potekhin SA. Scanning microcalorimetry in studying temperature-induced changes in proteins. Methods Enzymol. 1986;131:4–51. doi: 10.1016/0076-6879(86)31033-4. [DOI] [PubMed] [Google Scholar]
- 49.Privalov GP, Privalov PL. Problems and prospects in microcalorimetry of biological macromolecules. Methods Enzymol. 2000;323:31–62. doi: 10.1016/s0076-6879(00)23360-0. [DOI] [PubMed] [Google Scholar]
- 50.Marti-Renom MA, Stuart AC, Fiser A, Sanchez R, Melo F, Sali A. Comparative Protein Structure Modeling of Genes and Genomes. Annu Rev Biophys Biomol Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 51.Carson M. Ribbons 2.0. J Appl Crystallogr. 1991;24:958–961. [Google Scholar]
- 52.Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ. GROMACS: fast, flexible, and free. J Comput Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 53.Hess B. GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. J Chem Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 54.Jorgensen WL, Tirado-Rives J. The OPLS Force Field for Proteins: Energy Minimizations for Crystals of Cyclic Peptides and Crambin. J Am Chem Soc. 1988;110:1657–1666. doi: 10.1021/ja00214a001. [DOI] [PubMed] [Google Scholar]
- 55.Kaminski GA, Friesner RA, Tirado-Rives J, Jorgensen WL. Evaluation and Reparametrization of the OPLS-AA Force Field for Proteins via Comparison with Accurate Quantum Chemical Calculations on Peptides. J Phys Chem B. 2001;105:6474–6487. [Google Scholar]
- 56.Toukan K, Rahman A. Molecular-dynamics study of atomic motions in water. Physical Review B. 1985;31:2643–2648. doi: 10.1103/physrevb.31.2643. [DOI] [PubMed] [Google Scholar]
- 57.Berendsen HJC, Grigera JR, Straatsma TP. The Missing Term in Effective Pair Potentials. J Phys Chem. 1987;91:6269–6271. [Google Scholar]
- 58.Darden TA, York D, Pedersen L. Particle mesh Ewald: An N.log(N) method for Ewald sums in large systems. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 59.Hess B, Bekker H, Berendsen HJC, Fraaije JGEM. LINCS: A linear constraint solver for molecular simulations. J Comput Chem. 1997;18:1463–1472. [Google Scholar]