Abstract
Time-resolved fluorescence decay profiles of N-acetyl-L-tryptophan-amide (NATA) and tryptophan (Trp) dipeptides of the form Trp-X and X-Trp, where X is another aminoacyl residue, have been investigated using an ultraviolet upconversion spectrophoto fluorometer with time resolution better than 350 fs, together with a time correlated single photon counting apparatus on the 100ps to 20ns time scale. We analyzed the set of fluorescence decay profiles at multiple wavelengths using the global analysis technique. Nanosecond fluorescence transients for Trp dipeptides all show multiexponential decay, while NATA exhibits a monoexponential decay near 3 ns independent of pH. In the first 100 ps, a time constant for the water “bulk relaxation” around Trp, NATA and Trp dipeptides is seen near 1-2 ps, with an associated preexponential amplitude that is positive or negative depending on emission wavelength, as expected for a population-conserving spectral shift. The initial brightness (sub-ps) we measure for all these dipeptides is less than that of NATA, implying even faster (<200fs) intra-molecular (quasi) static quenching occurs within them. A new, third, ultrafast decay, bearing an exponential time constant of 20-30 ps with positive amplitude, has been found in many of these dipeptides. We believe it verifies our previous predictions of dipeptide QSSQ (“quasi static self quenching”) –the loss of quantum yield to sub-100ps decay process (Chen et al., Biochemistry, 1991, 30, 5184). Most important, this term is found in proteins as well (J.A.C.S., 2006, 128, 1214; Biophysical Journal 2008, 94, 546; 2009, 96, 46a), suggesting an ultrafast quenching mechanism must be common to both.
I. Introduction
Understanding peptide dynamics in aqueous solution is critical. Their functions depend strongly on their structures (and structural change) during biological processes such as protein folding, protein hydration, and protein-peptide recognition.1-4 In order to gain insight into the relationships among the solution environment, conformation changes and peptide function, Tryptophan (Trp) and other indole-containing compounds have been used as probes. Trp reveals structure and dynamics in its emission maximum and width, quantum yield, lifetime and anisotropy. Among amino acid residues, Trp possesses the highest UV extinction coefficient and highest quantum yield of emission.5-8 In a polar environment, Trp fluorescence is emitted from the singlet 1La state because ultrafast (<100 fs) internal conversion converts a mixture with 1Lb to 1La.9. Trp is very sensitive to the polarity and dynamics of the immediate environment; spectral energy and width depend strongly on exposure to water (and/or the polarity of the electrostatic environment inside protein).10,11 The ability to probe time resolved fluorescence of tryptophan, which, if not already present, can often be incorporated in a peptide without significant structural changes, opens a window into peptide/protein dynamics inaccessible to other biophysical techniques.12,13 Recently, interest in Trp has grown, as tunable ultrafast Ti: sapphire lasers have made it possible to monitor the molecular vibrational modes and femtosecond solvent relaxation. 14-16
It is well known that many peptides and proteins containing even a single Trp yield multiexponential fluorescence decays linked to different Decay Associated Spectra (DAS).17-19 The origin of these DAS is still controversial. Basically, three explanations have been proposed for complex fluorescence decays. First, these phenomena are most easily explained on the basis of ground state heterogeneity; e. g. the presence of several conformers, each with different fluorescence lifetimes.20,21 NMR-derived rotamer population data has thus been used to predict amplitudes.21,22 Further, single-Trp proteins in crystals provided angle-dependent preexponential factors that support a rotamer concept.23 Second, in addition to frank rotamers of the Trp side chain, microconformational states of peptides or proteins can create different local environments of the indole ring that constitute a source of ground-state heterogeneity,24 and third, relaxation of the peptide/protein matrix (or solvent water) surrounding the indole ring during the lifetime of the excited state will also produce a complex decay.25-27 The latter interpretation, driven by the preponderance in literature of shortlived bluer and longlived redder DAS, requires generalized solvent relaxation on the nanosecond timescale (in response to the increased dipole moment of the excited state).28 In this model, decay curves change character across the emission surface; rapid decay at the blue end of the fluorescence spectrum is replaced by a central region of simple exponential decay, and fast rising behavior is seen at the red end of the spectrum.14,15 A negative preexponential amplitude is thus generated at the red tail of spectrum that is always associated with a short lifetime (i.e. “short negative DAS”). The presence or absence of this negative term has become an important signature for relaxation, although mixtures of relaxation and heterogeneity processes might obscure it. The DAS recovered in the relaxation model should generally show a trend of longer decay times for the longer wavelength components.29
Most Trp fluorescence studies in peptides/proteins have been made with photomultipliers providing sub-nanosecond time resolution.30-32 In such previous work, analysis of lifetime and quantum yield data for Trp-X dipeptides provided evidence for quasi-static self quenching (QSSQ).33 Very short lifetimes (e.g., under 100ps) can be mistaken for scattered light or obscured by noise on the nanosecond scale, so they would be underrepresented or lost. Their presence was inferred from quantum yield defects.
In this work, extended time resolved fluorescence transients, including fluorescence decay kinetics and DAS, are collected from both an upconversion spectrophotofluorometer and a time correlated single photon counting (TCSPC) apparatus coupled to fs and ps laser sources. The full dynamics are dissected to reveal contributions from both the heterogeneous environs of the peptides and the evolving solvent shell. Recently, Larsen et al.34 also found a 16 ps fluorescence component for Trp in a small 22-mer peptide, and they proposed that this lifetime originated from a rotamer of Trp, which reinforces the conclusions of this paper.
II. Experimental Section
Upconversion spectrophotofluorometer
The experimental setup has been partly described elsewhere.14 Briefly, a mode-locked Ti: sapphire laser was used to generate a 400 mW pulse train with a typical pulse duration of 120 fs at a repetition rate of 80MHz. After seeding a Ti:sapphire regenerative amplifier, the amplified infrared pulses at 885nm each had an energy of ~160 J and an autocorrelation pulse width of 350 fs at a repetition rate of 5 kHz. They were frequency-doubled and tripled to generate ultraviolet excitation (295 nm) with an average power of 30 mW. The UV beam (pump pulse) was then separated from the infrared beam (885nm, probe pulse), and blue beam (doubled) by two dichroic mirrors, and the average power for the excitation of the sample was carefully attenuated to less than 1mW to avoid photodegradation, saturation, hole burning, and other undesirable effects. The sample was placed into “sandwich” cells with a path length of 1 mm that were, in turn, mounted in a delrin disk for attachment to a chopper motor and continuously spun so the tangential velocity of sample through the beam was greater than 1 m/s. The residual infrared pulse was retroreflected from a hollow cube corner on a computer-controlled stage, and this variably delayed pulse was used as a sampling probe pulse for the upconversion process. The fluorescence emission was collected by a pair of parabolic mirrors and focused into a BBO crystal, and the upconversion signal was produced from 232nm to 280 nm via type I sum frequency generation with the sampling pulse in the crystal. To reject the strong background signals (infrared laser, remnant UV and fluorescence) accompanying the upconverted signal, a non-collinear configuration was set up between infrared laser and fluorescence. The polarization of the excitation beam (295 nm) was chosen by a motor-controlled zero-order half-wave plate. Since polarizations of laser pulses and fluorescence were strictly determined by the orientation of nonlinear crystals,35 no extra linear polarizer was necessary in our experiments (in the terms of normal fluorometers, G=1.). The upconverted fluorescence was detected by a monochromator with a bandwidth of 0.5nm and a solar blind photomultiplier tube. Amplified signals were discriminated and then recorded by a Ortec 994 gated single photon counter The “lamp” (or instrument response) function under the current nonlinear geometry was around 400 fs as determined from the cross-correlation between UV-generated Raman scattering at 328 nm in water and the infrared laser. Instrument calibration with the linear fluorophore p-terphenyl yielded an initial anisotropy of 0.40+/−.01 and a single rotational correlation time of 41ps in cyclohexane (both from Sigma/Aldrich).
TCSPC apparatus
A tunable Spectra-Physics cavity dumped dye laser (3520) was synchronously pumped by a mode-locked DPSS (Vanguard) green laser, and dye laser output was doubled into the ultraviolet. For the present work, rhodamine 6G was used as the laser dye, and vertical excitation was employed at 295nm with pulses having a FWHM of 2ps. The fluorescence was recorded from 310nm to 405 nm through a magic angle polarizer by use of a cooled microchannel plate photomultiplier. Fluorescence decays for the samples and reference were measured to 1 × 104 counts in the peak channel. The instrumental response time was about 90ps, so the measurement of lifetimes of ~60ps and greater can be made in this instrument. A JYH10 monochromator with 1.5mm slit width was used to select the emission. Melatonin in water was used as a standard, for which separate experiments showed monoexponential decay and a lifetime of 5.4ns. Lifetimes were obtained by fitting the decay data to a multiple-exponential model, according to the weighted, least-squares method. Goodness of fit was assessed with the χ2R function.36 The fits for which data are given in this work yielded values of 1.01-1.2. For decay-associated spectra (DAS), time-resolved data were obtained at every 5 nm over the emission band from 310 nm to 405 nm. Alternating the sample with the scatterer, stepping of the emission monochromator, data collection, and transfer of data from the multichannel buffer to the computer was done automatically. In the analysis of the multiple curves obtained for a DAS, they were all fit to the same multiexponential model:37 I(λ, t) = ∑αi(λ) · e−t/τi.
Steady-state absorption and fluorescence spectra were characterized with a diode array spectrophotometer (HP 8452A) and Fluorolog-3 spectrophotofluorometer (SPEX), respectively.
Samples
N-acetyl-L-tryptophan-amide (NATA), L-Trp, and the Trp dipeptides (Trp-Leu, Trp-Ala, Trp-Phe, Trp-Gly, Leu-Trp, Val-Trp, Gly-Trp) were purchased from Sigma-Aldrich chemical Co. They were used without further purification. The solutions were prepared in a 10 mM sodium acetate buffer at pH 5.2 or sodium borate buffer at pH 9.3 using distilled deionized water. A typical concentration of the dipeptides in water for upconversion was 1 mM. All samples were made at the room temperature. A fresh sample solution was prepared for each time-resolved measurement.
III. Results
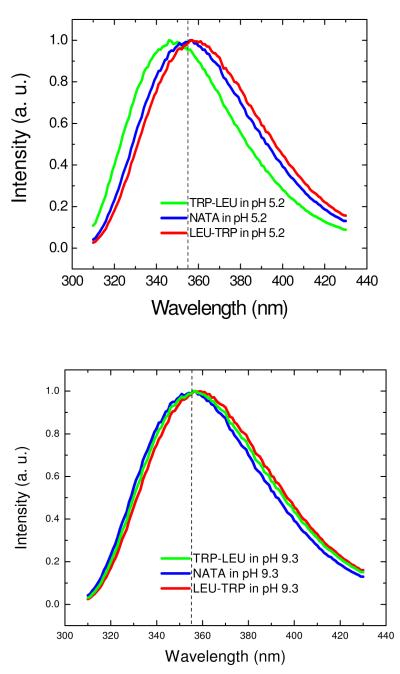
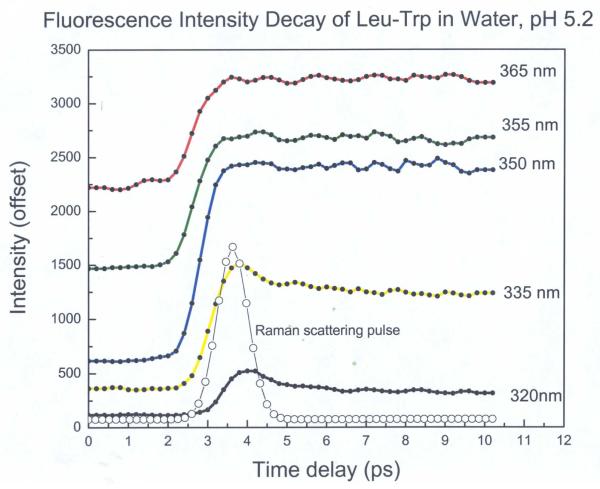
All Trp compounds studied here had a typical indole absorption band at a peak around 280nm (data not shown). Figures 1A, 1B show the steady state fluorescence spectra (295nm excitation) of NATA and Trp dipeptides (Trp-Leu, Leu-Trp) in water at pH 5.2 and 9.3, respectively. Several features should be noted: First, the spectrum of NATA (~355 nm) does not depend on pH. Second, the dipeptides show red (Leu-Trp) or blue (Trp-Leu) shifts relative to NATA (355nm) when pH is 5.2. For example, the peak for Trp-Leu at pH 5.2 is shifted by 11 nm to the blue compared with that of Leu-Trp. However, when pH is 9.3, the spectrum maximum matches that of NATA, indicating the identity of X does not govern spectral position when pH is high (anionic form). This is in accord with prior observations 33 and recent simulations.45 A series of upconversion transients for Leu-Trp (taken with magic angle excitation geometry) at various emission wavelengths is shown in Fig. 2. Clearly, there is a rapid process (corresponding to a 1-2ps exponential) that is depleting the blue side of the spectrum and creating a corresponding rising term on the red edge. This 1-2ps term is seen in all picosecond Trp studies and is ascribable to the dipolar relaxation of bulk water in response to the larger dipole of Trp in the excited (1LA) singlet state.14-16 This term provides smaller amplitude contributions near the center of the spectrum (where its sign changes).
Figure 1.
Normalized steady state fluorescence spectrum of dipeptides in water at (A) pH 5.2 or (B) pH 9.3.
Figure 2.
Representative fluorescence intensity decay for Leu-Trp in water at different emission wavelengths after excitation at 295 nm within a 10 ps window.
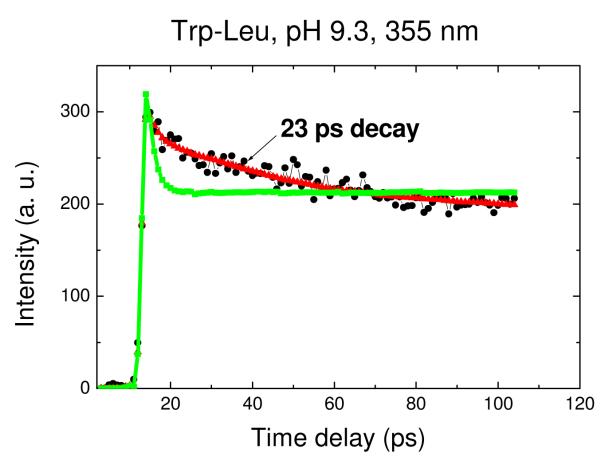
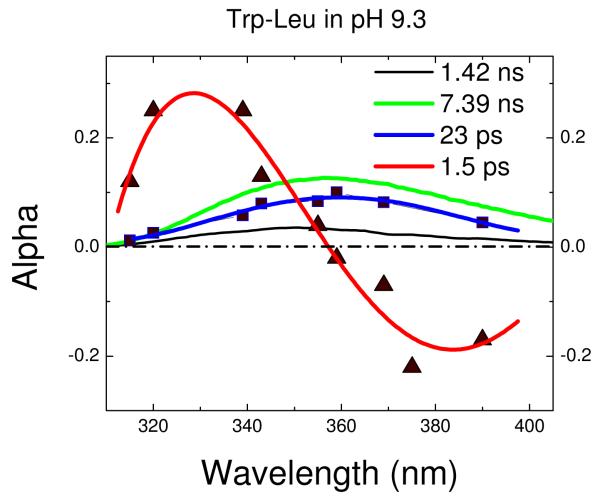
The upconverted transient for the central (~355nm) portion of the spectrum for Trp-Leu is shown in Fig. 3. It is clear that a biexponential fit is inferior to one including not only the ns mean lifetime (gleaned from TCSPC) and the 1-2 ps water relaxation term, but also a positive exponential of 20-30ps. To ascertain the origin of this term, one must collect a full decay surface. As we have previously shown for the proteins monellin and IIAGlc, the shape of the spectrum composed of preexponential amplitudes (DAS) can help distinguish heterogeneous vs. solvent relaxation mechanisms.16 The DAS from a full decay surface for Trp-Leu at pH 9.3 is shown in Fig. 4. Importantly, the DAS for the 1-2ps exponential has the “positive blue, negative red” signature of solvent relaxation while the DAS for 23ps is positive at all measured wavelengths (up to 400nm, where our detection system becomes inefficient). Further, the DAS shape does not suggest an imminent axis crossing; in fact, the 23ps DAS has about the same shape and width as those found by TCSPC (likely for heterogeneous conformers with distinct nanosecond decay times). The positive portion of a shortlived, solvent relaxation DAS that becomes negative in the red tail will necessarily be more narrow than the entirety of a positive-definite DAS arising from heterogeneity. One can never rule out the coexistence of some solvent relaxation in these spectra on similar timescales, but the data here do not require it.
Figure 3.
Representative fluorescence intensity decay for Trp-Leu in water within a 100 ps window. The ultrafast emission decay of Trp-Leu in pH 9.3 buffer was taken at 355 nm. The green line is an attempted biexponential fit yielding lifetimes of 1.6 ps and ~6 ns with amplitudes of 0.21 and 0.13, respectively (χ2R = 2.3). The triexponential fit yields lifetimes of 1.6 ps, 23 ps, and ~6 ns with amplitudes of 0.10, 0.05 and 0.19, respectively; only the triexponential has acceptable fitting error (χ2R = 1.1).
Figure 4.
Decay associated spectra of Trp-Leu in water. The DAS for terms other than bulk water relaxation (1-2ps) are positive-definite and have similar spectral widths.
We have compiled transients and DAS for several commercially available nonpolar Trp dipeptides, both in Trp-X and X-Trp configurations. In all cases, the 1-2ps DAS changes sign with wavelength while the 20-30ps term remains positive even at the red tail. Thus, the 1-2ps term represents bulk water relaxation and the 20-30ps term represents a rapidly decaying subpopulation (quenching mechanism yet to be determined). The bulk relaxation process reduces the energy of, but does not depopulate, the singlet (shifting bluer emitters to the red side). Hence it has no first order influence on the quantum yield. The 20-30ps decay term, however, contributes to the previously measured (and predicted) QSSQ of the peptides where it is found.
We have compiled distillates of these dipeptide decay characteristics in Table I and II. We define the parameter δ as the ratio of the sum of amplitudes of all terms with lifetimes greater than 2ps to all those above 100ps. In other words, δ is the ratio of molecular population that can be observed decaying in this new ultrafast measurement to that observable by TCSPC alone. The same ratios could be expected for any method limited by the intrinsic jitter of a photomultiplier detector (even though a few virtuoso TCSPC and phase/modulation measurements have been made down to ~40ps). We have studied the confidence limits on δ by examining the relationship between the fitting error (reduced chi-squared, χ2R) and excursions in the fixed amplitude of the 20-30ps term (all other parameters free). The F-test for such a system with ~90 degrees of freedom sets bounds at an approximately 10% χ2R increase; thus we expect (averaging multiple runs) that tabulated δ are accurate to approximately +/− 6%.
Table I.
Apparent and Corrected Radiative Lifetimes for Dipeptides at 350 nm, pH 5.2
| Dipeptides And Standard |
Qrel | <τ> | δ | IB I |
IB II |
“τrad”= <τ>/Qrel |
τ’rad = τrad/δ |
τcorr = τ’rad*IB |
|
|---|---|---|---|---|---|---|---|---|---|
| I | II | ||||||||
| NATA | 1.0 | 2.96 ns | 1.0 | 1.0 | 1.0 | 2.96 | 2.96 | 2.96 | 2.96 |
| L-Trp | 0.73 | 2.71 ns | 1.0 | 0.82 | 0.80 | 3.71 | 3.71 | 3.04 | 2.97 |
| Trp-Ala | 0.381 | 1.33 ns | 1.15 | 0.82 | 0.80 | 4.01 | 3.49 | 2.86 | 2.80 |
| Trp-Phe | 0.331 | 1.60 ns | 1.06 | 0.90 | 0.74 | 4.83 | 4.56 | 4.10 | 3.36 |
| Trp-Gly | 0.335 | 1.38 ns | 1.0 | 0.76 | 0.74 | 4.12 | 4.12 | 3.13 | 3.10 |
| Trp-Leu | 0.404 | 2.24 ns | 1.22 | 0.67 | 0.67 | 5.54 | 4.50 | 3.02 | 3.02 |
| Leu-Trp | 0.215 | 1.18 ns | 1.0 | 0.54 | 0.54 | 5.49 | 5.49 | 2.96 | 2.96 |
| Val-Trp | 0.237 | 1.18 ns | 1.14 | 0.90 | 0.75 | 4.98 | 4.36 | 3.92 | 3.27 |
| Gly-Trp | 0.144 | 0.81 ns | 1.28 | 0.77 | 0.77 | 5.63 | 4.39 | 3.38 | 3.38 |
Table II.
Apparent and Corrected Radiative Lifetimes for Dipeptides at 350 nm, pH 9.3
| Dipeptides And Standard |
Qrel | <τ> | δ | IB I |
IB II |
“τrad” = <τ>/Qrel |
τ’rad = τrad/δ |
τcorr = τ’rad*IB |
|
|---|---|---|---|---|---|---|---|---|---|
| I | II | ||||||||
| NATA | 0.99 | 2.94 ns | 1.0 | 1.01 | 0.99 | 2.97 | 2.97 | 2.97 | 2.91 |
| L-Trp | 1.19 | 5.59 ns | 1.0 | 0.82 | 0.70 | 4.70 | 4.70 | 3.85 | 3.29 |
| Trp-Ala | 1.36 | 6.80 ns | 1.18 | 0.82 | 0.80 | 5.0 | 4.24 | 3.48 | 3.39 |
| Trp-Phe | 1.09 | 5.96 ns | 1.08 | 0.98 | 0.72 | 5.47 | 5.06 | 4.96 | 3.59 |
| Trp-Gly | 1.18 | 5.60 ns | 1.0 | 0.90 | 0.71 | 4.75 | 4.75 | 4.28 | 3.39 |
| Trp-Leu | 1.198 | 6.04 ns | 1.34 | 0.77 | 0.77 | 5.04 | 4.54 | 3.49 | 3.49 |
| Leu-Trp | 0.563 | 3.54 ns | 1.0 | 0.82 | 0.52 | 6.28 | 6.28 | 5.15 | 3.27 |
| Val-Trp | 0.958 | 4.73 ns | 1.16 | 0.75 | 0.73 | 4.94 | 4.26 | 3.20 | 3.07 |
| Gly-Trp | 0.302 | 1.50 ns | 1.05 | 0.69 | 0.69 | 4.97 | 4.73 | 3.26 | 3.26 |
Examining Table I and II, it appears the presence or absence of the 20-30ps QSSQ term does not follow a predictable pattern vis-à-vis pH or Trp-X vs. X-Trp. On the other hand, it is present in a subset of these hydrophobic dipeptides but not in NATA or Trp itself, at either zwitterionic or anionic pH. This comports with prior evidence for specific configurations of the peptide bond as an important source of ultrafast quenching.37
We also sought to quantify the remaining fraction of the molecules that were quenched in subpicosecond events and/or, in truly static fashion, the ground state. This can be accomplished by recalculating <t> 33 with carefully normalized ps and ns measurements, or more simply, by meaning “Initial Brightness” (IB). If a time-resolved emission curve contains all exponential terms (i.e. none were below the instrument resolution) and no ground state molecules were selectively sequestered, a measurement at the peak of the curve would essentially be the sum of amplitudes times the radiative rate (and some other instrumental factors). Thus, equimolar (OD 295) solutions of compounds sharing the same radiative rate will show the same IB unless they have unresolved QSSQ. For our instrument, only events faster than ~350fs will be unrecorded in an initial brightness measurement.
In Table I and II, we use the same natural (radiative) lifetime surrogate, <τ>/QREL, used by Chen et al.33 to survey QSSQ across the range of peptides. A minor exception is that we choose NATA as the Q=1 standard, since it is essentially pH independent while Trp and the peptides have differing pKa values. As a reminder, these are apparent radiative lifetime ratios only; the fact that they exceed the value for a standard like NATA is what provides evidence of unreconciled QSSQ. The underlying principle we use is that spectra in the same approximate location for the same molecule should share a common true radiative rate.
In Table I and II, we show how each peptide which has a quantum yield defect relative to <τ> (e.g., QSSQ) is affected by both 20-30ps and subpicosecond events. The accompanying bar graphs (Fig.5 and 6) show how the initially elevated <τ>τ> τ>/QREL levels, when multiplied by first δ and then IB factors, lead to essentially constant <τ>τ> τ>/QREL values that agree with the standards (within error).
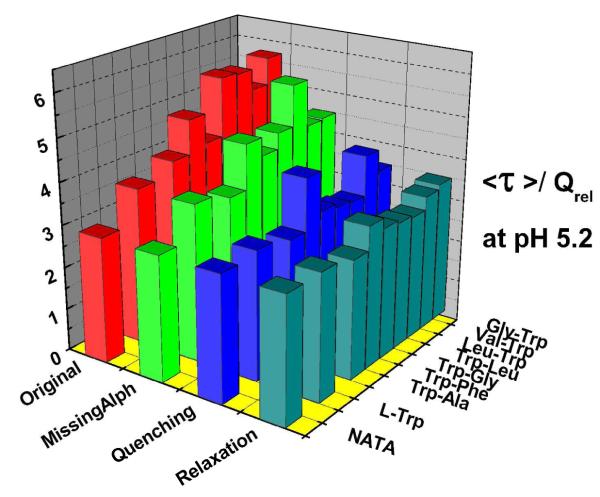
Figure 5.
<τ>τ> τ>/QREL values of Trp dipeptides at pH 5.2.
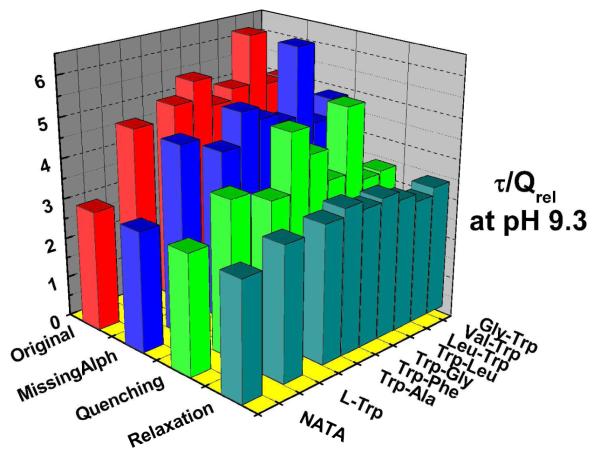
Figure 6.
<τ>τ> τ>/QREL values of Trp dipeptides at pH 9.3.
The 1-2ps solvent relaxation DAS seen for all of these compounds provides a small but important contribution to our IB measurements taken at 350nm; the final row of bars in Figs. 5 and 6 represent a correction to the IB factor that reduces it by the ratio (all amplitudes except 2ps/ all amplitudes). This extracts and excludes the presumed solvent relaxation term from IB. This presumes the 2ps term is a population conserving relaxation (if the 2ps decay term were entirely quenching rather than relaxation, the uncorrected IB term would be appropriate). The fact that the <τ>τ> τ>/QREL levels are both more consistent and closer to standards for the corrected IB approach suggests, again, that most of this 1-2ps term is “zero-sum” wrt yield and therefore arises from relaxation. The symmetry of the 2ps DAS wrt the abscissa also support that view.
The final reconciliation of <τ>τ> τ>/QREL values in Fig.5 and 6 leads to values consistent with NATA. Thus we can generally ascribe the QSSQ previously identified by Chen et al.32 to two ultrafast processes: either 20-30ps or sub-ps processes that deplete the population prior to conventional measurements.
IV. DISCUSSION
The role of water in time resolved fluorescence of Trp has long been controversial. Clearly, an increased Trp dipole moment in the excited state, inside a macromolecule dissolved in a polar solvent like water, cannot be immune to generalized relaxation. Every Trp analog, peptide or protein we have studied possesses the strong 1-2ps transient with “blue positive/red negative” DAS that are signatory of excited state reaction).39 Bulk water relaxation near 1-2ps is known from a variety of measurements.14-16
What is more difficult to ascertain is the role, if any, played by slow relaxation in the macromolecular matrix that strongly couples to water 40 or simpler processes like water desorption 41 in the period of a few ps to ~100ps. If one focuses on the TDFSS (Time Dependent Fluorescent Stokes Shift) alone, it is impossible to discern relaxation from heterogeneity. Only a study of DAS, perhaps reinforced with TDFSS spectral width evolution profiles, can identify which mechanism is dominant. 16 Further peeling the 10-200ps time span apart into mixed relaxation and heterogeneity terms 42 can only be done with outside information (e.g., ad hoc, by postulating that terms faster than ca. 200ps have pure relaxation origins).
In contrast, we have shown that exponential terms near 16ps can have origins in heterogeneity in protein;16 here we show that even simple dipeptides display the same ultrafast quenching decay terms. One might expect that dipeptides would display a much simpler set of slow conformational fluctuations than complex polypeptides in folded configurations. Thus it is likely that the agreement between peptides and proteins about a 20--30ps quenching term is due to a common quenching mechanism. A plethora of other evidence points to the peptide bond as a potent quencher of Trp in agreement with our observations.43,44
It is not clear why the QSSQ proceeds at ~20ps in only a subset of these peptides; all have subpicosecond quenching as well. Further, NATA possesses the equivalent of two bonds but does not suffer the quench. It will require a rather large series of QM-MM simulations upon a variety of seeded peptide structures to learn whether only certain rotamers of both indole and the nonpolar sidechain lead to charge-transfer proximity with the peptide bond (or perhaps other quenching moieties). The fact that Trp also exhibits IB below the IB for NATA suggests that, at least for sub-ps decay, other (non peptide bond) quenching processes are still possible. As a final aside, we note for completeness that an atypical sharp reduction in kr upon relaxation can, in limited cases, also mimic a quenching. 39
V. Conclusions
The ultrafast emission of NATA and Trp dipeptides in water has been studied with femtosecond time resolution. Trp in many of these dipeptides exhibits a multiexponential fluorescence decay including a 20-30ps term in addition to the water relaxation of 1-2ps. This multiexponentiality is apparently associated with ground-state heterogeneity and may arise from different rotamers of Trp or the ‘X’ sidechains. Experiments on constrained system will be needed to confirm this view.44 The corresponding DAS (decay associated spectra) are positive throughout the measurement range. Since these ultrafast components are not detected in conventional time-resolved or phase/modulation experiments, their presence might frustrate the matching of ns decay populations to either nmr-derived or computational predictions for conformers. Accounting for QSSQ may thus improve our understanding of polypeptides. On a more challenging level, this range of ultrafast quenching rates provides signposts for QM-MM simulation of Trp and its environment. 45 Also, we must leave open the possibility that some lifetimes could be surrogates for rates of internal motions that quench certain subpopulations. In any case, heterogeneity provides the dominant source of ultrafast QSSQ.
Acknowledgment
This research was supported by the Intramural Research Program of the NIH, NHLBI. We also thank Profs. Patrik Callis, Dmitri Toptygin, and Ludwig Brand for helpful discussions about Trp photophysics.
References
- 1.Frauenfelder H, Sligar S, Wolynes P. Science. 1991;254:1598. doi: 10.1126/science.1749933. [DOI] [PubMed] [Google Scholar]
- 2.Carerri G, Fasella P, Gratton E. Annu. Rev. Biophys. Bioeng. 1979;8:69. doi: 10.1146/annurev.bb.08.060179.000441. [DOI] [PubMed] [Google Scholar]
- 3.Santos J, Sica M, Marino Buslje C, Garrote A, Ermacora M, Delfino J. Biochemistry. 2009;48:595. doi: 10.1021/bi801969w. [DOI] [PubMed] [Google Scholar]
- 4.Hicks MR, Damianoglou A, Rodger A, Dafforn T. J. Molecular Biology. 2008;383:358. doi: 10.1016/j.jmb.2008.07.091. [DOI] [PubMed] [Google Scholar]
- 5.Beechem JM, Brand L. Annu. Rev. Biochem. 1985;54:43. doi: 10.1146/annurev.bi.54.070185.000355. [DOI] [PubMed] [Google Scholar]
- 6.Lakowicz JR. Principles of Fluorescence Spectroscopy. second edition Kluwer Academic/Plenum Publishers; New York: 1999. [Google Scholar]
- 7.Pan C, Callis P, Barkley M. J. Phys. Chem. B. 2006;110:7009. doi: 10.1021/jp056164p. [DOI] [PubMed] [Google Scholar]
- 8.Callis PR. Methods Enzymol. 1997;278:113. doi: 10.1016/s0076-6879(97)78009-1. [DOI] [PubMed] [Google Scholar]
- 9.Shen X, Knutson JR. Chem. Phys. Lett. 2001;339:191. [Google Scholar]
- 10.Li W, Li H, Zhang G, Bin Chao J, Ling L, Shuang S, Dong C. Journal of Photochemistry and Photobiology A–Chemistry. 2008;197:389. [Google Scholar]
- 11.Kamath D, Kartha B, Mahato K. Spectrochimica ACTA part A-molecular and biomolecular spectroscopy. 2008;70:187. doi: 10.1016/j.saa.2007.06.041. [DOI] [PubMed] [Google Scholar]
- 12.Kao P, Chen K, Lin S, Chang L. Journal of Peptide Science. 2008;14:342. doi: 10.1002/psc.943. [DOI] [PubMed] [Google Scholar]
- 13.Nemec N, Pande H, Qin S, Urbauer B, Tan S, Moe D, Tatulian A. Biochemistry. 2006;45:12448. doi: 10.1021/bi061440r. [DOI] [PubMed] [Google Scholar]
- 14.Shen X, Knutson JR. J. Phys. Chem. B. 2001;105:6260. [Google Scholar]
- 15.Qiu W, Li T, Zhang L, Yang Y, Kao Y, Wang Li., Zhong D. Chemical Physics. 2008;350:154. [Google Scholar]
- 16.Xu J, Toptygin D, Graver K, Albertini R, Savtchenko R, Meadow N, Roseman S, Callis P, Brand L, Knutson J. J. Am. Chem Soc. 2006;128:1214. doi: 10.1021/ja055746h. [DOI] [PubMed] [Google Scholar]
- 17.Rayner D, Szabo AG. Can J. Chem. 1978;56:743. [Google Scholar]
- 18.Knutson JR, Walbridge DG, Brand L. Biochemistry. 1982;21:4671. doi: 10.1021/bi00262a024. [DOI] [PubMed] [Google Scholar]
- 19.Stortelder A, Buijs J, Bulthuis J, van der Vies S, Gooijer C, van der Zwan G. Journal of Photochemistry and Photobiology B-Biology. 2005;78:53. doi: 10.1016/j.jphotobiol.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 20.Szabo A, Rayner D. J. Am. Chem Soc. 1980;102:554. [Google Scholar]
- 21.Ross JA, Wyssbrod HR, Porter RA, Schwartz GP, Michaels CA, Laws WR. Biochemistry. 1992;31:1585. doi: 10.1021/bi00121a002. [DOI] [PubMed] [Google Scholar]
- 22.Mcmahon LP, Yu HT, Vela MA, Morales GA, Shui L, Fronczek FR, McLaughlin ML, Barkley MD. J. Phys Chem B. 1997;101:3269. [Google Scholar]
- 23.Dahms T, Szabo A. Biophysical Journal. 1995;69:569. doi: 10.1016/S0006-3495(95)79930-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.She M, Dong W, Umeda P, Cheung H. Biophysical Journal. 1997;73:1042. doi: 10.1016/S0006-3495(97)78137-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vincent M, Gallay J, Demchenko AP. J. Phys Chem. 1995;99:14931. [Google Scholar]
- 26.Toptygin D, Brand L. Chem. Phys. Lett. 2000;322:496. [Google Scholar]
- 27.Toptygin D, Gronenborn A, Brand L. J. Phys Chem B. 2006;110:26292. doi: 10.1021/jp064528n. [DOI] [PubMed] [Google Scholar]
- 28.Vincent M, de Foresta B, Gallay J. Biophysical Journal. 2005;88:4337. doi: 10.1529/biophysj.104.057497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lakowicz JR. Photochemistry and Photobiology. 2000;72:421. doi: 10.1562/0031-8655(2000)072<0421:OSRIP>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kamal J, Behere D. Journal of Biological Inorganic Chemistry. 2002;7:273. doi: 10.1007/s007750100294. [DOI] [PubMed] [Google Scholar]
- 31.Neyroz P, Menna C, Polverini E, Masotti L. Journal of Biological Chemistry. 1996;271:27249. doi: 10.1074/jbc.271.44.27249. [DOI] [PubMed] [Google Scholar]
- 32.Russo A, Brand L. Journal of Fluorescence. 1999;9:333. [Google Scholar]
- 33.Chen RF, Knutson JR, Ziffer H, Porter D. Biochemistry. 1991;30:5184. doi: 10.1021/bi00235a011. [DOI] [PubMed] [Google Scholar]
- 34.Larsen OFA, van Stokkum IHM, Pandit A, van Grondelle R, van Amerongen H. J. Phys. Chem. B. 2003;107:3080. [Google Scholar]
- 35.Xu J, Knutson J. In: Ultrafast Fluorescence Spectroscopy via Upconversion: Applications to Biophysics, Methods in Enzymology. Brand Ludwig, Johnson Michael L., editors. Volume 450. Academic Press; 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Badea MG, Brand L. Methods Enzymol. 1979;61:378. doi: 10.1016/0076-6879(79)61019-4. [DOI] [PubMed] [Google Scholar]
- 37.Knutson JR, Beechem JM, Brand L. Chem. Phys. Lett. 1983;102:501. [Google Scholar]
- 38.Chen Y, Liu B, Yu H, Barkley M. J. Am. Chem. Soc. 1996;118:9271. [Google Scholar]
- 39.Xu J, Tcherkasskaya O, Gronenborn A, Callis P, Toptygin D, Gleason F, Brand L, Knutson J. Biophysical Journal. 2009;96:46a. [Google Scholar]
- 40.Mark P, Nilsson L. J. Phys Chem B. 2002;106:9440. [Google Scholar]
- 41.Peon J, Pal SK, Zewail AH. PNAS. 2002;99:10964. doi: 10.1073/pnas.162366099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim J, Lu W, Qiu W, Wang L, Caffrey M, Zhong D. J. Phys Chem B. 2006;110:21994. doi: 10.1021/jp062806c. [DOI] [PubMed] [Google Scholar]
- 43.Callis P, Vivian J. Chem. Phys. Lett. 2003;369:409. [Google Scholar]
- 44.Adams P, Chen Y, Ma K, Zagorski M, Snnichsen F, McLaughlin M, Barkley M. J. Am. Chem. Soc. 2002;124:9278. doi: 10.1021/ja0167710. [DOI] [PubMed] [Google Scholar]
- 45.Tusell J, Callis P. Biophysical Journal. 2009;96:590a. [Google Scholar]