Abstract
O-linked N-acetylglucosamine (O-GlcNAc) is a dynamic, reversible monosaccharide modifier of serine and threonine residues on intracellular protein domains. Crosstalk between O-GlcNAcylation and phosphorylation has been hypothesized. Here, we identified over 1750 and 16,500 sites of O-GlcNAcylation and phosphorylation from murine synaptosomes, respectively. In total, 135 (7%) of all O-GlcNAcylation sites were also found to be sites of phosphorylation. Although many proteins were extensively phosphorylated and minimally O-GlcNAcylated, proteins found to be extensively O-GlcNAcylated were almost always phosphorylated to a similar or greater extent, indicating the O-GlcNAcylation system is specifically targeting a subset of the proteome that is also phosphorylated. Both PTMs usually occur on disordered regions of protein structure, within which, the location of O-GlcNAcylation and phosphorylation is virtually random with respect to each other, suggesting that negative crosstalk at the structural level is not a common phenomenon. As a class, protein kinases are found to be more extensively O-GlcNAcylated than proteins in general, indicating the potential for crosstalk of phosphorylation with O-GlcNAcylation via regulation of enzymatic activity.
O-GlcNAcylation, the addition of a single sugar (β-N-acetylglucosamine) to serine and threonine residues on intracellular domains of proteins, is a regulated, reversible post-translational modification. The O-GlcNAcylation state of proteins is responsive to numerous cellular stimuli, including nutrient levels and stress. The addition of this post-translational modification is catalyzed by a single enzyme known as uridine diphosphate-N-acetylglucosamine:peptide β-N-acetylglucosaminyltransferase, referred to as O-GlcNAc-transferase (OGT)1, and it is removed by a single enzyme known as O-glycoprotein 2-acetamido-2-deoxy-β-d-glucopyranosidase, referred to as O-GlcNAcase (OGA) (1). Both of these enzymes are highly expressed in the brain. The physiological roles of protein O-GlcNAcylation may be particularly important in the central nervous system (2, 3). OGT is present in dendrites and axon terminals; it is also associated with microtubules (4). Neuron-specific deletion of OGT results in neonatal lethality due in part to abnormal neuronal development and motor deficits (5).
Because O-GlcNAcylation modifies serine and threonine side chains, there is the potential for interplay between the function(s) of this moiety and those of phosphorylation. Over 1000 proteins have been identified as O-GlcNAc modified. Although the majority of these are also phosphorylated (6), the implications of this are unclear given that the majority of all cellular proteins are probably phosphorylated. In most cases, the exact site of O-GlcNAc modification within a protein is still unknown. Traditional biochemical analysis has revealed numerous proteins that have been shown to be both phosphorylated and O-GlcNAcylated, including c-Myc, nitric oxide synthase, RNA polymerase II, synapsin I, tau, and amyloid precursor protein (reviewed in (6). In cell culture, modulation of the global levels of phosphorylation is accompanied by changes in O-GlcNAcylation levels of many proteins, and vice versa (7, 8). However, the specific sites involved have not been reported (6). Furthermore, pharmacological inhibition of a kinase causes a complex response, with an increase in O-GlcNAcylation of some proteins and a decrease in others (9). A mechanistic explanation of results from these experiments is lacking.
Cross talk has been proposed at the cellular level based on experiments inhibiting or promoting either O-GlcNAc or phosphorylation levels directly and examining what effect these treatments have on levels of the other PTM. For example, cells have been treated with lithium to inhibit GSK-3 (9), resulting in identification of 10 and 19 proteins with increased and decreased levels of O-GlcNAcylation, respectively. Inhibition of the O-GlcNAcase in NIH-3T3 cells resulted in elevated O-GlcNAcylation levels, which in turn was correlated with increases or decreases in over 50% of the measured phosphorylation sites (7). Overexpression of the O-GlcNAc transferase resulted in decreased phosphorylation in 17% of phosphorylation sites and increased phosphorylation at 7% of sites (10). Interpretation of these studies is complicated by the fact that the stimulation approaches were nonphysiological and resulted in widespread changes in cellular physiology. For example, if overexpression of OGT altered activity of a kinase, all phosphorylation changes, however far downstream, might be interpreted as evidence for crosstalk between the PTM systems. In general, these studies have not established unambiguously that individual protein molecules showing an increase in O-GlcNAcylation also had a change in phosphorylation (or vice versa).
Driven by advances in affinity chromatography and the development of several generations of even more powerful tandem mass spectrometers (11, 12), our knowledge of the complexity and extent of cellular phosphorylation is still growing dramatically. In contrast, our knowledge of O-GlcNAcylation is progressing slowly, because of less robust enrichment methodologies and lack of suitable, broadly applicable and sensitive mass spectrometric methodologies.
Early tandem mass spectrometric studies required relatively large amounts of individual proteins and were successful in the assignment of sites of O-GlcNAc modifications in α-crystallin (13) as well as of both O-GlcNAc and phosphorylation sites in isolated serum response factor (14). These studies on O-GlcNAcylation employed traditional collision-induced dissociation (CID); however, this common internal energy deposition technique suffered from the instability of the sugar group upon vibronic energy deposition in the gas phase (15). Therefore, although O-GlcNAcylated peptides could be identified readily by the shift in their molecular weights by 203 Da, the assignment of the exact site of modification within a peptide required care in adjustment of minimal collision energy. Later, using the widely available CID MS technique, derivatization strategies such as beta-elimination/Michael addition were pursued to alleviate the site assignment problem (16, 17). Recent work has addressed the need for O-GlcNAc-specific peptide enrichment based on both lectin chromatography2 (15, 18, 19) and derivatization of O-GlcNAcylated peptides with biotinylated reagents (10, 20). The recent electron capture and electron transfer dissociation (ECD and ETD, respectively) techniques allow dissociation of peptide backbone linkages without causing elimination of O-GlcNAc-moieties from O-GlcNAcylated peptides. Thus, the formation of O-GlcNAc-containing sequence ion series permits unambiguous site localization without the need for derivatization (15).
In the present work, we established a workflow that permits the combined detection and determination of O-GlcNAcylation and phosphorylation sites on proteins in the same biological sample. Using this approach, we extensively characterized mouse synaptosome preparations. We identified 6621 proteins, including 1750 sites of O-GlcNAcylation and 16,500 sites of phosphorylation. We estimate that 19 and 63% of synaptosome proteins are O-GlcNAcylated and phosphorylated, respectively. These results permit the first statistically robust analyses regarding crosstalk between O-GlcNAcylation and phosphorylation at the structural level. In addition, kinases are more frequently targets of OGT than proteins in general, demonstrating crosstalk at the catalytic level.
EXPERIMENTAL PROCEDURES
Preparation of Mouse Synaptic Membranes
Synaptic membrane samples were purified at 4 °C, as described previously (21) in the presence of the O-GlcNAcase inhibitor PUGNAc (Toronto Research Chemicals, North York, ON, Canada) and a mixture of phosphatase inhibitors throughout the preparation. Briefly, brains from adult mice (strain C57BL/6J) were dissected; the cerebellum was removed and the brains immediately frozen in liquid nitrogen. Material from several animals was combined prior to the biochemical purification. The brain tissue was homogenized in a sucrose buffer containing phosphatase inhibitors (1 mm Na3VO4, 1 mm NaF, 1 mm Na2MoO4, 4 mm sodium tartrate, 100 nm fenvalerate, 250 nm okadaic acid) and 20 μm PUGNAc, and cleared by centrifugation. Ten milliliters of buffer was used per gram of brain. The membranous fraction was layered on a sucrose density and fractionated by centrifugation. Synaptic membranes were collected at the 1.0–1.2 M interface and harvested by centrifugation.
Digestion of Synaptosome Samples
Thirty milligrams of synaptosome was resuspended in 1 ml buffer containing 50 mm ammonium bicarbonate, 6 m guanidine hydrochloride 6× Phosphatase Inhibitor Cocktails I and II (Roche), and 20 μm PUGNAc (Tocris). The mixture was incubated for one hour at 57 °C with 2 mm Tris(2-carboxyethyl)phosphine hydrochloride to reduce cysteine side chains, these side chains were then alkylated with 4.2 mm iodoacetamide in the dark for 45 min at 21 °C. The mixture was diluted sixfold with ammonium bicarbonate to a final ammonium bicarbonate concentration of 100 mm and 1:50 (w/w) modified trypsin (Promega, Madison, WI) was added. The pH was adjusted to 8.0 and the mixture was digested for 12 h at 37 °C. The digests were desalted using a C18 Sep Pak cartridge (Waters, Milford, MA) and lyophilized to dryness using a SpeedVac concentrator (Thermo Electron, San Jose, CA).
Preparation of the Lectin Weak Affinity Chromatography Column
Three hundred micrograms of POROS Al resin (Applied Biosystems) was reacted with 25 mg of WGA per the manufacturer's instructions. Briefly, 10 mm bicine, pH 7.5 was used as the reaction buffer and 5 mg/ml sodium cyanoborohydride was added along with 200 μl of 2 m sodium sulfate. The mixture was rotated at 21 °C for 24 h. The resin was spun down and washed with 10 ml bicine, then quenched with 10 ml of a 200 mm Tris acetate buffer, pH 7.5 and 200 μl sodium cyanoborohydride (100 mg/ml). The resin was then packed into a 2 × 250 mm stainless steel column.
Enrichment of GlcNAcylated Peptides Using a WGA Column
Peptides were resuspended in 50 μl buffer A (100 mm Tris pH 7.5, 150 mm NaCl, 2 mm MgCl2, 2 mm CaCl2, 5% acetonitrile). Peptides were run over the column at 125 μl/min. GlcNAcylated peptides eluted as an unresolved smear on the right side of the flow through tail peak. After 1.3 ml, an additional 100 μl of 20 mm GlcNAc in buffer A was injected to elute any remaining peptides. A GlcNAc-enriched fraction was collected between ∼1.3 and 3.7 ml. To decrease the chance of overloading the column each 10 mg portion was split into two 5 mg samples and run separately and the GlcNAc enriched fractions were combined subsequently. For subsequent rounds of enrichment, the pooled fractions were run together in a similar fashion as before. In subsequent rounds, the GlcNAc-enriched fraction was also collected from 1.3 ml to 3.7 ml.
Enrichment of Phosphorylated Peptides Using Titanium Dioxide
Peptides were resuspended in 250 μl buffer B1 (1% trifluoroacetic acid, 20% acetonitrile). The samples were run at 80 μl/min in buffer B1 over an analytical guard column with a 62 μl packing volume (Upchurch Scientific, Oak Harbor, WA) packed with 5 μm titanium dioxide beads (GL Sciences, Tokyo, Japan) (22). The column was rinsed with H2O, then eluted with 3 × 250 μl saturated KH2PO4 followed by 3 × 250 μl 5% phosphoric acid. A switching valve was used to direct these elutions onto a C18 macrotrap peptide column (Michrom Bioresources, Auburn, CA). The peptides were washed with H2O then eluted with 50% acetonitrile, and this solution was lyophilized to dryness using a SpeedVac concentrator.
High pH Reverse Phase Chromatography
High pH RP chromatography was performed using an ÄKTA Purifier (GE Healthcare, Piscataway, NJ) equipped with a 1 × 100 mm Gemini 3μ C18 column (Phenomenex, Torrance, CA). Individual GlcNAc-enriched or phospho-enriched fractions loaded onto the column in 1% buffer A (20 mm NH4FA, pH 10). Buffer B consisted of buffer A with 50% acetonitrile. The gradient went from 1% B to 21% B over 1.1 ml, to 62% B over 5.4 ml, and then directly to 100% B. The flow rate was 80 μl/min. Twenty fractions were collected and dried down using a SpeedVac concentrator. One milligram of the GlcNAc- and phospho-depleted flow-through material was separated by high pH reverse phase to collect 60 fractions.
Mass Spectrometry Analysis
All peptides were analyzed on an LTQ Orbitrap Velos equipped with a nano-Acquity UPLC. GlcNAc-enriched fractions were analyzed using electron transfer dissociation (ETD). Phospho-enriched fractions were analyzed using collision activated dissociation (CID). Nonmodified peptides were analyzed using HCD. Peptides were eluted using a 90-min gradient. MSMS peaklists were extracted using the PAVA program (23). Data was searched against the UniProt Mus Musculus database (downloaded January 11, 2011; 72932 entries) using Protein Prospector (version 5.10.0). To this database, a randomized version was concatenated to allow determination of false discovery rates. The cleavage specificity was set to “trypsin,” allowing for one missed cleavage. Carbamidomethylation of cysteine residues was set as a fixed modification. Acetylation of protein amino termini, oxidation of methionine residues, pyrolization of amino-terminal glutamine residues, and loss of protein terminal methionine residues were set as variable modifications. For the main GlcNAc search, HexNAc modification of serine, threonine, and asparagines was set as variable modifications. For the phosphorylation search, phosphorylation of serine, threonine and tyrosine was set as variable modifications. Data was searched initially with a 20 ppm tolerance of the parent ion, 0.6 Da tolerance of MS/MS measured in the ion trap (CID and ETD) and 20 ppm tolerance for HCD MS/MS. The precursor mass tolerance was then recalibrated on a file by file basis based upon the mass accuracy of high scoring peptides. Final precursor mass tolerances were between 10 and 13 ppm.
Subsequent searches were carried out to find those peptides simultaneously modified by both PTMs. The search parameters were as above, this time allowing for phosphorylation of serine and threonine residues and HexNAc modification of serine/threonine residues (ETD data), or HexNAc modification with neutral loss (CID data). Peptides simultaneously bearing both modifications were accepted if the corresponding peptide had been found with at least one of the PTMs and the peptide expectation value was below 0.01.
Searches were also conducted allowing the addition of 283.04 Da, corresponding to addition of protein glycosyl phosphorylation on a single residue (24). Peptides with both modifications (or potential glycosyl phosphorylation) were accepted if the corresponding peptide had been found bearing at least one of the PTMs and the peptide expectation value was below 0.01. PTMs were considered positively localized to a single residue if they possessed a SLIP score greater than or equal to six (corresponding to a local false localization rate of less than 5%) (25).
For the resulting output, the corresponding UniGene name, gene, and entry numbers were appended (http://www.ncbi.nlm.nih.gov/unigene). UniProt entries were grouped by their corresponding UniGene genes and redundant peptides within a gene group were removed.
For the nonmodified peptide identifications, a peptide expectation value threshold ≤ 0.01 was used. A protein was considered positively identified if the most confident peptide for that protein had an expectation value ≤ 1e – 7. This resulted in the identification of 6621 UniGene entries and 60,421 unique peptides. At this threshold, the decoy database contained six entries and eight unique peptides (protein FDR = 0.097%, peptide FDR = 0.013%).
O-GlcNAcylation and O-GalNAcylation both increase the mass of the modified peptide by the same amount (203.08 Da), and therefore these two PTMs are indistinguishable in the mass spectrometer. While O-GlcNAcylation occurs almost exclusively on intracellular protein regions, the extracelluar domain of Notch is O-GlcNAcylated (26). Peptides were assigned as ambiguous between O-GalNAcylated or O-GlcNAcylated based upon their annotation in UniProt (downloaded April 2011) as located in extracelluar or luminal regions, without further corrections (27). In the case of transmembrane proteins, UniProt topological information was used when available to determine protein extracellular and cytosolic regions. Proteins and protein regions not annotated as extracellular or luminal were assumed to be cytosolic.
Calculations of Expected versus Observed Frequencies
The expected versus observed crosstalk between the two types of PTMs was determined in three different contexts. In all cases, the analysis was restricted to peptides where the site of modification was assigned with a false localization probability of less than 5% (25). (1) For crosstalk at a single residue, we restricted the entire analysis to the portion of each protein predicted to be in disordered regions in order to maximize the probability that all the serine and threonine residues we were considering were accessible for modification. Of the 135 observed alternatively modified residues, 96 were on disordered regions. For each protein, we then counted the number of times a residue was observed to be both O-GlcNAcylated and phosphorylated in different experiments. For that protein, we then calculated the number of residues expected to be alternatively modified by chance as n * rg * rp, where n represented the number of serine and threonine residues on that protein and rg and rp were the rates of O-GlcNAcylation and phosphorylation, respectively for the same protein (calculated as the number of each modification over the total number of serine and threonine residues). The total expected number of alternatively modified residues was determined by summing across all proteins and this was compared with the observed value using χ2 evaluation (2). For crosstalk within sequence proximity level, we compared the observed versus expected values for the number of times an O-GlcNAcylation event was observed at a distance of n residues from a phosphorylation, for different values of n along the protein sequence. Thus, for each O-GlcNAcylation, we counted the number of phosphorylations at distance n to create a distribution of observed distances. Expected distances were calculated as n * rp limiting the serine and threonine residues to those also at distance n. Values of n were binned in intervals of five to create a larger sample size. Expected values were compared with observed values at each bin interval using χ2 (3). For crosstalk at the three-dimensional proximity level, we compared the expected and observed values for the number of times an O-GlcNAcylation was observed within n Å of a phosphorylation, for different values of n. Calculations proceeded as in (2). Analysis was limited to those modifications falling in a solved structure or good quality comparative model of the protein. In all alternative modification analysis, we limited the serine and threonine residues to those falling in disordered regions only.
Structural Analysis of PTMs
For proteins with an experimentally determined atomic structure or a “good quality” (defined as a model being evaluated with a GA341 score of > 0.8 (28) comparative model in ModBase (29)), secondary structure assignments for each residue were computed by DSSP (30). For proteins without structural information, secondary structure was predicted from sequence using PSIPRED (31). For all proteins, disorder was predicted from sequence using the DISOPRED algorithm with the default 5% false positive threshold parameter. The median length of disordered regions was 28 residues, in line with expected values (32). To assess the distribution of PTMs with respect to protein kinase domains, ATP binding sites, and proton acceptor sites, the corresponding amino acid ranges and positions were obtained from Uniprot (uniprot.org). If this information was not annotated for a given entry (as was the case for some TrEMBL entries), the homologous Swiss-Prot entry was used.
Protein-Protein Docking
Known OGT substrate proteins with an experimentally determined atomic structure or comparative models in ModBase (29) with greater than 85% sequence identity (to limit to models likely to be of near-native quality) were docked to Chain A of the open OGT structure (PDB ID 3PE4) using PatchDock (http://bioinfo3d.cs.tau.ac.il/PatchDock/) (33). Twenty-six O-GlcNAcylation sites from 20 substrate proteins (B1ATI9-T34, O88935-T262, P01942-S53, P05064-S354, P12787-S100, P14094-S160, P18760-S8, P60710-S199;-S365, P62874-S136, P68134-S241, P70365-T642, P97315-S192, Q3UHK5-S45;-S559;-S650, Q80X68-T454, Q8K4S1-S2139, Q8VC88-S159, Q9CWF2-T285, Q9CZ13-T217, Q9D1Q6-T367;-S380;-S381, Q9JKV1-S211;-S213) were docked to a truncated version, residues 467–1031, of Chain A of the open OGT structure, which excludes the TPR domain. Two distance constraints were applied to each of the substrate proteins. Atom 1462 in the 3PE4 structure, the nitrogen atom on the catalytic H498 of OGT, was required to be within 13.5 Å of the oxygen atom on the serine or threonine to be O-GlcNAcylated. Atom 11140 in the 3PE4 structure, an oxygen atom on the α-phosphate of UDP, was required to be within 13.0 Å of the nitrogen atom on the serine or threonine residue to be O-GlcNAcylated. A clustering RMSD of 4.0 Å and the default complex type were used. Using the link provided on the PatchDock results page, the best 100 solutions were further automatically refined using FireDock (34). The following substrates had O-GlcNAcylation sites that were not successfully docked: A2AA49; O08599; P08249; P14094; P48962; P56480; P60710; P62874; P68134; P68369; Q03265; Q80X68; Q91VR2; Q9CWF2; Q9CWS0; Q9JKV1.
RESULTS
Abundance of O-GlcNAcylation and Phosphorylation
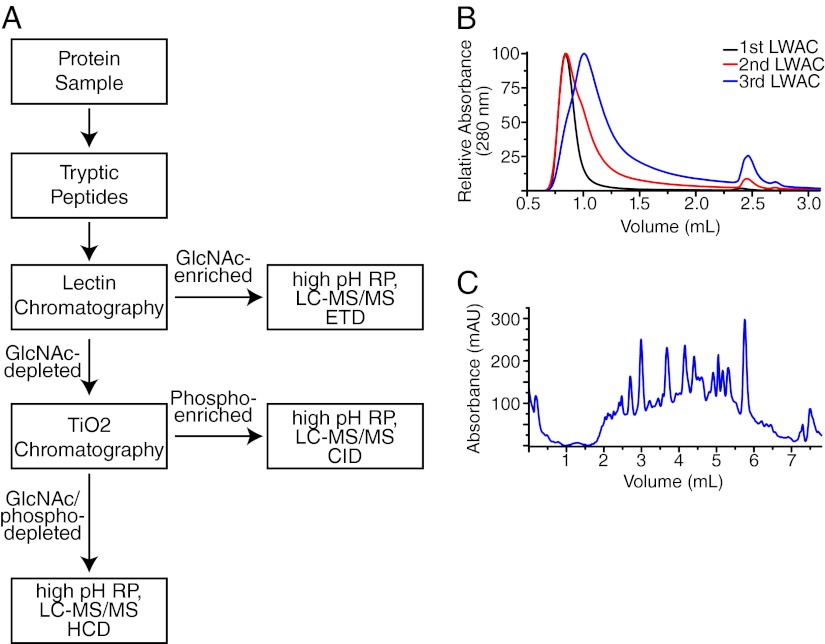
We developed a workflow to sequentially enrich O-GlcNAcylated and phosphorylated peptides from tryptic digests of mouse synaptosomes, which also allowed for analysis of the protein content from the PTM-depleted sample (Fig. 1A). O-GlcNAcylated peptides were isolated using three rounds of lectin weak affinity chromatography (LWAC) (Fig. 1B), yielding a final pool containing ∼30% O-GlcNAcylated peptides. Phosphorylated peptides were isolated using an automated TiO2-based enrichment step (supplemental Fig. S1). Then these two PTM-enriched fractions as well as the final unbound fraction (containing nonmodified peptides) were fractionated using high pH reverse phase chromatography (Fig. 1C). All fractions were analyzed on an LTQ-Orbitrap Velos mass spectrometer using electron transfer dissociation (ETD) for O-GlcNAc peptides and collisional dissociation (CID or HCD) for phospho- and other peptides. Interpretation of these mass spectral analyses resulted in the identification of 2434 distinct O-GlcNAcylated- and 23,206 phosphorylated peptides, respectively. These assignments correspond to over 1750 unique sites of O-GlcNAcylation and 16,500 unique sites of phosphorylation. Additional analysis of the PTM-depleted peptide fraction identified 60,422 peptides from 6621 proteins, all at global false discovery rates (FDRs) of less than 1% (supplemental Tables S1–S3). As we have previously reported, our enrichment technique using the lectin wheat germ agglutinin (WGA) also enriches for N-GlcNAcylated peptides (15). In our current analysis, we found 546 N-HexNAcylated peptides (supplemental Table S4). Although WGA has been reported to be selective for GlcNAcylated peptides and proteins (35), we have identified 181 peptides in the WGA-enriched fractions that are potentially O-GalNAcylated based upon sub-cellular localization of the modification (supplemental Table S5).
Fig. 1.
Combined analysis for O-GlcNAcylation, phosphorylation and proteins. A, Workflow schematic for the serial analysis of O-GlcNAcylation, phosphorylation, and protein content. Synaptosome preparations were digested with trypsin, and the resulting peptides were enriched for GlcNAcylation using lectin weak affinity chromatography. The GlcNAc-depleted peptides were next enriched for phosphorylated peptides using TiO2 chromatography. The final GlcNAc/phospho depleted fractions as well as the individual PTM-enriched fractions were each fractionated using high pH reverse phase chromatography. Individual fractions were then analyzed on an Orbitrap Velos. O-GlcNAc-enriched peptides were analyzed by ETD, phosphorylated peptides by CID, and nonmodified peptides by HCD. B, The UV trace during three sequential runs of lectin chromatography. In each case, an aliquot of GlcNAc (eluting at 2.5 ml) was used to release tightly binding peptides. Peptides were collected as a single fraction starting at 1.2 ml, desalted and re-run for a total of three rounds. The UV shift to later elution times corresponded with a subsequent enrichment in the percentage or peptides that were O-GlcNAc-modified. C, The UV trace of the high pH reverse phase gradient for the final O-GlcNAc-enriched fraction. Similar gradients were used to fractionate all peptides prior to analysis by LC-MS/MS. See also supplemental Fig. S1.
Efficiencies of PTM Detection
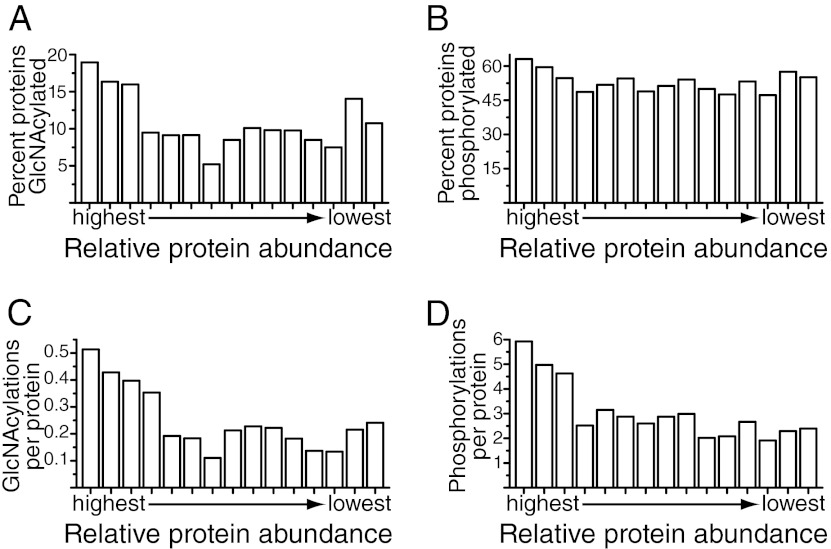
A major factor affecting whether or not a given peptide is detected in a proteomic study is its relative abundance (36). To estimate how efficiently we identified sites of O-GlcNAcylation and phosphorylation within the synaptosome preparation, we took advantage of our concurrent in-depth analysis of proteins from the same sample. The 6621 identified proteins were divided into bins based upon their relative abundance as determined by calculating an exponentially modified protein abundance index (emPAI) value for each protein (37). We then calculated the percentage of proteins in each bin that were either O-GlcNAcylated or phosphorylated. For the most abundant proteins, we identified 19 and 63% of them to be O-GlcNAcylated and phosphorylated, respectively (Figs. 2A, 2B). Proteins present at lower abundance were substantially less likely to be identified as O-GlcNAcylated (an average of 9.8% for the 12 lowest bins). For phosphorylation, this decrease was more modest. For 52% of the proteins in the 12 lowest bins, at least one site of phosphorylation was identified. Proteins in the most abundant bin had an average of 0.51 and 5.9 sites of O-GlcNAcylation and phosphorylation, respectively (Figs. 2C, 2D). The average number of sites identified per protein dropped off significantly with decreased protein abundance for both PTMs.
Fig. 2.
Prevalence of O-GlcNAcylation and phosphorylation. A, B, Comparison of the percentage of proteins found to be O-GlcNAcylated or phosphorylated as a function of relative protein abundance. The total number of unique nonphosphorylated and non-O-GlcNAcylated peptides per protein was used as an estimate of protein abundance. Only proteins identified with at least three peptides were used in analysis for this figure. The ability to detect that a protein is O-GlcNAcylated increased with protein abundance. Nineteen and 63% of the highest abundance proteins were O-GlcNAcylated and phosphorylated, respectively. C, D Comparison of the average number of modification sites as a function of relative protein abundance. The number of identified PTM-sites per protein increased with protein abundance. An average of 0.5 and 5.9 sites of O-GlcNAcylation and phosphorylation per protein was observed for the most abundant proteins. Note: because the proteins bassoon and piccolo were O-GlcNAcylated to a much higher extent than other proteins, they excluded from the calculations in 2C, where they would occur in the third most abundant protein group. Including them would raise the average number of O-GlcNAcylations per protein in this group from 0.40 to 1.33.
Overall, this finding suggests that although we were able to identify large numbers of both PTMs, we are not identifying all PTM-modified peptides present in the sample, particularly not those originating from lower abundance proteins. This is most likely because of the fact that when a given analysis does not identify all the components in a mixture, there is a strong bias toward acquiring MS/MS on higher abundance components. Based upon the average modifications per protein for the most abundant and thoroughly characterized proteins, we now can postulate the existence of at least 3400 O-GlcNAcylation sites and 39,000 phosphorylation sites for the 6621 proteins identified in our synaptosome preparation. Using the same rationale, we estimated that we identified ∼51 and 42% of the O-GlcNAcylation and phosphorylation sites in our sample, respectively.
Mass Spectrometric Characterization of PTM-Modified Peptides
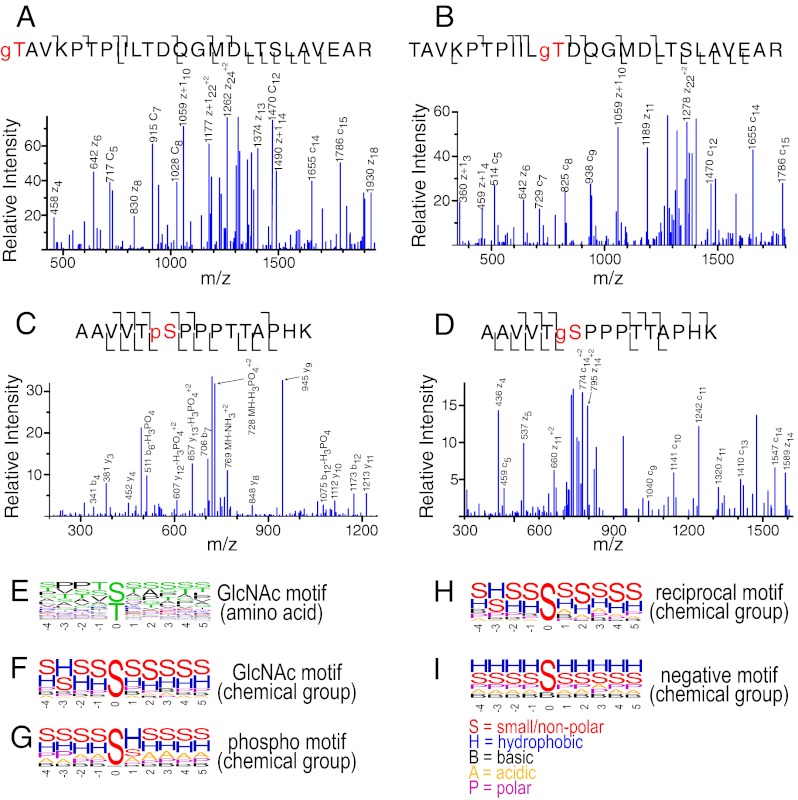
Multiple sites of O-GlcNAcylation were found close in protein primary sequence. We observed 130 instances where the same peptide sequence was found singly O-GlcNAcylated at different residues. Figs. 3A and 3B show MS/MS spectra of two O-GlcNAc positional site isomers of the peptide sequence, TAVKPTPIILTDQGMDLTSLAVEAR, from the protein bassoon. We observed 439 instances of peptides containing multiple sites of O-GlcNAcylation. An example of such a peptide, SVTDTALPGQSSGPFYSPR, modified at serine 1 and threonine 3, is shown in supplemental Fig. S2A. The presence of a relatively bulky O-GlcNAc moiety carboxyl terminal to an arginine or lysine residue may decrease tryptic cleavage efficiency, but these data indicated such cleavage events proximal to an O-GlcNAcylation site are not strictly prevented.
Fig. 3.
PTM-modified MS/MS spectra and motif analysis. Peptides from the O-GlcNAc enrichment were analyzed using ETD, whereas those from the phosphopeptides enrichment were analyzed using CID. A, B, The peptide TAVKPTPIILTDQGMDLTSLAVEAR O-GlcNAcylated at the first or 11th amino acid, respectively. C, D, An example of the peptide AAVVTSPPPTTAPHK phosphorylated or O-GlcNAcylated at the serine in the sixth position, respectively. E, The weblogo motif for an alignment of O-GlcNAcylation sites identified in this study. F–I, show a weblogo motif where individual amino acids are grouped by chemical property. “S” designates small/nonpolar (A, G, S, T); “A” designates acidic (D, E); “B” designates basic (H, K, R); “H” designates hydrophobic (C, R, I, L, M, P, V, W); and “P” designates polar (N, Q, Y). The chemical property weblogos for the O-GlcNAc motif, phospho motif, alternatively modified sites found modified by both PTMs and the background/negative distribution (those serine/threonine residues not found modified) are shown. See also supplemental Fig. S2.
Overall, we observed 135 peptide pairs where one version was O-GlcNAcylated and the other phosphorylated at the same residue. Figs. 3C and 3D show MS/MS spectra of two alternatively modified versions of the peptide AAVVTSPPPTTAPHK from the protein α-adducin, either phosphorylated or O-GlcNAcylated at serine 6. In addition, we found 66 peptide sequences that were simultaneously modified by both phosphorylation and O-GlcNAcylation (supplemental Table S6). One such example is the peptide RASQp(SS)LESSTGPSYgSR, shown in supplemental Fig. S2C. Supplemental Fig. S2B shows an example of an N-GlcNAcylated peptide with the sequence LNGTDPIVAADSKR from the Prolow-density lipoprotein receptor-related protein 1, modified at aspargine 2.
PTM Sequence Motifs
Previous analyses, based on a significantly smaller scale O-GlcNAcylated peptide data set, suggested a PVXS/T motif for substrates of OGT (18). Although this motif is found in a subset of modified peptides in this study, the majority of our O-GlcNAcylation sites fit this motif poorly. In fact, less than 20% of our modified peptides display this motif. Fig. 3E shows a weblogo representation of the amino acid residues surrounding the modified serine/threonine residue. There is a moderate preference for a proline residue either two or three residue positions amino-terminal to the site of modification (the –2 and –3 positions). There is also a slight preference for valine residues at either –1 or –3 positions. Overall, O-GlcNAc appears to be targeted toward regions on substrates rich in serine/threonine residues, as evidenced by an increased frequency of detection of these residues within the five residue region around the modification site that we examined. Such a preference for serine/threonine rich stretches may help explain our finding 439 multiply O-GlcNAc-modified peptides. In summary, these observations suggest that OGT does not recognize the exact primary sequence on substrates.
To investigate motifs within our phosphorylation data set, we used Motif-X to search for over-represented patterns (38). We find that a total of 56 distinct submotifs show statistically significant overrepresentation (supplemental Table S7). To examine the motifs, we grouped amino acid residue types by chemical property (e.g. small hydrophobic, charged and polar side chains) as shown in Figs. 3F–3I. When grouped by chemical property, the most prevalent amino acid residue types present around the site of O-GlcNAcylation are small or nonpolar residues, indicating existence of a hydrophobic residue at the –3 position. Overall, phosphorylation sites in our data set have a similar preference for small or nonpolar residues. In addition, because of the prevalence of proline-directed kinases in the mammalian kinome, there was an increased probability of having a hydrophobic residue at the +1 position. Finally, we examined those serine/threonine residues showing comodification by both PTMs. This subset had a motif most similar to that of the overall O-GlcNAcylation motif. We compared these motifs to the population of serine/threonine residues not found to be PTM-modified. Hydrophobic residues are most prevalent at all positions immediately surrounding these serine/threonine residues (Fig. 3I).
O-GlcNAcylation and Phosphorylation on PTM-Modifying Enzymes
We identified one site of O-GlcNAcylation on OGT, on the carboxy-terminal tail. However we did not identify any phosphorylation on OGT despite the protein being present at relatively abundant levels (28 unique peptides identified) in our synaptosome preparation. Six unique phosphorylation sites on OGT from mitotically active cells have been previously identified (39). On the O-GlcNAcase, we identified two sites of phosphorylation and no sites of O-GlcNAcylation. We identified 280 and 87 proteins in Gene Ontology annotated with the protein kinase (GO:0004672) and phosphatase activities (GO:0004721), respectively. Kinases were more frequently phosphorylated than proteins in the full data set (66% versus 48%, p < 3.8 × 10−11, hypergeometric distribution). Kinases were also more frequently O-GlcNAcylated than proteins in the full data set (16% versus 10%, p < 3.6 × 10−4, hypergeometric distribution). Protein phosphatases however, were not found to be PTM-modified at rates different from the overall data set (52% phosphorylated and 8% O-GlcNAcylated). This evidence supports the notion that O-GlcNAcylation crosstalks with phosphorylation via OGT's regulation of a substantial subset of kinases.
To examine in more detail the manner in which OGT activity may modify kinase behavior, we looked at the localization of O-GlcNAcylation sites on the 46 modified kinases with existing structural information in UniProt. In particular, we asked how closely sites of O-GlcNAcylation mapped to the proton acceptor and ATP binding sites as well as the protein kinase catalytic domain as a whole. Of the 131 O-GlcNAcylated peptides mapping to these 46 proteins, in only two instances was a site of O-GlcNAcylation located within the ∼250 residue kinase catalytic domain (1.5%). In contrast, 134 of the 480 phosphorylated peptides (28%) mapped to the protein kinase domain of kinases. Therefore, while protein kinase domains themselves are subject to extensive modification by phosphorylation, they are minimally O-GlcNAcylated.
With respect to the two kinase domains that were modified, in the first instance (CaMKII α) the O-GlcNAcylation site was more than 100 residues from either the proton or ATP acceptor site. In the second instance (CaMKII β), the O-GlcNAcylation site was 41 residues from the proton acceptor site. Although it is possible that O-GlcNAc may be modifying kinase regulatory domains, it does not appear to be directly affecting kinase catalytic ability or the ability of the catalytic domain to interact with substrate proteins. For several CaMKII isoforms, we observed O-GlcNAcylation of threonine 306/307 (in α and δ, respectively). Upon auto-phosphorylation, this residue inhibits calmodulin binding to CaMKII (40). The phosphorylation state of the adjacent threonine 305 and threonine 306 residues in CaMKII α modulates LTP (41). We detected 11 sites of phosphorylation on CaMKII α, but did not identify phosphopeptides corresponding to threonine 306 or 307. The PTM's of CaMKII α are mapped on its homology-based structural model in supplemental Fig. S3.
Modeling OGT Interactions With Its Substrates
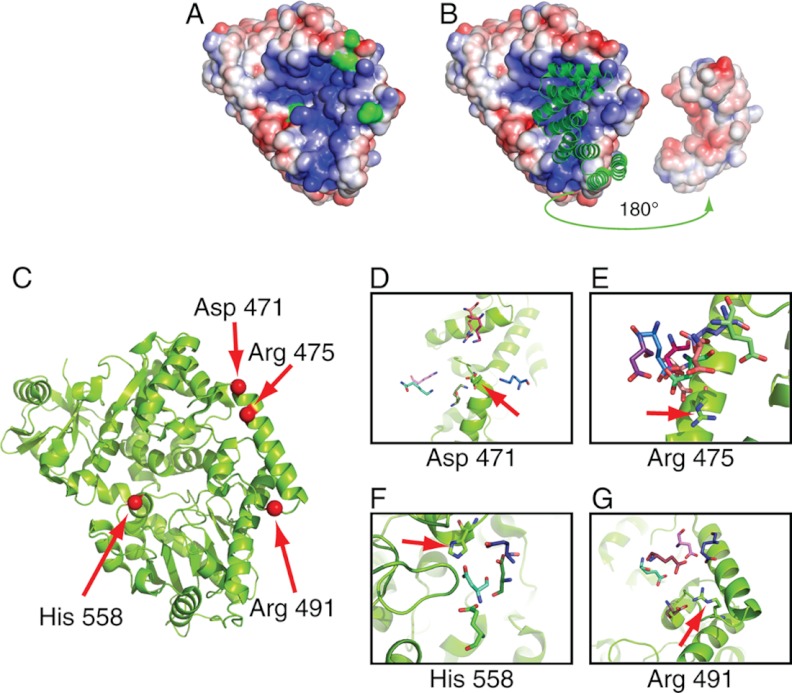
A recent solved crystallographic structure of human OGT in complex with an O-GlcNAcylated peptide and a tetratricopeptide repeat (TPR) domain gave insight into possible factors mediating interaction specificity with the enzyme and its substrates (42). First, the contacts between the enzyme and peptide were made primarily through the peptide main-chain atoms, indicating that peptide side-chain identity does not play a strong role in substrate recognition (Fig. 3E). The possibility exists that this specificity is determined through enzyme-substrate interactions distal to the active site. Second, the TPR domain appears to restrict substrate access to the active site, leading to speculation that the TPR domain swings out in a hinge motion prior to OGT-substrate complex formation. We examined the catalytic face of OGT with the TPR domain removed and observed a large basic patch that encompasses the catalytic site of OGT (Fig. 4A). A complementary acidic patch is present on the proximal TRP domain that interacts with this basic patch (Fig. 4B). The existence of these complementary electrostatic regions was not reported in the original crystal structure manuscript. To determine possible exosite contacts, and examine the possible role of the TPR domain, we applied computational docking methods to model the conformations of 46 O-GlcNAcylated sites identified in our study (from 29 O-GlcNAcylated proteins); the proteins we selected all had solved structures or high quality comparative models (Experimental Procedures). Two docking runs were performed; the first incorporated the TPR domain as observed in the solved structure of the OGT complex, and the second did not.
Fig. 4.
Structural and docking analysis of OGT with a series of known substrates. A, The catalytic domain of OGT as visualized with the surface electrostatic potential visualized using the APBS Tools plugin for Pymol (65). The peptide binding grove exists within a basic patch on the protein surface (blue). Four residues on the catalytic face that made frequent contacts with docked substrates are colored green. B, The TPR domains positioned against the catalytic domain. In the left side, the TPR domains are illustrated green in ribbon format. On the right side, the TPR domain is rotated 180° and shown as the charge distribution along the surface. An acidic patch can be seen (red) which would complement the basic patch on the catalytic domain in the closed confirmation. The protein-protein interface between OGT and each docked substrate was examined. Charged OGT surface residues were identified along with oppositely charged substrate residues in which their charged moiety was found within 10Å of and oriented toward the charged OGT residue. C, Four residues on OGT that were found to make frequent contacts with a range of substrates are shown. The positions of these four residues are shown in red on the OGT catalytic domain. D, aspartate 471 shown potentially interacting residues from six substrates (Q9D1Q6-Lys106; P18760-Lys22; Q61423-Arg28; Q8OX68-Lys393; Q9CQA3-Lys139; Q8K4S1-Lys2216). E, arginine 475 shown potentially interacting residues from eight substrates (P18760-Asp17; P62874-Glu172; Q3UHK5-Glu228; Q9CQA3-Asp50; Q9D1Q6-Asp324; Q9CZ13-Glu157; Q3UHK5-Asp640; Q8K4S1-Asp2194). F, histidine 558 shown potentially interacting residues from four substrates (Q80X68-Asp455; P97450-Asp88; Q9CZ13-Glu157; P62874-Glu138). G, arginine 491 shown potentially interacting residues from five substrates (P14094-Asp270; Q8K4S1-Glu2171; P12787-Asp60; Q3UHK5-Glu221; Q9D1Q6-Glu290). See also supplemental Fig. S3.
The docking protocol attempted to model the native conformation of a complex, assuming that complex formation does not significantly alter the three-dimensional structure of either component. Only one modified protein (cysteine and glycine-rich protein 1, P97315) was able to dock with OGT when the TPR domain was included. However, on removal of the TPR -domain, 26 O-GlcNAcylated sites docked successfully. This observation suggests that it is indeed necessary for the TPR domain to “swing out” prior to complex formation. Furthermore, the inability of the remaining 20 substrates to dock indicates that the bona fide enzyme:substrate complex in these instances requires conformational changes of the substrate and/or OGT for O-GlcNAcylation to occur.
Using the docked conformations of these 26 substrates, we searched for interactions between OGT side-chains and residues structurally conserved across multiple substrates. Although we observed no fully conserved interactions, we identified four charged patches conserved across subsets of substrates that interact with oppositely charged residues on the enzyme (Figs. 4C–4G).
Secondary Structural Elements Involved in Modification Of Substrate Proteins
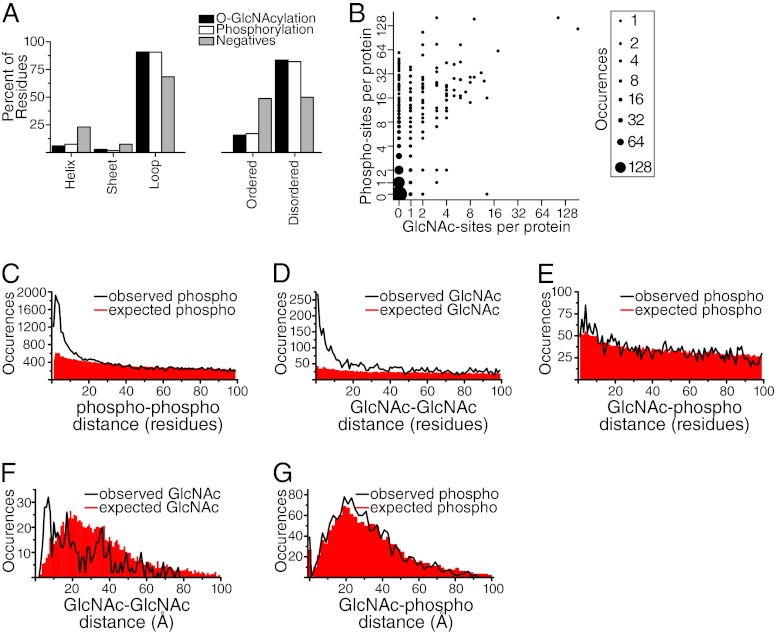
To gain insight into what secondary structure elements may be needed for O-GlcNAcylation and phosphorylation, we determined the frequency with which they appeared on loops, α-helices, and β-sheets. The secondary structure states of all residues in a solved structure or a good quality comparative model were assigned by the Define Secondary Structure of Proteins (DSSP) program (30). For substrate proteins without known or modeled structures, the secondary structure states were predicted by the PSIPRED program (31). Relative to the distribution of these structural elements in general, both O-GlcNAcylation and phosphorylation moieties were enriched within loops and relatively less prevalent within sheets and helices (Fig. 5A). For both PTMs, the site of modification occurred on loops for ∼90% of the sample. We then calculated to what extent the two PTMs were found in ordered versus disordered regions of protein structure. Both phosphorylation and O-GlcNAcylation were ∼sixfold more likely to occur on disordered rather than ordered regions of protein structure (Fig. 5A), in agreement with a previous observation for phosphorylation (43).
Fig. 5.
Relationship between O-GlcNAcylation and phosphorylation sites. A, A structural analysis of O-GlcNAcylation and phosphorylation sites compared with a background list of nonmodified serine and threonine residues. Both PTMs showed a propensity to occur on loop structures. In addition, both were significantly enriched in disordered protein regions. B, A comparison of the number of identified O-GlcNAcylation sites versus phosphorylation sites per protein. The analysis was restricted to the 919 most abundant proteins (based upon emPAI index), where our ability to identify PTMs was highest. The size of each data-point is proportional to the square root of occurrences. Note: There were 2060 proteins at the 0-0 data-point. For clarity, this data-point was given a size of 196 rather than 2060. C, An analysis of phosphorylation distribution with respect to primary sequence. For proteins found multiply phosphorylated, the distance along the primary sequence between pairs of phosphorylation sites was calculated. Phosphorylation sites show a propensity to cluster within regions of protein primary sequence. D, An analysis of O-GlcNAcylation distribution with respect to primary sequence. For proteins found multiply O-GlcNAcylated, the distance along the primary sequence between pairs of O-GlcNAcylation sites was calculated. O-GlcNAcylation sites show a propensity to cluster within regions of protein primary sequence. E, An analysis of the relationship between O-GlcNAcylation and phosphorylation with respect to primary sequence. For proteins found to be modified by both types of PTMs, the distance along the primary sequence from sites of O-GlcNAcylation to sites of phosphorylation was calculated. This distribution was not significantly different from the overall distribution of serine and threonine residues. F, An analysis of the relationship between O-GlcNAcylation sites with respect to three-dimensional distance for multiply O-GlcNAcylated proteins. For those proteins with solved structures or high quality comparative models, we took each site of O-GlcNAcylation and calculated the density of additional O-GlcNAcylation sites at various distances. O-GlcNAcylation sites showed a propensity to co-occur within 10 Å of other O-GlcNAcylation sites. G, An analysis of the relationship between O-GlcNAcylation and phosphorylation with respect to three-dimensional distance. This indicates that within alternatively modified proteins, phosphorylation was not more likely to be localized near O-GlcNAcylation than chance alone.
O-GlcNAcylation and Phosphorylation on Individual Proteins
To investigate how these two PTMs might be coupled at the level of individual proteins, we examined the number of phosphorylation sites per protein as a function of the number of O-GlcNAcylation sites per protein (Fig. 5B). Phosphorylated proteins were significantly more likely to be also O-GlcNAcylated than non-phosphorylated proteins (25.6% versus 4.8%, p < 1.2 × 10−18, hypergeometric distribution). There is a weak correlation between the frequencies of these two PTMs (r2 = 0.25). Interestingly, the vast majority of proteins partitioned to the top left half (i.e. above the diagonal with a phospho:O-GlcNAc ratio of one). The number of O-GlcNAc sites per protein was approximately equal to the minimum number of phosphorylation sites per protein, particularly when the number of O-GlcNAc sites was greater than two. However, for many proteins we observed extensive phosphorylation and only a limited number of O-GlcNAcylation sites. In contrast, in only a single instance did we observe a heavily O-GlcNAc-modified protein that was not also heavily phosphorylated.
The only protein that appeared heavily O-GlcNAcylated (13 sites) with no observed phosphorylation was CCR4-NOT transcription complex subunit 1. CCR4-NOT was among the 20% most abundant proteins in our preparation as estimated by the emPAI value. As a transcription factor, this protein is likely to partition between the nucleus and cytoplasm (for a review see (44)). Regulation of gene transcription is a protein functional class known to be preferentially O-GlcNAcylated (45). Only one site of phosphorylation on CCR4-NOT has been reported (46). Because only a minor fraction of this protein was likely present in our synaptosome preparation, it is possible that analysis of a total cell lysate (rather than of a specific compartment) would reveal additional sites of CCR4-NOT phosphorylation.
Substrates of OGT in general appear to consist of a subset of kinase targets. The fact that the extent of potential protein O-GlcNAcylation tracks with the extent of potential protein phosphorylation is consistent with OGT and serine/threonine kinases using similar mechanisms to target substrates. For example, a portion of OGT has been reported to occur in complex with catalytic subunits of protein phosphatase 1 (47).
Sites that are Modified by Either PTM
As noted above, in 135 instances, we observed phosphorylation and O-GlcNAcylation of the same residue, representing 8% of the identified O-GlcNAcylation sites found in this study. This number of alternatively modified sites suggests coupling between these two PTM systems; however, given the extensive number of phosphorylation and O-GlcNAcylation sites we identified in this study, it is expected that both PTMs would map to the same amino acid residue at some frequency by chance alone. If these two PTM systems have evolved to crosstalk functionally, the observed frequency with which the same residue was found modified by both PTMs should substantially exceed the frequency predicted by chance alone. We assumed that all serine and threonine residues on disordered regions were accessible to enzymatic modification (which was not necessarily the case for serine and threonine residues in ordered protein regions). We therefore limited our analysis to disordered regions. Approximately 50% of all serine and threonine residues are in disordered regions, and 96 of our 135 observed alternatively modified sites mapped on disordered regions (Fig. 5A). For a given protein, the number of alternatively modified sites expected by chance alone is modeled as the rates of phosphorylation and O-GlcNAcylation on disordered regions for that protein multiplied by the total number of disordered serine and threonine residues. Summing the expected alternatively modified residues across all proteins in our data set resulted in 96.4 sites of alternative modification expected by chance alone. Therefore, although both PTMs are preferentially targeted to disordered regions of protein structure, within these disordered regions, we find no increased propensity for modification at the same residue above chance.
Relationship Between the Two PTMs With Respect to Primary Structure
Spatial proximity between sites of O-GlcNAcylation and phosphorylation has been posited as a mechanism for crosstalk (6), whereby addition of one PTM will impact on the odds of the other via electrostatic or steric factors. If an organism has evolved to use such a mechanism, sites of O-GlcNAcylation should display an increased propensity to be localized in spatial proximity to sites of phosphorylation. We tested this hypothesis using two different representations of spatial proximity. The first computes proximity as the number of co-occurrences within a given number of residues along the protein sequence (“sequence proximity”) and the second computes it as the number of co-occurrences within a given three-dimensional distance in solved substrate structures (“three-dimensional proximity”; next section). For each, we compared the number of observed co-occurrences within different distances to the number expected by chance alone.
To calculate the expected number of co-occurrences in sequence proximity for a given protein and residue distance cutoff, we multiplied the phosphorylation rate for that protein by the number of serine and threonine residues within the distance cutoff of each observed O-GlcNAcylation, and summed the results for all observed O-GlcNAcylations in that protein. We found that there was essentially no increase in the number of observed phosphorylations at close distances (i.e. fewer than ten residues) in sequence proximity to O-GlcNAcylation sites relative to that expected by chance alone (Fig. 5E). We compared this distribution to the propensity of phosphorylation sites to cluster within a protein. We calculated the expected versus observed number of phosphate-phosphate co-occurrences at different distances in sequence proximity using the same approach as for phosphorylation - O-GlcNAcylation and detected a significant enrichment (Fig. 5C) similar to that reported previously (48–50). We then examined the same enrichment of observed versus expected co-occurrences of pairs of O-GlcNAcylation modifications (Fig. 5D) and detected a similar propensity to cluster. While there is a slight increase in the number of observed phosphorylation - O-GlcNAcylation co-occurrences relative to that expected by chance, it is significantly less than the robust increase in co-occurrences found within each type, indicating there is not strong evolutionary pressure for crosstalk through sequence proximity between O-GlcNAcylation and phosphorylation. In addition, our observation of 66 peptide sequences simultaneously bearing both PTMs (supplemental Table S6) demonstrates that electrostatic/steric factors do not strictly prohibit simultaneous alternative modification by both PTMs nearby in primary sequence distance.
Relationship Between the Two PTMs With Respect to Tertiary Structure
Although sequence proximity between PTM pairs is a proxy for the underlying three-dimensional distance, we could not calculate this value for all pairs since most of our OGT substrates lack solved angstrom level structures. Nevertheless, of the 285 proteins with both types of PTMs, 52 had such a structure or comparative model available covering both sites of modification. For these, we counted the number of observed phosphorylated residues within a given three-dimensional distance of each O-GlcNAcylated residue, using shells of increasing radii. We compared this observed value to that expected by chance, using the phosphorylation rates and total number of serine and threonine residues at the same distance, as was done in the sequence proximity analysis. We found that there was no enrichment for co-occurrences relative to that expected by chance (Fig. 5G). We compared this result to the enrichment for pairs of O-GlcNAc modifications in three-dimensional proximity. Here, we found that observed pairs of these modifications are more likely to co-occur than expected by chance at less than 10Å (Fig. 5F). This result supports the one determined from the sequence proximity analysis, indicating that there is little to no evolutionary pressure for crosstalk of these two types of PTMs within either type of proximity.
DISCUSSION
O-GlcNAcylation is a Widespread Post-Translational Modification
Previous investigations of protein O-GlcNAcylation have been limited in scope and in particular have lacked analogous characterization of phosphorylation for modified proteins occurring in the same biological preparations. Our identification of 1750 O-GlcNAcylation sites provides a 20-fold increase in the number of sites previously identified from any sample with endogenous levels of O-GlcNAcylation.
Our extensive O-GlcNAcylation coverage of both proteins modified and sites occupied, coupled with identification of over 16,500 phosphorylation sites, allowed us to systematically characterize O-GlcNAc distribution on synaptic proteins, model the interaction of target sites with OGT, and address potential crosstalk between these two post-translational modifications. Our increased coverage was mainly attributed to three factors: (1) the use of more sensitive mass spectrometry (an Orbitrap Velos equipped with ETD fragmentation), (2) high pH fractionation of the O-GlcNAc-enriched fractions prior to LC-MS/MS, and (3) improved efficiency of the lectin-enrichment step. The primary improvements in the lectin-enrichment step were the switch from an agarose-immobilized lectin to one immobilized on POROS resin (51) carried out in three rounds of enrichment at 4 °C.
Comparison With Chemoenzymatic Strategies for O-GlcNAc Analysis
An alternative approach for the enrichment of O-GlcNAc-modified peptides involves the chemoenzymatic addition of biotinylated GalNAc to O-GlcNAc using an engineered version of galactosyl transferase (20). This approach was used previously in combination with ETD to identify 141 sites of O-GlcNAcylation from mitotic spindle and midbodies (10). A paper was published while this current manuscript was in review extending the chemoenzymatic strategy to identify 458 sites (52). Although our study identified several-fold more O-GlcNAc peptides, it is hard to compare the selectivity and sensitivity of the two approaches, because the biological samples studied and amounts employed were different and because the previous experiment was carried out in a cell line overexpressing OGT, thus elevating the level of modification above endogenous levels. A possible shortcoming of the chemoenzymatic approach is less than 100% efficiency of the enzymatic incorporation of the tag and/or photocleavage from the resin. Despite the high coverage of O-GlcNAcylation at the spindle, only three of the 138 (2.1%) modified peptides reported previously (10) were doubly O-GlcNAcylated. In contrast, 439 of the 2,434 peptides (18%) in the current study were found to be multiply O-GlcNAcylated. The chemoenzymatic approach may bias against multiply modified peptides (e.g. by limited enzyme efficiencies). In contrast, our chromatographic approach may bias in favor of multiply modified peptides because such peptides may be more efficiently separated from the flow-through peak during lectin-enrichment.
Physiological Role of O-GlcNAcylation in the Brain
O-GlcNAcylation plays a critical role in neuronal biology. Neuron-specific knock-out of OGT leads to early postnatal death, which suggests a role for this enzyme in essential pathways (5). Both OGT and OGA are enriched at synapses (2, 3). In addition, O-GlcNAc has been implicated in a diverse set of neuronal processes, such as axonal branching and LTP at CA3/CA1 hippocampal synapses (53, 54).
We examined potential biological functions of O-GlcNAc using gene ontology analysis (http://amigo.geneontology.org). For this analysis, we used a background consisting of proteins in our data set not found to be O-GlcNAcylated and of a similar abundance distribution to the O-GlcNAcylated proteins. Consistent with O-GlcNAc modifications occurring on a large percentage of proteins, there were no GO categories in which O-GlcNAcylated proteins were significantly (greater than 50%) enriched. This finding suggests that modification by O-GlcNAc regulates a wide range of biological processes in synaptic regions of the brain.
The protein bassoon is extensively modified by both O-GlcNAcylation and phosphorylation (185 and 117 sites, respectively). Bassoon is a core presynaptic active zone protein, and participates in targeting of cargo to distal axons as a component of piccolo-bassoon transport vesicles. The binding of bassoon to dynein light chain is thought to regulate transport of these vesicles along microtubules (55). Bassoon contains three functional dynein light chain-binding motifs. We identified O-GlcNAcylation sites within two of these motifs, while none of them were found to be phosphorylated. O-GlcNAcylation of these motifs may disrupt interactions between dynein and bassoon, indicating a potential role for O-GlcNAcylation in regulation of vesicular transport.
A fraction of CaMKII α is O-GlcNAcylated at threonine residue 306. Auto-phosphorylation at both threonine residue 306 and the adjacent threonine residue 305 reduced the sensitivity of CaMKII to stimulation by CaM (inhibitory auto-phosphorylation). Genetic mutations of these residues affected spatial learning and synaptic plasticity in vivo. Inhibitory auto-phosphorylation of CaMKII controls PSD association, plasticity, and learning (56). Our observation of O-GlcNAcylation at threonine residue 306, suggests that mutation of this threonine to alanine (phospho-null mutation) needs more careful interpretation because this mutation eliminates the possibility of either O-GlcNAcylation or phosphorylation. The strong phenotype shown by mice carrying the phospho-mimicking allele (T306D) confirmed the importance of phosphorylation at this residue, but leaves the role of O-GlcNAcylation unclear. Our failure to find phosphopeptides corresponding to threonine residues 305/306 despite identifying over 16,500 phosphorylation sites suggests that the phosphorylation stoichiometry of these residues is low. Proteomic analyses (such as ours) typically rely on data-dependent acquisition of MS/MS spectra, which is biased toward the identification of abundant peptides. We cannot therefore rule out the possibility that phosphorylation at threonine residues 305/306 was present at some low level and that we failed to identify these sites because of chance. This example highlights the future need to determine the absolute occupancy of alternative site-specific PTMs as an important step toward understanding their respective functional roles for all sites that are affected by both PTMs.
The microtubule associated protein tau is extensively phosphorylated (for a review on tau PTMs, see (57) and has been reported to be modified by O-GlcNAc (58), giving rise to the hypothesis that O-GlcNAc may be regulating levels of tau phosphorylation (59). We identified 112 unique tau peptides, including 67 phosphopeptides mapping to 46 sites of phosphorylation. However, we did not find any peptides corresponding to O-GlcNAcylation on tau. This finding calls into question whether endogenous synaptic tau in healthy young animals is sufficiently O-GlcNAcylated to significantly modulate its phosphorylation site occupancies.
O-GlcNAcylation Sites are Often Found in Local Clusters
Our data set allowed for the first time a determination of whether or not O-GlcNAcylation sites are randomly distributed or occur in local clusters on the primary protein sequence or within three-dimensional distance. Our data strongly support the presence of local clusters of O-GlcNAcylation sites on the primary sequence with a distribution that is akin to but distinct from the one described for serine/threonine phosphorylation sites (48–50). Although members of clusters must originate from the catalysis of the single OGT, there remains a remote possibility that different OGT coproteins are involved in the O-GlcNAcylation of the adjacent sites; more likely explanations include a tendency of OGT to progressively modify a substrate in a concerted manner (perhaps via a tendency of OGT itself to directly bind O-GlcNAcylated substrates), and a promiscuous transfer of the high energy GlcNAc group to nearby nucleophilic serine and threonine residues on the docked substrate (60). Our lectin-based enrichment scheme may lead to an overrepresentation of multiply O-GlcNAcylated peptides, which in turn could lead to an overestimation of the extent to which O-GlcNAcylated sites are clustered. However, repeating the analysis in Figs. 5D and 5F using only singly O-GlcNAcylated peptides resulted in a consistent observation of clustering with respect to both primary sequence distance and three-dimensional spatial proximity. It appears that Ser/Thr kinases and OGT share a similar tendency to cluster their respective modifications. As a result, future investigations on the functional role of individual O-GlcNAcylation sites will need to account for the fact that O-GlcNAcylation may occur locally in a cluster and that O-GlcNAcylation at a distinct site may not be necessary and sufficient to cause a biological effect, nor may a O-GlcNAcylation-null mutation at a single site within a cluster be sufficient to test for a phenotype.
PTMs Can Potentially Crosstalk at Multiple Levels
Crosstalk between the two types of PTMs can occur via three distinct yet potentially simultaneous mechanisms. A: Crosstalk at the level of substrate where addition of one PTM directly regulates the ability of an enzyme to add the second PTM (structural crosstalk). B: Crosstalk at the enzymatic level where one type of PTM modifies enzymes responsible for the addition/removal of the second PTM, thus regulating their activity (catalytic crosstalk). C: Crosstalk where addition of one PTM changes the subcellular localization of the substrate and thereby regulates the ability of substrate to be modified by the second PTM (localization crosstalk) (61).
In this study, we identified 135 instances of individual serine and threonine residues alternatively modified by both O-GlcNAcylation and phosphorylation. Previously, fewer than 20 such instances have been reported in the literature (6). The scope of our analysis allowed us to demonstrate that the frequency of alternatively modified sites was essentially equal to the frequency expected by chance alone. Thus, our data suggests that there is no common evolutionary pressure to increase local structural crosstalk. Nevertheless, for these 135 instances addition of O-GlcNAc will compete with addition of phosphate, and vice versa. Biological relevance of this competition requires that the levels of modification be sufficiently high, in order to significantly alter the concentration of unmodified protein. Although we did not measure absolute modification levels, recent reports have examined these values for both PTMs on a range of proteins (62, 63). An examination of O-GlcNAcylation occupancy at the protein level for seven proteins showed a range of 2 to 100%, although the occupancies at individual sites for multiply modified proteins will likely be lower. Wu and colleagues calculated phosphorylation occupancies for over 5000 yeast phosphorylation sites. These values varied from 1 to 100%, with a median phosphorylation occupancy value of ∼25%. Based upon these results, it would appear that basal occupancies for both PTMs are in a range where moderate increases in one PTM may or may not result in a significant decrease in the substrate availability of the other PTM.
In addition to demonstrating that kinases and OGT do not preferentially target the same residues, we also show that the distribution of these two post-translational modifications do not cocluster on proteins. Therefore, it does not appear to be a general principle that addition of one type of PTM on a protein inhibits the addition of the other (via electrostatic or steric effects). Nevertheless, our results leave open the possibility of positive interactions between these two PTMs, whereby addition of one PTM increases the rate of addition of the other. For example, addition of phosphorylation may create a docking site used by OGT to GlcNAcylate a phosphorylated protein. Such a mechanism may explain our observation in Fig. 5B that the extent to which a protein may be GlcNAcylated is a function of the extent to which that protein is phosphorylated. Regulation of this sort has recently been proposed for GlcNAcylation of phosphorylated CREB (64). Importantly, if substrate phosphorylation creates a docking site (or conformational change) enabling recognition by an OGT complex, the resulting site of GlcNAcylation may not be in close proximity to the phosphorylation site. Our analysis relied on identification of peptides after tryptic cleavage. Although we identified 66 peptides simultaneously modified by both PTMs, digestion of the proteins likely resulted in many cases were nearby O-GlcNAc and phosphorylation sites ended up on different tryptic peptides. As such, our current data set does not readily allow us to determine the extent to which one type of PTM might potentially act as a primer for a second modification.
It has recently been reported that O-GlcNAc and phosphorylation levels are of similar abundance at spindles and midbodies (10). However, without controlling for differential detection efficiency of the two PTMs, it is difficult to make such claims with a high degree of confidence. When we attempted to account for this effect (Figs. 2C, 2D), we observed 11-fold more sites of phosphorylation than O-GlcNAcylation in synaptosomes. Although different subcellular compartments will undoubtedly have different ratios of these two PTMs at subsets of proteins, our results encompass measurements for over 6000 proteins.
In summary, our large-scale analysis of O-GlcNAcylation and phosphorylation revealed three key findings. First, O-GlcNAcylation occurs infrequently on proteins that are not phosphorylated, indicating that targeting of OGT does not extend beyond the set of substrates that has evolved for kinases. Second, similar to phosphorylation by serine/threonine kinases, O-GlcNAcylation also occurs in a clustered pattern, likely indicating similar evolutionary selection of sites or enzymatic mechanism for these two PTMs and their transferring enzymes. Third, although both O-GlcNAcylation and phosphorylation occur in clusters, these clusters do not colocalize. The localization of O-GlcNAcylation sites is statistically independent from localization of phosphorylation. This is consistent with OGT and kinases having independently evolved substrate site specificity. It also indicates that at the proteome level, there was no evolutionary advantage to promote local spatial crosstalk between these two PTMs.
Supplementary Material
Acknowledgments
We thank Ursula Pieper for comparative modeling of the modified proteins.
Footnotes
* This work was supported by the Biotechnology and Biological Sciences Research Council (to R. S.), by National Institutes of Health NIGMS 8P41GM103481 (to A.L.B.), the HHMI (to A.L.B, purchase of an Orbitrap Velos mass spectrometer), the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation (to A.L.B.) as well as by R01 GM083960, P01 AI091575, and U54 GM094662 (to A.S.). J.C.T was additionally supported by P50 GM081879 (to A.L.B, co-PI).
 This article contains supplemental Figs. S1 to S3, Tables S1 to S7 and Data.
This article contains supplemental Figs. S1 to S3, Tables S1 to S7 and Data.
2 New insight on GlcNAcylation sites from IRS2 and proteins from osteoblast cell lysates were also recently reported (L Ball, M Schilling, A Nagel, L Waller, S Comte-Walters, “Characterization of O-GlcNAc peptides by electron transfer dissociation MS/MS” Abstracts 9th Uppsala Conf on Electron Capture and Transfer Dissociation, Feb 2012, Charleston, SC USA).
1 The abbreviations used are:
- OGT
- O-GlcNAc-transferase
- DSSP
- Define Secondary Structure of Proteins
- ECD
- electron capture dissociation
- ETD
- electron transfer dissociation
- emPAI
- exponentially modified protein abundance index
- FDR
- false discovery rate
- HCD
- higher-energy C-trap dissociation
- LWAC
- lectin weak affinity chromatography
- OGA
- O-glycoprotein 2-acetamido-2-deoxy-β-d-glucopyranosidase, referred to as O-GlcNAcase
- O-GlcNAc
- O-linked N-acetylglucosamine
- PTM
- post-translational modification
- TPR
- tetratricopeptide repeat
- WGA
- wheat germ agglutinin.
REFERENCES
- 1. Hanover J. A., Krause M. W., Love D. C. (2010) The hexosamine signaling pathway: O-GlcNAc cycling in feast or famine. Biochim. Biophys. Acta 1800, 80–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cole R. N., Hart G. W. (2001) Cytosolic O-glycosylation is abundant in nerve terminals. J. Neurochem. 79, 1080–1089 [DOI] [PubMed] [Google Scholar]
- 3. Gao Y., Wells L., Comer F. I., Parker G. J., Hart G. W. (2001) Dynamic O-glycosylation of nuclear and cytosolic proteins: cloning and characterization of a neutral, cytosolic beta-N-acetylglucosaminidase from human brain. J. Biol. Chem. 276, 9838–9845 [DOI] [PubMed] [Google Scholar]
- 4. Akimoto Y., Comer F. I., Cole R. N., Kudo A., Kawakami H., Hirano H., Hart G. W. (2003) Localization of the O-GlcNAc transferase and O-GlcNAc-modified proteins in rat cerebellar cortex. Brain Res. 966, 194–205 [DOI] [PubMed] [Google Scholar]
- 5. O'Donnell N., Zachara N. E., Hart G. W., Marth J. D. (2004) Ogt-Dependent X-Chromosome-Linked Protein Glycosylation Is a Requisite Modification in Somatic Cell Function and Embryo Viability. Mol. Cell. Biol. 24, 1680–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hart G. W., Slawson C., Ramirez-Correa G., Lagerlof O. (2011) Cross Talk Between O-GlcNAcylation and Phosphorylation: Roles in Signaling, Transcription, and Chronic Disease. Annu. Rev. Biochem. 80, 825–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang Z., Gucek M., Hart G. W. (2008) Cross-talk between GlcNAcylation and phosphorylation: site-specific phosphorylation dynamics in response to globally elevated O-GlcNAc. Proc. Natl. Acad. Sci. U.S.A. 105, 13793–13798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Griffith L. S., Schmitz B. (1999) O-linked N-acetylglucosamine levels in cerebellar neurons respond reciprocally to pertubations of phosphorylation. Eur. J. Biochem. 262, 824–831 [DOI] [PubMed] [Google Scholar]
- 9. Wang Z., Pandey A., Hart G. W. (2007) Dynamic interplay between O-linked N-acetylglucosaminylation and glycogen synthase kinase-3-dependent phosphorylation. Mol. Cell Proteomics 6, 1365–1379 [DOI] [PubMed] [Google Scholar]
- 10. Wang Z., Udeshi N. D., Slawson C., Compton P. D., Sakabe K., Cheung W. D., Shabanowitz J., Hunt D. F., Hart G. W. (2010) Extensive Crosstalk Between O-GlcNAcylation and Phosphorylation Regulates Cytokinesis. Sci. Signal. 3, ra2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Choudhary C., Mann M. (2010) Decoding signalling networks by mass spectrometry-based proteomics. Nat. Rev. Mol. Cell Biol. 11, 427–439 [DOI] [PubMed] [Google Scholar]
- 12. Thingholm T. E., Jensen O. N., Larsen M. R. (2009) Analytical strategies for phosphoproteomics. Proteomics 9, 1451–1468 [DOI] [PubMed] [Google Scholar]
- 13. Chalkley R. J., Burlingame A. L. (2001) Identification of GlcNAcylation sites of peptides and alpha-crystallin using Q-TOF mass spectrometry. J. Am. Soc. Mass Spectrom. 12, 1106–1113 [DOI] [PubMed] [Google Scholar]
- 14. Chalkley R. J., Burlingame A. L. (2003) Identification of novel sites of O-N-acetylglucosamine modification of serum response factor using quadrupole time-of-flight mass spectrometry. Mol. Cell Proteomics 2, 182–190 [DOI] [PubMed] [Google Scholar]
- 15. Chalkley R. J., Thalhammer A., Schoepfer R., Burlingame A. L. (2009) Identification of protein O-GlcNAcylation sites using electron transfer dissociation mass spectrometry on native peptides. Proc. Natl. Acad. Sci. U.S.A. 106, 8894–8899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wells L., Vosseller K., Cole R. N., Cronshaw J. M., Matunis M. J., Hart G. W. (2002) Mapping sites of O-GlcNAc modification using affinity tags for serine and threonine post-translational modifications. Mol. Cell Proteomics 1, 791–804 [DOI] [PubMed] [Google Scholar]
- 17. Vosseller K., Hansen K. C., Chalkley R. J., Trinidad J. C., Wells L., Hart G. W., Burlingame A. L. (2005) Quantitative analysis of both protein expression and serine / threonine post-translational modifications through stable isotope labeling with dithiothreitol. Proteomics 5, 388–398 [DOI] [PubMed] [Google Scholar]
- 18. Vosseller K., Trinidad J. C., Chalkley R. J., Specht C. G., Thalhammer A., Lynn A. J., Snedecor J. O., Guan S., Medzihradszky K. F., Maltby D. A., Schoepfer R., Burlingame A. L. (2006) O-linked N-acetylglucosamine proteomics of postsynaptic density preparations using lectin weak affinity chromatography and mass spectrometry. Mol. Cell. Proteomics 5, 923–934 [DOI] [PubMed] [Google Scholar]
- 19. Myers S. A., Panning B., Burlingame A. L. (2011) Polycomb repressive complex 2 is necessary for the normal site-specific O-GlcNAc distribution in mouse embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 108, 9490–9495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Khidekel N., Ficarro S. B., Clark P. M., Bryan M. C., Swaney D. L., Rexach J. E., Sun Y. E., Coon J. J., Peters E. C., Hsieh-Wilson L. C. (2007) Probing the dynamics of O-GlcNAc glycosylation in the brain using quantitative proteomics. Nat. Chem. Biol 3, 339–348 [DOI] [PubMed] [Google Scholar]
- 21. Trinidad J. C., Specht C. G., Thalhammer A., Schoepfer R., Burlingame A. L. (2006) Comprehensive identification of phosphorylation sites in postsynaptic density preparations. Mol. Cell. Proteomics 5, 914–922 [DOI] [PubMed] [Google Scholar]
- 22. Pinkse M. W., Uitto P. M., Hilhorst M. J., Ooms B., Heck A. J. (2004) Selective isolation at the femtomole level of phosphopeptides from proteolytic digests using 2D-NanoLC-ESI-MS/MS and titanium oxide precolumns. Anal. Chem. 76, 3935–3943 [DOI] [PubMed] [Google Scholar]
- 23. Guan S., Price J. C., Prusiner S. B., Ghaemmaghami S., Burlingame A. L. (2011) A data processing pipeline for mammalian proteome dynamics studies using stable isotope metabolic labeling. Mol. Cell. Proteomics 10, M111.010728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Graham M. E., Thaysen-Andersen M., Bache N., Craft G. E., Larsen M. R., Packer N. H., Robinson P. J. (2011) A Novel Post-translational Modification in Nerve Terminals: O-Linked N-Acetylglucosamine Phosphorylation. J. Proteome Res. 10, 2725–2733 [DOI] [PubMed] [Google Scholar]
- 25. Baker P. R., Trinidad J. C., Chalkley R. J. (2011) Modification site localization scoring integrated into a search engine. Mol. Cell. Proteomics 10, M111.008078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Matsuura A., Ito M., Sakaidani Y., Kondo T., Murakami K., Furukawa K., Nadano D., Matsuda T., Okajima T. (2008) O-linked N-acetylglucosamine is present on the extracellular domain of notch receptors. J. Biol. Chem. 283, 35486–35495 [DOI] [PubMed] [Google Scholar]
- 27. Thalhammer A., Trinidad J. C., Burlingame A. L., Schoepfer R. (2009) Densin-180: revised membrane topology, domain structure and phosphorylation status. J. Neurochem. 109, 297–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Melo F., Sánchez R., Sali A. (2002) Statistical potentials for fold assessment. Protein Sci. 11, 430–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pieper U., Webb B. M., Barkan D. T., Schneidman-Duhovny D., Schlessinger A., Braberg H., Yang Z., Meng E. C., Pettersen E. F., Huang C. C., Datta R. S., Sampathkumar P., Madhusudhan M. S., Sjölander K., Ferrin T. E., Burley S. K., Sali A. (2011) ModBase, a database of annotated comparative protein structure models, and associated resources. Nucleic Acids Res. 39, D465–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kabsch W., Sander C. (1983) Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22, 2577–2637 [DOI] [PubMed] [Google Scholar]
- 31. Jones D. T. (1999) Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292, 195–202 [DOI] [PubMed] [Google Scholar]
- 32. Schlessinger A., Schaefer C., Vicedo E., Schmidberger M., Punta M., Rost B. (2011) Protein disorder–a breakthrough invention of evolution? Curr. Opin. Struct. Biol. 21, 412–418 [DOI] [PubMed] [Google Scholar]
- 33. Duhovny D., Nussinov R., Wolfson H. J. (2002) Efficient Unbound Docking of Rigid Molecules. in Algorithms in Bioinformatics (Guigó R., Gusfield D., eds.) pp. 185–200, Springer Berlin Heidelberg, Berlin, Heidelberg [Google Scholar]
- 34. Andrusier N., Nussinov R., Wolfson H. J. (2007) FireDock: Fast interaction refinement in molecular docking. Proteins 69, 139–159 [DOI] [PubMed] [Google Scholar]
- 35. Nagata Y., Burger M. M. (1974) Wheat germ agglutinin. Molecular characteristics and specificity for sugar binding. J. Biol. Chem. 249, 3116–3122 [PubMed] [Google Scholar]
- 36. Liu H., Sadygov R. G., Yates J. R., 3rd (2004) A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 76, 4193–4201 [DOI] [PubMed] [Google Scholar]
- 37. Shinoda K., Tomita M., Ishihama Y. (2010) emPAI Calc—for the estimation of protein abundance from large-scale identification data by liquid chromatography-tandem mass spectrometry. Bioinformatics 26, 576–577 [DOI] [PubMed] [Google Scholar]
- 38. Schwartz D., Gygi S. P. (2005) An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat. Biotechnol. 23, 1391–1398 [DOI] [PubMed] [Google Scholar]
- 39. Olsen J. V., Vermeulen M., Santamaria A., Kumar C., Miller M. L., Jensen L. J., Gnad F., Cox J., Jensen T. S., Nigg E. A., Brunak S., Mann M. (2010) Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal 3, ra3. [DOI] [PubMed] [Google Scholar]
- 40. Hudmon A., Schulman H. (2002) Structure-function of the multifunctional Ca2+/calmodulin-dependent protein kinase II. Biochem. J. 364, 593–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lucchesi W., Mizuno K., Giese K. P. (2011) Novel insights into CaMKII function and regulation during memory formation. Brain Res. Bull. 85, 2–8 [DOI] [PubMed] [Google Scholar]
- 42. Lazarus M. B., Nam Y., Jiang J., Sliz P., Walker S. (2011) Structure of human O-GlcNAc transferase and its complex with a peptide substrate. Nature 469, 564–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Iakoucheva L. M., Radivojac P., Brown C. J., O'Connor T. R., Sikes J. G., Obradovic Z., Dunker A. K. (2004) The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 32, 1037–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Collart M. A. (2003) Global control of gene expression in yeast by the Ccr4-Not complex. Gene 313, 1–16 [DOI] [PubMed] [Google Scholar]
- 45. Jackson S. P., Tjian R. (1988) O-glycosylation of eukaryotic transcription factors: Implications for mechanisms of transcriptional regulation. Cell 55, 125–133 [DOI] [PubMed] [Google Scholar]
- 46. Tang L. Y., Deng N., Wang L. S., Dai J., Wang Z. L., Jiang X. S., Li S. J., Li L., Sheng Q. H., Wu D. Q., Li L., Zeng R. (2007) Quantitative phosphoproteome profiling of Wnt3a-mediated signaling network: indicating the involvement of ribonucleoside-diphosphate reductase M2 subunit phosphorylation at residue serine 20 in canonical Wnt signal transduction. Mol. Cell. Proteomics 6, 1952–1967 [DOI] [PubMed] [Google Scholar]
- 47. Wells L., Kreppel L. K., Comer F. I., Wadzinski B. E., Hart G. W. (2004) O-GlcNAc transferase is in a functional complex with protein phosphatase 1 catalytic subunits. J. Biol. Chem. 279, 38466–38470 [DOI] [PubMed] [Google Scholar]
- 48. Yachie N., Saito R., Sugahara J., Tomita M., Ishihama Y. (2009) In silico analysis of phosphoproteome data suggests a rich-get-richer process of phosphosite accumulation over evolution. Mol. Cell. Proteomics 8, 1061–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schweiger R., Linial M. (2010) Cooperativity within proximal phosphorylation sites is revealed from large-scale proteomics data. Biol. Direct 5, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Moses A. M., Hériché J.-K., Durbin R. (2007) Clustering of phosphorylation site recognition motifs can be exploited to predict the targets of cyclin-dependent kinase. Genome Biol. 8, R23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Afeyan N. B., Gordon N. F., Mazsaroff I., Varady L., Fulton S. P., Yang Y. B., Regnier F. E. (1990) Flow-through particles for the high-performance liquid chromatographic separation of biomolecules: perfusion chromatography. J. Chromatogr. 519, 1–29 [DOI] [PubMed] [Google Scholar]
- 52. Alfaro J. F., Gong C. X., Monroe M. E., Aldrich J. T., Clauss T. R., Purvine S. O., Wang Z., Camp D. G., 2nd, Shabanowitz J., Stanley P., Hart G. W., Hunt D. F., Yang F., Smith R. D. (2012) Tandem mass spectrometry identifies many mouse brain O-GlcNAcylated proteins including EGF domain-specific O-GlcNAc transferase targets. Proc. Natl. Acad. Sci. U.S.A. 109, 7280–7285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Francisco H., Kollins K., Varghis N., Vocadlo D., Vosseller K., Gallo G. (2009) O-GlcNAc post-translational modifications regulate the entry of neurons into an axon branching program. Dev Neurobiol 69, 162–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tallent M. K., Varghis N., Skorobogatko Y., Hernandez-Cuebas L., Whelan K., Vocadlo D. J., Vosseller K. (2009) In vivo modulation of O-GlcNAc levels regulates hippocampal synaptic plasticity through interplay with phosphorylation. J. Biol. Chem. 284, 174–181 [DOI] [PubMed] [Google Scholar]
- 55. Fejtova A., Davydova D., Bischof F., Lazarevic V., Altrock W. D., Romorini S., Schöne C., Zuschratter W., Kreutz M. R., Garner C. C., Ziv N. E., Gundelfinger E. D. (2009) Dynein light chain regulates axonal trafficking and synaptic levels of Bassoon. J. Cell Biol. 185, 341–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Elgersma Y., Fedorov N. B., Ikonen S., Choi E. S., Elgersma M., Carvalho O. M., Giese K. P., Silva A. J. (2002) Inhibitory autophosphorylation of CaMKII controls PSD association, plasticity, and learning. Neuron 36, 493–505 [DOI] [PubMed] [Google Scholar]
- 57. Martin L., Latypova X., Terro F. (2011) Post-translational modifications of tau protein: implications for Alzheimer's disease. Neurochem. Int. 58, 458–471 [DOI] [PubMed] [Google Scholar]
- 58. Arnold C. S., Johnson G. V., Cole R. N., Dong D. L., Lee M., Hart G. W. (1996) The microtubule-associated protein tau is extensively modified with O-linked N-acetylglucosamine. J. Biol. Chem. 271, 28741–28744 [DOI] [PubMed] [Google Scholar]
- 59. Liu F., Iqbal K., Grundke-Iqbal I., Hart G. W., Gong C. X. (2004) O-GlcNAcylation regulates phosphorylation of tau: a mechanism involved in Alzheimer's disease. Proc. Natl. Acad. Sci. U.S.A. 101, 10804–10809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhao P., Viner R., Teo C. F., Boons G. J., Horn D., Wells L. (2011) Combining High-Energy C-Trap Dissociation and Electron Transfer Dissociation for Protein O-GlcNAc Modification Site Assignment. J. Proteome Res. 10, 4088–4104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hunter T. (2007) The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol. Cell 28, 730–738 [DOI] [PubMed] [Google Scholar]
- 62. Rexach J. E., Rogers C. J., Yu S. H., Tao J., Sun Y. E., Hsieh-Wilson L. C. (2010) Quantification of O-glycosylation stoichiometry and dynamics using resolvable mass tags. Nat. Chem. Biol. 6, 645–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wu R., Haas W., Dephoure N., Huttlin E. L., Zhai B., Sowa M. E., Gygi S. P. (2011) large-scale method to measure absolute protein phosphorylation stoichiometries. Nat. Methods 8, 677–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Rexach J. E., Clark P. M., Mason D. E., Neve R. L., Peters E. C., Hsieh-Wilson L. C. (2012) Dynamic O-GlcNAc modification regulates CREB-mediated gene expression and memory formation. Nat. Chem. Biol. 8, 253–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. (2001) Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.