Abstract
Regenerative tissues such as the skin epidermis, the intestinal mucosa or the hematopoietic system are organized in a hierarchical manner with stem cells building the top of this hierarchy. Somatic stem cells harbor the highest self-renewal activity and generate a series of multipotent progenitors which differentiate into lineage committed progenitors and subsequently mature cells. In this report, we applied an in-depth quantitative proteomic approach to analyze and compare the full proteomes of ex vivo isolated and FACS-sorted populations highly enriched for either multipotent hematopoietic stem/progenitor cells (HSPCs, LinnegSca-1+c-Kit+) or myeloid committed precursors (LinnegSca-1−c-Kit+). By employing stable isotope dimethyl labeling and high-resolution mass spectrometry, more than 5000 proteins were quantified. From biological triplicate experiments subjected to rigorous statistical evaluation, 893 proteins were found differentially expressed between multipotent and myeloid committed cells. The differential protein content in these cell populations points to a distinct structural organization of the cytoskeleton including remodeling activity. In addition, we found a marked difference in the expression of metabolic enzymes, including a clear shift of specific protein isoforms of the glycolytic pathway. Proteins involved in translation showed a collective higher expression in myeloid progenitors, indicating an increased translational activity. Strikingly, the data uncover a unique signature related to immune defense mechanisms, centering on the RIG-I and type-1 interferon response systems, which are installed in multipotent progenitors but not evident in myeloid committed cells. This suggests that specific, and so far unrecognized, mechanisms protect these immature cells before they mature. In conclusion, this study indicates that the transition of hematopoietic stem/progenitors toward myeloid commitment is accompanied by a profound change in processing of cellular resources, adding novel insights into the molecular mechanisms at the interface between multipotency and lineage commitment.
Multipotent hematopoietic stem cells (HSCs)1 are of primary importance for our body due to their unique capacity to replenish all cell types of the blood system (1–3). HSCs are extremely rare and reside in the bone marrow of the trabecular bones. They possess the capability to self-renew and give rise to multipotent progenitors (MPPs), a transient amplifying precursor pool, which also conserves multi-lineage potential ((4) and Fig. 1A). The HSCs and MPPs together form the hematopoietic stem/progenitor cell (HSPC) fraction, which is distinguished as negative for mature lineage-markers and positive for the stem-cell markers Sca-1 and c-Kit (LS+K). MPPs commit to progenitors with more restricted cell fate potential, either myeloid committed progenitors (Lineage−, Sca-1−, c-Kit+, LS−K) or lymphoid progenitors, which ultimately give rise to mature effector cells of the blood, like erythrocytes or neutrophils (4). Two key characteristics of early hematopoiesis are the maintenance of multipotency in the most immature HSPC compartment and the commitment to either myeloid or lymphoid fate, which coincides with loss of multipotency and the onset of more specialized function-specific protein repertoires (5, 6). Surprisingly, despite intense research efforts over the last decades, the molecular basis for both multipotency and commitment, phenomena of fundamental importance for the development of all organisms and tissues, remains poorly understood (7). Notably, with respect to malignant aberration of the hematopoietic system, there is extensive debate about the compartment of origin for leukemias (8–10). Thus, an improved understanding of differentiation under homeostatic conditions is an important prerequisite to specify the molecular roots of leukemias. Following this aim, the greatest obstacle has been the lack of global protein data on early hematopoietic cell fractions. Application of proteomic techniques to stem cell populations has long been hampered by scarcity of cells, as well as the sensitivity of the mass spectrometric instrumentation (11). Recently, several large-scale quantitative proteomic studies have investigated various aspects of stem cell differentiation (12, 13). Although some attempts have been made to distinguish the proteome of hematopoietic immature cells (14), no comprehensive characterization has yet been described. Using the most recent generation of instruments, biochemical and software tools, we set out to characterize the global protein signatures that are essential for multipotency and commitment in the hematopoietic system. In this study, we present an in-depth quantitative proteomic analysis of primary multipotent progenitor cells and early committed cells of the mouse. Combined with extensive bioinformatic analysis, our results provide strong evidence that multipotency and differentiation of hematopoietic stem and progenitor cells involves fundamental changes in the processes of cytoskeleton organization, metabolism, and immune response. In addition, our data point to gene-regulatory proteins that potentially participate in the control of hematopoietic multipotency and commitment. Based on our results, we discuss possible implications for a new concept of early hematopoiesis.
Fig. 1.
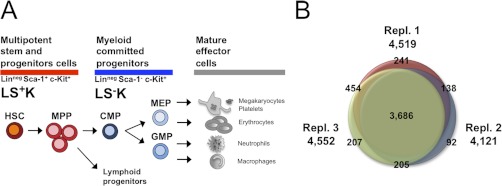
Quantitative proteomic analysis of hematopoietic stem and progenitor cells. A, Early hematopoiesis. Multipotent hematopoietic stem cells (HSCs) give rise to multipotent progenitors (MPPs), which commit either to myeloid specified progenitors (CMP; common myeloid progenitors; GMPs: granulocyte-macrophage progenitors, and MEPs: megakaryocyte/erythrocyte progenitors) or to lymphoid specified progenitors with limited cell fate. Cell fractions can be highly purified by FACS using specific surface markers for Lineage (Lin), Sca-1, and c-Kit, distinguishing multipotent progenitor cells (LS+K) from myeloid committed (LS−K) cells. B, Overlap of quantified proteins in three biological replicates. The Venn diagram displays number of proteins quantified in each of the three replicates and their overlap. In total 5023 proteins were quantified.
EXPERIMENTAL PROCEDURES
Animals
Eight- to 12-week-old female C57BL/6 mice purchased from Harlan Laboratories (The Netherlands) were used throughout the study. All mice were maintained in the animal facility at DKFZ, Heidelberg, Germany, under specific pathogen free (SPF) conditions and kept in individually ventilated cages (IVC). Animal procedures were performed according to protocols approved by the German authorities, Regierungspräsidium Karlsruhe (Nr. Z110/02).
Fluorescence-activated Cell Sorting: Staining and Sorting
Bone marrow was isolated from hind legs (femur, tibia), hips (ilium), and backbone (vertebra). Muscle, connective tissue, and spinal cord were removed, bones were crushed in Roswell Park Memorial Institute media/2% fetal bovine serum (Invitrogen, Carlsbad, CA) using mortar and pestle. Single cell-suspensions were made by flushing through a 40 μm filter mesh. Cell numbers and viability were determined using a ViCell Counter (Beckman Coulter, Brea, CA). To deplete lineage-positive cells, total bone marrow was stained for 30 min with a combination of the following monoclonal rat antibodies directed against mature cell specific lineage markers: anti-CD4 (clone GK1.5), anti-CD8a (53.6.7), anti-CD11b (M1/70), anti-B220 (RA3.6B2), anti-Gr1 (RB6.8C5) and anti-Ter119 (Ter119). Labeled cells were incubated for 20 min with polyclonal sheep anti-rat IgG coated magnetic Dynabeads (Invitrogen, Hercules, CA) at a ratio 2:1 beads to cell and depleted using a magnet, enriching for the lineage-negative (Linneg) cell fraction. Beads were washed twice with RPMI/2% fetal bovine serum to harvest residual cell fractions. Centrifugation steps were carried out at 1500 rpm and 4 °C for 5 min (5810r, Eppendorf, Hamburg, Germany). To specify multipotent and myeloid progenitor fractions, the Linneg fraction was stained for 30 min using the following rat monoclonal fluorochrome-coupled antibodies: anti-CD4 (GK1.5), anti-CD8a (53.6.7), anti-CD11b (M1/70), anti-B220 (RA3–6B2), anti-Gr1 (RB6.8C5), and anti-Ter119 (Ter119), all phycoerythrobilin-cyanine dye 7 (PE-Cy7) conjugated; anti-CD117/c-Kit (2B8)- PE; anti-Sca-1 (D7)-Alexa700. Lineage antibodies for depletion of mature effector cells were purified in our laboratory according to standard protocols. Monoclonal antibody conjugates were purchased from eBioscience (San Diego, CA) or BioLegend (San Diego, CA). All antibodies were titrated prior to use. FACS analyses were performed on a LSR II or LSR Fortessa cell analyzer (Becton Dickinson, San Jose, CA). Cell sorting was performed on a FACS Aria I or FACS Aria II (Becton Dickinson, San Jose, CA) at the DKFZ Flow Cytometry Service Unit, Heidelberg, Germany. The following sort parameters were used: 70 μm nozzle; 15,000 evt/s.; 70 psi. Data were analyzed using BD FACSDiva (Becton Dickinson, San Jose, CA) and FlowJo software (Tree Star, Ashland, OR). For proteome analysis, LS+K cells and LS−K cells were obtained by sorting LinnegSca-1+c-Kit+ and LinnegSca-1+c-Kit−, respectively, in biological triplicate (supplemental Fig. S2). Sorted cells were collected into ice-cold PBS, centrifuged and snap-frozen as dry pellets using liquid nitrogen. Frozen samples were stored at −20 °C until further usage.
Protein Extraction and Digestion
Cell pellets corresponding to 106 FACS-sorted cells were lysed with 0.1% RapiGest (Waters) in 50 mm ammonium bicarbonate, then heated at 90 °C for 5 min, followed by sonication for 20 min. Cell debris were removed by centrifugation and the sample was concentrated with a Microcon YM-10 centrifugal filter unit (Millipore). After reduction of disulfide bonds with 5 mm dithiotreitol for 30 min at 56 °C and alkylation of cysteines with 10 mm iodoacetamide for 60 min at room temperature in the dark, the proteins were digested overnight with sequencing grade modified trypsin (Promega) at 37 °C. The reaction was stopped by adding trifluoroacetic acid to a final concentration of 0.5% (v/v), and RapiGest was precipitated by further incubation at 37 °C for 20 min. Following centrifugation the supernatants were collected and protein digests were stored at −20 °C until further use.
Peptide Stable Isotope Labeling and Fractionation
Protein digests were dimethyl labeled on column as previously described with slight modifications (15). Briefly, SepPak C18 cartridges (Waters) were washed with acetonitrile and conditioned with 0.1% (v/v) formic acid. Acidified samples were loaded and washed with 0.1% formic acid. Samples were labeled by flushing the columns with labeling reagent (light, intermediate or heavy using CH2O (Fisher) + NaBH3CN (Fluka), CD2O (Isotec) + NaBH3CN or 13CD2O (Isotec) + NaBD3CN (Isotec), respectively). After washing with 0.1% formic acid, labeled peptides were eluted with 80% (v/v) acetonitrile/0.05% (v/v) formic acid. Samples were mixed in 1:1 ratio based on total peptide amount, determined by analyzing an aliquot of the labeled samples on regular liquid chromatography (LC)-MS runs and comparing overall peptide signal intensities. Samples were dried by vacuum centrifugation, reconstituted in IPG rehydration buffer and fractionated according to manufacturer's instructions using pH 3–10 IPG strips and 3100 OFFGEL fractionator (Agilent). The 12 fractions resolved were acidified and desalted with C18 Stagetips (Empore 3 m) (16). Peptide samples were dried by vacuum centrifugation and stored at −20 °C until further use.
LC-Electrospray Ionization (ESI)-Tandem MS (MS/MS) Analysis
Peptides were separated using the nanoACQUITY UltraPerformance LC system (Waters) fitted with a trapping column (nanoAcquity Symmetry C18, 5 μm, 180 μm x 20 mm) and an analytical column (nanoAcquity BEH C18, 1.7 μm, 75 μm × 200 mm). The outlet of the analytical column was coupled directly to an LTQ Orbitrap Velos (Thermo Fisher Scientific) using a Proxeon nanospray source. The mobile phases for LC separation were 0.1% (v/v) formic acid in LC-MS grade water (solvent A) and 0.1% (v/v) formic acid in acetonitrile (solvent B). Peptides were first loaded with a constant flow of solvent A at 15 μl/min onto the trapping column. Trapping time was 2 min. Subsequently, peptides were eluted via the analytical column at a constant flow of 300 nl/min. During the elution step, the percentage of solvent B increased in a linear fashion from 3% to 7% in 10 min, then increased to 25% in 100 min and finally to 40% in a further 10 min. The peptides were introduced into the mass spectrometer via a Pico-Tip Emitter 360 μm OD × 20 μm ID; 10 μm tip (New Objective) and a spray voltage of 2.1 kV was applied. The capillary temperature was set at 230 °C. Full scan spectra from m/z 300 to 1700 at resolution 30,000 (profile mode) were acquired in the Orbitrap MS. The filling time was set at maximum of 500 ms with limitation of 106 ions. The most intense ions (up to 15) from the full scan MS were selected for fragmentation in the ion trap. Normalized collision energy of 40% was used, and the fragmentation was performed after accumulation of 3 × 104 ions or after filling time of 100 ms for each precursor ion (whichever occurred first). MS/MS data was acquired in centroid mode. Only multiply charged (2+, 3+) precursor ions were selected for MS/MS. The dynamic exclusion list was restricted to 500 entries with maximum retention period of 30 s and relative mass window of 10 ppm. In order to improve the mass accuracy, a lock mass correction using a background ion (m/z 445.12003) was applied. The data associated with this manuscript may be downloaded from ProteomeCommons.org Tranche as “Proteomic cornerstones of hematopoietic stem cell differentiation” using the following hash:
L4Cn7Gpxl+YAGrlzanOPc0FwvYxuJyb/BnuW6hZF9crXLooWehbMDkjJXc44QFjroay2TpoZ3GLyNxNflROtAVqMwhsAAAAAABLjeA==.
Protein Identification and Quantification
MS raw data files were processed with MaxQuant (version 1.0.13.13) (17). Enzyme specificity was set to trypsin/P and a maximum of two missed cleavages were allowed. Cysteine carbamidomethylation and methionine oxidation were selected as fixed and variable modifications, respectively. The derived peak list was searched using the in-built Andromeda search engine (version 1.0.13.13) in MaxQuant against the International Protein Index (IPI) mouse database version 3.68 containing 56,729 proteins to which 265 frequently observed contaminants as well as reversed sequences of all entries had been added. Initial maximal allowed mass tolerance was set to 20 ppm for peptide masses, followed by 6 ppm in the main search, and 0.5 Dalton for fragment ion masses. The minimum peptide length was set to six amino acid residues and three labeled amino acid residues were allowed. A 1% false discovery rate (FDR) was required at both the protein level and the peptide level. In addition to the FDR threshold, proteins were considered identified if they had at least one unique peptide. The protein identification was reported as an indistinguishable “protein group” if no unique peptide sequence to a single database entry was identified.
Bioinformatic Analysis
Statistical analysis was performed using the Limma package in R/Bioconductor (18, 19). In order to capture the effect that the statistical spread of unregulated proteins is much more focused for highly abundant proteins than for low abundance ones (17), proteins quantified in all three replicates were grouped into bins based on their summed peptide intensities reported by MaxQuant. After fitting a linear model to the data, an empirical Bayes moderated t test was used and p values were adjusted for multiple testing with Benjamini and Hochberg's method. Proteins with an adjusted p value lower than 0.05 were considered to be differentially expressed between LS+K and LS−K cells.
Protein classification was performed using PANTHER classification system (20), and membrane proteins predicted using the TMHMM algorithm (21). Gene Ontology (GO) enrichment analysis was performed using the functional annotation tool of MetaCore (GeneGo Inc., (22)). To investigate interactions between differentially expressed proteins, unsupervised network analysis was performed using STRING (v9.0; (23)). The network was then visualized in Cytoscape (24). Nodes were re-arranged after manual Gene Ontology curation (UniProt, (25)) and extensive literature search according to functional classification and STRING interaction scores.
Confirmation by FACS
For FACS experiments on multipotent and myeloid sub-populations, the following rat monoclonal fluorochrome-coupled antibodies were used: anti-Tfrc (R17217)- PE; anti-CD169 (3D6.112)- fluorescein isothiocyanate (FITC); anti-Ly6c (HK1.4)- allophycocyanin (APC)- Cy7; anti-CD4 (GK1.5), anti-CD8a (53.6.7), anti-CD11b (M1/70), anti-B220 (RA3–6B2), anti-Gr1 (RB6.8C5), and anti-Ter119 (Ter119), all PE-Cy7 conjugated; anti-CD117/c-Kit (2B8)- APC-Cy7; anti-Sca-1 (D7)- APC; anti-CD34 (RAM 34)- FITC or Alexa700; anti-CD150 (TC15–12F12.2)- PE-cyanine dye 5; anti-CD48 (HM48–1)- PacificBlue; anti-CD16/32 (93)- PacificBlue, and anti-CD127 (A7R34)- APC. For labeling of myeloid progenitor subpopulations expressing FcγRII/III, the blocking antibody 24G2 (anti-CD16/32-FcRγ) was omitted and staining was carried out in RPMI/2% fetal bovine serum. Monoclonal antibody conjugates were purchased from eBioscience (San Diego, CA) or BioLegend (San Diego, CA) except for anti-CD169 (AbD Serotec, Oxford, UK). The following rat isotype controls were used: IgG2a- FITC, IgG2a- APC-Cy7 (all Biolegend, San Diego, CA) and IgG2a- PE (eBioscience, San Diego, CA).
Cytospins and Immunofluorescence
For immunocytochemical stainings, Linneg cells (enriched using magnetic beads as described above) were fixed using Cytofix/Cytoperm (Becton Dickinson, San Jose, CA) for 10 min at 4 °C and subsequently FACS-sorted, as described above. Sorted cells were spun on SuperFrost Plus slides (Menzel, Braunschweig, Germany) using the Cytospin 4 Cytocentrifuge (Thermo Fisher Scientific, Waltham, MA). Slides were incubated with 10% donkey serum (Abcam, Cambridge, UK) for 30 min to block unspecific binding. Next, slides were incubated with primary antibodies in 10% donkey serum overnight at 4 °C. The following polyclonal primary antibodies were used (all Santa Cruz, Santa Cruz, CA): rabbit IgG anti-Pml (H-238; sc-5621; dilution 1:50), goat IgG anti-MYL3 (G-12; sc-49054; 1:50), rabbit anti-Padi4 (M-70; sc-98990; 1:50), goat anti-RIG-I (l-15; sc-48931; 1:250), and goat anti-RIG-I (C-15; sc-48929; 1:250). After a wash with PBS, slides were incubated with secondary antibodies in 2.5% donkey serum for 2 h at room temperature. The following polyclonal secondary antibodies were used at 1:200 (all Invitrogen, Hercules, CA): Alexa568 donkey anti-goat IgG (A-11057); Alexa488 donkey anti-rabbit IgG (A-21206). Controls using only rabbit IgG isotype control (Imgenex, San Diego, CA) or goat IgG isotype control (R&D Systems, Minneapolis, MN) or secondary antibodies did not show staining (data not shown). For costaining with cytoskeletal F-actin, slides were incubated with Alexa488 phalloidin (Invitrogen, Hercules, CA; A-12379; 1:2) for 2 h at room temperature. After washing with PBS, cells were embedded using ProLong® embedding solution (Invitrogen, Hercules, CA) with DAPI for staining of nuclei. Confocal microscopy was carried out using a LSM700 microscope and ZEN software (both Zeiss, Göttingen, Germany) at the DKFZ Light Microscopy Facility, Heidelberg, Germany.
RESULTS
Strategy
In the present study, we elucidated the proteome differences between multipotent LS+K cells and myeloid committed LS−K progenitor cells in the mouse hematopoietic system (Fig. 1A). The strategies used in the study are illustrated in supplemental Fig. S1. In three biological replicates, proteins extracted from 106 FACS-sorted primary LS+K and LS−K cells (supplemental Fig. S2) were digested with trypsin. Subsequently, stable isotopes were introduced by dimethyl labeling, using label swaps for replicates. In order to maximize protein identification, sample complexity was reduced by peptide fractionation using isoelectric focusing. The resulting twelve peptide fractions were analyzed by high-resolution nano-LC-MS/MS. In total 564,779 MS/MS spectra were assigned to peptides at a FDR below 1%, leading to the confident identification of a total of 5139 protein groups with on average seven unique peptides per protein (supplemental Table S1, S7, and S8) and an average absolute mass deviation of 0.60 ppm for the identified peptide masses. To the best of our knowledge, this coverage is the largest achieved by any proteome analysis of hematopoietic stem and progenitor cells. Remarkably, when comparing our data set with a previous report on similar cell fractions identifying 1263 proteins (14), we found an overlap of 1023 proteins and identified 4116 proteins in addition. Most importantly, our data includes many proteins that are known to be of low abundance or that are difficult to detect by mass spectrometry, such as 336 transcription factors (based on PANTHER protein classification) and 491 membrane proteins (predicted by TMHMM to contain transmembrane helices). Notably, these classes comprise proteins important for multipotent progenitors and stem cells (e.g. CD34, FLT3, EVI1, or c-Kit) or myeloid committed progenitors (e.g. GATA1, GFI1, C/EBPalpha), demonstrating the sensitivity and relevance of our data (supplemental Table S1).
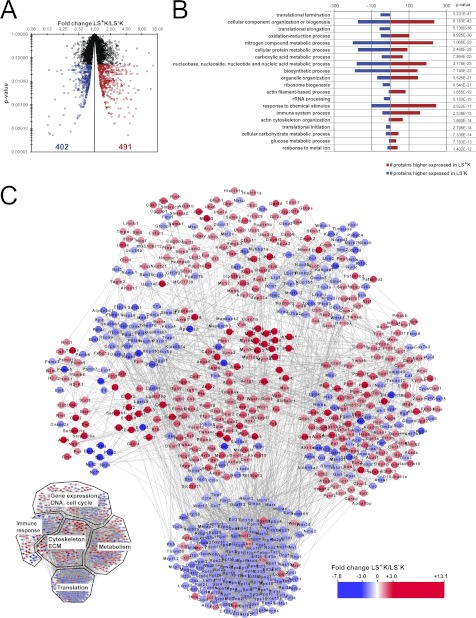
In the three replicates, 4519, 4121, and 4552 proteins were quantified (in total 5023 proteins) with an overlap of 3686 proteins quantified in all three replicates (Fig. 1B, supplemental Table S1). The accuracy of quantification was supported by the high number of ratio counts (on average 45 ratio counts per protein; supplemental Fig. S3A), and 86% of the protein ratios showed a variability below 50% (supplemental Fig. S3B). Biological variability was higher than technical variability (supplemental Fig. S4), indicating that our approach to perform biological triplicate experiments combined with rigorous statistics was essential to account for biological variability. Statistical analysis of these 3686 proteins revealed 893 proteins that are differentially expressed with statistical significance (p < 0.05), with fold changes ranging between 1.2 and 13.1, clearly distinguishing LS+K from LS−K cells (Fig. 2A; supplemental Fig. S3C; supplemental Table S2). Of these, 491 proteins were higher expressed in LS+K cells, and 402 proteins were higher expressed in LS−K cells (Fig. 2A). In order to confirm the MS quantitative data further, we used flow cytometry as an independent technique (supplemental Fig. S5). We examined expression of three selected differentially expressed surface proteins, transferrin receptor 1 (Tfrc), Ly6c and sialoadhesin (CD169), which have been studied in hematopoietic cells before, but without a direct comparison of expression in LS+K and LS−K cells (26–28). Confirmatory, Tfrc (MS fold change −3.2) and Ly6c (MS fold change −5.7) were detected by FACS in LS−K with 5–10-fold higher expression compared with LS+K (supplemental Fig. S5). In accordance with the MS results (fold change +2.9), CD169 showed higher expression in LS+K (supplemental Fig. S5), and interestingly no significant difference between immature HSCs (LS+K CD150+CD48−) and more mature MPPs (LS+K CD150−CD48+) (supplemental Fig. S5; (29)). In summary, the FACS measurements confirmed the MS quantifications, further demonstrating the robustness of the data.
Fig. 2.
Differential protein expression between multipotent and myeloid committed progenitors. A, Significantly changed proteins. After statistical test and correction of p value for multiple testing, 893 proteins showed a significant (p < 0.05) change of expression in the LS+K to LS−K transition. 491 proteins were higher expressed in LS+K cells, and 402 proteins were higher expressed in LS−K cells. B, Enriched biological processes of differentially expressed proteins. Significantly changed proteins were mapped onto biological processes according to Gene Ontology classification system. The presented biological processes are sorted according to the p value. To show diverse processes enriched in the data, redundant or highly similar terms were removed. Within each biological process, number of proteins with a higher expression in LS+K (red) and LS−K (blue) are shown. C, Protein-protein interaction network of differentially expressed proteins. An interaction network was built based on the 893 significantly differentially expressed proteins (p < 0.05) using STRING. Proteins higher expressed in LS+K cells are shown in red and proteins higher expressed in LS−K cells are shown in blue.
Classification of Proteomic Differences Between LS+K and LS−K cells
Subsequent bioinformatic analysis was focused on the 3686 proteins quantified in all replicates. To provide an overview of the functional roles of the differentially expressed proteins in a biological context, the 893 proteins were annotated based on GO biological processes assignments (Fig. 2B). The most prominent over-represented biological processes in multipotent progenitor cells included actin cytoskeleton organization, glucose metabolic process, and immune system process, whereas translation was clearly over-represented in the myeloid committed cells. Protein interactions between and within the affected processes are displayed in an interaction network, which also revealed a high number of proteins of gene regulation to be differentially expressed between the two cell states (Fig. 2C). These evident effects on several fundamental and essential cellular processes demonstrate the profound difference between multipotent LS+K and myeloid committed LS−K cells. In order to get a deeper insight into the key proteome differences, the protein expression profiles within these biological processes were evaluated, as detailed below.
A Marked Difference in Expression of Proteins that Regulate Translation in LS+K to LS−K Cell Transition
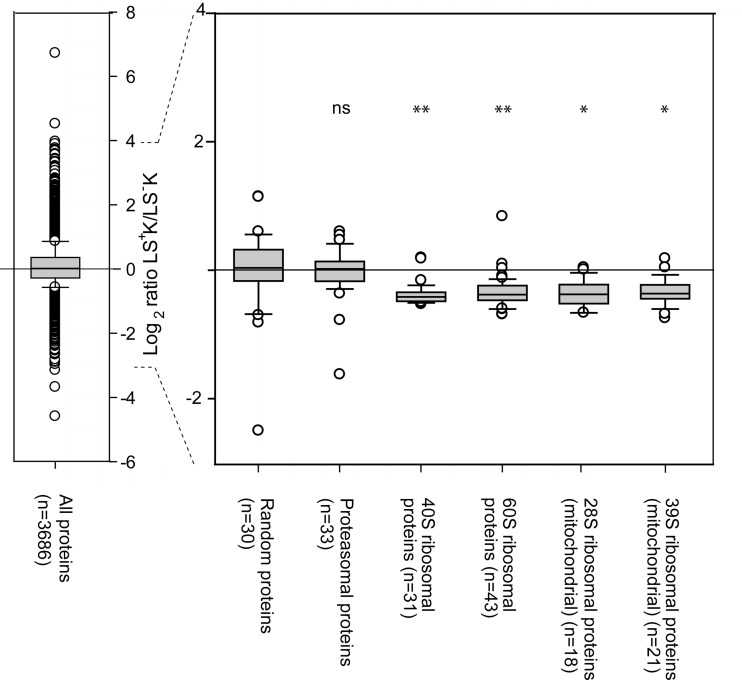
The vast majority of the proteins involved in translation initiation, elongation and termination were found to be lower expressed in LS+K cells in comparison to LS−K cells (supplemental Table S3). For example, of the 47 quantified eukaryotic translation initiation factors, 43 showed an average fold change of lower expression in LS+K cells. Similarly, 25 of 31 proteins of the small 40S subunit complex and 28 of 43 proteins of the large 60S subunit complex were significantly lower expressed in LS+K cells (supplemental Table S3). Because most proteins showed a significant but small fold change, we next investigated the statistical significance of this collective behavior. Both 40S and 60S ribosomal complexes (with average fold change –1.3, respectively) showed a statistically significant down-regulation compared with a randomly selected size-matched set of proteins (Fig. 3). In addition, the mitochondrial ribosomal protein complexes were also found to be down-regulated in a concerted manner (Fig. 3). For comparison, the proteins of the proteasome complex, for which a similar number of proteins were quantified as for the ribosomal complexes, did not show any collective regulation (Fig. 3). This demonstrates that our quantification method is highly accurate, allowing the detection of even such small changes. Together, the data strongly suggest that LS+K cells have a lower translation activity than LS−K, which may be a reflection of decreased protein turnover or the previously described lower proliferation rate (30). Notably, the multipotent progenitor cells showed a higher expression of numerous proteins known as internal ribosome entry segment trans-acting factors (ITAFs) (La ribonucleoprotein (fold change +1.2), polypirimidine tract binding protein (fold change +1.2), and several heterogeneous nuclear ribonucleoproteins (supplemental Table S1)). These ITAFs are important in providing an alternative mechanism of translation initiation of certain mRNAs when conventional cap-dependent translation is reduced (31) (supplemental Table S1), raising the possibility that the translation of mRNAs coding for specific proteins is assured, whereas the overall translation activity is low in LS+K multipotent cells.
Fig. 3.
Ribosomal complexes show reduced expression in the LS+K to LS−K transition compared with the overall data set. Box plots show ratio distribution for all proteins quantified in three replicates (n = 3686), a random set of 30 proteins (to show a similar-sized group of proteins as the ribosomal and proteasomal proteins), all quantified proteasomal proteins (n = 33), all quantified 40S ribosomal proteins (n = 31), all quantified 60S ribosomal proteins (n = 43), all quantified 28S mitochondrial ribosomal proteins (n = 18), and all quantified 39S mitochondrial ribosomal proteins (n = 21). Student's t test was performed to evaluate if the protein ratios of the groups of proteasomal and ribosomal proteins were significantly different from the the 30 random proteins. * p < 0.05; ** p < 0.005; ns = not significant (p > 0.05).
Multipotent Progenitor Cells Have a Distinct Metabolic Profile of Glycolytic Phenotype
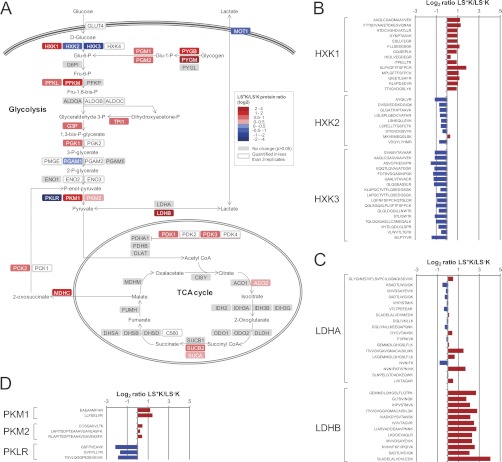
Many enzymes regulating energy metabolism were found to be differentially expressed between the multipotent progenitor cells and myeloid committed progenitors (supplemental Table S4). Within the glycolytic pathway, enzymes catalyzing all the steps from glucose to pyruvate were quantified and revealed a distinct glycolytic phenotype, with a solid differential expression pattern of several of the isoforms catalyzing the same step (Fig. 4A). More specifically, with confident quantitative values for most of the unique peptide sequences of the hexokinase isoforms, hexokinase 1 was found to be higher expressed in LS+K cells (fold change +2.0), whereas hexokinase 2 and 3 were lower (fold change –1.6 and –2.1, respectively) (Fig. 4B). Hexokinase catalyzes the first step of the glycolysis and the selective expression of its isoforms may be an important factor in directing the glucose to metabolism through the various potential downstream pathways. The type 1 isoform is thought to primarily function in a catabolic role, whereas the type 2 and 3 are thought to have an anabolic role (32). A high expression of hexokinase 1 in LS+K cells therefore suggests an introduction of glucose into the glycolytic pathway for energy (ATP) generation in these cells, whereas in LS−K cells, the higher expression of hexokinase 2 and 3 may provide the produced Glu-6-P for glycogen synthesis or metabolism via the pentose phosphate pathway for lipid synthesis. Interestingly, also glycogen phosphorylase B and M, as well as phosphoglucomutase-1 and −2 were all found to be higher expressed in the multipotent LS+K cells (Fig. 4A), indicating an additional fuel of the glycolysis via glycogen in LS+K cells and further suggesting opposing direction of metabolites in the two cell fractions at this step.
Fig. 4.
A, Differential expression of proteins involved in glycolysis and TCA cycle. A pathway map was built based on Metacore GeneGO pathway maps of glycolysis and glyconeogenesis. Proteins higher expressed in LS+K cells are shown in red and proteins higher expressed in LS−K cells are shown in blue. Quantified but not significantly differentially expressed proteins (p > 0.05) are shown in gray. B, Peptide ratios for unique peptides of isoforms of hexokinase. Average values from two or three replicates are shown. C, Peptide ratios for unique peptides of subunits of lactate dehydrogenase. Average values from two or three replicates are shown. D, Peptide ratios for unique peptides of isoforms of pyruvate kinase. Average values from two or three replicates are shown.
An isoform shift was also evident for the enzymes of the last step of the glycolysis, i.e. the pyruvate kinases. The isozymes M1 and M2 were both higher expressed in the multipotent progenitor cells (fold change +2.4 and +1.2, respectively), in contrast to the R/L form, which was strongly higher expressed in the myeloid committed progenitors (fold change –4.2) (Fig. 4D). In addition, we found that Pml, a negative regulator of pyruvate kinase M2 (33), showed a significantly higher expression in LS+K cells (fold change +2.3 which we confirmed by immunocytochemistry (supplemental Fig. S6A)). This result is in line with earlier observations that a low-activity form of pyruvate kinase M2 directs pyruvate away from the mitochondria to lactate formation in tumor cells and proliferating cells, suggesting a similar scenario in LS+K cells (34). This observation was further supported by the quantification of many of the proteins involved in energy metabolism downstream of pyruvate (Fig. 4A), including all enzymes of the tricarboxylic acid (TCA) cycle. While the large majority of them were not differentially expressed, proteins at the interaction modules between glycolysis and the TCA cycle, i.e. the pyruvate dehydrogenase kinase (PDK1 and PDK3), were found to be higher expressed in LS+K cells (Fig. 4). This protein phosphorylates and thereby inactivates pyruvate dehydrogenase, preventing the import of pyruvate into the mitochondrial matrix (35). Surprisingly, of the two subunits A and B of lactate dehydrogenase, the enzyme that governs pyruvate-lactate conversion and substrate supply to mitochondria, we found LDH-B to be strongly higher expressed in the multipotent progenitor cells (fold change +4.3), whereas LDH-A was not differentially expressed (Fig. 4C). LDH-A is generally suggested to favor the reaction of pyruvate to lactate, and it has recently been suggested that a suppression of LDH-B has a critical role in a metabolic shift toward aerobic glycolysis, by activating glycolysis and inhibiting mitochondrial respiration (36). The observed shift toward lower proportions of the B subunit during differentiation thereby suggests a favoring of pyruvate to lactate conversion in LS−K cells. In addition to the metabolic enzymes, the monocarboxylate transporter 1 (MOT1), which is responsible for the cell's uptake of exogenous lactate and pyruvate (37), was lower expressed in LS+K cells (fold change −1.9). Notably, also proteins of lipid and fatty acid metabolism were found to have a profoundly different expression between the multipotent progenitor cells and the myeloid committed cells (e.g. acyl-CoA thioesterase 2, acyl-CoA thioesterase 11, AMPK gamma2; fold change +2.9, +3.5 and +3.3, respectively (supplemental Table S2)), further demonstrating that a distinct switch in the metabolic profile occurs during hematopoietic differentiation.
Cytoskeleton and Extracellular Matrix Proteins Build a Specific Signature in Multipotent Progenitor Cells
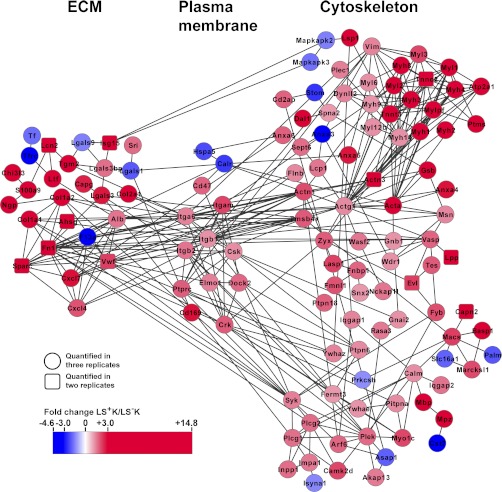
Our data analysis revealed a highly interconnected group of 88 proteins involved in the dynamics of the cytoskeleton that showed strong up-regulation in the LS+K multipotent cells (Fig. 2, Fig. 5, and supplemental Table S5). This network (Fig. 5) was centered around alpha-actin-1 and gamma actin, which are known to be key proteins for cell shape and motility (38, 39). The actin network was linked to a cluster of 14 myosin proteins strongly up-regulated in LS+K (fold-change +1.3 to +13.0; Fig. 5 and supplemental Table S5). This cluster included the ubiquitous Myosin II proteins MYL6, MYH9, and MYL12b, which are thought to play a central role in cell adhesion, migration and division in non-muscle cells (40). To gain insight into the subcellular localization of the differentially expressed myosins, MYL3 was examined by immunocytochemistry on cytospins of LS−K and LS+K cells (supplemental Fig. S6B). Notably, MYL3 showed strong and exclusive detection at the cell periphery of multipotent LS+K cells (Fig. 6B), suggesting a possible role in plasma membrane dynamics (38). In addition, we found 48 actin accessory proteins (e.g. gelsolin, vasodilator-stimulated phosphoprotein (VASP), lymphocyte-specific protein 1 (Fig. 5 and supplemental Table S5)), which are known to modulate organization, polymerization and movement of actin filaments (41–43). Interestingly, we found thymosin beta-4 (Tmsb4x) to be higher expressed in LS+K cells. This protein has been shown to modulate multiple processes related to extracellular matrix (ECM) function, including induction of metallo-proteinases, chemotaxis, angiogenesis as well as inhibition of inflammation and bone marrow stem cell proliferation (44, 45). Tmsb4x might exert similar roles in LS+K and thus contribute to maintenance of the multipotent state. Consistently, the actin-network was linked by alpha-actinin-1, alpha-actinin-3, and Tmsb4x via four integrins (integrin alpha 6, beta-1, beta-2, and alpha-M) to a cluster of 30 components of the ECM, 24 of them strongly up-regulated in the LS+K (+1.6 to +14.8-fold; Fig. 5 and supplemental Table S5). These ECM components included both structural proteins (collagen alpha-1(I) chain, -2(I) chain, and -1(II) chain) and functional proteins like receptors, cytokines, or peptidase inhibitors (e.g. galectin-3, CXCL7, neutrophilic granule protein; Fig. 5). As potential downstream mediators, our network analysis highlighted tyrosine-protein phosphatases (tyrosine-protein phosphatase C receptor-type (Ly-5), protein-tyrosine phosphatase non-receptor type 6 and 18), tyrosine-protein kinases (CSK and SYK; (46)) and adapter proteins (proto-oncogene C-crk, dedicator of cytokinesis protein 2; (47, 48) and Fig. 5). Furthermore, components of InsP3-signaling (phospholipase C-gamma-1 and 2, inositol monophosphatase 1, inositol polyphosphate 1-phosphatase) and PKC-signaling (pleckstrin, myosin-Ic, ADP-ribosylation factor 6) were found to be up-regulated in LS+K (Fig. 5). In addition, Marcks, Marcksl, and Basp1, known regulators of adhesion and membrane dynamics (49, 50), were found to be two- to fourfold higher expressed in LS+K (Fig. 5). Taken together, our data demonstrate that an extensive network of cytoskeleton and extracellular matrix proteins is elevated in multipotent progenitor cells, which may promote adhesion, membrane dynamics and chemotaxis in the multipotent state.
Fig. 5.
Differential expression of cytoskeletal and extracellular matrix proteins. An interaction network was built based on differentially expressed proteins (p < 0.05) of cytoskeleton and extracellular matrix (according to GO-classification) using STRING. Based on proteins quantified in three replicates (circular nodes), highly related proteins quantified in two replicates were implemented (square nodes). Proteins higher expressed in LS+K cells are shown in red and proteins higher expressed in LS−K cells are shown in blue.
Fig. 6.
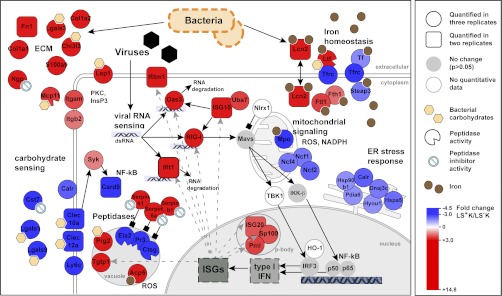
Differential expression of proteins involved in innate immune response. The illustration depicts self-protective mechanisms of multipotent and myeloid committed progenitors. Proteins were grouped according to GO-classification and literature (see main text). Proteins higher expressed in LS+K cells are shown in red and proteins higher expressed in LS−K cells are shown in blue.
Distinct Self-protective Signatures in multipotent LS+K and Myeloid LS−K Cells at Steady-state
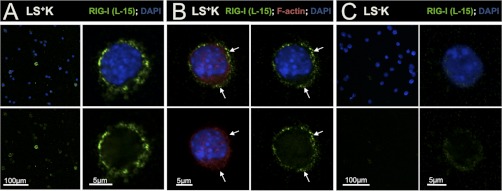
We found a large diversity of self-protective mechanisms to be differentially regulated between multipotent and myeloid committed progenitors under homeostatic conditions (Fig. 2, Fig. 6, and Table I). The multipotent defense signature included a diverse repertoire of extracellular matrix proteins known to exert antimicrobial functions (e.g. chitinase-3-like protein 3, neutrophilic granule protein, mast cell protease-11; Fig. 6). Further, several members of the family of serine protease inhibitors (Serpins b1, b6a, h1) were higher expressed in LS+K (+2.9 to +3.4-fold; Fig. 6 and Table I). Serpins are known to counterbalance cytoplasmic inflammatory proteases e.g. in neutrophils (51), suggesting such a role in LS+K cells. Strikingly, 2′-5′ oligoadenylate synthetase 3 (Oas3), retinoic acid-inducible gene 1 (RIG-I, also known as DDX58) and Ifit1, three different cytoplasmic sensors for viral RNA (52–54), were strongly up-regulated in the multipotent state (Oas3 + 3.9, RIG-I +4.2, and Ifit1 + 3.2 times higher in LS+K; Table I). Whereas binding of foreign RNA to Oas3 or Ifit1 leads to RNA degradation (52, 54), activation of the RIG-I-like helicase (RLH) RIG-I induces a complex signaling cascade, which triggers antiviral response by activation of type-I interferons (55). Consistently, we found downstream RLH-signaling components and interferon-stimulated proteins to be induced in the multipotent population (e.g. Uba7, Pml, Sp100, Stat1, and Tgtp1; Table I). To investigate subcellular localization of RIG-I, we examined its expression in cytospins of LS−K and LS+K cells (Fig. 7). Interestingly, we found a strong peripheral expression in LS+K multipotent cells (Fig. 7A, 7B) but only weak signals in LS−K cells (Fig. 7C). Co-staining with F-actin revealed a localization of RIG-I in close apical proximity to the actin cortex but without overlap (Fig. 7B). These results were confirmed by an independent RIG-I antibody (C-15; data not shown). Five proteins involved in antibacterial iron regulation were found to be induced in the LS+K population (Lcn2, Ltf, Fth, Ftl1, and Fpn1; (56, 57)). Further, several proteins participating in ROS signaling and secretion of pro-inflammatory cytokines were up-regulated in the multipotent state (e.g. tartrate-resistant acid ATPase +5.3-fold and adenosine deaminase +2.1-fold; (58, 59)). Notably, also components of the adaptive immune response showed up-regulation in LS+K cells, including the “don't-eat-me” signal CD47 (+1.8-fold; (60)) and the B-cell immunity receptor Igh-6 (+9.9-fold). Taken together, our analyses indicate a robust and diverse self-protective signature in multipotent LS+K cells at steady-state conditions. In contrast, myeloid committed LS−K cells showed a different pattern of defense-related proteins, reflecting distinct self-protective mechanisms (Fig. 6 and Table I). Strikingly, three neutrophil serine proteases (elastase-2, proteinase 3 and cathepsin G) were found in our data among the top-5 changed proteins (fold change –7.1 to –7.9; Table I). Such proteases serve for bacterial killing in specialized lysosomal granules and are known to be specifically expressed in early myeloid progenitors (61). Further, C-type lectins (Clec10a, Clec12a), which are involved in sensing of bacterial carbohydrates (62) and their downstream mediator Card9 (63) were pronounced in the myeloid compartment. Notably, numerous ER stress response proteins that participate in unfolded protein response and immunomodulatory functions (e.g. calreticulin, Dnajc3, heat shock 70kDa protein 5; Fig. 2, Fig. 6 and Table I; (64)) were found higher expressed in the myeloid LS−K cells. In comparison to LS+K cells, other proteins involved in iron homeostasis (STEAP3, serotransferrin and transferrin receptor 1 (Tfrc); (65, 66)) and ROS signaling (myeloperoxidase, neutrophil NADPH oxidase factor 1, 2, and 4; (67, 68)) showed elevated expression in the myeloid state, demonstrating a distinct profile compared with the multipotent progenitors. Notably, our FACS-analysis revealed differential expression of Tfrc in subpopulations of LS−K, with a higher expression in common myeloid progenitors (CMPs) and megakaryocyte/erythrocyte progenitors (MEPs), and intermediate expression in granulocyte/macrophage progenitors (GMPs) (supplemental Fig. S5). All together, our data demonstrate a self-protective signature in multipotent cells at steady-state, which is distinct from the myeloid committed profile with respect to proteins and mechanisms involved.
Table I. List of differentially expressed proteins between LS+K and LS−K involved in immune response.
(* = quantified in two replicates)
| Uniprot ID | Gene Name | Protein Name | Biological Process/Molecular Function | Fold change LS+K/LS−K | Adjusted p value | # of peptides |
|---|---|---|---|---|---|---|
| Extracellular matrix | ||||||
| P29699 | Ahsg* | Alpha-2-HS-glycoprotein | Acute phase response; peptidase inhibitor activity | 5.42 | – | 2 |
| O35744 | Chi3l3 | Chitinase-3-like protein 3 | Bacterial carbohydrate sensing; chemotaxis | 4.85 | 0.00 | 7 |
| P11087 | Col1a1 | Collagen alpha-1(I) chain | Structural constituent; cell-substrate adhesion | 6.99 | 0.00 | 5 |
| Q01149 | Col1a2 | Collagen alpha-2(I) chain | Structural constituent; cell-substrate adhesion | 6.51 | 0.02 | 6 |
| P28481 | Col2a1 | Collagen alpha-1(II) chain | Structural constituent; cell-substrate adhesion | 5.63 | 0.04 | 6 |
| Q9Z126 | Cxcl4 | Chemokine (C-X-C motif) ligand 4 | Cytokine activity; chemotaxis | 2.07 | 0.03 | 3 |
| Q9EQI5 | Cxcl7 | Chemokine (C-X-C motif) ligand 7 | Cytokine activity; chemotaxis | 2.95 | 0.02 | 1 |
| P11276 | Fn1* | Fibronectin | Acute phase response; cell adhesion | 4.95 | – | 11 |
| P16110 | Lgals3 | Galectin-3 | Lectin; IgE binding | 2.84 | 0.04 | 4 |
| B1AQJ6 | Lgals3bp | Galectin-3 binding protein | Cell adhesion; integrin-signaling | 1.70 | 0.03 | 2 |
| Q6RUT2 | Mcp11 | Mast cell protease-11 | Peptidase activity | 2.54 | 0.02 | 3 |
| O08692 | Ngp | Neutrophilic granule protein | Defense response; peptidase inhibitor activity | 4.05 | 0.03 | 7 |
| P31725 | S100a9 | Protein S100-A9 | Bacterial carbohydrate sensing; antimicrobial | 3.22 | 0.01 | 4 |
| P07214 | Sparc* | SPARC | Cell growth; proliferation; Ca2+ | 14.79 | – | 1 |
| P21981 | Tgm2 | Transglutaminase-2 | GPCR signaling; cell adhesion; Ca2+ | 6.00 | 0.00 | 8 |
| Q8CIZ8 | Vwf* | von Willebrand factor | Focal adhesion; ECM-receptor interaction | 2.31 | – | 1 |
| O89098 | Cst7 | Cystatin-7 | Peptidase inhibitor activity | −3.96 | 0.01 | 2 |
| Q8BH61 | F13a | Coagulation factor XIII A chain | Blood coagulation; peptide cross-linking | −4.56 | 0.00 | 15 |
| P16045 | Lgals1 | Galectin-1 | Cell-cell adhesion; myoblast differentiation | −1.97 | 0.00 | 7 |
| B1AQR8 | Lgals9 | Galectin-9 | Lectin | −1.50 | 0.00 | 11 |
| Carbohydrate sensing | ||||||
| Q62230 | Cd169 | Sialoadhesin | Bacterial carbohydrate sensing; cell adhesion | 2.94 | 0.00 | 41 |
| Q61735 | Cd47 | Leukocyte surface antigen CD47 | Cell adhesion; response to bacterium | 1.84 | 0.01 | 4 |
| Q91V08 | Clec2d* | C-type lectin domain family 2 member D | Carbohydrate sensing; C-type lectin; differentiation | 2.30 | – | 3 |
| P01873 | Igh-6 | Ig mu chain C region membrane-bound form | Antigen binding; activation of MAPK activity | 9.95 | 0.03 | 13 |
| P19973 | Lsp1 | Lymphocyte-specific protein 1 | Chemotaxis; PKC signaling; leukocyte migration | 4.38 | 0.00 | 8 |
| Q61878 | Prg2 | Bone marrow proteoglycan | Carbohydrate sensing; C-type lectin; cytotoxin | 3.19 | 0.01 | 6 |
| Q9JL95 | Prg3* | Proteoglycan 3, Isoform 1 | Carbohydrate sensing; C-type lectin; cytotoxin | 7.68 | – | 1 |
| Q09200 | B4galnt1 | Beta-1,4 N-acetylgalactosaminyltransferase 1 | Bacterial carbohydrate sensing | −3.83 | 0.00 | 11 |
| A2AIV8 | Card9* | Caspase recruitment domain family, member 9 | C-type lectin coupling; NF-κB activation | −4.40 | – | 5 |
| A9XX86 | Clec10a* | Macrophage galactose-type C-type lectin 2 | Bacterial carbohydrate sensing | −3.11 | – | 1 |
| Q504P2 | Clec12a* | C-type lectin domain family 12 member A | Bacterial carbohydrate sensing | −3.55 | – | 3 |
| P09568 | Ly6c | Lymphocyte antigen 6C2 | −5.74 | 0.00 | 3 | |
| A9XX86 | Mgl2* | Macrophage galactose N-acetyl-galactosamine specific lectin 2 | Carbohydrate sensing; C-type lectin | −3.11 | – | 1 |
| Peptidases/peptidase inhibitors | ||||||
| P70202 | Lxn | Latexin | Metalloprotease inhibitor activity | 2.07 | 0.01 | 3 |
| Q9D154 | Serpinb1 | Serine proteinase inhibitor, clade B, member 1 | Peptidase inhibitor activity | 2.94 | 0.00 | 17 |
| Q3U3L3 | Serpinb6a | Serine proteinase inhibitor, clade B, member 6a | Peptidase inhibitor activity | 3.41 | 0.03 | 2 |
| O08804 | Serpinb6b* | Serine peptidase inhibitor, clade B, member 6b | Peptidase inhibitor activity | 1.96 | – | 2 |
| O08797 | Serpinb9* | Serine proteinase inhibitor, clade B, member 9 | Peptidase inhibitor activity | 4.95 | – | 6 |
| P19324 | Serpinh1 | Serine proteinase inhibitor, clade H, member 1 | Peptidase inhibitor activity | 3.12 | 0.04 | 5 |
| P28293 | Ctsg | Cathepsin G | Endopeptidase activity | −7.09 | 0.00 | 15 |
| Q3UP87 | Ela2 | Elastase-2 | Endopeptidase activity | −7.43 | 0.00 | 12 |
| Q61096 | Prtn3 | Proteinase 3 | Endopeptidase activity | −7.85 | 0.00 | 7 |
| Antiviral response | ||||||
| Q3U5Q7 | Cmpk2 | UMP-CMP kinase 2, mitochondrial; | LPS response; IFN response | 2.34 | 0.01 | 9 |
| Q9Z0E6 | Gbp2 | Interferon-induced guanylate-binding protein 2 | GTPase activity | 3.55 | 0.00 | 9 |
| Q80SU7 | Gvin1 | Interferon-induced very large GTPase 1 | GTPase activity | 3.45 | 0.00 | 34 |
| Q3U5K8 | Ifit1* | Interferon-induced tetratricopeptide repeats 1 | Viral ppp-RNA binding; IFIT complex | 3.24 | – | 4 |
| Q8R2S7 | Ifitm1* | Interferon induced transmembrane protein 1 | Control cell growth; anti-proliferative | 3.87 | – | 1 |
| Q9D8C4 | Ifi35 | Interferon-induced 35 kDa protein homolog | 1.47 | 0.03 | 5 | |
| Q61179 | Irf9* | Interferon regulatory factor 9 isoform 1 | Transcription regulation | 1.93 | – | 4 |
| Q0GUM2 | Irgb10* | Interferon-gamma-inducible p47 GTPase | Host resistance | 2.15 | – | 3 |
| Q60766 | Irgm1* | Immunity-related GTPase family M protein | Cell motility; adhesion; autophagy | 1.80 | – | 3 |
| Q64339 | Isg15* | Interferon-stimulated protein 15 | RLH signaling; Ubl conjugation pathway | 3.89 | – | 2 |
| Q9JL16 | Isg20* | Interferon-stimulated gene 20 kDa protein | DNA/RNA metabolism; exonuclease activity | 2.03 | – | 5 |
| B9EIU4 | Oas3 | 2'-5' oligoadenylate synthetase 3 | Viral RNA binding; OAS/RNase L pathway | 3.88 | 0.03 | 4 |
| Q60953 | Pml | Probable transcription factor PML | Antiviral defense; apoptosis; transcription regulation | 2.34 | 0.00 | 22 |
| Q6Q899 | RIG-I | Retinoic acid-inducible gene 1 protein | RLH signaling; viral RNA/DNA binding; | 4.16 | 0.00 | 17 |
| Q60710 | Samhd1 | SAM domain- and HD domain-containing 1 | Hydrolase activity | 2.67 | 0.00 | 15 |
| O35892 | Sp100 | Nuclear autoantigen Sp-100 | PML body; DNA binding | 2.10 | 0.05 | 8 |
| P42225 | Stat1 | Signal transducer and activator of transcription 1 | 1.79 | 0.01 | 15 | |
| P42227 | Stat3 | Signal transducer and activator of transcription 3 | 1.43 | 0.03 | 11 | |
| Q3T9E4 | Tgtp1 | T-cell specific GTPase 1 | Response to virus; GTPase activity | 3.16 | 0.00 | 10 |
| Q9DBK7 | Uba7 | Ubiquitin-activating enzyme E1-like | RLH signaling; ISG15 activating enzyme activity | 2.36 | 0.01 | 10 |
| Iron homeostasis | ||||||
| Q9JHI9 | Fpn1* | Ferroportin-1 | Iron ion transport; apoptosis | 2.15 | – | 2 |
| P09528 | Fth1 | Ferritin heavy chain | Iron ion storage | 1.75 | 0.01 | 7 |
| Q3THE6 | Ftl1 | Ferritin light chain 1 | Iron ion transport; oxidoreductase activity | 2.37 | 0.01 | 11 |
| Q60842 | Lcn2* | Lipocalin-2 | Iron ion transport;siderophore transport; apoptosis | 4.49 | – | 3 |
| P08071 | Ltf | Lactotransferrin | Iron ion transport; Peptidase activity; antimicrobial | 4.82 | 0.02 | 8 |
| Q8CI59 | Steap3 | Metalloreductase STEAP3 | Iron ion uptake; apoptosis | −2.47 | 0.03 | 6 |
| Q921I1 | Tf | Serotransferrin | Iron ion transport; cell proliferation | −1.73 | 0.02 | 24 |
| Q62351 | Tfrc | Transferrin receptor protein 1 | Iron ion uptake; osteoclast differentiation | −3.18 | 0.00 | 11 |
| ROS signaling | ||||||
| Q05117 | Acp5 | Tartrate-resistant acid ATPase | Cytokinesis; antibactericial; H2O2 repression | 5.26 | 0.02 | 1 |
| P03958 | Ada | Adenosine deaminase | Amplification ROS; adenosine deaminase activity | 2.07 | 0.00 | 11 |
| Q9EQ20 | Ald6a1 | Aldehyde dehydrogenase family 6 member A1 | Aldehyde dehydrogenase (NAD) activity; differentiation | 2.05 | 0.02 | 6 |
| Q3TFC7 | Ald7a1 | Aldehyde dehydrogenase family 7 member A1 | Aldehyde dehydrogenase (NAD) activity | 2.73 | 0.02 | 6 |
| Q64669 | Nqo1 | NAD(P)H dehydrogenase [quinone] 1 | Response to oxidative stress | 2.03 | 0.04 | 4 |
| P11247 | Mpo | Myeloperoxidase | Response to ROS; microbicidal activity; | −7.49 | 0.00 | 45 |
| Q3UBI5 | Ncf1 | Neutrophil NADPH oxidase factor 1 | Bacterial killing; superoxide-generating oxidase | −2.02 | 0.01 | 11 |
| O70145 | Ncf2 | Neutrophil NADPH oxidase factor 2 | Bacterial killing; superoxide-generating oxidase | −2.28 | 0.01 | 11 |
| Q3TBC6 | Ncf4 | Neutrophil NADPH oxidase factor 4 | Bacterial killing; superoxide-generating oxidase | −2.01 | 0.01 | 7 |
| ER stress response | ||||||
| P14211 | Calr | Calreticulin | Carbohydrate binding; Lectin; protein stabilization | −2.38 | 0.00 | 29 |
| Q91YW3 | Dnajc3 | DnaJ homolog subfamily C member 3 | Proteolysis | −2.01 | 0.00 | 12 |
| P20029 | Hspa5 | Heat shock 70 kDa protein 5 | Proteolysis | −2.16 | 0.00 | 43 |
| P08113 | Hsp90b1 | Heat shock protein 90kDa beta | Proteolysis; unfolded protein binding | −1.80 | 0.00 | 38 |
| Q99LP6 | Grpel1 | GrpE like protein 1 | Protein folding | −1.63 | 0.03 | 7 |
| Q9JKR6 | Hyou1 | Hypoxia up-regulated protein 1 | Response to hypoxia | −1.83 | 0.00 | 34 |
| Q921X9 | Pdia5 | Protein disulfide-isomerase A5 | Response to stress; cell redox homeostasis | −1.77 | 0.01 | 7 |
| Q922R8 | Pdia6 | Protein disulfide isomerase A6 | Cell redox homeostasis | −1.98 | 0.00 | 23 |
Fig. 7.
RIG-I is differentially expressed between multipotent and myeloid committed progenitors. A, Expression and subcellular localization of the viral nucleic acid sensor RIG-I was examined on cytospins of LS+K. B, Costaining of RIG-I with F-actin (phalloidin) on LS+K cells revealed a distinct, non-matching localization of RIG-I more peripheral than F-actin. C, Expression and subcellular localization of RIG-I on cytospins of LS−K cells.
Gene-regulatory Proteins Are Differentially Expressed Between Multipotent and Myeloid Cells
We found a high number of proteins involved in regulation of gene expression to be differentially expressed between LS+K and LS−K cells (Fig. 2C and supplemental Table S6). These included transcription factors or co-factors such as Pml (+2.3-fold), ERG (+2.4-fold), and homeobox protein Meis1 (+2.7-fold), which were up-regulated in multipotent LS+K cells and on the contrary, Myb and Baz1a, which were higher expressed in myeloid LS−K cells (–1.9-fold and –2.0-fold, respectively; supplemental Table S6). Further, we found brain acid soluble protein 1 (Basp1), a corepressor shown to cooperate with WT1 in the regulation of hematopoietic differentiation (69) and antagonist to Myc (70) to be 4.5-fold increased in LS+K cells (supplemental Table S6). In addition, several proteins involved in epigenetic control of transcription were found to be strongly up-regulated in the multipotent state (supplemental Table S6). Peptidylarginine deiminase 4 (Padi4), a potent regulator of histone deimination and deacetylation (71) was 6.1-fold higher expressed in LS+K cells. In immunocytochemical stainings of LS+K cells, Padi4 showed a strong expression with nuclear pattern, whereas in myeloid LS−K cells, only weak expression was detected (supplemental Fig. S6C). These results are in line with observations that Padi4 is expressed in immature hematopoietic cells and potentially involved in tumorigenesis (72). Finally, we found Methyl-CpG-binding protein 2 (MeCp2), a powerful transcriptional repressor involved in DNA methylation and histone deacetylation (73) to be fourfold up-regulated in LS+K cells. Interestingly, Mecp2 has been shown to be involved in down-regulation of HIF-1A in multipotent CD34+ hematopoietic progenitors (74). Taken together, our data show that proteins at different levels of gene regulation are differentially expressed between LS+K and LS−K cells, which suggests their participation in the control of hematopoietic multipotency and commitment.
DISCUSSION
In this study, we aimed to compare global protein expression of multipotent hematopoietic stem/progenitor cells (HSPCs; LS+K) and myeloid committed progenitors (LS−K). The quantitative proteomics data (5023 quantified proteins) represents an important resource for advances in our understanding of the molecular basis of multipotency and commitment in the hematopoietic system.
Progress in stem cell biology research has proposed that specific metabolic properties control self-renewal and differentiation capacities (75–78). However, evidence for the hematopoietic stem cells remains elusive. It was therefore of great value to find a marked difference in the metabolic profile between LS+K and LS−K cells. Most importantly, key players regulating the metabolic fate of glucose and pyruvate were found differentially expressed. A clear shift in isoforms of hexokinase and pyruvate kinase as well as in the lactate dehydrogenase subunits collectively point to a remodeling of the cellular energetic infrastructure in the LS+K to LS−K transition. Surprisingly, the data suggests a difference between HSPCs and other stem cells during differentiation for some of the metabolic enzymes, e.g. in the expression of the hexokinase isoforms. In contrast to embryonic stem cells and induced pluripotent stem cells, which have been reported to maintain high glycolytic flux by high levels of hexokinase 2 (76), our data show that hexokinase 2 as well as the isoform 3 are higher expressed in the differentiated cells of the hematopoietic lineage. Instead, hexokinase 1 has an elevated expression in the multipotent progenitor cells, indicating that the different stem cells have different metabolic properties.
Many findings in our data (e.g. higher expression of pyruvate kinase M2, Pml, and pyruvate dehydrogenase kinase as well as lower expression of MOT1) strongly suggest that the pyruvate generated from glycolysis is redirected away from mitochondrial oxidative phosphorylation to generate lactate in the multipotent progenitor cells. In fact, the metabolic profile of LS+K cells resembles very much that seen in tumor cells and proliferating cells, involving a switch in the amount glucose metabolized by oxidative phosphorylation and aerobic glycolysis (Warburg effect) (79). The redirection of pyruvate away from the mitochondria to lactate formation is also observed for cells when oxygen is limited (79). Since HSPCs cells are known to have a lower proliferation rate than myeloid committed progenitors (30), the metabolic profile we observe for LS+K cells cannot be explained by a difference in proliferation rate, but rather suggests a more hypoxic environment for the multipotent progenitor cells, in line with a recent report for long-term hematopoietic stem cells (75). In addition to the findings on metabolism, our data on translational proteins supports a low-oxygen environment of the multipotent progenitors. In accordance to previous reports (14) our results solidly point to a lower translation activity in LS+K cells than in their myeloid successors, as is observed in response to stress, such as hypoxia (80). In such cases, an inhibition by phosphorylation of specific translation initiation factors is normally seen (81). Although we cannot exclude that this also occurs in LS+K cells, a different phenotype is evident, with a decreased expression of a majority of the translation initiation factors as well as other proteins involved in translation. A lower translation rate in the multipotent progenitor cells may also be because of a lower proliferation rate of these cells (30), and therefore the possible cause of low translation activity remains to be determined.
Strikingly, we found a large network of cytoskeleton proteins coupled to a distinct ECM repertoire up-regulated in multipotent LS+K cells. This network could exert possible roles in multiple processes including adhesion dynamics regulated at the actin cortex, cell-cell or cell-matrix interactions, chemotactic sensing of the environment, anchoring of signaling components as receptors to e.g. lipid rafts and enhanced motility (blebs and lamellipodia; (38)). The data strongly suggest that such processes are critical in the multipotent state. Notably, pseudopod morphologies were described previously for immature hematopoietic cells (82). The observed expression pattern is in accordance to a recently described biochemical interaction between pleckstrin, several ECM components and actin cytoskeleton (83). Expansion of these concepts on the basis of functional experiments will be of high interest. This can be investigated by e.g. challenging primary LS+K cultures in migration assays or by exposure to different ECM components and chemoattractants.
The crucial task of protecting the stem cell compartment in the hematopoietic system was long thought to be exerted exclusively by mature effector cells as e.g. neutrophils (84). It is a recent notion only that HSCs and multipotent progenitors themselves can be activated via feedback-loops in response to stress (5, 85). In addition, they seem to have intrinsic machinery for defense against attacking bacterial or viral pathogens (86). The robust self-protective signatures of LS+K and LS−K presented in this study, for the first time enable us to dissect the utilization of distinct immune mechanisms along differentiation on a comprehensive protein level. Strikingly, with three prominent cytoplasmic RNA sensors (Oas3, RIG-I, and Ifit1) higher expressed in the LS+K population, our results point to a crucial role of cytoplasmic dsRNA sensing in the multipotent state and contrast with recent reports of an attenuated dsRNA signaling as a general characteristic of pluripotency in hESCs (87). Notably, all three sensors are thought to be modulated in a feedback loop by the type-I interferon response (Fig. 6; (54, 88)), which represents a potential regulatory module. Of note, RIG-I is involved in diverse cellular functions including myeloid differentiation induced by retinoic acid (89), leukemogenesis (90) and control of apoptosis (91). Similarly, the other interferon-stimulated proteins we found to be regulated could exert various cellular roles. Accessibility of extracellular iron is another key strategy to limiting pathogens (92) and is indicated by our data to be important for multipotent LS+K cells. Interestingly, the iron transport proteins lipocalin-2 and lactotransferrin are also shown to serve other functionalities, like the sensing of bacterial carbohydrates (56, 57).
Our results suggest that multipotent progenitors employ a highly specific ECM repertoire, which is possibly an immediate sensor and at the same time a first line barrier against invading pathogens. Even the highly regulated actin cytoskeleton can be seen as a physical barrier, which, equipped with a repertoire of self-protective proteins, may ensure fast and specific acute immune response. On the other hand, myeloid committed progenitors seem to utilize a lectin-based detection of bacteria and lysosomal bacterial killing by granular serine proteases. Further, unfolded protein response in the ER seems to be myeloid-specific. In summary, our results indicate a switch of self-protective mechanisms at commitment from the multipotent to the myeloid committed state, which might reflect an elevated protective machinery for multipotent cells justified by their enormous value for the body. In addition, differences in ECM repertoires might reflect adaptations to specific microenvironments of multipotent versus myeloid committed cells in the bone marrow. Here, subtle changes might even correspond to sub-population-specific phenotypes, e.g. stem cell niche factors of the HSC fraction.
We found essential stem cell maintenance proteins like Pml and Meis1 (75, 93) to be higher expressed in multipotent LS+K cells. Importantly, several other gene-regulatory proteins like Basp1, Padi4, and Mecp2 were differentially expressed. These might be involved in up-stream regulation of the affected cellular processes and therefore of primary importance for biology of HSPCs. Accordingly, we will focus on the analysis of these gene-regulatory proteins on more refined HSC cell populations.
Taken together, we found that processes of cytoskeleton remodeling, energy metabolism, translation, and immune response are critical in characterizing the hematopoietic transition from multipotent progenitor cells to myeloid committed cells. In particular, the novel finding that these cells display a switch in proteins involved in self-protective mechanisms, suggests the exciting possibility that specific, and so far unrecognized, mechanisms are involved in protecting these precious cells as they mature. Collectively, our study has indicated that the progression from hematopoietic progenitors to myeloid committed cells is accompanied by a profound change in their proteomes, adding novel insights into the molecular mechanisms regulating multipotency and differentiation in the hematopoietic system. We expect these results to be very helpful to direct future research and functionally define the described phenomena in LS+K and LS−K cells, and to establish related phenotypes in more refined compartments of multipotent and myeloid committed cells.
Acknowledgments
Support by the EMBL Proteomics Core Facility, DKFZ Flow Cytometry Service Unit and DKFZ Light Microscopy Facility is gratefully acknowledged. We would like to thank Drs. Nina Cabezas, Marieke Essers, Joanna Kirkpatrick and Kiran Raosaheb Patil for comments on the manuscript.
Footnotes
* This work was supported in part by the Netherlands Organisation for Scientific Research (NWO).
 This article contains supplemental Tables S1 to S8 and Figs. S1 to S6.
This article contains supplemental Tables S1 to S8 and Figs. S1 to S6.
1 The abbreviations used are:
- HSC
- Hematopoietic stem cell
- APC
- Allophycocyanin
- Cy7
- Cyanine dye 7
- ECM
- Extracellular matrix
- FDR
- False discovery rate
- LS+K
- Lineage negative, Sca-1 positive, c-Kit positive
- LS−K
- Lineage negative, Sca-1 negative, c-Kit positive
- MPP
- Multipotent progenitor cell
- PE
- Phycoerythrobilin
- ROS
- Reactive oxygen species
- TCA
- Tricarboxylic acid.
REFERENCES
- 1. Till J. E., McCulloch C. E. (1961) A direct measurement of the radiation sensitivity of normal mouse bone marrow cells. Radiation Res. 14, 213–222 [PubMed] [Google Scholar]
- 2. Purton L. E., Scadden D. T. (2007) Limiting factors in murine hematopoietic stem cell assays. Cell Stem Cell 1, 263–270 [DOI] [PubMed] [Google Scholar]
- 3. Wilson A., Oser G. M., Jaworski M., Blanco-Bose W. E., Laurenti E., Adolphe C., Essers M. A., Macdonald H. R., Trumpp A. (2007) Dormant and self-renewing hematopoietic stem cells and their niches. Ann. NY Acad. Sci. 1106, 64–75 [DOI] [PubMed] [Google Scholar]
- 4. Weissman I. L., Shizuru J. A. (2008) The origins of the identification and isolation of hematopoietic stem cells, and their capability to induce donor-specific transplantation tolerance and treat autoimmune diseases. Blood 112, 3543–3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Trumpp A., Essers M., Wilson A. (2010) Awakening dormant haematopoietic stem cells. Nature Rev. Immunol. 10, 201–209 [DOI] [PubMed] [Google Scholar]
- 6. Graf T., Enver T. (2009) Forcing cells to change lineages. Nature 462, 587–594 [DOI] [PubMed] [Google Scholar]
- 7. Enver T., Pera M., Peterson C., Andrews P. W. (2009) Stem cell states, fates, and the rules of attraction. Cell Stem Cell 4, 387–397 [DOI] [PubMed] [Google Scholar]
- 8. Passegue E., Jamieson C. H., Ailles L. E., Weissman I. L. (2003) Normal and leukemic hematopoiesis: are leukemias a stem cell disorder or a reacquisition of stem cell characteristics? Proc. Natl. Acad. Sci. U. S. A. 1, 11842–11849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jamieson C. H. (2008) Chronic myeloid leukemia stem cells. Hematology Am. Soc. Hematol. Educ. Program 436–442 [DOI] [PubMed] [Google Scholar]
- 10. Becker M. W., Jordan C. T. (2011) Leukemia stem cells in 2010: current understanding and future directions. Blood Rev. 25, 75–81 [DOI] [PubMed] [Google Scholar]
- 11. Reiland S., Salekdeh G. H., Krijgsveld J. (2011) Defining pluripotent stem cells through quantitative proteomic analysis. Expert Rev. Proteomics 8, 29–42 [DOI] [PubMed] [Google Scholar]
- 12. Rigbolt K. T., Prokhorova T. A., Akimov V., Henningsen J., Johansen P. T., Kratchmarova I., Kassem M., Mann M., Olsen J. V., Blagoev B. (2011) System-wide temporal characterization of the proteome and phosphoproteome of human embryonic stem cell differentiation. Sci. Signaling 4, rs3. [DOI] [PubMed] [Google Scholar]
- 13. Van Hoof D., Muñoz J., Braam S. R., Pinkse M. W., Linding R., Heck A. J., Mummery C. L., Krijgsveld J. (2009) Phosphorylation dynamics during early differentiation of human embryonic stem cells. Cell Stem Cell 5, 214–226 [DOI] [PubMed] [Google Scholar]
- 14. Spooncer E., Brouard N., Nilsson S. K., Williams B., Liu M. C., Unwin R. D., Blinco D., Jaworska E., Simmons P. J., Whetton A. D. (2008) Developmental fate determination and marker discovery in hematopoietic stem cell biology using proteomic fingerprinting. Mol. Cell. Proteomics 7, 573–581 [DOI] [PubMed] [Google Scholar]
- 15. Boersema P. J., Raijmakers R., Lemeer S., Mohammed S., Heck A. J. (2009) Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat. Protoc. 4, 484–494 [DOI] [PubMed] [Google Scholar]
- 16. Rappsilber J., Mann M., Ishihama Y. (2007) Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896–1906 [DOI] [PubMed] [Google Scholar]
- 17. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 18. Gentleman R. C., Carey V. J., Bates D. M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., Hornik K., Hothorn T., Huber W., Iacus S., Irizarry R., Leisch F., Li C., Maechler M., Rossini A. J., Sawitzki G., Smith C., Smyth G., Tierney L., Yang J. Y., Zhang J. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5, R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Smyth G. K. (2004) Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Statistical Appl. Gen. Mol. Biol. 3, Article3 [DOI] [PubMed] [Google Scholar]
- 20. Mi H., Guo N., Kejariwal A., Thomas P. D. (2007) PANTHER version 6: protein sequence and function evolution data with expanded representation of biological pathways. Nucleic Acids Res. 35, D247–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580 [DOI] [PubMed] [Google Scholar]
- 22. Nikolsky Y., Ekins S., Nikolskaya T., Bugrim A. (2005) A novel method for generation of signature networks as biomarkers from complex high throughput data. Toxicol. Lett. 158, 20–29 [DOI] [PubMed] [Google Scholar]
- 23. Szklarczyk D., Franceschini A., Kuhn M., Simonovic M., Roth A., Minguez P., Doerks T., Stark M., Muller J., Bork P., Jensen L. J., von Mering C. (2011) The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 39, D561–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., Ideker T. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jain E., Bairoch A., Duvaud S., Phan I., Redaschi N., Suzek B. E., Martin M. J., McGarvey P., Gasteiger E. (2009) Infrastructure for the life sciences: design and implementation of the UniProt website. BMC Bioinformatics 10, 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dong H. Y., Wilkes S., Yang H. (2011) CD71 is selectively and ubiquitously expressed at high levels in erythroid precursors of all maturation stages: a comparative immunochemical study with glycophorin A and hemoglobin A. Am. J. Surgical Pathol. 35, 723–732 [DOI] [PubMed] [Google Scholar]
- 27. Chow A., Lucas D., Hidalgo A., Méndez-Ferrer S., Hashimoto D., Scheiermann C., Battista M., Leboeuf M., Prophete C., van Rooijen N., Tanaka M., Merad M., Frenette P. S. (2011) Bone marrow CD169+ macrophages promote the retention of hematopoietic stem and progenitor cells in the mesenchymal stem cell niche. J. Exp. Med. 208, 261–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wölfler A., Danen-van Oorschot A. A., Haanstra J. R., Valkhof M., Bodner C., Vroegindeweij E., van Strien P., Novak A., Cupedo T., Touw I. P. (2010) Lineage-instructive function of C/EBPalpha in multipotent hematopoietic cells and early thymic progenitors. Blood 116, 4116–4125 [DOI] [PubMed] [Google Scholar]
- 29. Kiel M. J., Yilmaz O. H., Iwashita T., Yilmaz O. H., Terhorst C., Morrison S. J. (2005) SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell 121, 1109–1121 [DOI] [PubMed] [Google Scholar]
- 30. Passegué E., Wagers A. J., Giuriato S., Anderson W. C., Weissman I. L. (2005) Global analysis of proliferation and cell cycle gene expression in the regulation of hematopoietic stem and progenitor cell fates. J. Exp. Med. 202, 1599–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. López-Lastra M., Rivas A., Barria M. I. (2005) Protein synthesis in eukaryotes: the growing biological relevance of cap-independent translation initiation. Biol. Res. 38, 121–146 [DOI] [PubMed] [Google Scholar]
- 32. Wilson J. E. (2003) Isozymes of mammalian hexokinase: structure, subcellular localization and metabolic function. J. Exp. Biol. 206, 2049–2057 [DOI] [PubMed] [Google Scholar]
- 33. Shimada N., Shinagawa T., Ishii S. (2008) Modulation of M2-type pyruvate kinase activity by the cytoplasmic PML tumor suppressor protein. Genes Cells 13, 245–254 [DOI] [PubMed] [Google Scholar]
- 34. Christofk H. R., Vander Heiden M. G., Harris M. H., Ramanathan A., Gerszten R. E., Wei R., Fleming M. D., Schreiber S. L., Cantley L. C. (2008) The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature 452, 230–233 [DOI] [PubMed] [Google Scholar]
- 35. Kolobova E., Tuganova A., Boulatnikov I., Popov K. M. (2001) Regulation of pyruvate dehydrogenase activity through phosphorylation at multiple sites. Biochem. J. 358, 69–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kim J. H., Kim E. L., Lee Y. K., Park C. B., Kim B. W., Wang H. J., Yoon C. H., Lee S. J., Yoon G. (2011) Decreased lactate dehydrogenase B expression enhances claudin 1-mediated hepatoma cell invasiveness via mitochondrial defects. Exp. Cell Res. 317, 1108–1118 [DOI] [PubMed] [Google Scholar]
- 37. Poole R. C., Halestrap A. P. (1993) Transport of lactate and other monocarboxylates across mammalian plasma membranes. Am. J. Physiol. 264, C761–782 [DOI] [PubMed] [Google Scholar]
- 38. Ridley A. J. (2011) Life at the leading edge. Cell 145, 1012–1022 [DOI] [PubMed] [Google Scholar]
- 39. Pollard T. D., Cooper J. A. (2009) Actin, a central player in cell shape and movement. Science 326, 1208–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Park I., Han C., Jin S., Lee B., Choi H., Kwon J. T., Kim D., Kim J., Lifirsu E., Park W. J., Park Z. Y., Kim do H., Cho C. (2011) Myosin regulatory light chains are required to maintain the stability of myosin II and cellular integrity. Biochem. J. 434, 171–180 [DOI] [PubMed] [Google Scholar]
- 41. Silacci P., Mazzolai L., Gauci C., Stergiopulos N., Yin H. L., Hayoz D. (2004) Gelsolin superfamily proteins: key regulators of cellular functions. Cell. Mol. Life Sci. 61, 2614–2623 [DOI] [PubMed] [Google Scholar]
- 42. Dominguez R., Holmes K. C. (2011) Actin structure and function. Ann. Rev. Biophys. 40, 169–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Petri B., Kaur J., Long E. M., Li H., Parsons S. A., Butz S., Phillipson M., Vestweber D., Patel K. D., Robbins S. M., Kubes P. (2011) Endothelial LSP1 is involved in endothelial dome formation, minimizing vascular permeability changes during neutrophil transmigration in vivo. Blood 117, 942–952 [DOI] [PubMed] [Google Scholar]
- 44. Huff T., Muller C. S., Otto A. M., Netzker R., Hannappel E. (2001) beta-Thymosins, small acidic peptides with multiple functions. Int. J. Biochem. Cell Biol. 33, 205–220 [DOI] [PubMed] [Google Scholar]
- 45. Husson C., Cantrelle F. X., Roblin P., Didry D., Le K. H., Perez J., Guittet E., Van Heijenoort C., Renault L., Carlier M. F. (2010) Multifunctionality of the beta-thymosin/WH2 module: G-actin sequestration, actin filament growth, nucleation, and severing. Ann. NY Acad. Sci. 1194, 44–52 [DOI] [PubMed] [Google Scholar]
- 46. Mócsai A., Ruland J., Tybulewicz V. L. (2010) The SYK tyrosine kinase: a crucial player in diverse biological functions. Nat. Rev. Immunol. 10, 387–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Feller S. M. (2001) Crk family adaptors-signalling complex formation and biological roles. Oncogene 20, 6348–6371 [DOI] [PubMed] [Google Scholar]
- 48. Cabodi S., del Pilar Camacho-Leal M., Di Stefano P., Defilippi P. (2010) Integrin signalling adaptors: not only figurants in the cancer story. Nat. Rev. Cancer 10, 858–870 [DOI] [PubMed] [Google Scholar]
- 49. Larsson C. (2006) Protein kinase C and the regulation of the actin cytoskeleton. Cell. Signal. 18, 276–284 [DOI] [PubMed] [Google Scholar]
- 50. Sheetz M. P., Sable J. E., Dobereiner H. G. (2006) Continuous membrane-cytoskeleton adhesion requires continuous accommodation to lipid and cytoskeleton dynamics. Ann. Rev. Biophys. Biomol. Structure 35, 417–434 [DOI] [PubMed] [Google Scholar]
- 51. Benarafa C., LeCuyer T. E., Baumann M., Stolley J. M., Cremona T. P., Remold-O'Donnell E. (2011) SerpinB1 protects the mature neutrophil reserve in the bone marrow. J. Leukocyte Biol. 90, 21–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bréhin A. C., Casadémont I., Frenkiel M. P., Julier C., Sakuntabhai A., Desprès P. (2009) The large form of human 2′,5′-Oligoadenylate Synthetase (OAS3) exerts antiviral effect against Chikungunya virus. Virology 384, 216–222 [DOI] [PubMed] [Google Scholar]
- 53. Hornung V., Ellegast J., Kim S., Brzózka K., Jung A., Kato H., Poeck H., Akira S., Conzelmann K. K., Schlee M., Endres S., Hartmann G. (2006) 5′-Triphosphate RNA is the ligand for RIG-I. Science 314, 994–997 [DOI] [PubMed] [Google Scholar]
- 54. Pichlmair A., Lassnig C., Eberle C. A., Gorna M. W., Baumann C. L., Burkard T. R., Burckstümmer T., Stefanovic A., Krieger S., Bennett K. L., Rulicke T., Weber F., Colinge J., Muller M., Superti-Furga G. (2011) IFIT1 is an antiviral protein that recognizes 5′-triphosphate RNA. Nat. Immunol. 12, 624–630 [DOI] [PubMed] [Google Scholar]
- 55. Loo Y. M., Gale M., Jr. (2011) Immune signaling by RIG-I-like receptors. Immunity 34, 680–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Flo T. H., Smith K. D., Sato S., Rodriguez D. J., Holmes M. A., Strong R. K., Akira S., Aderem A. (2004) Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature 432, 917–921 [DOI] [PubMed] [Google Scholar]
- 57. Devireddy L. R., Hart D. O., Goetz D. H., Green M. R. (2010) A mammalian siderophore synthesized by an enzyme with a bacterial homolog involved in enterobactin production. Cell 141, 1006–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bune A. J., Hayman A. R., Evans M. J., Cox T. M. (2001) Mice lacking tartrate-resistant acid phosphatase (Acp 5) have disordered macrophage inflammatory responses and reduced clearance of the pathogen, Staphylococcus aureus. Immunology 102, 103–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kälvegren H., Fridfeldt J., Bengtsson T. (2010) The role of plasma adenosine deaminase in chemoattractant-stimulated oxygen radical production in neutrophils. Eur. J. Cell Biol. 89, 462–467 [DOI] [PubMed] [Google Scholar]
- 60. Jaiswal S., Jamieson C. H., Pang W. W., Park C. Y., Chao M. P., Majeti R., Traver D., van Rooijen N., Weissman I. L. (2009) CD47 is upregulated on circulating hematopoietic stem cells and leukemia cells to avoid phagocytosis. Cell 138, 271–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Korkmaz B., Moreau T., Gauthier F. (2008) Neutrophil elastase, proteinase 3 and cathepsin G: physicochemical properties, activity and physiopathological functions. Biochimie 90, 227–242 [DOI] [PubMed] [Google Scholar]
- 62. Osorio F., Reis e Sousa C. (2011) Myeloid C-type lectin receptors in pathogen recognition and host defense. Immunity 34, 651–664 [DOI] [PubMed] [Google Scholar]
- 63. Kerrigan A. M., Brown G. D. (2011) Syk-coupled C-type lectins in immunity. Trends Immunol. 32, 151–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Peters L. R., Raghavan M. (2011) Endoplasmic reticulum calcium depletion impacts chaperone secretion, innate immunity, and phagocytic uptake of cells. J. Immunol. 187, 919–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kaplan J. (2002) Mechanisms of cellular iron acquisition: another iron in the fire. Cell 111, 603–606 [DOI] [PubMed] [Google Scholar]
- 66. Pan X., Tamilselvam B., Hansen E. J., Daefler S. (2010) Modulation of iron homeostasis in macrophages by bacterial intracellular pathogens. BMC Microbiol. 10, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Tsuruta T., Tani K., Hoshika A., Asano S. (1999) Myeloperoxidase gene expression and regulation by myeloid cell growth factors in normal and leukemic cells. Leukemia Lymphoma 32, 257–267 [DOI] [PubMed] [Google Scholar]
- 68. El-Benna J., Dang P. M., Gougerot-Pocidalo M. A., Marie J. C., Braut-Boucher F. (2009) p47phox, the phagocyte NADPH oxidase/NOX2 organizer: structure, phosphorylation and implication in diseases. Exp. Mol. Med. 41, 217–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Goodfellow S. J., Rebello M. R., Toska E., Zeef L. A., Rudd S. G., Medler K. F., Roberts S. G. (2011) WT1 and its transcriptional cofactor BASP1 redirect the differentiation pathway of an established blood cell line. Biochem. J. 435, 113–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Hartl M., Nist A., Khan M. I., Valovka T., Bister K. (2009) Inhibition of Myc-induced cell transformation by brain acid-soluble protein 1 (BASP1). Proc. Natl. Acad. Sci. U. S. A. 106, 5604–5609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Cuthbert G. L., Daujat S., Snowden A. W., Erdjument-Bromage H., Hagiwara T., Yamada M., Schneider R., Gregory P. D., Tempst P., Bannister A. J., Kouzarides T. (2004) Histone deimination antagonizes arginine methylation. Cell 118, 545–553 [DOI] [PubMed] [Google Scholar]
- 72. Chang X., Fang K. (2010) PADI4 and tumourigenesis. Cancer Cell Int. 10, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Adkins N. L., Georgel P. T. (2011) MeCP2: structure and function. Biochem. Cell Biol. 89, 1–11 [DOI] [PubMed] [Google Scholar]
- 74. Walczak-Drzewiecka A., Ratajewski M., Pulaski L., Dastych J. (2010) DNA methylation-dependent suppression of HIF1A in an immature hematopoietic cell line HMC-1. Biochem. Biophys. Res. Commun. 391, 1028–1032 [DOI] [PubMed] [Google Scholar]
- 75. Simsek T., Kocabas F., Zheng J., Deberardinis R. J., Mahmoud A. I., Olson E. N., Schneider J. W., Zhang C. C., Sadek H. A. (2010) The distinct metabolic profile of hematopoietic stem cells reflects their location in a hypoxic niche. Cell Stem Cell 7, 380–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Varum S., Rodrigues A. S., Moura M. B., Momcilovic O., Easley C. A., 4th., Ramalho-Santos J., Van Houten B., Schatten G. (2011) Energy metabolism in human pluripotent stem cells and their differentiated counterparts. PloS One 6, e20914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Folmes C. D., Nelson T. J., Martinez-Fernandez A., Arrell D. K., Lindor J. Z., Dzeja P. P., Ikeda Y., Perez-Terzic C., Terzic A. (2011) Somatic oxidative bioenergetics transitions into pluripotency-dependent glycolysis to facilitate nuclear reprogramming. Cell Metab. 14, 264–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Suda T., Takubo K., Semenza G. L. (2011) Metabolic regulation of hematopoietic stem cells in the hypoxic niche. Cell Stem Cell 9, 298–310 [DOI] [PubMed] [Google Scholar]
- 79. Vander Heiden M. G., Cantley L. C., Thompson C. B. (2009) Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Liu L., Simon M. C. (2004) Regulation of transcription and translation by hypoxia. Cancer Biol. Therapy 3, 492–497 [DOI] [PubMed] [Google Scholar]
- 81. Proud C. G. (2005) eIF2 and the control of cell physiology. Seminars Cell Develop. Biol. 16, 3–12 [DOI] [PubMed] [Google Scholar]
- 82. Francis K., Palsson B., Donahue J., Fong S., Carrier E. (2002) Murine Sca-1(+)/Lin(-) cells and human KG1a cells exhibit multiple pseudopod morphologies during migration. Exp. Hematol. 30, 460–463 [DOI] [PubMed] [Google Scholar]
- 83. Baig A., Bao X., Haslam R. J. (2009) Proteomic identification of pleckstrin-associated proteins in platelets: possible interactions with actin. Proteomics 9, 4254–4258 [DOI] [PubMed] [Google Scholar]
- 84. Mantovani A., Cassatella M. A., Costantini C., Jaillon S. (2011) Neutrophils in the activation and regulation of innate and adaptive immunity. Nat. Rev. Immunol. 11, 519–531 [DOI] [PubMed] [Google Scholar]
- 85. Essers M. A., Offner S., Blanco-Bose W. E., Waibler Z., Kalinke U., Duchosal M. A., Trumpp A. (2009) IFNalpha activates dormant haematopoietic stem cells in vivo. Nature 458, 904–908 [DOI] [PubMed] [Google Scholar]
- 86. King K. Y., Goodell M. A. (2011) Inflammatory modulation of HSCs: viewing the HSC as a foundation for the immune response. Nat. Rev. Immunol. 11, 685–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Chen L. L., Yang L., Carmichael G. G. (2010) Molecular basis for an attenuated cytoplasmic dsRNA response in human embryonic stem cells. Cell cycle 9, 3552–3564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Kim M. J., Hwang S. Y., Imaizumi T., Yoo J. Y. (2008) Negative feedback regulation of RIG-I-mediated antiviral signaling by interferon-induced ISG15 conjugation. J. Virol. 82, 1474–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Zhang N. N., Shen S. H., Jiang L. J., Zhang W., Zhang H. X., Sun Y. P., Li X. Y., Huang Q. H., Ge B. X., Chen S. J., Wang Z. G., Chen Z., Zhu J. (2008) RIG-I plays a critical role in negatively regulating granulocytic proliferation. Proc. Natl. Acad. Sci. U. S. A. 105, 10553–10558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Liu T. X., Zhang J. W., Tao J., Zhang R. B., Zhang Q. H., Zhao C. J., Tong J. H., Lanotte M., Waxman S., Chen S. J., Mao M., Hu G. X., Zhu L., Chen Z. (2000) Gene expression networks underlying retinoic acid-induced differentiation of acute promyelocytic leukemia cells. Blood 96, 1496–1504 [PubMed] [Google Scholar]
- 91. Ishibashi O., Ali M. M., Luo S. S., Ohba T., Katabuchi H., Takeshita T., Takizawa T. (2011) Short RNA Duplexes Elicit RIG-I-Mediated Apoptosis in a Cell Type- and Length-Dependent Manner. Sci. Signal. 4, ra74. [DOI] [PubMed] [Google Scholar]
- 92. Wang L., Cherayil B. J. (2009) Ironing out the wrinkles in host defense: interactions between iron homeostasis and innate immunity. J. Innate Immun. 1, 455–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Ito K., Bernardi R., Morotti A., Matsuoka S., Saglio G., Ikeda Y., Rosenblatt J., Avigan D. E., Teruya-Feldstein J., Pandolfi P. P. (2008) PML targeting eradicates quiescent leukaemia-initiating cells. Nature 453, 1072–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]