Abstract
Here we describe a systematic structure-function analysis of the human ubiquitin (Ub) E2 conjugating proteins, consisting of the determination of 15 new high-resolution three-dimensional structures of E2 catalytic domains, and autoubiquitylation assays for 26 Ub-loading E2s screened against a panel of nine different HECT (homologous to E6-AP carboxyl terminus) E3 ligase domains. Integration of our structural and biochemical data revealed several E2 surface properties associated with Ub chain building activity; (1) net positive or neutral E2 charge, (2) an “acidic trough” located near the catalytic Cys, surrounded by an extensive basic region, and (3) similarity to the previously described HECT binding signature in UBE2L3 (UbcH7). Mass spectrometry was used to characterize the autoubiquitylation products of a number of functional E2-HECT pairs, and demonstrated that HECT domains from different subfamilies catalyze the formation of very different types of Ub chains, largely independent of the E2 in the reaction. Our data set represents the first comprehensive analysis of E2-HECT E3 interactions, and thus provides a framework for better understanding the molecular mechanisms of ubiquitylation.
Ubiquitin (Ub)1 is a conserved polypeptide that is covalently conjugated to other proteins in a reversible manner, to alter their function in a variety of ways. Ub conjugation (ubiquitylation) is a highly regulated process, consisting of a sequential series of E1-E2-E3 activation, conjugation, and ligation reactions. An E1 enzyme must first activate a mature Ub polypeptide via the formation of a high-energy thiol-ester bond with the Ub carboxyl-terminal Gly residue. The activated Ub polypeptide is then transferred to a Cys residue of an E2 conjugating protein. Finally, via an E3 ligase, the Ub polypeptide is covalently conjugated to a target protein (reviewed in 1–5).
Monoubiquitylation (i.e. the conjugation of a single Ub molecule to a target protein) has been implicated in a number of biological processes including transcriptional control, endocytosis, plasma membrane receptor recycling, and DNA damage signaling (6, 7). However, Ub itself contains seven lysine residues, all of which can be ubiquitylated to form polyubiquitin oligomers, or Ub “chains” (2–5, 8). Ub chains of varying lengths and linkage types can confer very different biological outcomes to a targeted protein substrate. For example, the best-characterized function of Ub chains (in this case, consisting of at least four Ub polypeptides linked via K48) is the targeting of a protein substrate for 26S proteasome-dependent degradation (9). By contrast, K63-linked Ub chains play roles in the DNA damage response, epsin-mediated endocytosis and aggresome formation (10, 11).
Although only two human Ub E1 activating proteins have been identified (12), 40 E2s (including both active E2 proteins and inactive E2 variants) are encoded in the human genome (Table I). All E2 proteins share a conserved “core” ubiquitin conjugating (UBC) domain of ∼150 amino acid residues, and many E2s possess additional N- and/or C-terminal protein sequences that can govern intracellular localization, confer regulatory properties, or provide specificity for interactions with particular E3 ligases (13, 14).
Table I. E2 (including both active and inactive E2 variants) and HECT domain protein information. Indicated are gene name, aliases, protein length, PDBID code (if structure has been solved) and NCBI protein and gene accession numbers. “Protein Note” indicates whether we were able to produce soluble full length or core recombinant protein. “Ub loading” indicates the ability of each recombinant E2 protein to form a thio-ester bond with Ub, in our assay.
| Gene Name | Synonyms | Prot Len | PDB Code | Prot Acc # | Gene Acc # | Protein Note | Ub-loading | Function |
|---|---|---|---|---|---|---|---|---|
| UBE2A | UBC2, HHR6A, RAD6A | 152 | NP_003327 | NM_003336 | full length | Yes | Ubiquitylation | |
| UBE2B | UBC2, HR6B, HHR6B, RAD6B | 152 | 1jas | NP_003328 | NM_003337 | full length | Yes | Ubiquitylation |
| UBE2C | UBCH10 | 179 | 1i7k | NP_008950 | NM_007019 | full length | Yes | Ubiquitylation |
| UBE2D1 | SFT, UBCH5, UBC4/5, UBCH5A | 147 | 2c4p | NP_003329 | NM_003338 | full length | Yes | Ubiquitylation |
| UBE2D2 | UBCH5B, UBC4 | 147 | 2esk | NP_003330 | NM_003339 | full length | Yes | Ubiquitylation |
| UBE2D3 | UBC4/5, UBCH5C | 147 | 1×23 | NP_003331 | NM_003340 | full length | Yes | Ubiquitylation |
| UBE2D4 | HBUCE1 | 147 | 3eb6 | NP_057067 | NM_015983 | full length | Yes | Ubiquitylation |
| UBE2E1 | UBCH6 | 193 | 3bzh | NP_003332 | NM_003341 | full length | Yes | Ubiquitylation |
| UBE2E2 | UBCH8, FLJ25157 | 201 | 1y6l | NP_689866 | NM_152653 | full length | Yes | Ubiquitylation |
| UBE2E3 | UBCH9, UBCM2 | 207 | NP_006348 | NM_006357 | full length | Yes | Ubiquitylation | |
| UBE2F | NCE2 | 185 | 2edi | NP_542409 | NM_080678 | full length | No | Neddylation |
| UBE2G1 | UBE2G | 170 | 2awf | NP_003333 | NM_003342 | full length | Yes | Ubiquitylation |
| UBE2G2 | UBC7 | 165 | 2cyx | NP_003334 | NM_003343 | full length | Yes | Ubiquitylation |
| UBE2H | UBC8, UBCH, UBCH2, E2-20K | 183 | 1yh6 | NP_003335 | NM_003344 | full length | Yes | Ubiquitylation |
| UBE2I | UBC9 | 158 | 1a3s | NP_003336 | NM_003345 | full length | No | Sumoylation |
| UBE2J1 | UBC6p, CGI-76, NCUBE1, HSPC153 | 318 | NP_057105 | NM_016021 | core | Yes | Ubiquitylation | |
| UBE2J2 | NCUBE2, PRO2121 | 275 | 2f4w | NP_919296 | NM_194315 | core | Yes | Ubiquitylation |
| UBE2K | HIP2, LIG, UBC1 | 200 | 1yla | NP_005330 | NM_005339 | full length | Yes | Ubiquitylation |
| UBE2L3 | E2-F1, l-UBC, UBCH7, UbcM4 | 154 | 1fbv, 1c4z | NP_937800 | NM_198157 | full length | Yes | Ubiquitylation |
| UBE2L6 | RIG-B, UBCH8, MGC40331 | 153 | 1wzw | NP_004214 | NM_004223 | full length | No | ISGylation |
| UBE2M | UBC12, hUbc12, UBC-RS2 | 183 | 1y8x | NP_003960 | NM_003969 | full length | No | Neddylation |
| UBE2N | UBCH-BEN, UBC13, MGC8489 | 152 | 2c2v | NP_003339 | NM_003348 | full length | Yes | Ubiquitylation |
| UBE2NL | 153 | NP_001013007 | NM_001012989 | not available | ND | |||
| UBE2O | E2–230K, FLJ12878, KIAA1734 | 1292 | NP_071349 | NM_022066 | insoluble | ND | ||
| UBE2Q1 | GTAP, UBE2Q, NICE-5, PRO3094 | 422 | 2qgx | NP_060052 | NM_017582 | core | Yes | Ubiquitylation |
| UBE2Q2 | 375 | 1zuo | NP_775740 | NM_173469 | full length | Yes | Ubiquitylation | |
| UBE2QL | FLJ25076, LOC134111 | 161 | NP_001138633 | NM_001145161.1 | full length | Yes | Ubiquitylation | |
| CDC34 | UBE2R1, E2-CDC34 | 236 | 2ob4 | NP_004350 | NM_004359 | full length | Yes | Ubiquitylation |
| UBE2R2 | UBC3B, CDC34B | 238 | NP_060281 | NM_017811 | full length | Yes | Ubiquitylation | |
| UBE2S | E2-EPF | 225 | 1zdn | NP_055316 | NM_014501 | full length | Yes | Ubiquitylation |
| UBE2T | PIG50, HSPC150 | 197 | 1yh2 | NP_054895 | NM_014176 | full length | Yes | Ubiquitylation |
| UBE2U | MGC35130, RP4-636O23.1 | 226 | 1yrv | NP_689702 | NM_152489 | core | Yes | |
| UBE2W | FLJ11011 | 151 | 2a7l | NP_060769 | NM_018299 | full length | Yes | Ubiquitylation |
| UBE2Z | HOYS7, FLJ13855, USE1 | 246 | NP_075567 | NM_023079 | full length | No | ||
| BIRC6 | BRUCE, APOLLON, FLJ13726, KIAA1289 | 4829 | 3ceg | NP_057336 | NM_016252 | core | ND |
Ub E3 ligases facilitate the transfer of Ub from an activated E2 to a substrate protein or another Ub molecule. Members of the RING (really interesting new gene) type E3 family recruit activated E2∼Ub complexes to substrates, resulting in direct Ub transfer from the E2 to the target (2), whereas members of the HECT (homologous to E6-AP carboxyl terminus) domain E3 family form a thiol-ester linkage with Ub prior to its transfer to a target protein (15, 16). The human genome encodes hundreds of RING type E3s, but only 28 HECT domain-containing E3 ligases (16). E3s clearly demonstrate specificity for subsets of E2 proteins, and different E2-E3 combinations can generate different types of Ub chains (e.g. 17–20). However, the molecular determinants involved in E2-HECT E3 interaction specificity are not well defined, the physical properties associated with processivity (i.e. the synthesis of long versus short Ub chains) are not well understood, and the molecular mechanisms involved in the specification of different types of Ub linkages remain cryptic.
Because of the modest affinity of most E2-E3 interactions, techniques such as co-immunoprecipitation have been largely unsuccessful in the characterization of functional E2-E3 pairs. Although yeast two hybrid screening has identified a number of putative E2-E3 functional interactions (e.g. 21, 22), this methodology does not provide information concerning processivity or Ub chain linkage types generated by each pair. In vitro E2-E3 ubiquitylation reactions performed with purified proteins must be used to obtain this type of information.
Functional interactions between a number of RING domain E3s and a smaller number of E2 proteins have been investigated previously (e.g. 17–20), but no large-scale study has focused on the HECT E3 ligases. Here, we present the first comprehensive human E2-HECT E3 structure function analysis.
MATERIALS AND METHODS
Plasmids
Human E2 open reading frames were amplified by PCR from various templates (see NM numbers in Table I), and inserted using the Infusion system (BD Biosciences) into a pET28 vector with 6xHis-tag and a thrombin or TEV-cleavage site located upstream of the cDNA insert. Human HECT domain proteins (Table I) and Ub were similarly cloned. The library of E. coli E2 expression clones will be made available through Addgene and/or other nonprofit sources.
Protein Purification, Size-Exclusion Chromatography and Crystallization
Proteins were expressed in E. coli BL21 (DE3) Gold (Stratagene, La Jolla, CA) grown in TB medium in the presence of 50 μg/ml kanamycin at 37 °C to an OD600 of 4–5, induced with 2 mm isopropyl-1-thio-d-galactopyranoside and further grown for 16–18 h at 15 °C. Recombinant E2 and HECT domain proteins were purified using standard metal-affinity chromatography with TALON resin (Stratagene), according to manufacturer's instructions. All proteins for the Ub assays were dialyzed against 20 mm Tris-Cl pH 8.0, 150 mm NaCl, 10% glycerol, 2 mm dithiothreitol and stored at −80 °C. For crystallization of the E2 proteins, the N-terminal 6xHis-tag was removed by incubation with thrombin (1 unit/mg protein, 2 h at 21–23 °C). Proteins were further purified by gel filtration on a HighLoad 16/60 Superdex 200 column (GE Healthcare); fractions containing the target protein were pooled and concentrated by ultrafiltration using an Amicon Ultra centrifugal filter with 10 kDa cutoff (Millipore, Billerica, MA) to a final protein concentration of 20–40 mg/ml.
Structure Determination
E2 protein crystals were grown at 18 °C using the hanging drop method. Suitable crystals were immersed in well solution supplemented with cryoprotectant prior to dunking and storage in liquid nitrogen. Diffraction data were collected either in-house using an FRE system or at synchrotron beamlines at the Argonne Photon Source or the Cornell High Energy Synchrotron Source. Data were processed with HKL-2000 (23), the XDS program package (24), or D*TREK (25). All structures, other than that of UBE2H, UBE2Q2 and BIRC6, were solved by molecular replacement methods using AMoRE(26), Phaser (27) or MOLREP (28). The structure of UBE2H, UBE2Q2 and BIRC6 were solved by single wavelength anomalous diffraction using either the program BnP (http://www.hwi.buffalo.edu/BnP/) or SOLVE (29). In some cases automatic model building was carried out using either ARP/wARP (30) or RESOLVE (31). Iterative manual model building was conducted using the graphics program Coot (32) combined with refinement with REFMAC5 (33), CNS1.1 (34), or PHENIX (35). Extensive use was also made of the CCP4 program suite (65). All structures have been deposited and statistics can be found in supplemental Table S1.
Autoubiquitylation and E2 Loading Assays
E2 loading assays were carried out in a volume of 10 μl containing 1 μg E1, 1 μg of E2, and 5 μg of 6xHis tagged Ub in a buffer consisting of 10 mm HEPES pH7.5, 100 mm NaCl, 40 μm ATP and 2 mm MgCl2. Reaction mixtures were incubated for 10 min at 30 °C, stopped by the addition of non-reducing SDS-PAGE sample buffer, separated by 4–20% gradient SDS-PAGE gels (Invitrogen), and visualized by Western blotting, using a mouse monoclonal antibody directed against the 6xHis epitope tag (Qiagen, Valencia, CA), an HRP-conjugated goat-mouse secondary (GE), and ECL (BioRad, Hercules, CA).
Autoubiquitylation reactions (in the presence of E3s) were performed in a volume of 20 μl in a buffer of 50 mm Tris pH 7.6, 5 mm MgCl2, 2 mm ATP, and 2 mm dithiothreitol, containing E1 (50 ng), E2 (100 ng), ubiquitin (5 μg), and E3 (6xHis tagged HECT domain proteins, 0.5 μg). After incubation at 30 °C for 90 min, reactions were stopped by the addition of SDS-PAGE sample buffer and resolved on 7% SDS-PAGE gels. In the absence of E3, reaction conditions were 50 mm Tris pH 7.6, 50 mm NaCl, 50 mm KCl, 10 mm MgCl2, 5 mm ATP and 0.1 mm dithiothreitol, with 100 ng E1, E2 (200 ng of UBE2R1 or 1 μg of UBE2K) and 5 μg Ub. Reactions were incubated at 30 °C for 3 h. Ubiquitylated proteins were evaluated by Western blotting using monoclonal antibodies directed against 6xHis (Qiagen), as above.
Analysis of E2 Surface Charge Characteristics
Electrostatic potential distributions of a representative group of 32 human E2 conjugating enzymes were evaluated using the AESOP (Analysis of Electrostatic Similarities Of Proteins) framework. The coordinates of the x-ray crystal structures of UBE2C (PDBID: 1I7K.A), UBE2D1 (PDBID: 2C4P.A), UBE2D2 (PDBID: 2ESK.A), UBE2D3 (PDBID: 1X23.D), UBE2E1 (PDBID: 3BZH.A), UBE2E2 (PDBID: 1Y6L.C), UBE2G1 (PDBID: 2CYX.B), UBE2G2 (PDBID: 2CYX.B), UBE2H (PDBID: 2Z5D.A), UBE2K (PDBID: 1YLA.B), UBE2L3 (PDBID: 1C4Z.D), UBE2S (PDBID: 1ZDN.A), UBE2T (PDBID: 1YH2.A), and UBE2U (PDBID: 1YRV.A) were used for surface charge analysis (36). An NMR ensemble was used for UBE2B (PDBID: 1JAS.A/21 models). All of the ensemble models were included in the analysis, however only one model is shown for UBE2B in the dendrograms, as they clustered together. To obtain structural representatives for human E2 conjugating enzymes that lack an experimental structure, homology modeling was performed to generate seven models, using SwissModel. Template models were selected based on percent sequence identity and domain coverage, as follows: UBE2R2: 2OB4.A - 79.2% seq id, residues 9–181; UBE2R1: 2OB4.A - 90.8% seq id, residues 9–181; UBE2J2: 2F4W.B - 94.4% seq id, residues 11–171; UBE2J1: 2F4W.B - 36.1% seq id, residues 9–151; UBE2E3: 1Y7L.C - 97.9% seq id, residues 60–207; UBE2D4: 2OXQ.B - 95.9% seq id, residues 1–147; UBE2A: 1JAS.A - 96.0% seq id, residues 1–151. Structural models (homology models and experimentally determined models) were inspected using Chimera, and extraneous atoms removed (e.g. HIS-tag, water molecules, other proteins/peptides, and residues that extended beyond the E2 core domain).
Structural models were prepared for electrostatic potential calculations by determining partial charges at a pH of 7.6 and van der Waals radii using PDB2PQR (37) with the PARSE (38) forcefield. Electrostatic potentials were calculated using the linearized Poisson Boltzmann equation,
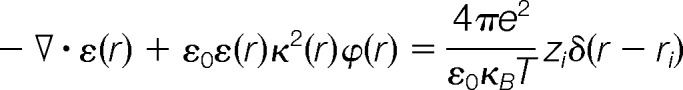 |
where r represents discrete grid point positions within and around the protein, ε(r) is the dielectric coefficient, ε0 is the vacuum permittivity, κ(r) is the ion accessibility function, ϕ(r) is the electrostatic potential, e is the electron charge, κB is the Boltzmann constant, T is the temperature, and z is the unit or partial charge located at position δ(r − rr) (39). The Adaptive Poisson-Boltzmann Solver (APBS) software package calculates electrostatic potential by embedding each E2 enzyme in a grid, and solves the Poisson-Boltzmann equation to determine electrostatic potential at each grid point based on assigned charge, dielectric coefficient, and ion accessibility at that position (40). The dielectric surface was defined using a sphere probe with a radius of 1.4 Å, and ion accessibility surface was defined using a sphere probe with a radius of 2.0 Å. To ensure a proper comparison, all of the E2 enzymes were superimposed and grid dimensions (129 × 97 × 97 points) were selected to fully enclose each enzyme when the calculated isopotential contour surfaces were plotted at ±1kbT/e. Electrostatic potentials were visualized using the molecular graphics software Chimera (41).
Comparison of the spatial distributions of electrostatic potentials of the E2 enzymes were performed by generating a similarity distance matrix according to the metric:
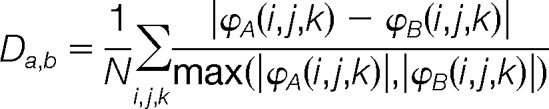 |
where ϕA(i,j,k) and ϕB(i,j,k) are electrostatic potentials of proteins A and B, respectively, at a common grid point (i,j,k), and N the number of grid points. This method implies that proteins having a distance of 0 have identical spatial distributions of electrostatic potentials, whereas those having a distance of 2 have completely different electrostatic potential spatial distributions.
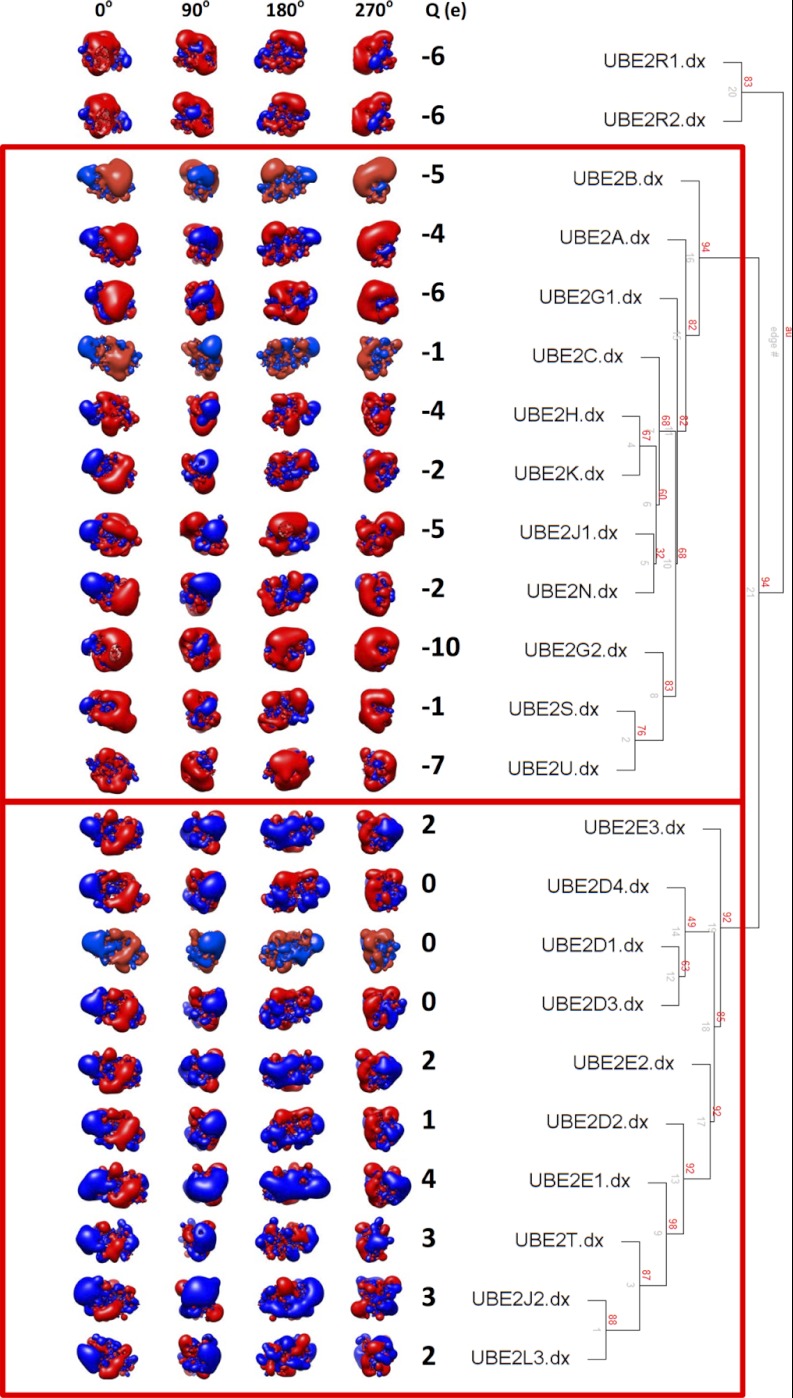
Agglomerative hierarchical clustering of the similarity distance matrix was performed with a euclidean metric and average linkage using the pvclust library (42, 43), which performs average-linkage hierarchical clustering while assessing its uncertainty through bootstrap resampling and providing “approximately unbiased” (AU) probability values (p values) and “bootstrap probability” (BP) values. Our analysis was performed using 1,000,000 bootstrap replications. The hierarchical clustering dendrogram generated by pvclust displays AU p values (in red) calculated using multiscale bootstrap resampling, edge numbers (in gray), and a red box outlining clusters strongly supported by data (i.e. edges with a multiscale bootstrap resampling AU p value greater than 90%). The AU p value represents the frequency that a particular cluster appears in the bootstrap replicates. A discrete cluster having high probability (AU > 0.9; edge 21) excludes UBE2R1 and UBE2R2 from the other 21 E2 enzymes, which are further divided into two high probability clusters (AU > 0.9; edges 16 and 19). These two high probability clusters correspond to a net negative charged cluster (UBE2B, UBE2A, UBE2G1, UBE2C, UBE2H, UBE2K, UBE2J1, UBE2N, UBE2G2, UBE2S, and UBE2U) and a net positive charged cluster (UBE2E3, UBE2D4, UBE2D1, UBE2D3, UBE2E2, UBE2D2, UBE2E1, UBE2T, UBE2J2, and UBE2L3). In addition to the calculation and clustering of electrostatic potential distribution for the full 23 domains, additional calculations were performed on subsets of residues within the E3-binding interface, and two ubiquitin-binding interfaces.
Mass Spectrometry
Autoubiquitylation reactions were scaled-up threefold for mass spectrometric analysis, and subjected to 4–12% gradient SDS-PAGE. Gels were stained with Coomassie brilliant blue for visualization, and the region containing proteins migrating at >125kDa was processed as in (44) for mass spectrometry. The digested peptide mixture was subjected to nLC-ESI-MS/MS, performed using an Orbitrap Velos instrument (Thermo Fisher Scientific) coupled to a Proxeon nanoHPLC system (Odense, Denmark). Reaction products from each E2-E3 pair were analyzed twice. Analytical columns were prepared in-house from 10 cm fused silica capillaries (75 μm inner diameter; InnovaQuartz, Phoenix, AZ) and packed with C18 coated silica particles (300Å pore size, 5 μm particle size; Michrom Bioresources, Auburn, CA). Peptides were first injected onto a 2 cm (100 μm inner diameter) C18 precolumn, and chromatographic separation was achieved using a 120 min gradient, from 100% buffer A (5% acetonitrile with 0.1% formic acid) to 40% buffer B (95% acetonitrile with 0.1% formic acid) running at a constant flow rate of 250 nl/min. The mass spectrometer was operated in data-dependent acquisition mode: one survey (400–1800 m/z) MS scan (at 60,000 resolution) was performed, and the forty most intense ions were chosen for fragmentation using collision-induced dissociation in the ion trap. Target ions for which two previous collision-induced dissociation scans had been collected (within 30 s) were dynamically excluded for 60 s. Thermo .raw files were converted to the .mzXML format with ReadW software v.3.5.1 (45), and data were searched using both; (a) automated database search software X!Tandem (46, 47) against the Homo sapiens ENSEMBL Genome Reference Consortium assembly GRCh37 database (75,126 entries), and (b) spectral matching against our previously published Ub/Ubl spectral library (48), supplemented with additional consensus spectra derived from commercial (Boston Biochem) di-ubiquitin K6, K27, K29, and K33 linkages. Search parameters for X!Tandem specified a parent MS tolerance of 10 ppm and an MS/MS fragment ion tolerance of 0.4 Da, with up to two missed cleavages allowed for trypsin. A +114.0429 Da modification of lysine was specified as a variable search parameter to identify the ubiquitin-derived diglycine motif. Oxidation of methionine (+15.995) and deamidation of Gln (+0.985) were also allowed as variable modifications. A GPM expect score of –2 was used as a cutoff, corresponding to a calculated false discovery rate of 0.80%. SpectraST (49, 50) was used for spectral matching, with a dot product of ≥0.7 used as a cutoff, corresponding to a calculated false discovery rate of 0.66%. We have uploaded all files relevant to MS analysis of in vitro autoubiquitylation assays to Tranche, with the associated hash tag
L6k/4UcFa9AEIRTwa7vNg7NiQ/gBNiHYZK1Z0yY/oH6b9reBLNl3E1XU2iYztfVVHQN6Lf7DBQbv+ZKDHI1RxZg52esAAAAAAAAudw==.
To correct for inherent differences in Ub linkage signal intensity, Proteome Discoverer (Thermo, Ver. 1.3) was used to search and analyze three replicate MS analyses of trypsin digested equimolar mixes of all seven di-ubiquitin polypeptides (Boston Biochem). Files were searched using the Sequest search algorithm and the ipi.HUMAN.v3.83 database, with a 10 ppm precursor mass tolerance and 0.5 Da fragment mass tolerance. Oxidation of M, deamidation of N or Q, and ubiquitylation of lysines (+114.043) were also allowed. Sequest results were analyzed using the Percolator algorithm, with a target FDR of 0.01. Precursor ion area detection was enabled, using a precision of 4ppm, and the AUC for each Ub linkage was calculated (for all observed charge states). A “detection bias” ratio was then calculated as the ratio of the AUC for each linkage type with respect to the linkage type with the lowest AUC, K29. An average detection bias ratio was calculated from the three replicate MS runs, and this conversion ratio applied to all data (supplemental Table S2).
RESULTS
E2 evolutionary relationships
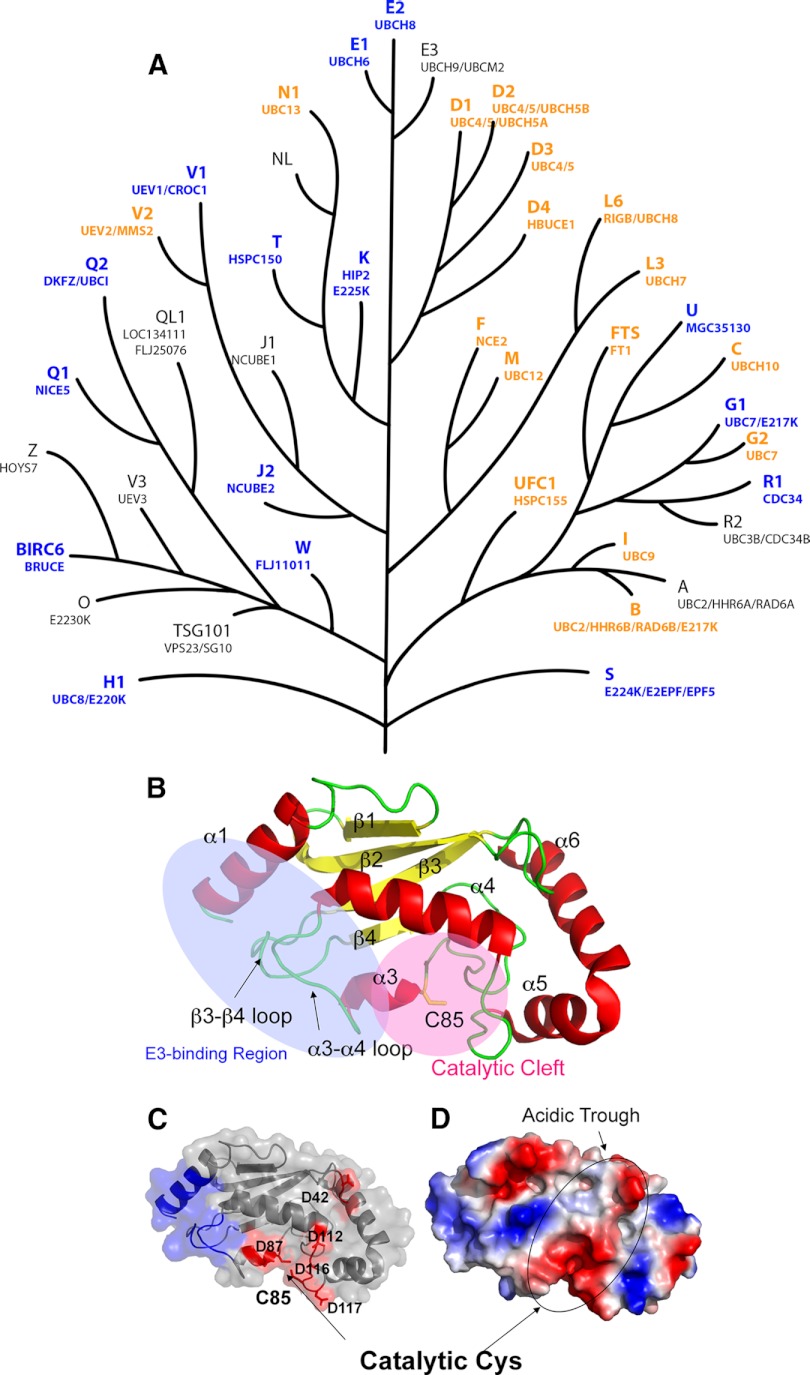
Based on the presence of a conserved catalytic domain of ∼150 residues (domain ID #CD00195), we constructed a phylogenetic model depicting putative evolutionary relationships among the human E2 proteins (Fig. 1A). Interestingly, although this approach successfully grouped closely related E2s (e.g. the UBE2D and UBE2E subfamilies), the E2s that mediate the conjugation of ubiquitin-like proteins (Ubls) are not readily distinguished from those that mediate Ub conjugation (see also the very similar phylogenetic tree in 12). For example, UBE2I (Ubc9), the E2 for the Ubl SUMO (small ubiquitin-related modifier; 51, 52), is grouped with two known Ub E2s, UBE2A and UBE2B. Similarly, UBE2M and UBE2F, E2s for the Ubl NEDD8 (neural precursor cell expressed, developmentally down-regulated 8; 53), are placed on the same evolutionary branch as UBE2L3, a Ub E2 (54). Overall sequence similarity and putative evolutionary relationships thus appear to have somewhat limited value in predicting E2 functional properties.
Fig. 1.
The UBC domain. A, Dendrogram of human UBC domains, depicting putative evolutionary relationships. A phylogenetic tree was generated from a cladogram derived from a ClustalW2 alignment of the minimal UBC fold. Nodal distances and relationships have been modified for clarity. NCBI gene nomenclature is shown above in larger font, and aliases below. Protein structures solved in this study are colored blue, and previously published structures are depicted in orange. B, Ribbon diagram of UBE2D1 (PDBID: 2C4P). Helices are labeled as α1-α6, and strands as β1-β4. Helix α2 is not observed in UBE2D1, but contained in the structures of UBE2Q1 (2QGX) and UBE2Q2 (1ZUO), and is located between β2 and β3. The E3 ligase binding region (blue) and the catalytic cleft (pink) encompassing Cys85 at the active site are also indicated. C, A surface and ribbon representation of UBE2D1, with the E3-binding region colored in blue. Acidic residues on the negatively charged surface are also indicated. D, An electrostatic surface representation of UBE2D1 in the same orientation. Locations of the acidic trough and catalytic Cys residue are indicated.
Construction of an E2 Library
To better understand the human E2s, we constructed a recombinant E2 protein library. Full-length and “core” (UBC domain) versions of the 40 H. sapiens E2 proteins were cloned into a 6xHis expression vector (Table I). Using standard methods, both core and full-length polypeptides for 29 different E2s were successfully expressed in E. coli, and purified to homogeneity. The remaining full-length proteins were not expressed or were insoluble, but we were able to express the core domains of eight of the remaining E2s (Table I). These 66 purified E2 proteins (covering 37 of the 40 UBC domains in the human genome) form the basis of our library, representing the most complete collection of recombinant human E2 enzymes available to date.
A conserved E2 structural core
Although the structures of a number of E2s have been solved (Table I), those from several of the main branches of the E2 evolutionary tree (Fig. 1A) have not been previously characterized. To better understand E2 structure-function relationships, we determined high-resolution three-dimensional structures of 15 additional human E2 core domains (Table I, supplemental Fig. S1, and supplemental Table S1). These structures double the number of solved E2 UBC domains, and, combined with those that have been previously characterized, provide nearly complete structural coverage of the human E2 core.
As expected, most of the new structures display a canonical E2 fold, composed of a four stranded, anti-parallel curled β-sheet surrounded on three sides by α-helical segments. The core domains of these E2s share a remarkable degree of similarity in three-dimensional structure, with an average root mean square deviation (RMSD) of 2Å over aligned Cα atoms of 145 residues (relative to UBE2D1), despite sequence identity as low as 15% (supplemental Fig. S2). Interestingly, the RMSD values of the ubiquitin-like protein E2s (UBE2I, UBE2F and UBE2M) are also ∼2.0Å, indicating that the core polypeptide backbone structure does not diverge significantly for these conjugating enzymes. Alternative features such as surface properties or additional structural elements must therefore be important for specificity.
Highly conserved areas in the UBC fold include (Figs. 1B–1D); (1) the catalytic cleft, encompassing the catalytic Cys in the β4–310 loop, (2) the E3 binding region, encompassing helix α1 and the β3-β4 and α3-α4 loops, and (3) the central β sheet composed of β2, β3 and β4 in all E2 structures. Less conserved areas of the UBC fold include the surface-exposed region between the E3 ligase binding site and the catalytic cleft, and helices 5 and 6, when present.
An E2-HECT E3 Functional Screen
Our entire E2 panel was next subjected to an in vitro “Ub loading” assay, to assess the ability of each conjugating enzyme to form a thio-ester∼Ub intermediate. In the presence of Ub, ATP and the E1 enzyme Uba1, we found that 26 of the recombinant human E2s were capable of loading Ub (supplemental Fig. S3). This result is in accordance with previous studies (e.g. 12), with one minor difference; UBE2U was able to load Ub (albeit weakly) in our assay. Our Ub loading screen also revealed that three E2s, UBE2K, UBE2R1 (Cdc34), and UBE2R2, do not require the presence of an E3 protein to carry out Ub conjugation (i.e. the formation of an isopeptide bond with the epsilon amine group of a lysine residue in one of the proteins in the reaction) under our standard in vitro reaction conditions. As expected, we did not observe Ub loading for the known ubiquitin-like protein E2s UBE2I, UBE2F, UBE2M, and UBE2L6, nor the inactive Ub E2 variants (UEVs, which lack a catalytic Cys) TSG101, TMEM189, UBE2V1, UBE2V2, or AKTIP. UBE2Z also had no activity in our assay, consistent with a previous report demonstrating that this E2 is loaded specifically by an alternative Ub E1, UBA6 (12) (not tested here).
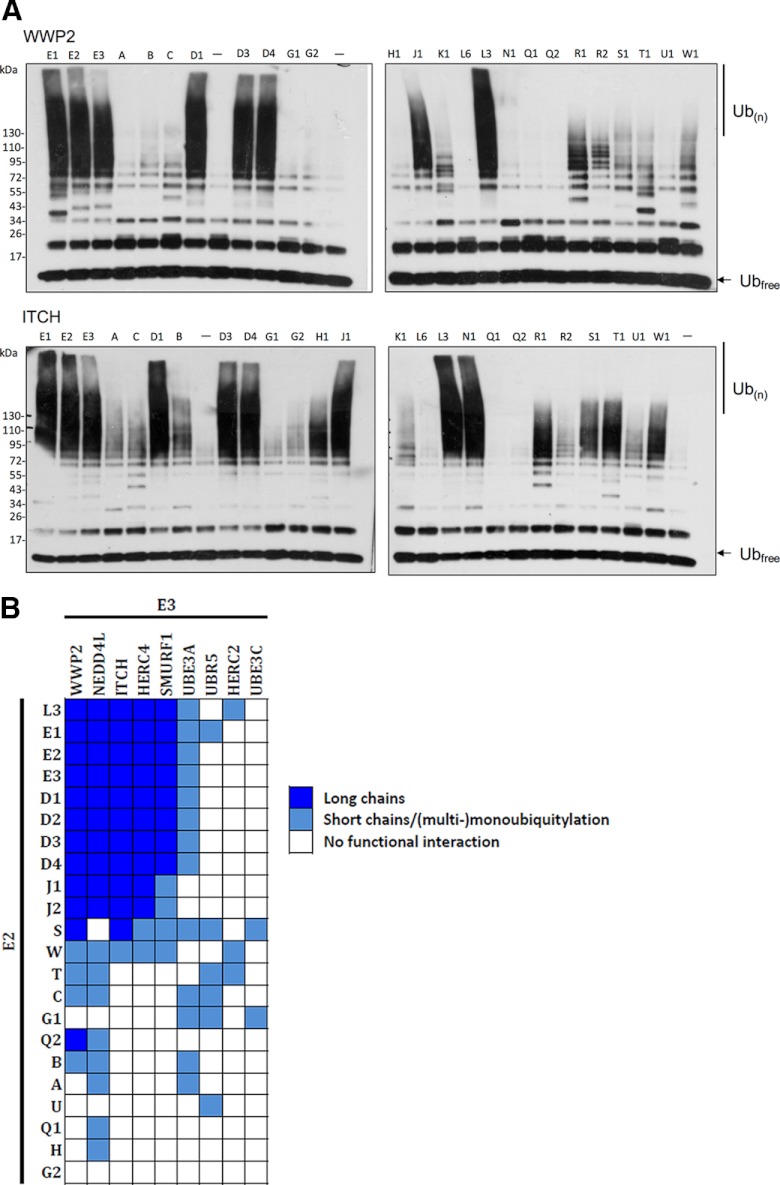
Functional interactions between the entire set of Ub loading E2s and a number of HECT E3 ligases were next characterized. Nine HECT domains (Table I), including representatives of the NEDD4 subfamily (NEDD4L, ITCH, SMURF1, and WWP2), the HERC subfamily (HERC2 and HERC4), and other more distantly related HECT domains (UBR5, UBE3A and UBE3C) were expressed as 6xHis fusion proteins in E. coli, purified to homogeneity, and tested in an autoubiquitylation assay with each of the 26 Ub-loading E2 proteins. After 90 min at 30 °C, reaction mixtures were resolved via SDS-PAGE, and ubiquitylated products were detected by Western blotting (Fig. 2A and supplemental Fig. S4).
Fig. 2.
Autoubiquitylation reactions. A, Autoubiquitylation was performed in the presence of recombinant E1, ATP, ubiquitin and E2. Reactions were subjected to SDS-PAGE and anti-Ub Western analysis. Shown is a representative screen (with the HECT E3 domains of WWP2 and ITCH), typical of results obtained in three independent experiments. Each lane represents an individual autoubiquitylation reaction with E2 indicated at top. (-) indicates negative control, lacking E2 protein. The locations of unconjugated and oligomeric His-Ub are indicated to the right of each Western blot. B, A heat map depicting Ub chain-building activity of each of the 234 E2-E3 pairs in in vitro autoubiquitylation reactions. Dark blue indicates long Ub chains (>125 kDa), light blue indicates short chains or (multi-) monoubiquitylation, and white indicates no functional interaction. E2s and E3s are hierarchically clustered according to activity.
Based on Ub adduct migration in SDS-PAGE, we classified each of the 234 E2-E3 autoubiquitylation reactions into one of three groups: (1) those containing long Ub chains (MW >125 kDa), (2) those containing shorter Ub oligomers or (multi-)monoubiquitylation (MW <125 kDa), and (3) those with no apparent products (Fig. 2B). UBE2L3, UBE2D1–4, UBE2E1–3, and UBE2J1–2 displayed functional interactions with all of the NEDD4 subfamily HECT domains, as well as with HERC4 and UBE3A (E6-AP). A majority of these interactions were highly productive, resulting in the synthesis of long Ub chains. UBE2S was able to catalyze the synthesis of long chains with WWP2 and ITCH, and short chains or multimonoubiquitylation with several other E2s. UBE2C, UBE2G1, UBE2T, and UBE2W also functionally interacted with several of the HECT domains to produce short chains or multimonoubiquitylation. UBE2A, UBE2B, UBE2H, UBE2Q1, and UBE2U displayed only low levels of activity in our screen, and no interactions were observed with UBE2G2. These data, for the first time, define a human E2-HECT functional interaction landscape.
In a few cases, we observed significant differences in activity between the core and full-length versions of an E2. For example, the core domains of UBE2J1 and UBE2W displayed increased activity compared with their full-length counterparts in some autoubiquitylation reactions. Conversely, the full-length UBE2S protein was more active in some reactions than the core domain. These data suggest that additional sequences outside the core can act as positive or negative regulatory domains and/or provide additional E3 specificity.
E2 Surface Charge is Associated With Ub Chain-Building Processivity
To identify physicochemical characteristics that confer Ub chain building activity and functional interactions with HECT domains, surface properties of each E2 structure were next analyzed. Charge, dielectric coefficient and ion accessibility were calculated for each E2 protein, and the Poisson-Boltzmann equation was used to determine electrostatic potential at >1,200,000 grid points/structure (Fig. 3, see Methods for further detail). Interestingly, the majority of the highly active E2s identified in our HECT autoubiquitylation screen displayed an overall net positive or neutral charge, whereas the remaining E2s displayed a net negative charge. Closer inspection revealed that most of the highly active E2s possess a clearly defined “acidic trough” adjacent to the catalytic cysteine residue (corresponding to residues Asp42, Asp87, Asp112, Asp116, and Asp117 in UBE2D1, located in the α5 and β2-β3 loop; Figs. 1B–1D), surrounded by extensive basic regions. Indeed, agglomerative hierarchical clustering of distance matrix values grouped the majority of the active E2s, based on the spatial distribution of electrostatic potential within 6Å of the ubiquitin donor interaction interface (corresponding to residues E51, N97, R101, D102, C118, I121, S127, E139, and Y141 in UBE2S, the entire region encompassing a total of 53 aa). The most active E2s in HECT-containing reactions thus share a high degree of similarity specifically on the Ub donor binding surface, along with a higher net charge, and these properties clearly differentiate them from other E2 proteins. By contrast, the surface electrostatic potentials of the E3 binding site and Ub acceptor region (supplemental Fig. S5) were not good predictors of activity in our assays.
Fig. 3.
left - Spatial distributions of electrostatic potentials of human E2 protein structures; red = acidic, blue = basic at ± 1kBT/e. Four different orientations around the vertical axis are shown, along with net charge (Q(e)) of the full structure. right - An agglomerative hierarchical clustering dendrogram was generated using pvclust, calculated based on the spatial distribution of electrostatic potential within 6Å of the ubiquitin donor interaction interface. Bootstrap probability values (red), and edge numbers (gray) are indicated.
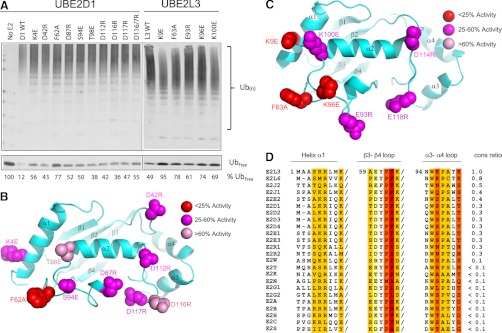
Several previous reports have indicated that E2 acidic residues may contribute to catalysis. Asp117 and the nearby HPN (where N = Asn77) motif in UBE2D2 were shown to assist catalysis by stabilizing the E2-Ub oxyanion intermediate (55). Other negatively charged residues adjacent to the catalytic cleft may also contribute to the binding of the positively charged residues in the region surrounding the carboxy-terminus of Ub (56). To test whether the acidic trough plays a role in HECT-mediated ubiquitylation, we generated a series of UBE2D1 proteins in which individual amino acids in this region were converted to basic residues. As a measure of E2 activity, the amount of free (unconjugated) Ub remaining after a standard autoubiquitylation reaction with the ITCH HECT domain was monitored (Fig. 4A). Wild type (WT) UBE2D1 incorporated ∼90% of the Ub in the reaction mix into conjugates within 90 min (compare lane 1, no E2, to lane 2). Consistent with previous findings, mutation of residues important for E2-E3 interactions (K4 and F62) greatly attenuated Ub conjugation (lanes 3 and 5). Conversion of individual UBE2D1 acidic trough residues to basic amino acids (D42R, D87R, D112R, D116R, and D117R; see Fig. 4B for residue locations) also resulted in markedly lower activity in this assay (lanes 4, 6, 9, 10, and 11). Similar results were observed with other HECT domains (data not shown). These data are consistent with an important role of the acidic residues surrounding the catalytic Cys (as was recently also observed for UBE2S; 17).
Fig. 4.
Defining residues important for E2-HECT function. A, Mutational analysis of the UBE2D1 acidic trough and UBE2L3 HECT binding domain. (top) E2 proteins bearing single point mutations (as indicated) were assayed in a standard autoubiquitylation assay. Percentage unconjugated (free) Ub is indicated below each reaction. (bottom) B, UBE2D1 ribbon diagram highlighting the locations of each of the mutated residues in the E2 structure, color coded according to magnitude of effect on Ub conjugation in vitro. C, UBE2L3 ribbon diagram highlighting locations of mutated residues. D, Alignment of the previously defined UBE2L3 HECT binding domain with other Ub-loading human E2 sequences. Residues previously found to play a role in HECT binding are highlighted. Column color corresponds to the change in binding energy observed when this residue was mutated (39). A “conservation score” for each E2 was assigned as follows; identical residues were assigned a score according to binding energy, where: red residues = +4, orange = +3, mustard = +2, light yellow = +1; conserved mutations at the same location (e.g. R to K, or S to T) were assigned half scores. Similar amino acids (e.g. aliphatic, charged or small amino acids) were assigned a score of 0. Dramatically altered amino acids at the same position were penalized with a negative score corresponding to the binding energy of each UBE2L3 amino acid. Raw scores were summed for each E2, and conservation score determined by dividing by the E2L3 score of 32.
A HECT Binding Signature Correlates with Ub Chain-Building Activity
Although most E2s can catalyze ubiquitylation with RING E3 ligases, UBE2L3 is preferentially utilized by HECT E3s (54, 57). The HECT domain protein E6-AP (UBE3A) selectively interacts with UBE2L3 and UBE2L6 with a Kd of ∼5 μm, and binds to other E2s with reduced affinity (54). The UBE2L3 residues responsible for binding to the E6-AP HECT domain were previously identified on helix α1, β3-β4 loop 4, and α3-α4 loop 7. We aligned this region through all of the active Ub E2s in our screen, and scored them for potential HECT binding affinity, based on the binding energies associated with a number of critical UBE2L3 residues (54) (Figs. 4C and 4D). Notably, the active E2s highlighted in our HECT screen also scored highest in this analysis, suggesting that these proteins possess structural features more favorable for HECT domain binding than other human E2 proteins. A higher degree of structural similarity to the E6-AP binding domain of UBE2L3 was thus an excellent predictor of Ub chain building activity in in vitro reactions with HECT domains.
To test whether this signature is important for E2 activity in HECT-containing autoubiquitylation reactions, we generated a series of UBE2L3 proteins in which individual residues in the E6-AP HECT interaction domain were mutated, and assessed their activity in vitro with the ITCH E3 ligase HECT domain (Fig. 4A). UBE2L3 proteins mutated at residues in the E6-AP binding motif (K9E, F63A, E93R, K96E, and K100E; see Fig. 4C for residue locations) displayed a consistent decrease in activity with ITCH. These data suggest that the affinity of the E2 protein for HECT domains also plays an important role in HECT-mediated ubiquitylation.
Characterization of Ub Chain Linkages in E2 - HECT Reactions
Finally, we utilized mass spectrometry to characterize the Ub chain linkages generated by a number of functional E2-E3 pairs. Autoubiquitylation reaction products were separated via SDS-PAGE, and proteins migrating at >125 kDa were subjected to trypsin proteolysis. The resulting peptides were analyzed using nanoflow liquid chromatography-electrospray ionization-tandem mass spectrometry (nLC-ESI-MS/MS). Ub-Ub linkages were identified using two methods: (1) standard database searching, with the inclusion of a +114.0429 Da mass shift (corresponding to the Ub tryptic GG remnant on lysine) as a variable modification (58), and (2) spectral matching, using a Ub/Ubl spectral library that we recently developed, and which now contains consensus spectra derived from all seven Ub chain linkage types (59, 48).
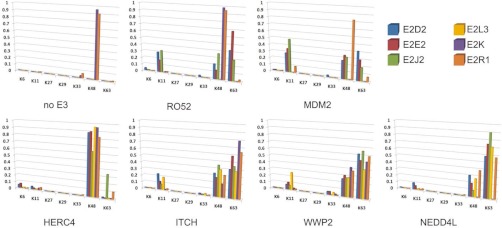
When UBE2D2, UBE2E3 or UBE2J2 were combined with the RING E3s Mdm2 or Ro52, a mix of Ub linkages was generated, in which K63 ≈ K48 ≈ K11 ≫ all other linkage types (Fig. 5 and supplemental Table S2). UBE2L3 was not functional with either of these RING proteins. In the presence of the same E2s, the HECT domains derived from ITCH, WWP2, and NEDD4L also catalyzed the formation of a mix of K63-, K48- and K11-linked Ub products, where rank order of abundance was K63 > K48 > K11 ≫ all other linkage types (Fig. 5 and supplemental Table S2). In contrast, the HERC4 HECT domain gave rise to a very different spectrum of Ub chains with the same set of E2s, synthesizing Ub oligomers that were markedly enriched in K48 linkages (K48 ≫ K63 > all other linkages). Thus, very different linkage types were observed in reactions containing the same E2, but different HECT E3s.
Fig. 5.
Ub linkage analysis. Autoubiquitylation reactions were subjected to SDS-PAGE, and reaction products analyzed by mass spectrometry (see Methods for details). Ub linkage composition for each E2-E3 reaction are presented as a fraction of the total number of linkages detected. See Legend inset for color codes.
To further delineate the role of HECT domains in Ub linkage specification, UBE2R1 (Cdc34) and UBE2K, both of which have been previously demonstrated to generate Ub chains enriched in K48 linkages (60, 61), were analyzed in reactions containing; (1) no E3, (2) the RING E3s Mdm2 or Ro52, and (3) HECT domains derived from WWP2, ITCH, NEDD4L, and HERC4. Both of these E2s catalyzed the synthesis of Ub oligomers highly enriched in K48 linkages in the absence of E3, or when combined with RING E3s (Fig. 5 and supplemental Table S2). A similar linkage distribution was observed in reactions containing HERC4. However, when UBE2R1 or UBE2K were combined with NEDD4 subfamily HECT E3s (n.b. NEDD4L was inactive with UBE2K), the composition of Ub linkages in the reactions were similar to those observed with UBE2D2, UBE2E3, UBE2J2, and UBE2L3; i.e. mixed chain products, with significant levels of K63 and K48 linkages, along with a low level of K11 linked Ub oligomers (Fig. 5 and supplemental Table S2).
Consistent with earlier studies (62, 63), our data thus confirm that; (1) HECT domains can govern linkage composition in autoubiquitylation reactions, largely independent of E2 linkage specificity, and (2) different HECT domains display different linkage preferences. Specifically, we found that HERC4 catalyzes the synthesis of Ub oligomers enriched in K48 linkages, whereas the NEDD4 subfamily HECT domains NEDD4L, ITCH and WWP2 synthesize Ub chains consisting primarily of K63, K48, and K11 linkages.
DISCUSSION
Structural genomics of the human E2 family confirmed that the UBC fold is stable through a large range of sequence variations, and highlighted an “acidic trough” near the catalytic cysteine, surrounded by extensive basic regions. This surface area is likely involved in a number of important roles in Ub/Ubl transfer. The area of the trough near the catalytic Cys specifically bonds with and desolvates the nucleophilic epsilon amino group, contributing to the activation of the substrate lysine (64). A previous study demonstrated that mutation of two Asp residues in this region (D100A and D127A) of UBE2I (Ubc9) significantly affects the transfer of SUMO from UBE2I to a target lysine (64). The area of the trough farther from the nucleophilic cysteine may be involved in recruiting and/or positioning the Ub/Ubl polypeptide during catalysis; in both the SUMO-UBE2I-RanGAP1 and the yeast UBC8-Ub (PDBID:1FXT) structures, this patch of the E2 mediates direct contact with the Ub/Ubl C terminus (56, 64). Consistent with this model, we found that mutation of acidic residues in the UBE2D1 trough decreased its activity in HECT domain-containing reactions. These observations do not extend to all E2s, however. For example, UBE2R1 was highly active in our autoubiquitylation assays, yet displays very different surface charge characteristics. Other structural elements apparently confer a similar function in this case. Previous reports have indicated that a long acidic loop in UBE2R1 plays an essential role in the assembly of polyubiquitin chains (60).
We also found that similarity to the previously defined HECT binding region of UBE2L3 was a good predictor of activity for all E2s in HECT-containing in vitro reactions (Fig. 4). Eletr and Kuhlman (54) first defined the regions of UBE2L3 involved in binding to the HECT E6-AP (UBE3A), and the NEDD4L-UBE2D2 (55) cocrystal structure highlighted the same region. We found that the E2s sharing the highest similarity with UBE2L3 in this region also displayed higher levels of Ub chain building activity in autoubiquitylation assays, with a number of different HECT domains. Mutational analysis of the HECT binding signature in UBE2L3 suggested that this motif likely plays an important role in Ub chain-building activity with all HECT proteins.
Both NEDD4L and E6-AP contact the E2 protein via helices H7 and H8 (55). However, E6-AP appears to make more extensive interactions with UBE2L3 via the loop between helix H7 and S5. Interestingly, E6-AP and HERC4 possess similar E2 binding sites, whereas NEDD4L, WWP2, SMURF1 and ITCH diverge in this region. This could explain the variation in chain linkage types synthesized by the NEDD4 subfamily versus HERC4 with the same E2 proteins, if the more robust HERC4 binding to E2s stabilizes the E2-E3 pair in a conformation that favors the formation of K48-linked Ub oligomers. Consistent with this model, HERC4-containing autoubiquitylation reactions were also less productive overall than NEDD4 subfamily-containing interactions with the same set of E2s (supplemental Fig. S4, and see Ub linkage counts in supplemental Table S2), suggesting that the E2 off rate for HERC4 could be slower. Further study will be required to test this model.
Previous publications have suggested that HECT E3s determine linkage specificity in autoubiquitylation reactions (62, 63), because they possess a catalytic Cys residue. However, many different E2-E3 interaction models can be envisioned. For example, (1) once a single Ub molecule is transferred to the HECT Cys residue, Ub chains could be built upon it by E2s. In this case, the E2 would specify the linkage type. Or, (2) Ub chains built by one or more E2s could be transferred en bloc to a HECT before conjugation to a target. Linkage specificity would be entirely determined by the E2 in this model. Or, (3) if an E2 interacts strongly with a HECT E3, it is possible that it could influence chain types produced by the E2-E3 pair. Unlike any other study that we are aware of, here we challenged several different HECT domains with two E2s that synthesize Ub chains specifically enriched in K48 linkages (UBE2R1/Cdc34 and UBE2K (19, 63)). We found that the chain types generated in HECT-containing autoubiquitylation reactions were quite different than those observed in RING-containing reactions (or in reactions lacking an E3) with these E2s, indicating that the HECT domain is the primary determinant of Ub-Ub linkages in this context.
Recent work from the Rape laboratory indicated that residues in UBE2S coordinate with residues of Ub itself to produce K11-specific Ub chain linkages (17). This interaction involves the Ub β1, β2 and β4 strands (K11 is in β2) interacting with the active site and helix α3 of UBE2S. Three side chains in UBE2S α3 (E131, R135 and E139) participate in hydrogen or salt bonds with Ub. To assess whether other E2s may function in a similar manner, we aligned all known E2 structures, and analyzed conservation in this region (supplemental Fig. S6). Notably, no other E2s possess similar residues at the α3 sites that make contact with Ub in UBE2S. Structural analyses therefore supports our current understanding that K11 specificity (at least using a mechanism like UBE2S) is a trait likely pertinent only to UBE2S.
Finally, it is important to note that functional E2-HECT pairs that can only extend Ub chains on mono-ubiquitylated targets, or those that require one or more cellular co-factors, would not be detected in our assay. Nevertheless, our new set of E2 core structures, combined with the construction of a nearly complete E2 protein library, a comprehensive E2-HECT functional interaction screen, and a mass spectrometry-generated Ub linkage data set, represents an important resource for better understanding the structural properties that mediate E2-HECT interactions, processivity, and the synthesis of different types of Ub chain linkages.
Supplementary Material
Acknowledgments
We thank F. MacKenzie, C. Butler-Cole, and Y. Li for cloning, along with A. –C. Gingras for critical reading of the manuscript. Results shown in this report are derived from work performed at Argonne National Laboratory, beamlines SBC 19-BM, GM/CA CAT 23-ID-B, IMCA-CAT and at the Advanced Photon Source. Argonne is operated by U Chicago Argonne, LLC, for the U.S. Department of Energy, Office of Biological and Environmental Research under contract DE-AC02–06CH11357. GM/CA CAT has been funded in whole or in part with Federal funds from the National Cancer Institute (Y1-CO-1020) and the National Institute of General Medical Science (Y1-GM-1104). Use of the IMCA-CAT beamline 17-BM was supported by the companies of the Industrial Macromolecular Crystallography Association through a contract with Hauptman-Woodward Medical Research Institute. Additional data collection was carried out at Cornell High Energy Synchrotron Source Beamline F-1. CHESS is supported by the NSF & NIH/NIGMS via NSF award DMR-0225180, and the MacCHESS resource is supported by NIH/NCRR award RR-01646.
Footnotes
* This work was supported by the Canadian Cancer Society, the Ontario Ministry of Health Research and Long-term Care, and the Structural Genomics Consortium, a registered charity (number 1097737) that receives funds from the Canadian Institutes of Health Research, the Canadian Foundation for Innovation, Genome Canada through the Ontario Genomics Institute, GlaxoSmithKline, Karolinska Institutet, the Knut and Alice Wallenberg Foundation, the Ontario Innovation Trust, the Ontario Ministry for Research and Innovation, Merck & Co., Inc., the Novartis Research Foundation, the Swedish Agency for Innovation Systems, the Swedish Foundation for Strategic Research and the Wellcome Trust. Molecular graphics images were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco. JHH, RD and TS were funded by CIHR Student Fellowship Awards, CHA holds the Canada Research Chair in Structural Proteomics and BR holds the Canada Research Chair in Proteomics and Molecular Medicine, with funding from the Canada Foundation for Innovation, the Ontario Innovation Trust and the Ontario Ministry of Research and Innovation.
 This article contains supplemental Figs. S1 to S6 and Tables S1 and S2.
This article contains supplemental Figs. S1 to S6 and Tables S1 and S2.
1 The abbreviations used are:
- Ub
- ubiquitin
- UBC
- ubiquitin conjugating
- HECT
- homologous to E6-AP carboxyl terminus.
REFERENCES
- 1. Schwartz A. L., Ciechanover A. (2009) Targeting proteins for destruction by the ubiquitin system: implications for human pathobiology. Annu. Rev. Pharmacol. Toxicol. 49, 73–96 [DOI] [PubMed] [Google Scholar]
- 2. Deshaies R. J., Joazeiro C. A. (2009) RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78, 399–434 [DOI] [PubMed] [Google Scholar]
- 3. Finley D. (2009) Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 78, 477–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pickart C. M., Eddins M. J. (2004) Ubiquitin: structures, functions, mechanisms. Biochim. Biophys. Acta 1695, 55–72 [DOI] [PubMed] [Google Scholar]
- 5. Pickart C. M., Fushman D. (2004) Polyubiquitin chains: polymeric protein signals. Curr. Opin. Chem. Biol. 8, 610–616 [DOI] [PubMed] [Google Scholar]
- 6. Saksena S., Sun J., Chu T., Emr S. D. (2007) ESCRTing proteins in the endocytic pathway. Trends Biochem. Sci 32, 561–573 [DOI] [PubMed] [Google Scholar]
- 7. Mosesson Y., Yarden Y. (2006) Monoubiquitylation: a recurrent theme in membrane protein transport. Isr. Med. Assoc. J. 8, 233–237 [PubMed] [Google Scholar]
- 8. Ye Y., Rape M. (2009) Building ubiquitin chains: E2 enzymes at work. Nat. Rev. Mol. Cell Biol. 10, 755–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Thrower J. S., Hoffman L., Rechsteiner M., Pickart C. M. (2000) Recognition of the polyubiquitin proteolytic signal. EMBO J. 19, 94–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Acconcia F., Sigismund S., Polo S. (2009) Ubiquitin in trafficking: the network at work. Exp. Cell Res. 315, 1610–1618 [DOI] [PubMed] [Google Scholar]
- 11. Chin L. S., Olzmann J. A., Li L. (2010) Parkin-mediated ubiquitin signalling in aggresome formation and autophagy. Biochem. Soc Trans 38, 144–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jin J., Li X., Gygi S. P., Harper J. W. (2007) Dual E1 activation systems for ubiquitin differentially regulate E2 enzyme charging. Nature 447, 1135–1138 [DOI] [PubMed] [Google Scholar]
- 13. Wenzel D. M., Stoll K. E., Klevit R. E. (2011) E2s: structurally economical and functionally replete. Biochem. J. 433, 31–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pickart C. M. (2001) Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70, 503–533 [DOI] [PubMed] [Google Scholar]
- 15. Bernassola F., Karin M., Ciechanover A., Melino G. (2008) The HECT family of E3 ubiquitin ligases: multiple players in cancer development. Cancer Cell 14, 10–21 [DOI] [PubMed] [Google Scholar]
- 16. Rotin D., Kumar S. (2009) Physiological functions of the HECT family of ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 10, 398–409 [DOI] [PubMed] [Google Scholar]
- 17. Wickliffe K. E., Lorenz S., Wemmer D. E., Kuriyan J., Rape M. (2011) The mechanism of linkage-specific ubiquitin chain elongation by a single-subunit E2. Cell 144, 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jin L., Williamson A., Banerjee S., Philipp I., Rape M. (2008) Mechanism of ubiquitin-chain formation by the human anaphase-promoting complex. Cell 133, 653–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Christensen D. E., Brzovic P. S., Klevit R. E. (2007) E2-BRCA1 RING interactions dictate synthesis of mono- or specific polyubiquitin chain linkages. Nat. Struct. Mol. Biol 14, 941–948 [DOI] [PubMed] [Google Scholar]
- 20. Ordureau A., Smith H., Windheim M., Peggie M., Carrick E., Morrice N., Cohen P. (2008) The IRAK-catalysed activation of the E3 ligase function of Pellino isoforms induces the Lys63-linked polyubiquitination of IRAK1. Biochem. J. 409, 43–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Markson G., Kiel C., Hyde R., Brown S., Charalabous P., Bremm A., Semple J., Woodsmith J., Duley S., Salehi-Ashtiani K., Vidal M., Komander D., Serrano L., Lehner P., Sanderson C. M. (2009) Analysis of the human E2 ubiquitin conjugating enzyme protein interaction network. Genome Res. 19, 1905–1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. van Wijk S. J., de Vries S. J., Kemmeren P., Huang A., Boelens R., Bonvin A. M., Timmers H. T. (2009) A comprehensive framework of E2-RING E3 interactions of the human ubiquitin-proteasome system. Mol. Syst. Biol. 5, 295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Otwinowski M., Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 24. Kabsch W. (2010) Xds. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pflugrath J. W. (1999) The finer things in X-ray diffraction data collection. Acta Crystallogr. D Biol. Crystallogr. 55, 1718–1725 [DOI] [PubMed] [Google Scholar]
- 26. Navaza J. (2001) Implementation of molecular replacement in AMoRe. Acta Crystallogr. D Biol. Crystallogr. 57, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 27. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vagin A., Teplyakov A. (2010) Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 66, 22–25 [DOI] [PubMed] [Google Scholar]
- 29. Terwilliger T. C., Berendzen J. (1999) Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 55, 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Morris R. J., Perrakis A., Lamzin V. S. (2003) ARP/wARP and automatic interpretation of protein electron density maps. Methods Enzymol. 374, 229–244 [DOI] [PubMed] [Google Scholar]
- 31. Terwilliger T. C. (2000) Maximum-likelihood density modification. Acta Crystallogr. D Biol. Crystallogr. 56, 965–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 33. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 34. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 35. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Berman H. M., Westbrook J., Feng Z., Gilliland G., Bhat T. N., Weissig H., Shindyalov I. N., Bourne P. E. (2000) The Protein Data Bank. Nucleic Acids Res. 28, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dolinsky T. J., Czodrowski P., Li H., Nielsen J. E., Jensen J. H., Klebe G., Baker N. A. (2007) PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 35, W522–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sitkoff D., Sharp K. A., Honig B. (1994) Accurate calculation of hydration free energies using macroscopic solvent models. J. Phys. Chem. 98, 1978–1988 [Google Scholar]
- 39. Davis M. E., McCammon J. A. (1990) Electrostatics in biomolecular structure and dynamics. Chem. Rev. 90, 509–521 [Google Scholar]
- 40. Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. (2001) Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem, 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 42. Shimodaira H. (2002) An approximately unbiased test of phylogenetic tree selection. System. Biol. 51, 492–508 [DOI] [PubMed] [Google Scholar]
- 43. Shimodaira H. (2004) Approximately unbiased test of regions using multistep-multiscale bootstrap resampling. Ann. Statist. 32, 2616–2641 [Google Scholar]
- 44. Lallemand-Breitenbach V., Jeanne M., Benhenda S., Nasr R., Lei M., Peres L., Zhou J., Zhu J., Raught B., de Thé H. (2008) Arsenic degrades PML or PML-RARalpha through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat. Cell. Biol. 10, 547–555 [DOI] [PubMed] [Google Scholar]
- 45. Pedrioli P. G., Eng J. K., Hubley R., Vogelzang M., Deutsch E. W., Raught B., Pratt B., Nilsson E., Angeletti R. H., Apweiler R., Cheung K., Costello C. E., Hermjakob H., Huang S., Julian R. K., Kapp E., McComb M. E., Oliver S. G., Omenn G., Paton N. W., Simpson R., Smith R., Taylor C. F., Zhu W., Aebersold R. (2004) A common open representation of mass spectrometry data and its application to proteomics research. Nat. Biotechnol. 22, 1459–1466 [DOI] [PubMed] [Google Scholar]
- 46. Craig R., Beavis R. C. (2004) TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20, 1466–1467 [DOI] [PubMed] [Google Scholar]
- 47. Beavis R. C. (2006) Using the global proteome machine for protein identification. Methods Mol. Biol. 328, 217–228 [DOI] [PubMed] [Google Scholar]
- 48. Srikumar T., Jeram S. M., Lam H., Raught B. (2010) A ubiquitin and ubiquitin-like protein spectral library. Proteomics 10, 337–342 [DOI] [PubMed] [Google Scholar]
- 49. Lam H., Deutsch E. W., Eddes J. S., Eng J. K., King N., Stein S. E., Aebersold R. (2007) Development and validation of a spectral library searching method for peptide identification from MS/MS. Proteomics 7, 655–667 [DOI] [PubMed] [Google Scholar]
- 50. Lam H., Deutsch E. W., Eddes J. S., Eng J. K., Stein S. E., Aebersold R. (2008) Building consensus spectral libraries for peptide identification in proteomics. Nat. Methods 5, 873–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Johnson E. S. (2004) Protein modification by SUMO. Annu. Rev. Biochem. 73, 355–382 [DOI] [PubMed] [Google Scholar]
- 52. Melchior F. (2000) SUMO–nonclassical ubiquitin. Annu. Rev. Cell Dev. Biol. 16, 591–626 [DOI] [PubMed] [Google Scholar]
- 53. Gong L., Yeh E. T. (1999) Identification of the activating and conjugating enzymes of the NEDD8 conjugation pathway. J. Biol. Chem. 274, 12036–12042 [DOI] [PubMed] [Google Scholar]
- 54. Eletr Z. M., Kuhlman B. (2007) Sequence determinants of E2-E6AP binding affinity and specificity. J. Mol. Biol. 369, 419–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kamadurai H. B., Souphron J., Scott D. C., Duda D. M., Miller D. J., Stringer D., Piper R. C., Schulman B. A. (2009) Insights into ubiquitin transfer cascades from a structure of a UbcH5B ∼ ubiquitin-HECT(NEDD4L) complex. Mol. Cell 36, 1095–1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hamilton K. S., Ellison M. J., Barber K. R., Williams R. S., Huzil J. T., McKenna S., Ptak C., Glover M., Shaw G. S. (2001) Structure of a conjugating enzyme-ubiquitin thiolester intermediate reveals a novel role for the ubiquitin tail. Structure 9, 897–904 [DOI] [PubMed] [Google Scholar]
- 57. Wenzel D. M., Lissounov A., Brzovic P. S., Klevit R. E. (2011) UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids. Nature 474, 105–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kirkpatrick D. S., Denison C., Gygi S. P. (2005) Weighing in on ubiquitin: the expanding role of mass-spectrometry-based proteomics. Nat. Cell. Biol. 7, 750–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Jeram S. M., Srikumar T., Pedrioli P. G., Raught B. (2009) Using mass spectrometry to identify ubiquitin and ubiquitin-like protein conjugation sites. Proteomics 9, 922–934 [DOI] [PubMed] [Google Scholar]
- 60. Petroski M. D., Deshaies R. J. (2005) Mechanism of lysine 48-linked ubiquitin-chain synthesis by the cullin-RING ubiquitin-ligase complex SCF-Cdc34. Cell 123, 1107–1120 [DOI] [PubMed] [Google Scholar]
- 61. Haldeman M. T., Xia G., Kasperek E. M., Pickart C. M. (1997) Structure and function of ubiquitin conjugating enzyme E2–25K: the tail is a core-dependent activity element. Biochemistry 36, 10526–10537 [DOI] [PubMed] [Google Scholar]
- 62. Kim H. C., Huibregtse J. M. (2009) Polyubiquitination by HECT E3s and the determinants of chain type specificity. Mol. Cell. Biol. 29, 3307–3318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wang M., Pickart C. M. (2005) Different HECT domain ubiquitin ligases employ distinct mechanisms of polyubiquitin chain synthesis. EMBO J. 24, 4324–4333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yunus A. A., Lima C. D. (2006) Lysine activation and functional analysis of E2-mediated conjugation in the SUMO pathway. Nat. Struct. Mol. Biol. 13, 491–499 [DOI] [PubMed] [Google Scholar]
- 65. Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Cryst. D. 50, 760–763 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.