Abstract
Metabolomics is a powerful new technology that allows for the assessment of global metabolic profiles in easily accessible biofluids and biomarker discovery in order to distinguish between diseased and nondiseased status information. Deciphering the molecular networks that distinguish diseases may lead to the identification of critical biomarkers for disease aggressiveness. However, current diagnostic methods cannot predict typical Jaundice syndrome (JS) in patients with liver disease and little is known about the global metabolomic alterations that characterize JS progression. Emerging metabolomics provides a powerful platform for discovering novel biomarkers and biochemical pathways to improve diagnostic, prognostication, and therapy. Therefore, the aim of this study is to find the potential biomarkers from JS disease by using a nontarget metabolomics method, and test their usefulness in human JS diagnosis. Multivariate data analysis methods were utilized to identify the potential biomarkers. Interestingly, 44 marker metabolites contributing to the complete separation of JS from matched healthy controls were identified. Metabolic pathways (Impact-value≥0.10) including alanine, aspartate, and glutamate metabolism and synthesis and degradation of ketone bodies were found to be disturbed in JS patients. This study demonstrates the possibilities of metabolomics as a diagnostic tool in diseases and provides new insight into pathophysiologic mechanisms.
Metabolomics, an omic science in systems biology, is the comprehensive profiling of metabolic changes occurring in living systems (1). It attempts to capture global changes and overall physiological status in biochemical networks and pathways in order to elucidate sites of perturbations, and has shown great promise as a means to identify biomarkers of diseases (2, 3). One area of considerable interest in the field of metabolomics is the detection of potential biomarkers associated with diseases, and the metabolic profiling could provide global changes of endogenous metabolites of patients. Metabolomics is the study of metabolic pathways and the measurement of unique biochemical molecules generated in a living system. It could facilitate biomarker discovery by distinguishing between diseased and nondiseased patients. Biomarker metabolites can also be therapeutic targets (4). Detecting changes in metabolite concentrations reveals the range of biochemical effects induced by a disease condition. Metabolic profiling of urine is particularly attractive because urine collection is noninvasive, and urine contains metabolic signatures of many biochemical pathways. The advantages of urine include its noninvasive collection and wide availability, its low protein and cellular levels, and its richness in metabolites. Monitoring certain metabolite levels in urine fluid has become an important way to detect early stages in disease (5). Urinary metabolomic approaches have been used to screen for potentially earlier diagnostic and prognostic biomarkers of disease (6). Metabolite changes observed in diseased individuals as a primary indicator have become possible, and hence the measurement of metabolites have been an important part of clinical practice.
Traditional markers used in conventional clinical chemistry and histopathology methods are not region-specific and only increase significantly after substantial disease injury. Therefore, more sensitive markers of disease are needed. The ideal biomarkers will identify disease early, resulting in safer drugs. Metabolomics, is an emerging and powerful discipline, which has become a promising player in the disease arena, and its benefits have been demonstrated in diverse clinical areas (7–9). Recent methodology, whose aim for complete characterization of the entire metabolome regardless of molecular size, are distinguishable from traditional tests on one or two components. The metabolomics approach has substantial impact on the development of diagnostics, therapeutics, and drug development (10–12). Particularly, for the early detection of disease, highly sensitive and specific biomarkers as primary indicators in bio-fluids are relatively more useful because these can be used for nonbiopsy tests. By analyzing and verifying the specificity of early biomarkers of a disease, metabolomics enables us to better understand pathological processes and metabolic pathways. Compared with traditional diagnostic methods, even small changes of metabolites can help to detect early pathologic changes earlier. Metabolic profiling has also been used as a diagnostic tool in the setting of liver disease. Studies of metabolites from patient urine that were discovered using metabolomics technology have recently come into focus as possible biomarkers for liver disease (13–15).
Understanding syndromes is a core research to develop more efficient therapeutic strategies, classification, and diagnostic criteria for patients. Pharmacological studies and clinical practice have shown that patients with liver disease are commonly complicated by jaundice syndrome (JS)1 (16–21). JS, a common and fatal disease requiring early diagnosis and effective treatment, exhibits two subtypes of YangHuang (YAH; acute) and YinHuang (YIH; chronic). Current diagnostic methods, measuring levels of alanine aminotransferase and aspartate aminotransferase etc. in blood samples, have been the main diagnostic markers for JS and can be considered as the current “gold standard” for initial diagnosis and surveillance. However, the sensitivity of these markers is relatively low, they are poor prognostic indicators and they are not particularly effective in separating cases of JS from other non-JS disorders. Fortunately, the rapid development of metabolomics technology platforms has been used to explore the particular metabolites, diagnostic and prognostic biomarkers, and pathways of the syndrome to provide a methodological basis for a deeper understanding of the essence of the syndrome from the aspect of systems biology (22). Consequently, this article was designed to investigate a comprehensive metabolome of JS by ultra-performance liquid-chromatography/electrospray-ionization synapt high-definition mass spectrometry (UPLC-Q-TOF-HDMS) combined with pattern recognition methods and pathways analysis, in order to establish a specific metabolite phenotype and explore the diagnostic possibilities, define new potential biomarkers, and generate a better understanding of the pathophysiology.
MATERIALS AND METHODS
Ethical Statement
Written informed consents were obtained from all subjects. The experimental protocol were reviewed and approved by the Ethical Committee of Heilongjiang University of Chinese Medicine and was conducted according to the principles expressed in the Declaration of Helsinki.
Subjects
Patients were collected from the Hospital of Heilongjiang University of Chinese Medicine, China for the period between June 1, 2009 and December 30, 2010. JS (n = 20), YAH (n = 19), and YIH (n = 14) patients and control subjects (n = 12) were recruited in this study. The outcomes of Health Survey Questionnaire in patients with JS, YAH, YIH and the normal controls were assessed, and the related clinical information including gender, age, body mass index (BMI), basic syndromes of disease, and main parameters of liver makers were collected in supplemental Table S1. Exclusion criteria included nonsmoker, cancer, cardiac insufficiency, hepatosis, renal inadequacy, respiratory failure, alimentary tract hemorrhage, or other diseases that will affect the clinical observations and biological indicators. Then, a second set of JS (n = 10), YAH (n = 12), and YIH (n = 9) patients and control subjects (n = 10) were to be blindly selected and tested using our approach.
Chemicals and Reagents
Acetonitrile, HPLC grade, was obtained from Merck (Darmstadt, Germany); methanol (HPLC grade) was purchased from Fisher Scientific Corporation (Loughborough, UK); water was produced by a Milli-Q Ultra-pure water system (Millipore, Billerica, MA); formic acid was of HPLC grade, and obtained from Honeywell Company (Morristown, New Jersey); leucine enkephalin was purchased from Sigma-Aldrich (St. Louis, MO). All other reagents were HPLC grade.
Sample Preparation
The subjects were given insulated ice packs in which they were asked to store the urine samples immediately until they were received by the study investigator. On arrival at the laboratory, the samples were centrifuged at 10,000 rpm for 10 min at 4 °C to remove any solid debris. Fractions (500 ml) of the urine supernatants were then stored at –80 °C until UPLC-Q-TOF-HDMS analysis. Thawed urine samples were collected after centrifugation at 13,000 rpm for 10 min at 4 °C, and the supernatant was transferred to a 1.5 ml polypropylene tube, and then filtered through a syringe filter (0.22 μm), 5 μl of the supernatant were injected into the UPLC-Q-TOF-HDMS.
Metabolic Profiling
Chromatographic Conditions
The UPLC/MS analysis was carried out using a Waters ACQUITY ultra-performance liquid-chromatography (UPLC) system (Waters Corp., Milford, MA) coupled with time-of-flight mass spectrometry (TOF-MS) from Waters. Chromatography was carried out with an ACQUITY BEH C18 chromatography column (2.1 mm × 100 mm, 1.7 um). The column temperature was maintained at 45 °C, and then gradient mobile phase conditions was composed of phase A (water with 0.1% formic acid) and phase B (acetonitrile containing 0.1% formic acid). A Waters Acquity UPLC BEH C18 (2.1 mm i.d. × 100 mm ACQUITY) column packed with 1.7 mm beads was used to separate the molecules in the biofluids set. The gradient for the urine sample was as follows: 0–5 min, 1–25% B; 5–9 min, 25–50% B; 9–9.1 min, 50–99% B; 9.1–11 min, 99% B; 11–11.1 min, 99–1% B; 11.1–13 min, 1% B. The flow rate was 0.40 ml/min and 5 μl aliquot of each sample was injected onto the column. The eluent was introduced to the mass spectrometry directly without a split. To ensure the stability and repeatability of the UPLC-Q-TOF-HDMS systems, pooled quality control (QC) samples were prepared from 10 μl of each sample and analyzed together with the other samples. The QC samples were also inserted and analyzed in every 10 samples.
TOF-MS Conditions
Mass spectrometry and accurate mass acquisition was performed with a Waters QTOF Premier operating at positive-ion (ESI+) and negaitive-ion (ESI−) electrospray ionization (ESI) mode. The optimal capillary voltage was set at 3200 V, and cone voltage at 35 V. Nitrogen was used as the dry gas, the desolvation gas flow rate was set at 500 L/h, and cone gas flow was maintained at 50 L/h. The desolvation temperature was set at 350 °C, and source temperature at 110 °C. The scan time and interscan delay were set to 0.4 s and 0.1 s, respectively. MS data were collected in the full scan mode from m/z 50–1000. All the data were acquired using an independent reference lock mass via the LockSprayTM interface to ensure accuracy and reproducibility during the MS analysis. Leucine enkaphalin was used as the reference compound (positive ion mode ([M+H]+ = 556.2771) and [M-H]− = 554.2615) at a concentration of 0.2 ng/ml under a flow rate of 100 μl·min−1. The data were collected in the centroid mode, and the LockSpray frequency set at 10 s and averaged over 10 scans for correction.
Data Processing
All data were processed using the MarkerLynx application manager for MassLynx 4.1 software (Waters Corp). The UPLC/MS data are detected and noise-reduced in both the UPLC and MS domains such that only true analytical peaks are further processed by the software. A list of intensities of the peaks detected is then generated for the first chromatogram, using the Rt-m/z data pairs as identifiers. The resulting normalized peak intensities form a single matrix with Rt-m/z pairs for each file in the data set. All processed data of each chromatogram were normalized and pareto scaled, prior to multivariate statistical analysis. SPSS 13.0 for Windows was used for the statistical analysis. The data were analyzed using the Wilcoxon Mann-Whitney Test, with p < 0.05 set as the level of statistical significance.
Multivariate Data Analysis
Centroided and integrated raw mass spectrometric data were processed using MassLynx V4.1 and MarkerLynx software (Waters). The intensity of each ion was normalized with respect to the total ion count to generate a data matrix that consisted of the retention time, m/z value, and the normalized peak area. The multivariate data matrix was analyzed by EZinfo software (Waters). The unsupervised segregation was checked by principal components analysis using pareto-scaled data. From the loading plots of orthogonal projection to latent structures discriminate analysis (OPLS-DA), various metabolites could be identified as being responsible for the separation between control and JS groups, and were therefore viewed as differentiating metabolites. Potential markers of interest were extracted from the combining S- and VIP- plots that were constructed from the OPLS analysis, and markers were chosen based on their contribution to the variation and correlation within the data set. With the completion of the OPLS-DA analysis, we can try computational systems analysis with MetaboAnalyst data annotation approach (http://www.metaboanalyst.ca/MetaboAnalyst/faces/Home.jsp) (22) to distinguish between control and JS subjects. The Heatmap, implemented in MetaboAnalyst tool commonly used for unsupervised clustering, were constructed based on the potential candidates of importance, which were extracted with OPLS-DA analysis.
Biomarker Identification
Exact molecular mass data from redundant m/z peaks corresponding to the formation of different parent and product ions were first used to help confirm the metabolite molecular mass. MS/MS data analysis highlights neutral losses or product ions, which are characteristic of metabolite groups and can serve to discriminate between database hits. The MassFragment™ application manager (Waters MassLynx v4.1, Waters) was used to facilitate the MS/MS fragment ion analysis process by way of chemically intelligent peak-matching algorithms. The identities of the specific metabolites were confirmed by comparison of their mass spectra and chromatographic retention times with those obtained using commercially available reference standards. A full spectral library, containing MS/MS data obtained in the positive and negative ion modes, for all metabolites reported in this work is available on request from the authors. This information was then submitted for database searching, either in-house or using the online ChemSpider database (www.chemspider.com), MassBank (http://www.massbank.jp/), and MetFrag (http://msbi.ipb-halle.de/MetFrag/) data source. Pathway analysis and visualization using the KEGG (www.genome.jp/kegg/) pathway database was carried out using Metaboanalyst.
RESULTS
Urine Metabonomic Study of Patients
Urine metabolic profiling was established to explore important biomarkers and metabolic pathways related to JS and develop a prediction model for aided diagnosis of the disease. The representative UPLC-HDMS base peak ion (BPI) current chromatograms from positive and negative ion modes of human urine from the patients with JS and controls were compared visually (Data not shown). The stable BPI reflected the stability of UPLC-HDMS analysis and reliability of the metabolomic data. Low molecular mass metabolites could be well separated in the short time of 13 min because of the small particles (less than 1.7 um) of UPLC. Using the optimized UPLC-HDMS analysis protocol and subsequent processes, such as baseline correction, peak deconvolution, alignment, and normalization, we obtained a three-dimensional matrix, including data file name, retention time exact mass pair, and normalized peak areas. Overall 9986 retention time-exact mass pairs were determined in each sample profile. Although some differences could be visually noted among the three sets of the detail illustrated in the BPI chromatogram, more subtle changes could be found using a pattern recognition approach, such as PLS, OPLS-DA. Typically, the metabolic profiles of disease cases and controls are compared with the aim of identifying spectral features, and ultimately metabolites, which discriminate the classes.
Analysis of Metabolic Pattern
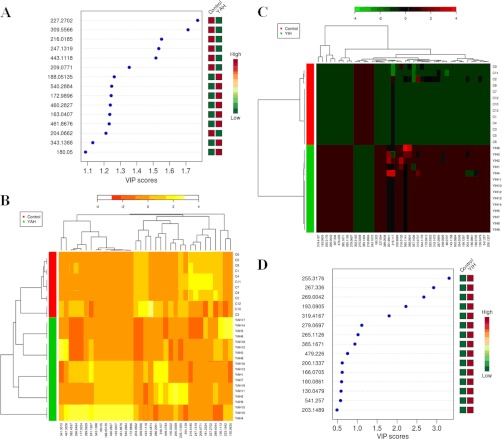
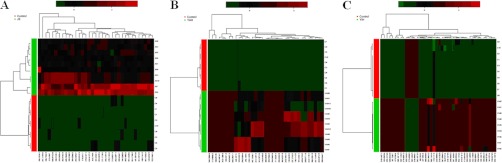
Using our metabolomics platform, the statistically important metabolites were studied. PCA was used first to investigate general interrelation between groups, including clustering and outliers among the samples. And then, PLS-DA was used to maximize the difference of metabolic profiles between control and JS groups and facilitate the detection of metabolites consistently present in the biological samples. Data were standardized using Pareto-scaling technique. The supervised OPLS-DA can improve biomarker discovery efforts and separate samples into two blocks was applied to obtain better discrimination between the control and JS groups. Score plots from the supervised OPLS-DA showed obvious separation between the JS, YAH, YIH groups and healthy group in both positive (Figs. 1A, 2A, and 3A) and negative ion modes (supplemental Figs. S1, S2, and S3), which suggests that urinary biochemical perturbation significantly occurs in JS and its subtype groups. Trajectory analysis of score plots (3-D) for the control and JS groups showed clear segregation (Fig. 1B). Additionally, the similar analysis based on subtype groups and control group did reveal clearer separation (Figs. 2B and 3B). From the corresponding the loading plots, the ions furthest away from the origin contribute significantly to be responsible for the separation between control and JS groups and may be therefore regarded as the differentiating metabolites for JS, YAH, YIH groups (Figs. 1C, 2C, and 3C). Combining the results of S- and VIP- plots from the OPLS analysis (Figs. 1D, 2D, and 3D), the UPLC-HDMS analysis platform provided the retention time, precise molecular mass, and MS/MS data for the structural identification of biomarkers. Finally, according to the variable importance in the projection (VIP >2), the variables (ions) were identified based on the metabolite identification strategy (listed in supplemental Tables S2–S4). The parallel analysis of the samples with MetaboAnalyst allows for the ability to verify that ions, which are identified through both ways (i.e. Heatmaps, OPLS-DA), are highly significant, as depicted through two completely different algorithms. Analysis of the control and JS samples utilizing MetaboAnalyst's data annotation tools revealed differences between the two groups. Figs. 4A and 5D showed top 15 significant features of the metabolite markers based the VIP projection. The heatmaps, commonly used for unsupervised clustering, were constructed based on the potential candidates of importance, which were extracted with OPLS-DA analysis. The parallel heatmap visualization (Figs. 4B and 5C) using Ward's method in computational systems analysis for the JS, YAH, YIH, and controls showed distinct segregation. To test the usefulness of the marker metabolites for diagnosis, a second set of JS (n = 10), YAH (n = 12), and YIH (n = 9) patients and control subjects (n = 10) were blindly selected and analyzed. Using our metabolomics platform, interestingly, the heatmap visualization (Fig. 6) using in computational systems analysis could be achieved for discrimination of all the patients.
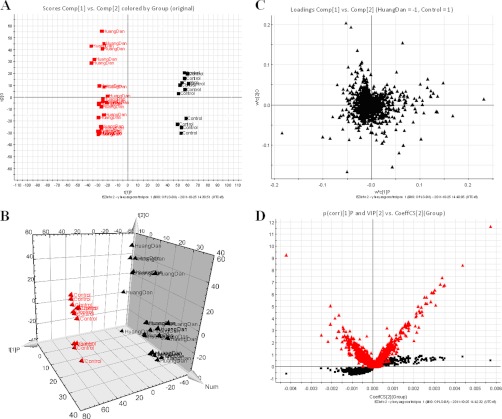
Fig. 1.
Metabolomic profiling of JS. A, OPLS-DA model results for JS group in positive mode. B, 3-D of OPLS-DA model for JS group. C, Loading plot of OPLS-DA of JS in positive mode. D, shows the combination of S- and VIP-score plots constructed from the supervised OPLS analysis of urine (ESI+ mode). Ions with the highest abundance and correlation in the JS group with respect to the controls are present on the upper far right hand quadrant, whereas ions with the lowest abundance and correlation in the JS group with respect to the control group are residing in the lower far left hand quadrant.
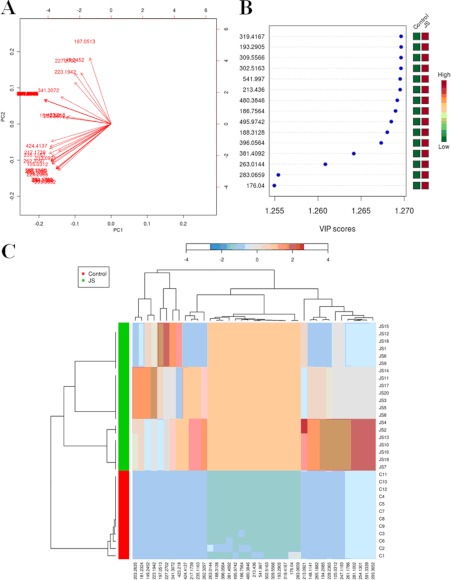
Fig. 2.
Analysis of the control and JS samples utilizing MetaboAnalyst's data annotation tools revealed differences between the two groups. A, Significance changes of the metabolite markers illustrated in the Biplot from PLS-plot. It can be readily observed that the concentrations of differential metabolites. B, Top 15 significant features of the metabolite markers based the VIP projection. C, Heat map visualization for the urine of JS. The heatmaps were constructed based on the potential candidates of importance, which were extracted with OPLS-DA analysis. Variable differences are revealed between the control and JS groups, with verified and known ions marked on the bottom corresponding to supplemental Table S2. Rows: samples; Columns: metabolites; Color key indicates metabolite expression value, blue: Lowest, red: highest.
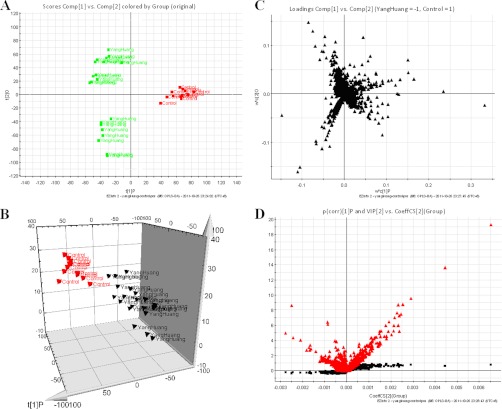
Fig. 3.
Metabolomic profiling of YAH. A, OPLS-DA model results for YAH group in positive mode. B, 3-D of OPLS-DA model for YAH group. C, Loading plot of OPLS-DA of YAH in positive mode. Combination of S- and VIP-score plots constructed from the supervised OPLS analysis of urine (ESI+ mode).
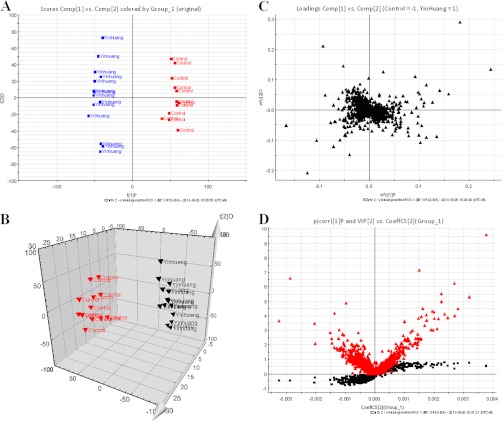
Fig. 4.
Metabolomic profiling of YIH. A, OPLS-DA model results for YIH group in positive mode. B, 3-D of OPLS-DA model for YIH group. C, Loading plot of OPLS-DA of YIH in positive mode. D, shows the combination of S- and VIP-score plots constructed from the supervised OPLS analysis of urine (ESI+ mode).
Fig. 5.
Systems analysis of Metabolomic alterations of the YAH, YIH and control samples with MetaboAnalyst's data annotation tools. A, Top 15 significant features of the metabolite markers based the VIP scores of OPLS-DA. B, Heat map visualization constructed based on the differential metabolites of importance for the urine of YAH. Heatmap represents unsupervised hierarchical clustering of groups (rows). Variable differences marked on the bottom corresponding to supplemental Table S3 are revealed between the control and YAH groups. Rows: samples; Columns: differential metabolites; Color key indicates metabolite expression value, red: Lowest, yellow: highest. C, Heat map visualization constructed based on the differential metabolites of importance for the urine of YIH. Variable differences marked on the bottom corresponding to supplemental Table S4 are revealed between the control and YIH groups. Rows: samples; Columns: differential metabolites; Color key indicates metabolite expression value, green: Lowest, red: highest. D, Top 15 significant features of the metabolite markers based the VIP scores.
Fig. 6.
Hierarchical clustering and diagnostic potential of JS metabolite composition. A, Unsupervised hierarchical clustering of JS metabolite profiles. B, Profiles of the signatures that distinguish YAH from control samples. C, Dendrogam obtained from unsupervised hierarchical clustering of metabolite profiles for YIH and control samples.
Biomarker Identification
The robust UPLC-HDMS analysis platform provides the retention time, precise molecular mass, and MS/MS data for the structural identification of biomarkers. Structure identification was performed according to their molecular ion masses and MS/MS product ion analysis compared with authentic standards or database resources. Some parameters, such as deviation from calculated mass (mDa or ppm), double bond equivalent (DBE), and i-fit value (the isotopic pattern of the selected ion) were used to evaluate the accuracy of possible formulas. Based on the relative intensities of the metabolites from the normalized spectrum, the combination of S- and VIP-score plots constructed from the supervised OPLS analysis of urine was used to reveal the significant differences of identified metabolites between the JS group and control group, as well as between the subtype groups and control group. The presumed molecular formula was searched in Chemspider, Human Metabolome Database, and other databases to identify the possible chemical constitutions, and MS/MS data were screened to determine the potential structures of the ions.
Here, the biomarker with Rt-m/z of 6.76–591.3339 in positive ion mode was detailed as an example to illustrate the identification process. Using a mass tolerance of 5mDa, C33H42N4O6 was located as the candidate because of its high mass accuracy and low i-fit value among the possible chemical formulas. And then, C33H42N4O6 was input in the KEGG ligand for possible compounds, and d-urobilinogen finally emerged, which was further confirmed by comparing it to its authentic standards. In this study, we performed UPLC/MS based metabolomic profiling in urine to detect potential biomarkers associated with JS patients. Finally, forty-four marker metabolites contributing to the complete separation of JS from matched healthy controls were identified, with epinephrine, deoxycytidine, methylglutarylcarnitine, etc. exhibiting the best combined classification performance (see supplemental Table S2). Forty endogenous metabolites contributing to a good separation between YAH and matched healthy controls were identified, with Kinetin, porphobilinogen, indoleacrylic acid, etc. showing the maximum combined classification performance (supplemental Table S3). Forty-nine metabolites contributing to the complete separation between YIH and matched healthy controls were identified, with 2-octenedioic acid, pyroglutamic acid, alpha-N-phenylacetyl-l-glutamine, etc. showing the best combined classification performance (supplemental Table S4).
Metabolic Pathway and Function Analysis
With pattern recognition analysis of metabolites, a clear separation of JS groups and healthy group was achieved. Metabolite profiling focuses on the analysis of a group of metabolites related to a specific metabolic pathway in biological states. More detailed analysis of the most relevant pathways and networks of JS were performed by Metaboanalyst that is a free, web-based tool that combines result from powerful pathway enrichment analysis involved in the conditions under study. Metaboanalyst, directed graph, uses the high-quality KEGG (www.genome.jp/kegg/) pathway database as the backend knowledgebase. Consequently, potential targets of metabolic pathway analysis (Impact-value ≥0.10) with Metaboanalyst revealed that the metabolites that were identified together are important for the host response to JS, and are responsible for d-glutamine and d-glutamate metabolism, synthesis and degradation of ketone bodies, alanine, aspartate, and glutamatemetabolism (supplemental Fig. S4A). The top three metabolic pathway of importance including vitamin B6 metabolism, tryptophan metabolism, arginine and proline metabolism were found to be disturbed in YAH patients (supplemental Fig. S4B). Distinct metabolic pathway analysis (Impact-value≥0.10), which were all related to YIH, are responsible for steroid hormone biosynthesis, primary bile acid biosynthesis, cysteine and methionine metabolism (supplemental Fig. S4C). The detailed construction of the d-glutamine and d-glutamate metabolism (supplemental Fig. S5A), vitamin B6 metabolism (supplemental Fig. S5B), primary bile acid biosynthesis (supplemental Fig. S5C) pathways with a higher score in humans were generated using the reference map by searching KEGG.
DISCUSSION
Systems biology is a general trend of contemporary scientific development (23). Identification of molecular markers and metabolic pathways has the potential to improve diagnostic, prognostication, and therapy of interest (24–27). The determination of metabolic profiles may probably provide a biomarker for the discrimination of diseases as well as differentiation of healthy volunteers from patients. Current diagnostic methods, measuring levels of alanine aminotransferase, and aspartate aminotransferase etc. in blood samples, have been the main diagnostic markers for JS and can be considered as the current “gold standard” for initial diagnosis and surveillance. However, the sensitivity of these markers is relatively low and it is difficult to get an outcome immediately. Hence, novel approaches for the detection of JS are urgently needed. The nontarget metabolomics provides a global view of the organism and can be used to monitor the dynamic metabolic alterations that occur in different pathological processes (28, 29).
Efficient diagnostic methods in clinical practice are needed for the metabolite biomarkers. Metabolomics is a powerful new technology that allows for the assessment of global metabolic profiles in easily accessible biofluids and biomarker discovery in order to distinguish between diseased and nondiseased status information (30). Offering a physiologically holistic, noninvasive platform, the emergent field of metabolomics has the potential to provide new diagnostic tools. Recent technological breakthroughs have provided researchers with the capacity to measure hundreds or even thousands of small-molecule metabolites in as little as a few minutes per sample, paving the way for studies ideally suited to complex diseases such as JS (31, 32). Early or subclinical diagnosis of JS would be helpful for prevention and treatment. Metabolomics is particularly suited to liver injury assessment, where urine is the most common sample made available for laboratory tests. Although gene and protein expression have been extensively profiled in human disease, little is known about the global metabolomic alterations that characterize disease. Multiple, complex molecular events characterize disease development and progression. Deciphering the molecular networks that distinguish disease from liver disease may lead to the identification of critical biomarkers for disease aggressiveness.
We utilized the metabolomics approach in a pilot study in urine samples of JS patients from control subjects. It was demonstrated that based on UPLC-MS measurements of urine samples with statistically significant multivariate models could be constructed to distinguish between diseased and nondiseased subjects. The global metabolic profiling and subsequent multivariate analysis clearly distinguished JS patients from matched controls. As shown in Figs. 1A and 1B, OPLS-DA revealed an evident and statistically significant separation between the JS and control samples. Results indicate that JS related metabolites play an essential role in glutamate metabolism, synthesis, and degradation of ketone bodies, alanine, and aspartate metabolism, which are tightly correlated with the genes of neurotransmitters, hormones, and cytokines in the metabolites interaction network. By using our metabolomics platform, 44 statistically important variables with VIP >2 were defined (supplemental Table S2). Interestingly, of the 44 distinct metabolites identified from these pathways, many are found in the various stages of progression of JS. Further study of these metabolites may facilitate the development of noninvasive biomarkers and more efficient therapeutic strategies for JS. Furthermore, vitamin B6 metabolism, tryptophan metabolism, arginine and proline metabolism was also the top function listed by Metaboanalyst for YAH patient. Significant changes associated with YAH disease, defined as metabolite changes in YAH versus controls were identified for 40 metabolites that are potential candidates for biomarkers. Additionally, steroid hormone biosynthesis, primary bile acid biosynthesis, cysteine and methionine metabolism were all related with YIH. It is noteworthy that 49 metabolites together are important for the host response to JS through target metabolism pathways.
We specifically introduced a new way to examining metabolome expression information of typical patients versus controls. A significant separation using Heatmap analysis was obtained for the difference between JS versus controls as well as between subtype patients versus controls. Computational systems analysis with MetaboAnalyst tool (Heatmap) provides a powerful approach to metabolic profiling of urine to differentiate patients from control subjects. The patients with JS were easily distinguished from control subjects by the Heatmap approach when using autoscaling methods. To determine whether these metabolic markers which were identified using our metabolomics platform, we performed a preliminary validation using a second set of patients and control subjects, which were blindly selected. Our results indicated that the model had a high sensitivity and specificity for the clinical JS diagnosis, as shown in Fig. 6. Targeted metabolite analysis studies have already shown that alterations of critical JS metabolic pathways, such as glutamate metabolism, synthesis and degradation of ketone bodies, alanine and aspartate metabolism, are strongly associated with JS development. Such changes are expected to be reflected in wider coverage metabolic profiles, which may in turn be explored for potential biomarkers for JS assessment and treatment. Recently, metabolic pathways incorporate complex networks of protein-based interactions and modifications, are usually considered to provide information on mechanisms of disease and have become a common and probably the most popular form of representing biochemical information for hypothesis generation and validation. These maps store a wide knowledge of complex molecular interactions and regulations occurring in the living organism in a simple and obvious way, often using intuitive graphical notation. Based on the KEGG, a detailed construction of the the d-glutamine and d-glutamate metabolism, vitamin B6 metabolism, primary bile acid biosynthesis pathways map with higher score is shown in supplemental Fig. S5. Our results indicate that these target pathways show the marked perturbations over the entire time-course of JS and could contribute to the development of JS. Furthermore, the more patients included and the detected metabolomic biomarkers make evaluation in further studies necessary before the significance of our results could be assured. Further investigations are also underway to clarify the precise pathogenesis why JS induced these results. Therefore, metabolomics suggests that there is a great potential for candidate metabolite biomarker discovery, metabolite signatures may also have the potential to be used as diagnostic biomarkers. Future research will focus on the discovery of additional biomarkers using metabolomics platforms and the validation of the explorative biomarkers. In addition, more effort will be directed to the biological interpretation: it will be investigated which pathways were involved in the biochemical changes associated with the onset, development, and progression of JS, and whether these changes are the same during onset and progression, or if different changes of biochemistry occur at the different stages of JS.
In summary, metabolomics provides a powerful approach to discover diagnostic and therapeutic biomarkers by analyzing global changes in an individual's metabolic profile. We here, for the first time, report a comprehensive analysis of metabolic patterns of JS and sub-types. We have identified some significantly changed metabolites associated with JS and specifically noted 44 metabolomic signatures in JS. Glutamate metabolism, synthesis, and degradation of ketone bodies, alanine and aspartate metabolism was found that the most altered functional pathway associated with JS according to ingenuity pathway analysis. The results not only indicated that metabolomic methods had sufficient sensitivity and specificity to distinguish JS from healthy controls, but also have the potential to be developed into a clinically useful diagnostic tool, and could also contribute to a further understanding of disease mechanisms. In conclusion, our findings suggested that metabonomics method would be helpful to establishing a suitable model for reasonably evaluating disease syndrome, exploring pathological mechanism of the syndrome, clarifying the relationships between the syndrome and related diseases.
Footnotes
* This work was supported by grants from the Key Program of Natural Science Foundation of State (Grant No. 90709019), the National Specific Program on the Subject of Public Welfare (Grant No. 200807014), National Key Subject of Drug Innovation (Grant No. 2009ZX09502–005), and National Program on Key Basic Research Project of China (Grant No. 2005CB523406).
 This article contains supplemental Figs. S1 to S5 and Tables S1 to S4.
This article contains supplemental Figs. S1 to S5 and Tables S1 to S4.
Author contributions: X.W., A.Z., and H.S. designed and performed the experiments and analyzed the raw data. X.W. and A.Z., wrote the manuscript. H.Y. and W.P. supervised the project. G.S., and Y.Y., X.Y., M.Z. collected urine. H.L., T.D., X.N. H.Z. and D.W. assisted in metabolomic experiment.
Competing financial interests: The authors declare no competing financial interests.
1 The abbreviations used are:
- JS
- Jaundice syndrome
- YAH
- YangHuang
- YIH
- YinHuang
- PCA
- principal components analysis
- OPLS-DA
- orthogonal projection to latent structures discriminate analysis.
REFERENCES
- 1. Nicholson J. K., Lindon J. C. (2008) Systems biology: Metabonomics. Nature 455, 1054–1056 [DOI] [PubMed] [Google Scholar]
- 2. Kim K., Aronov P., Zakharkin S. O., Anderson D., Perroud B., Thompson I. M., Weiss R. H. (2009) Urine metabolomics analysis for kidney cancer detection and biomarker discovery. Mol. Cell Proteomics 8, 558–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tohge T., Fernie A. R. (2010) Combining genetic diversity, informatics and metabolomics to facilitate annotation of plant gene function. Nat. Protoc. 5, 1210–1227 [DOI] [PubMed] [Google Scholar]
- 4. Arakaki A. K., Skolnick J., McDonald J. F. (2008) Marker metabolites can be therapeutic targets as well. Nature 456, 443. [DOI] [PubMed] [Google Scholar]
- 5. Cai Z., Zhao J. S., Li J. J., Peng D. N., Wang X. Y., Chen T. L., Qiu Y. P., Chen P. P., Li W. J., Xu L. Y., Li E. M., Tam J. P., Qi R. Z., Jia W., Xie D. (2010) A combined proteomics and metabolomics profiling of gastric cardia cancer reveals characteristic dysregulations in glucose metabolism. Mol. Cell. Proteomics 9, 2617–2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Veselkov K. A., Vingara L. K., Masson P., Robinette S. L., Want E., Li J. V., Barton R. H., Boursier-Neyret C., Walther B., Ebbels T. M., Pelczer I., Holmes E., Lindon J. C., Nicholson J. K. (2011) Optimized preprocessing of ultra-performance liquid chromatography/mass spectrometry urinary metabolic profiles for improved information recovery. Anal. Chem. 83, 5864–5872 [DOI] [PubMed] [Google Scholar]
- 7. Chadeau-Hyam M., Ebbels T. M., Brown I. J., Chan Q., Stamler J., Huang C. C., Daviglus M. L., Ueshima H., Zhao L., Holmes E., Nicholson J. K., Elliott P., De Iorio M. (2010) Metabolic profiling and the metabolome-wide association study: significance level for biomarker identification. J. Proteome Res. 9, 4620–4627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang T. J., Larson M. G., Vasan R. S., Cheng S., Rhee E. P., McCabe E., Lewis G. D., Fox C. S., Jacques P. F., Fernandez C., O'Donnell C. J., Carr S. A., Mootha V. K., Florez J. C., Souza A., Melander O., Clish C. B., Gerszten R. E. (2011) Metabolite profiles and the risk of developing diabetes. Nat. Med. 17, 448–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Beckonert O., Coen M., Keun H. C., Wang Y., Ebbels T. M., Holmes E., Lindon J. C., Nicholson J. K. (2010) High-resolution magic-angle-spinning NMR spectroscopy for metabolic profiling of intact tissues. Nat. Protoc. 5, 1019–1032 [DOI] [PubMed] [Google Scholar]
- 10. Clayton T. A., Lindon J. C., Cloarec O., Antti H., Charuel C., Hanton G., Provost J. P., Le Net J. L., Baker D., Walley R. J., Everett J. R., Nicholson J. K. (2006) Nicholson. Pharmaco-metabonomic phenotyping and personalized drug treatment, Nature 440, 1073–1077 [DOI] [PubMed] [Google Scholar]
- 11. Holmes E., Loo R. L., Stamler J., Bictash M., Yap I. K., Chan Q., Ebbels T., De Iorio M., Brown I. J., Veselkov K. A., Daviglus M. L., Kesteloot H., Ueshima H., Zhao L., Nicholson J. K., Elliott. P. (2008) Human metabolic phenotype diversity and its association with diet and blood pressure. Nature 453, 396–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sreekumar A., Poisson L. M., Rajendiran T. M., Khan A. P., Cao Q., Yu J., Laxman B., Mehra R., Lonigro R. J., Li Y., Nyati M. K., Ahsan A., Kalyana-Sundaram S., Han B., Cao X., Byun J., Omenn G. S., Ghosh D., Pennathur S., Alexander D. C., Berger A., Shuster J. R., Wei J. T., Varambally S., Beecher C., Chinnaiyan A. M. (2009) Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature 457, 910–914 [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 13. Yu K., Sheng G., Sheng J., Chen Y., Xu W., Liu X., Cao H., Qu H., Cheng Y., Li L. (2007) A metabonomic investigation on the biochemical perturbation in liver failure patients caused by hepatitis b virus. J. Proteome Res. 6, 2413–2419 [DOI] [PubMed] [Google Scholar]
- 14. Chen J., Wang W., Lv S., Yin P., Zhao X., Lu X., Zhang F., Xu G. (2009) Metabonomics study of liver cancer based on ultra performance liquid chromatography coupled to mass spectrometry with HILIC and RPLC separations. Anal. Chim. Acta 650, 3–9 [DOI] [PubMed] [Google Scholar]
- 15. Manna S. K., Patterson A. D., Yang Q., Krausz K. W., Idle J. R., Fornace A. J., Gonzalez F. J. (2011) UPLC-MS-based urine metabolomics reveals indole-3-lactic acid and phenyllactic acid as conserved biomarkers for alcohol-induced liver disease in the Ppara-null mouse model. J. Proteome Res. 10, 4120–4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tang X., Yang C., Gu Y., Song C., Zhang X., Wang Y., Zhang J., Hew C. L., Li S., Xia N., Sivaraman J. (2011) Structural basis for the neutralization and genotype specificity of hepatitis E virus. Proc. Natl. Acad. Sci. U.S.A. 108, 10266–102671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Friedman L. S., Gee M. S., Misdraji J. Case records of the Massachusetts General Hospital. Case 39–2010. (2010) A 19-year-old woman with nausea, jaundice, and pruritus. N. Engl. J. Med. 363, 2548–2557 [DOI] [PubMed] [Google Scholar]
- 18. Selmi C., Bowlus C. L., Gershwin M. E., Coppel R. L. (2011) Primary biliary cirrhosis. Lancet 377, 1600–1609 [DOI] [PubMed] [Google Scholar]
- 19. Tschopp S., Brücker R. (2003) Aching joints and jaundice. Lancet 361, 750. [DOI] [PubMed] [Google Scholar]
- 20. Arias I. M. (1998) New genetics of inheritable jaundice and cholestatic liver disease. Lancet 352, 82–83 [DOI] [PubMed] [Google Scholar]
- 21. Premawardhena A., Fisher C. A., Fathiu F., de Silva S., Perera W., Peto T. E., Olivieri N. F., Weatherall D. J. (2001) Genetic determinants of jaundice and gallstones in haemoglobin E beta thalassaemia. Lancet 357, 1945–1946 [DOI] [PubMed] [Google Scholar]
- 22. Wang X., Yang B., Sun H., Zhang A. (2012) Pattern recognition approaches and computational systems tools for ultra performance liquid chromatography-mass spectrometry-based comprehensive metabolomic profiling and pathways analysis of biological data sets. Anal. Chem. 84, 428–439 [DOI] [PubMed] [Google Scholar]
- 23. Bousquet J., Anto J. M., Sterk P. J., Adcock I. M., Chung K. F., Roca J., Agusti A., Brightling C., Cambon-Thomsen A., Cesario A., Abdelhak S., Antonarakis S. E., Avignon A., Ballabio A., Baraldi E., Baranov A., Bieber T., Bockaert J., Brahmachari S., Brambilla C., Bringer J., Dauzat M., Ernberg I., Fabbri L., Froguel P., Galas D., Gojobori T., Hunter P., Jorgensen C., Kauffmann F., Kourilsky P., Kowalski M. L., Lancet D., Pen C. L., Mallet J., Mayosi B., Mercier J., Metspalu A., Nadeau J. H., Ninot G., Noble D., Oztürk M., Palkonen S., Préfaut C., Rabe K., Renard E., Roberts R. G., Samolinski B., Schünemann H. J., Simon H. U., Soares M. B., Superti-Furga G., Tegner J., Verjovski-Almeida S., Wellstead P., Wolkenhauer O., Wouters E., Balling R., Brookes A. J., Charron D., Pison C., Chen Z., Hood L., Auffray C. (2011) Systems medicine and integrated care to combat chronic noncommunicable diseases. Genome Med. 3, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Catchpole G. S., Beckmann M., Enot D. P., Mondhe M., Zywicki B., Taylor J., Hardy N., Smith A., King R. D., Kell D. B., Fiehn O., Draper J. (2009) Hierarchical metabolomics demonstrates substantial compositional similarity between genetically modified and conventional potato crops. Proc. Natl. Acad. Sci. U.S.A. 102, 14458–14462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu J. Y., Li N., Yang J., Li N., Qiu H., Ai D., Chiamvimonvat N., Zhu Y., Hammock B. D. (2010) Metabolic profiling of murine plasma reveals an unexpected biomarker in rofecoxib-mediated cardiovascular events. Proc. Natl. Acad. Sci. U.S.A. 107, 17017–17022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Suhre K., Shin S. Y., Petersen A. K., Mohney R. P., Meredith D., Wägele B., Altmaier E., CARDIoGRAM, Deloukas P., Erdmann J., Grundberg E., Hammond C. J., de Angelis M. H., Kastenmüller G., Köttgen A., Kronenberg F., Mangino M., Meisinger C., Meitinger T., Mewes H. W., Milburn M. V., Prehn C., Raffler J., Ried J. S., Römisch-Margl W., Samani N. J., Small K. S, Wichmann H. E., Zhai G., Illig T., Spector T. D., Adamski J., Soranzo N., Gieger C., Assimes T. L., Deloukas P., Erdmann J., Holm H., Kathiresan S., König I. R., McPherson R., Reilly M. P., Roberts R., Samani N. J., Schunkert H., Stewart A. F. (2010) Human metabolic individuality in biomedical and pharmaceutical research. Nature 477, 54–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Anderson D. D., Quintero C. M., Stover P. J. (2011) Identification of a de novo thymidylate biosynthesis pathway in mammalian mitochondria. Proc. Natl. Acad. Sci. U.S.A. 108, 15163–15168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huang Z., Lin L., Gao Y., Chen Y., Yan X., Xing J., Hang W. (2011) Bladder cancer determination via two urinary metabolites: a biomarker pattern approach. Mol. Cell Proteomics 10, M111.007922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Deng W. J., Nie S., Dai J., Wu J. R., Zeng R. (2010) Proteome, phosphoproteome, and hydroxyproteome of liver mitochondria in diabetic rats at early pathogenic stages. Mol. Cell. Proteomics 9, 100–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stoop M. P., Coulier L., Rosenling T., Shi S., Smolinska A. M., Buydens L., Ampt K., Stingl C., Dane A., Muilwijk B., Luitwieler R. L., Sillevis Smitt P. A., Hintzen R. Q., Bischoff R., Wijmenga S. S., Hankemeier T., van Gool A. J., Luider T. M. (2010) Quantitative proteomics and metabolomics analysis of normal human cerebrospinal fluid samples. Mol Cell Proteomics 9, 2063–2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Naesens M., Sarwal M. M. (2010) Molecular diagnostics in transplantation. Nat. Rev. Nephrol. 6, 614–628 [DOI] [PubMed] [Google Scholar]
- 32. Chen T., Xie G., Wang X., Fan J., Qiu Y., Zheng X., Qi X., Cao Y., Su M., Wang X., Xu L. X., Yen Y., Liu P., Jia W. (2011) Serum and urine metabolite profiling reveals potential biomarkers of human hepatocellular carcinoma. Mol. Cell Proteomics 10, 110.004945 [DOI] [PMC free article] [PubMed] [Google Scholar]