Abstract
Annexin 1 (ANXA1), the first characterized member of the annexin superfamily, is known to bind or annex to cellular membranes in a calcium-dependent manner. Besides mediating inflammation, ANXA1 has also been reported to be involved in important physiopathological implications including cell proliferation, differentiation, apoptosis, cancer, and metastasis. However, with controversies in ANXA1 expression in breast carcinomas, its role in breast cancer initiation and progression remains unclear. To elucidate how ANXA1 plays a role in breast cancer initiation, we performed stable isotope labeling of amino acids in cell culture analysis on normal mammary gland epithelial cells from ANXA1-heterozygous (ANXA1+/−) and ANXA1-null (ANXA1−/−) mice. Among over 4000 quantified proteins, we observed 214 up-regulated and 169 down-regulated with ANXA1−/−. Bioinformatics analysis of the down-regulated proteins revealed that ANXA1 is potentially implicated in DNA damage response, whereas the analysis of up-regulated proteins showed the possible roles of ANXA1 in cell adhesion and migration pathways. These observations were supported by relevant functional assays. The assays for DNA damage response demonstrated an accumulation of more DNA damage with slower recovery on heat stress and an impaired oxidative damage response in ANXA1−/− cells in comparison with ANXA1+/− cells. Overexpressing Yes-associated protein 1 or Yap1, the most down-regulated protein in DNA damage response pathway cluster, rescued the proliferative response in ANXA1−/− cells exposed to oxidative damage. Both migration and wound healing assays showed that ANXA1+/− cells possess higher motility with better wound closure capability than ANXA1−/− cells. Knocking down of β-parvin, the protein with the highest fold change in the cell adhesion protein cluster, indicated an increased cell migration in ANXA1−/− cells. Altogether our quantitative proteomics study on ANXA1 suggests that ANXA1 plays a protective role in DNA damage and modulates cell adhesion and motility, indicating its potential role in cancer initiation as well as progression in breast carcinoma.
Annexin-1 (ANXA1), a 37-kDa protein, is a member of the family of Ca2+-dependent phospholipid-binding proteins. The diverse biological properties of the annexin family members are attributed to the variability in length and sequence of their N-terminal domains (1). Being the first characterized member of the annexin superfamily, ANXA1 has long been implicated to have anti-inflammatory properties, whereby it mediates the function of glucocorticoids (2), and as an inhibitor of phospholipase A2 activity (3). Its roles as a substrate for the epidermal growth factor receptor (EGFR)1 tyrosine kinase (4), modulator of the mitogen-activated protein kinase extracellular signal-regulated kinase pathway (5), as well as an “eat me” signal in apoptotic cells for phagocytes (6), establish ANXA1's involvement in important cellular regulatory pathways including cell proliferation, differentiation and apoptosis (7). This has driven recent research on ANXA1 toward the topic of carcinogenesis because any dysregulation in cellular regulatory pathways has the potential of leading to cancer.
There is accumulating evidence suggesting that ANXA1 could be playing critical roles in cancer. A first line of evidence comes from the observation that there is differential expression of ANXA1 in different cancers. ANXA1 has been shown to be lost in esophageal cancer (8), prostate cancer (9), and head and neck cancer (10) and overexpressed in hepatocarcinoma (11) as well as pancreatic cancer (12). The implication of ANXA1 in tumor growth and pathological angiogenesis, which are etiologies of cancer, has also been recently demonstrated in ANXA1-null mice upon subcutaneous injection of tumor cells, suggesting ANXA1 to be a tumor-induced vascular biomarker (13). There have been conflicting reports on the status of ANXA1 levels in breast carcinomas, with ductal carcinomas exhibiting a loss of ANXA1, whereas basal cell carcinomas express high levels of ANXA1. ANXA1 was reported as an important modulator for an epithelial-to-mesenchymal-like phenotypic switch via the transforming growth factor β signaling pathway (14). Furthermore, our recent study demonstrates that ANXA1 is required for constitutive NF-κB activity in basal cell carcinoma cell lines, which is of utmost importance to metastatic potential (15). Moreover, genomics approaches to studying molecular signatures associated with transformation and progression to breast cancer highlighted an up-regulation of ANXA1 in cellular transformation (16). However, its specific role in breast cancer initiation and progression remains unclear. Elucidating factors regulating normal mammary gland cell development is essential for our understanding of breast cancer. Here, we used a quantitative system wide approach to investigate the impact of ANXA1 in mammary gland cells from ANXA1-heterozygous and deficient mice. Stable isotope labeling of amino acids in cell culture (SILAC), deploying the in vivo incorporation of amino acids with substituted stable isotope-labeled amino acids (17) into cell culture, was employed for mass spectrometry-based quantitative proteomics. We quantified over 4000 proteins by SILAC and performed bioinformatics analysis for up- and down-regulated protein clusters. The analysis revealed that ANXA1 plays key roles in DNA damage responses, cell adhesion, and migration, which we further validated by using relevant functional assays.
EXPERIMENTAL PROCEDURES
Isolation and Culture of Primary Mammary Gland Epithelial Cells
Mammary glands were removed from C57 female ANXA1-heterozygous and ANXA1-null mice, minced to average fragment size of ∼1.5 mm3 using scalpel blades in serum-free Dulbecco's minimal essential medium with 0.01 mg/10 ml collagenase (Sigma) and incubated in a 37 °C water bath for 1 h with intermittent shaking. The fragments were then meshed through a 70-μm nylon filter, collected in the same medium before centrifugation at 80 × g for 5 min to remove fats. The pellet was washed with fresh medium without collagenase and centrifuged at 80 × g for 5 min before resuspending in complete SILAC medium and plating on culture dishes for the establishment of the individual cell lines. For SILAC culture, both the ANXA1+/− and ANXA1−/− mammary gland cells were adapted to SILAC DMEM (Thermo Scientific) containing either 13C- and 15N-labeled lysine and arginine (K8R10; “heavy”) (Cambridge Isotopes) or 12C- and 14N-labeled lysine and arginine (K0R0; “light”) (Sigma), 10% dialyzed FBS (Thermo Scientific), and 1% penicillin-streptomycin (Invitrogen) at 37 °C in a humidity-saturated 5% CO2 atmosphere.
Cell Lysate Preparation
SILAC-adapted cells were harvested by scraping on ice and lysed on ice for 20 min using 50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1% Nonidet P-40 with 1× Roche complete protease inhibitor mixture (Roche Applied Science), and 1× PhosSTOP phosphatase inhibitor (Roche Applied Science). Protein quantification was measured using the BCA assay kit (Pierce) before mixing equal amount of identically concentrated light and heavy SILAC-labeled ANXA1+/− and ANXA1−/− lysates.
Mass Spectrometry
Equal amounts of proteins from lysates of heavy and light labeled cells were mixed, and 80 μg of protein mixture was separated by one-dimensional 4–12% NuPage Novex Bis-Tris gel (Invitrogen), stained using the Colloidal Blue staining kit (Invitrogen) and digested with trypsin using published procedures (18). The samples were analyzed on an Orbitrap or Orbitrap XL (Thermo Fisher). Survey full scan MS spectra (m/z 300–1400) were acquired with a resolution of r = 60,000 at m/z 400, an AGC target of 1e6, and a maximum injection time of 500 ms. The 10 most intense peptide ions in each survey scan with an ion intensity of >2000 counts and a charge state of ≥2 were isolated sequentially to a target value of 1e4 and fragmented in the linear ion trap by collisionally induced dissociation using a normalized collision energy of 35%. A dynamic exclusion was applied using a maximum exclusion list of 500 with one repeat count, repeat, and exclusion duration of 30 s.
Identification and Quantification of Peptides and Proteins
The data were searched using Mascot (version 2.2; Matrix Science, London, UK) against International Protein Index mouse version 3.68 decoy database containing 52,288 proteins and 172 commonly observed contaminants. Database searches were performed with tryptic specificity allowing maximum two missed cleavages and two labeled amino acids as well as an initial mass tolerance of 7 ppm for precursor ions and 0.5 Da for fragment ions. Cysteine carbamidomethylation was searched as a fixed modification, and N-acetylation and oxidized methionine were searched as variable modifications. Labeled arginine and lysine were specified as fixed or variable modifications, depending on the prior knowledge about the parent ion. SILAC peptide and protein quantification was performed with MaxQuant version 1.0.13.13 (19, 20) using default settings. The maximum false discovery rates were set to 0.01 for both protein and peptide. The proteins were considered identified when supported by at least one unique peptide with a minimum length of six amino acids.
Western Blot Analysis
Equal amount of cell lysates were loaded onto NuPage Novex 4–12% Bis-Tris gel (Invitrogen) and transferred onto PVDF membrane (Bio-Rad). The following antibodies used for Western analyses were bought from Santa Cruz: anti-ANXA1, anti-caspase 3, anti-STAT1, anti-EGFR, anti-PAI1, and anti-TIMP2. Antibodies against MMP14 and p19ARF were from Abcam, p53 and Yap1 (Yes-associated protein 1) were from Cell Signaling Technologies, and β-actin was from Millipore.
RT-PCR Analysis
Total RNA was prepared using an RNeasy kit (Qiagen) and reverse transcribed using SuperScript first-strand synthesis system for RT-PCR (Invitrogen) according to the manufacturer's protocol. Gene-specific primer pairs used were as follows: β-actin (sense, 5′-GGACGACATGGAGAAAATCTG-3′; antisense, 5′-CAGCTCGTAGCTCTTCTCCA-3′), ANXA1 (sense, 5′-ATGGCAATGGTATCAGAATTCCTC AA-3′; antisense, 5′-TGTAAGGGCTTTTCTCAAGACTTC AT-3′), p53 (sense, 5′-CCAGAAGATATCCTGCCATCA-3′; antisense, 5′-TAGCTTATTGAGGGGAGGAGA-3′), p19ARF (sense, 5′-GTTGTTGAGGCTAGAGAGGAT-3′; antisense, 5′-TCGCACGATGTCTTGATGTCC-3′), STAT1 (sense, 5′-TCCCGTACAGATGTCCATGAT-3′; antisense, 5′-TGAACCAGCTTTGCAGCTGA-3′), Casp3 (sense, 5′-GGAATGTCATCTCGCTCTGGT-3′; antisense, 5′-CTTGTGCGCGTACAGCTTCAG-3′), EGFR (sense, 5′-AATGGACTTACAGAGCCATCC-3′; antisense, 5′-GGATGCCATCTTCTTCCACTT-3′), PAI1 (sense, 5′-TCAATGACTGGGTGGAAAGGC-3′; antisense, 5′-GGTCCACTTCAGTCTCCAGAG-3′), MMP14 (sense, 5′-CCACTTTGATTCTGCCGAGCC-3′; antisense, 5′-GCTTCGTCAAACACCCAGTGC-3′), TIMP2 (sense, 5′-CCGCAACAGGCGTTTTGCAAT-3′; antisense, 5′-TTCTCTTGATGCAGGCGAAGA-3′), and β-parvin (sense, 5′-GAG GTG ACT GAC TTA CAG GAA-3′; antisense, 5′-CTG CAC GGT GAC ATG CTC AG-3′). All reactions except for β-parvin follow a 2-min initial denaturation at 94 °C, and amplification conditions were as follows: 94 °C for 10 s, 55 °C for 30 s, and 68 °C for 45 s for 25 cycles, followed by an extension of 68 °C for 7 min. For β-parvin, a 3-min initial denaturation at 94 °C was followed by 94 °C for 30 s, 58 °C for 30 s, and 72 °C for 45 s for 25 cycles, and an extension of 68 °C for 7 min. PCR products were electrophoresed on 2% agarose gels stained with ethidium bromide. β-Actin was used as a normalizing control.
Heat Treatment
The cells were seeded in T75 tissue culture flasks 1 day prior to treatment. The flasks were sealed with parafilm and incubated in a water bath preheated to 45 °C for 30 min (heat-treated) or 37 °C for 30 min (control). The cells were harvested at 0-, 4-, and 24-h time points after treatment by trypsinization for comet assays.
Comet Assay
The assay was performed using a CometAssay® kit (Trevigen) according to the manufacturer's protocol. In summary, the cells were counted, mixed with low melting point agarose, spread onto Comet slides, and allowed to solidify before soaking in lysis solution in the dark for 30 min followed by 20 min in DNA unwinding buffer. The slides were loaded onto a gel electrophoresis tank with alkaline electrophoresis buffer and run at 25 V/300 mA in the dark. The samples were then washed in water, dehydrated in 70% ethanol, and dried at 37 °C. DNA was stained using SYBR Green in Tris-EDTA buffer (1:10,000), and slides were examined with an Axioplan 2 imaging fluorescence microscope (Carl Zeiss). Analysis of comets was performed using Comet Imager Software (Metasystems). The extent of DNA damage was expressed in terms of comet tail moments. 30 randomly selected cells were examined per sample.
Transfection Experiments
For knocking down of β-parvin, ANXA1−/− mammary gland cells were seeded onto 6-well plate and transfected using Dharmafect 1 according to the manufacturer's protocol with β-parvin small interfering RNA (si-β-parvin; Santa Cruz); mock cells simply treated with transfection reagent were used as the negative control. Upon 24 h post-transfection, the cells were washed twice with PBS and starved with serum-free medium overnight before cell adhesion assay. For overexpression of Yap1, pCDNA-Yap (His- and Xpress-tagged) was transfected in ANXA1−/− cells plated to ∼80% confluency using TurboFect in vitro transfection reagent (Fermentas) according to the manufacturer's protocol.
Cell Adhesion Assay
Prior to the assay, the underside of the porous translucent membranes in the cell culture inserts (Corning) were coated with 25 μg/ml of PureCol® collagen (Advanced BioMatrix). These inserts were then incubated overnight at 37 °C in a sterile condition and covered. 1 × 104 of the transfected cells in DMEM with 0.5% FBS were seeded into the upper chamber (with the collagen-coat on the underside) with 10% FBS in the lower chamber of the Transwell. The cells were incubated for another 24 h at 37 °C to allow for adhesion to and migration through the collagen-coated porous membrane on the underside of the cell culture insert. At the end of the assay, nonmigrated cells on the upper side of the insert (top chamber), as well as the migrated cells in the lower chamber, were carefully removed and counted. The collagen-coated underside of the membrane was washed twice with PBS and fixed with 10% formalin for 10 min at room temperature. After two washes with PBS, the migrated cells were permeabilized with 0.1% Tween 20 in PBS for 5 min at room temperature before staining with 500 μl of hematoxylin-2 (Richard-Allan Scientific) for 10 min at room temperature. The membrane was then washed in water by dipping it for 5 min before staining in 500 μl of eosin-Y alcoholic (Richard-Allan Scientific) for 2 min at room temperature. After three 5-min sessions in 95% alcohol, the cell culture inserts were then allowed to air dry upside down for 15 min at room temperature. The porous membranes were removed from the cell culture inserts with a razor blade and mounted on glass slides. The migrated cells on the underside of the collagen-coated membrane were visualized using light microscope (Zeiss) at 2.5× and 10× magnification.
Hydrogen Peroxide Treatment and Cell Proliferation (WST) Assay
24 h after transfection, 3 × 104 cells were seeded into 96-well before overnight treatment with 1 mm H2O2. Cell viability was measured using WST-1 assay (Roche Applied Science) at every 15-min interval.
Migration Assay
The cells were starved for 16 h prior to migration assay with serum-free DMEM. 2.5 × 105 cells in DMEM with 0.5% FBS were seeded into the upper chamber (8-μm membrane insert) with 10% FBS in the lower chamber of the Transwell (Corning) and incubated for 48 h under normal cell culture conditions. The migrated cells were then counted.
Wound Healing Assay
The cells were seeded into both reservoirs of the culture insert (Ibidi), allowed to grow to monolayer confluence before removal of the culture insert. The cells were fed with fresh DMEM, and the dishes were fixed on a motorized XY stage coupled to a Nikon Ti Eclipse with perfect focus system. The time-lapsed phase contrast images were acquired by a CoolSnap HQ2 (Photometrics). The images were selected and analyzed using Wimasis Image Analysis software (Wimasis GmbH).
Reactive Oxygen Species (ROS) Assay
The cells harvested by trypsinization were washed with PBS before incubation with 1 μm 5-(and-6)-chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate (Molecular Probes) for 20 min at 37 °C in the dark. The cells were then washed, resuspended with DMEM, and assessed for intracellular ROS by FACS analysis.
RESULTS
Relative Quantification of Proteins of ANXA1+/− and ANXA1−/− (Knockout) Murine Mammary Gland Cells by SILAC
We first wished to identify the functions of ANXA1 in mammary gland cells (nontumor) through quantitative profiling of proteins using SILAC. We adapted cell lines from murine mammary gland cells of ANXA+/− mouse (+/−) and from ANXA−/− mouse (−/−) in either media containing normal isotopes of l-lysine-(12C614N2) (K0) and l-arginine-(12C614N4) (R0) or media containing stable isotope l-lysine-(13C615N2) (K8) and l-arginine-(13C615N4) (R10). The equal amounts of cell lysates from light (K0R0) and heavy (K8R10) cells were mixed followed by separation of 80 μg of total protein on SDS-PAGE. Extracted peptides from in-gel trypsin-digested gel slices were subjected to LC Orbitrap-MS analysis. MS data were analyzed using MaxQuant software. The experiments were carried out in four replicates as two forward (heavy ANXA−/− cells versus light ANXA+/− cells) experiments and two reverse (heavy ANXA+/− cells versus light ANXA−/− cells) experiments (Fig. 1A). Each experiment contained two technical replicates for MS analysis. For initial data analysis, the combined raw data of two technical replicates of each experiment was used in MaxQuant analysis. We quantified ∼5000 proteins at least in one experiment (supplemental Table 1). The SILAC ratios of forward and reverse and biological replicates were well correlated (supplemental Fig. S1). Thus, raw data sets from forward experiments were compiled together for MaxQuant analysis to obtain final SILAC ratios for the forward experiment and the same analysis for the reverse experiment (Fig. 1B and supplemental Table 2). After removing contaminants and reverse assignments, we found 4108 proteins quantified in both forward and reverse experiments, which were used to categorize unchanged, up- and down-regulated proteins with ANXA1−/− as follows. First, proteins with contradictory ratio significance B (P) values from forward and reverse experiments were removed (e.g., proteins with ratio significance of less than 0.05 in the forward experiment, but that of more than 0.05 in the reverse experiment (and vice versa) were removed). From this filtered list “ratio significance B value of forward (PFwd) × ratio significance B value of reverse (PRev) <0.0025” was applied to select proteins with statistically significant changes (red and blue dots in Fig. 1C). To obtain biologically significant changes, a protein frequency (density) plot was generated using R, and the proteins below the 10% quantile were selected as down-regulated. Conversely, those above the 90% quantile were selected as up-regulated proteins (Fig. 1, B and C). We found that 10%/90% quantile matched to ∼2-fold changes in all experiments. Thus, up- or down-regulated proteins have at least 2-fold changes together with p < 0.05. This approach has the advantage that proteins with just below 2-fold changes but with statistically significant ratios were not eliminated. Through this approach, we found 214 up-regulated proteins and 169 down-regulated proteins (Fig. 1C) in ANXA1-deficient cells.
Fig. 1.
SILAC workflow and protein quantification analysis. A, a schematic workflow of the SILAC forward (Fwd) and reverse (Rev) experiments where ANXA1+/− and ANXA1−/− mammary gland cells were adapted in K0R0 and K8R10 (and vice versa). B, distribution of unchanged, up- and down-regulated proteins from merged Fwd and Rev experiments with high confidence ratios. Statistically significant changes in protein levels (unit by log2 ratio) between the two mammary gland cell lines are marked by the cutoffs shown in blue and red. C, a scatter plot showing the up-regulated (red), down-regulated (blue), and unchanged (black) protein clusters.
Analysis of Down-regulated Proteins in ANXA1-deficient Mammary Cells
Pathway Analysis
To assign preliminary functional relevance of the up- or down-regulated proteins, we performed bioinformatics analysis using GeneGo. GeneGo Map Folder analysis revealed that DNA damage response pathway proteins were enriched in down-regulated protein cluster with high significance value (<1e−6) (supplemental Fig. S2A). Thus, we focused on DNA damage response pathway for further analysis. In addition to the proteins in GeneGo DNA damage response pathway folder, we found two other proteins with statistically significant fold changes involved in this pathway through Gene Ontology/Kyoto Encyclopedia of Genes and Genomes pathway analysis (Table I). These proteins were found to play roles in DNA damage response pathways primarily through regulation of proliferation, transcription, caspase activation cascade, and apoptosis. We have confirmed that Yap1, p53, p19ARF, caspase-3, and STAT1 were down-regulated in ANXA1−/− cells by Western blot analysis (supplemental Fig. S2B). RT-PCR analysis of most of these proteins demonstrated that all protein expression levels correspond to their mRNA transcript levels. For both Western blot and RT-PCR analyses, β-actin was used as a loading and normalizing control.
Table I. Down-regulated proteins with ANXA1−/− involved in DNA damage response.
| Gene name | Normalized H/L |
P (forward) × P (reverse) | |
|---|---|---|---|
| Forward | Reverse | ||
| Yap1 | 0.09 | 6.34 | 3.20E-17 |
| Tp53 | 0.17 | 2.95 | 1.00E-07 |
| Eya3 | 0.27 | 3.27 | 1.03E-04 |
| Ptpn11 | 0.30 | 2.80 | 1.97E-05 |
| Bid | 0.31 | 2.74 | 8.95E-05 |
| Casp3 | 0.33 | 2.73 | 4.16E-05 |
| Prkar2b | 0.35 | 3.29 | 8.94E-06 |
| Cdkn2a | 0.36 | 3.96 | 1.72E-06 |
| Nmt2 | 0.36 | 2.17 | 2.43E-04 |
| Nfkb2 | 0.39 | 2.14 | 2.14E-03 |
| Set | 0.43 | 1.94 | 1.69E-04 |
| Stat1 | 0.45 | 3.38 | 2.69E-06 |
| Hmgb2 | 0.45 | 1.90 | 3.12E-04 |
| Lig1 | 0.46 | 2.07 | 1.38E-03 |
| Hsp90aa1 | 0.48 | 2.08 | 1.99E-04 |
| Hspa4 | 0.50 | 1.80 | 8.51E-04 |
Functional Validation
Pathway analysis of the SILAC data showed that ANXA1 plays important roles in DNA damage response and related pathways. To further confirm its involvement in the DNA damage response pathway, we performed a comet assay, which assesses the extent of DNA damage at various time points of recovery upon DNA damage. Comet tail moment (fraction of migrated DNA multiplied by some measure of tail length), indicative of the extent of damage, was analyzed using the Comet Imaging Software. The heat shock protein HSP70 is involved in the caspase cascade pathway (21), and because both HSP70 and HSP90 were down-regulated with ANXA1−/− cells, thermal stress was used to induce DNA damage (22) in both cell lines. As shown in Fig. 2A, the accumulation of damage in ANXA1−/− was higher than ANXA1+/− at both 4 and 24 h. Moreover, by the 24-h time point, more recovery was observed in ANXA1+/− whereas DNA damage was still obvious in ANXA1−/− cells (Fig. 2B). Interestingly, we also observed that there was a substantial difference in the mean tail moments in the control without any heat treatment at 0 h between the two mammary cell lines (Fig. 2C). In summary, we showed that ANXA1 indeed plays a protective role in the DNA damage response, because ANXA1−/− mammary gland cells not only accumulated more damage but recovered more slowly compared with ANXA1+/− cells after heat stress.
Fig. 2.
Heat stress responses of ANXA1−/−versus ANXA1+/− mammary gland cells and the effect of Yap1 on intrinsic ROS accumulation/damage in ANXA1−/−. A, comet assay was performed with the mammary gland cells with and without DNA damage (heat treatment). After 30 min of 45 °C (heat stress) or 37 °C (control), the cells were allowed to recover at the various time points for comet assay. DNA damage was measured as mean tail moment in the graph plotted. B, representatives of SYBER Green-stained comets 24 h after heat stress for both mammary gland cells. Comet Imager Software measures the tail moments of the comets as indicated by the sky blue trail. C, representative pictures from comet assays of the two mammary gland cells at the 0-h time point without any treatment (control) indicated a much higher mean tail moment (1.068) for the knockout than the ANXA1+/− (0.308). D–F, FACS analyses from the ROS assays. The intrinsic increase in ROS in the ANXA1-deficient is evidenced by the slight shift of the peak to the right relative to ANXA1+/− (D). No significant difference was observed between control and Yap-1-transfected cells (E and F). The data are representative of three repeats of the experiments.
To further verify the importance of ANXA1 in DNA damage functionally, a ROS assay was performed to confirm the difference in the cellular accumulation of ROS in ANXA1+/− and ANXA1−/− mammary gland cells. Fig. 2D shows a rightward shift of the ROS peak for ANXA1−/− cells compared with ANXA1+/− cells, indicative of more intrinsic ROS accumulation. This potentially could lead to more oxidative damage in the ANXA1−/− cells, further supporting the protective role of ANXA1 in mammary gland cells. To evaluate the relationship between down-regulated proteins in ANXA1−/− and DNA damage response pathway, we carried out both the comet and ROS assays with the most down-regulated protein in ANXA1−/− cells, Yap1 (Table I). Yap1 was overexpressed in ANXA1−/− cells in an attempt to see whether it is able to alleviate its ROS accumulation or intrinsic damage. Fig. 2 (E and F) shows that the ROS peaks for both mock and Yap1 totally overlapped with the ROS peak for ANXA1−/−. Moreover, the results of comet assays also show insignificant differences in the mean tail moments between the control ANXA1−/− and Yap-1-transfected ANXA1−/− cells (data not shown). Because Yap1 is unable to make any difference to the intrinsic ROS accumulation and damage, we assessed the lack of this DNA damage response phenotype in ANXA1−/− cells by the measurement of proliferation. Because oxidative insult to ANXA1−/− cells showed minimal proliferation response as compared with ANXA1+/− cells (Fig. 3A), we asked whether over- or re-expressing Yap1 would “rescue” this proliferation response. Overexpressing Yap1 in ANXA1−/− cells did not bring about any changes in the proliferation rate of the cells (Fig. 3B). However, Yap1 was able to bring a substantial reversal of the lack of DNA damage response phenotype in ANXA1−/− cells when the cells were exposed to oxidative damage (Fig. 3C), showing that down-regulated proteins in ANXA1−/− are key regulators in responding to DNA damage.
Fig. 3.
Oxidative stress responses of ANXA1−/− mammary gland cells with Yap1 overexpression. A, graph showing the cell proliferation of ANXA1+/− and ANXA1−/− cells with and without H2O2. The cells were treated with 1 mm H2O2 overnight, and the absorbance was measured over 2 h using WST reagent. B, Western blot showing the overexpression of mouse Yap1 in ANXA1−/− cells (left) and graph showing proliferation profile between ANXA1−/− and Yap1-transfected ANXA1−/− cells (right). The cells were transfected with pCDNA-Yap1 (mouse) for 24 h before seeding into 96-well for cell proliferation/viability assay by WST reagent. C, graph showing the rescued cellular proliferation response made by Yap1-transfected ANXA1−/− mammary gland cells. Transfected cells were treated overnight with 1 mm H2O2 before measurement. All of the data are representatives of triplicates of two repeats of the experiments.
Analysis of Up-regulated Proteins in ANXA1-deficient Mammary Cells
Pathway Analysis
Pathway analysis of up-regulated proteins clustered with GeneGo Map Folder analysis revealed that the tissue remodeling and wound repair pathway was enriched with a high significance value (<1e−5) among up-regulated protein cluster (supplemental Fig. S3A). To narrow down to more specific fundamental processes pertaining to tissue remodeling and wound repair pathway, we further analyzed subfolders associated with this pathway cluster. We identified cell adhesion and cell motility as major subclusters. Based on these outcomes, we searched the proteins involved in these pathways using GeneGo as well as Kyoto Encyclopedia of Genes and Genomes pathway. In this search we found several other proteins with statistically significant fold changes involved in these pathways (Table II). We confirmed that EGFR, MMP14, TIMP2, and PAI1 were up-regulated in the absence of ANXA1 by Western blot (supplemental Fig. S3B). Interestingly RT-PCR analysis of related genes demonstrated that MMP14 and TIMP2 were not affected at the mRNA level and that PAI1 was lower at the mRNA level. All other genes were consistent with protein expression. For both Western blot and RT-PCR analyses, β-actin was used as a loading and normalizing control.
Table II. Up-regulated proteins with ANXA1−/− involved in cell adhesion/motility.
| Gene name | Normalized H/L |
P (forward) × P (reverse) | |
|---|---|---|---|
| Forward | Reverse | ||
| Parvb | 16.46 | 0.26 | 1.68E-07 |
| Itga11 | 12.09 | 0.07 | 4.54E-13 |
| Col1a1 | 11.67 | 0.25 | 2.83E-11 |
| Col5a2 | 10.09 | 0.09 | 2.35E-12 |
| Tln2 | 8.24 | 0.11 | 1.24E-09 |
| Timp3 | 8.00 | 0.17 | 9.41E-07 |
| Col1a2 | 7.84 | 0.35 | 3.57E-08 |
| Lamb2 | 6.23 | 0.31 | 3.96E-05 |
| Mmp14 | 6.18 | 0.26 | 4.04E-06 |
| Serpine1 | 6.14 | 0.14 | 1.76E-07 |
| Tgfbr2 | 6.01 | 0.19 | 6.05E-07 |
| Il6st | 5.83 | 0.43 | 1.49E-05 |
| Sdc4 | 5.41 | 0.33 | 1.65E-04 |
| Lama4 | 5.13 | 0.25 | 5.89E-05 |
| Acta2 | 5.12 | 0.23 | 5.65E-06 |
| Tubb3 | 4.91 | 0.27 | 8.07E-06 |
| Timp2 | 4.89 | 0.22 | 6.00E-06 |
| Col3a1 | 4.81 | 0.48 | 1.67E-05 |
| Fbln2 | 4.53 | 0.29 | 6.14E-05 |
| Tgfb1i1 | 4.44 | 0.21 | 8.54E-06 |
| Figf | 4.32 | 0.16 | 5.08E-06 |
| Tgfbr3 | 4.26 | 0.29 | 1.50E-04 |
| Pdgfra | 4.15 | 0.24 | 2.01E-05 |
| Sh3kbp1 | 4.15 | 0.33 | 1.24E-04 |
| Jak1 | 4.15 | 0.37 | 2.45E-04 |
| Myl9 | 4.09 | 0.28 | 1.84E-05 |
| Egfr | 3.84 | 0.17 | 1.61E-06 |
| Pak3 | 3.32 | 0.34 | 1.30E-03 |
| Ncam1 | 3.16 | 0.31 | 7.29E-06 |
Functional Validation
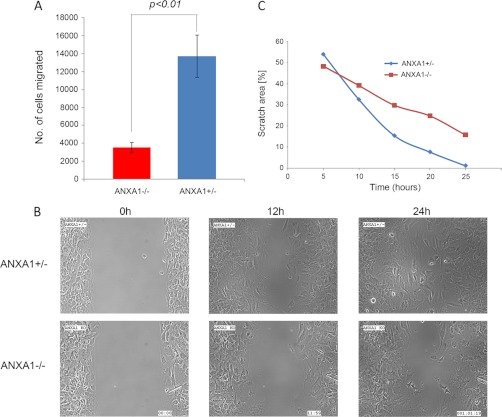
Pathway analysis revealed that ANXA1 is important in cell migration and adhesion because proteins involved in those pathways were up-regulated in ANXA1−/− cells. To verify this functional association, Transwell migration and wound healing assays were performed to compare the adhesive and migratory capabilities of these two cell lines, which we used to perform proteomics analysis. The Transwell migration assay demonstrated that the motility of ANXA1+/− was significantly higher (at least 3-fold) than the ANXA1−/− (Fig. 4A). Next, we used a culture insert for wound healing assay, where a nonbias cell-free gap of 500 ± 50 μm is produced as the “wound” when it is removed. Immediately after the removal of the culture insert, the cells were observed in real time under a video microscope by a CCD camera captured every 10 min. After 24 h, ANXA1+/− cells had achieved nearly complete wound closure (∼98%), whereas the wound in ANXA−/− cells was only ∼70% closed (Fig. 4, B and C, and supplemental Movie 1). Taken together, these results showed that ANXA1-expressing cells were able to migrate faster than ANXA1-deficient cells, implying that ANXA1 plays a positive role in cell migration and adhesion processes.
Fig. 4.
Migration and wound healing capabilities of ANXA1−/−versus ANXA1+/− mammary gland cells. A, graph showing an overall representation of four separate sets of migration assays. ANXA1+/− and ANXA1−/− mammary gland cells without any treatment were trypsinized for Transwell migration assays using 10% FBS in lower chamber for 48 h. B, representatives of real time snapshots of the directive migration or wound healing assay at different time points (from supplemental Movie 1). C, nearly complete closure of wound by ANXA1+/− mammary gland cells was observed at 24 h (left with 1.2% scratch area), whereas ANXA1−/− lagged with 15.8% scratched area as depicted.
For further verification, we tested up-regulated proteins in ANXA1−/− cells having direct roles in cell adhesion and migration. We selected β-parvin because it is the most up-regulated protein among those involved in adhesion processes (Table II) and performed both migration and wound healing assays for β-parvin knockdown cells (Fig. 5A). The Transwell migration assay demonstrated that the motility of the si-β-parvin in ANXA1−/− cells was higher than the mock control (Fig. 5B). The wound healing assay also supported the faster migratory capability of si-β-parvin in ANXA1−/− cells because it achieved near 100% wound closure, whereas the wound in mock control was only ∼80% closed by 12 h (Fig. 5, C and D, and supplemental Movie 2). Because β-parvin is a focal adhesion protein, we tested its effect on cell adhesion as described under “Experimental Procedures.” The results of these experiments showed an obvious abolition of collagen adhesion in cells with β-parvin knocked down in comparison with the mock control ANXA1−/− (Fig. 6A). To further ensure that this decrease in adhesiveness is a complement to the migratory capability of the cells, we counted the cells in both the upper and lower chambers. Fig. 6B shows no statistical significance in the number of cells left in the upper chamber across the mock, si-β-parvin, and ANXA1+/− or between si-β-parvin and ANXA1+/− cells. However, there is a significant difference in the number of cells in the lower chamber (p < 0.05) between the mock and si-β-parvin as well as mock and ANXA1+/− cells. This result clearly showed that the migratory ability of the ANXA1−/− cells was increased by knocking down of β-parvin, confirming the relevance of the up-regulation of proteins clustering in the cell adhesion/migration pathway to the phenotype of the ANXA1−/− cells.
Fig. 5.
Effect of β-parvin on migration and wound healing capabilities in ANXA1−/− mammary gland cells. A, RT-PCR showing the expression levels of β-parvin after knocking down in ANXA1−/− cells as compared with mock control. β-Actin was used as a normalizing control. B, graph showing an overall representation of two separate sets of migration assays. ANXA1−/− cells were mock-treated (control) or knocked down with β-parvin siRNA before trypsinized for Transwell migration assays using 10% FBS in lower chamber for 24 h. C, representatives of real time snapshots of the directive or wound healing assay at different time points for the mock control and si-β-parvin ANXA1−/− cells (from supplemental Movie 2). D, graph depicting nearly complete wound closure by si-β-parvin ANXA1−/− cells (left with 1% scratch area), whereas the mock control cells lagged with an 11% scratch area at 12 h.
Fig. 6.
Comparison of the cell adhesive/migratory abilities of mock-treated or antisense β-parvin-transfected ANXA−/− cells. A, hematoxylin and eosin staining of ANXA1−/− cells transfected with antisense β-parvin that have passed through the porous membrane and adhered to the collagen on the underside of the cell culture insert over 24 h as compared with mock and ANXA1+/− cells. Pictures (representatives of triplicates from two separate sets of the experiments) were taken under 2.5× and 10× magnification using a light microscope, showing an observable abolition of collagen adhesion by si-β-parvin-transfected ANXA1−/− cells as compared with the mock control. B, graph showing the total number of cells left in the upper chamber and those that have migrated through the collagen into the lower chamber after 24 h. No statistical significance was observed across the number of cells left in the upper chambers, whereas a statistical significance of p < 0.05 was observed between the mock and si-β-parvin and between the mock and ANXA1+/− cells but not between si-β-parvin and ANXA1+/− cells.
DISCUSSION
In this study, we sought to address the role of ANXA1 in mammary gland cells using SILAC-based mass spectrometry. Because proteins directly control almost all cellular processes and poor correlation between mRNA and corresponding protein abundance occurs because of post-transcriptional processing (23), information about proteome level changes is essential in deciphering biological mechanisms. With the recent major advances in the sensitivity and mass accuracy of mass spectrometry measurements, it is now possible to sequence thousands of peptides from complex mixtures in an automated manner and quantify thousands of proteins (comparable with mRNA quantification) using metabolic labeling of cells (SILAC). Here SILAC combined with bioinformatics allowed us to propose possible roles of ANXA1 through determination of changes in functionally relevant subproteomes. It usually would take extensive study and a long time to functionally characterize a given protein through hypothesis-based biological assays. However, by careful analysis of SILAC data and downstream pathways, we can propose direct and indirect roles of ANXA1 in cancer-related biological pathways of DNA damage-response, apoptosis, and cell migration in a much shorter time.
Importantly, we have used normal and nontumor mammary gland cells in our study to understand the importance of ANXA1 in normal cell function. The loss of ANXA1 expression in certain breast tumors points toward a possible functional importance of ANXA1 in tumor initiation. This led us to examine mammary gland cells from mice expressing ANXA1 and mice deficient in ANXA1. Further experiments on tumors from ANXA1+/− and ANXA1−/− mice will be useful for understanding the regulatory roles of ANXA1 in tumor growth and progression. Using bioinformatics analysis of ANXA1-regulated proteins, our initial finding that ANXA1 could positively regulate potent tumor suppressors provides clues of the regulatory effect of ANXA1 in tumor initiation. This can be a further mechanism through which the loss of ANXA1 can induce tumor initiation in mammary ductal carcinoma.
Through bioinformatics analysis of ANXA1-regulated proteins, we identified several down-regulated proteins in the DNA damage response pathway. This pathway is associated with various processes such as cell cycle, proliferation, apoptosis, etc. Yap1, which emerged as the most down-regulated proteins under DNA damage response category, has been shown to be not only able to induce apoptosis via stabilizing the well known tumor suppressor p53 upon DNA damage but also induce p53-regulated apoptosis-inducing protein 1 (p53AIP1), a p73 target gene (24, 25). Strikingly, another protein is p53, which plays vital roles in various important processes such as cell cycle checkpoints, apoptosis (26), transcriptional control (27), and DNA damage (28, 29). We observed a down-regulation of p53 in ANXA1−/− cells at the transcriptional level, implying that ANXA1 may regulate p53 transcription. Another important protein that is positively regulated by ANXA1 is p19ARF, the homologue of p14ARF in humans, which is another important tumor suppressor gene (30) regulating the cell cycle and p53 pathways (31). In addition, DNA ligase I, which is involved in base excision repair (32), is also down-regulated in ANXA1−/− mammary gland cells. The down-regulation of these important proteins involved in DNA damage response pathway suggests that ANXA1-deficient mammary gland cells may have a lack of responses to DNA damage that would compromise the intrinsic genome integrity of the cells. Indeed, our first functional study demonstrated that cells lacking ANXA1 accumulated more damage than ANXA1-expressing cells in response to heat, an inducer of DNA damage, supporting our pathway analysis. The observation that the recovery from DNA damage or DNA repair was also affected in ANXA1−/− cells is in line with our previous study on MCF-7 human breast carcinoma cells, where ANXA1 was shown to protect cells against heat-induced DNA damage (33). Our data also demonstrate that ANXA1 can positively regulate proteins involved in the caspase cascade and apoptosis. This indicates that ANXA1 can modulate the intrinsic apoptotic pathway that relies on the release of intracellular proapoptotic proteins to cleave caspases upon DNA damage (34). Bid, a member of the Bcl2 proapoptotic family, is cleaved to tBid by the initiator caspase-8 upon activation, myristoylated by myristoyl-CoA:protein N-myristoyltransferase before relocalization to mitochondria for the signaling of cytochrome c release (35) from the mitochondria, a characteristic of the intrinsic pathway (36, 37). Previous studies have shown that ANXA1 is a proapoptotic protein that can regulate tumor necrosis factor-related apoptosis-inducing ligand-induced cell death in follicular thyroid carcinoma cells through the regulation of Bcl-2/Bcl-XL-associated death promoter activity and translocation to the mitochondria (38). In addition, neutrophil apoptosis is induced by ANXA1 in a calcium-dependent manner and through the dephosphorylation of Bcl-2/Bcl-XL-associated death promoter (39). Our study functionally verified that ANXA1-expressing cells consistently exhibited lower ROS, and in the absence of ANXA1, there was an accumulation of cellular ROS. This may be linked to the greater DNA damage in unstimulated cells lacking ANXA1, because ROS is known to cause structural alterations to DNA, such as base pair mutations and deletions (40). In addition, ROS, as well as reactive nitrogen species, can modulate the activity of stress response genes. Because oxidative damage was also one of the pathways identified with the down-regulation of proteins in the ANXA1−/− mammary gland cells (data not shown), functional verification using a ROS assay was performed to compare the internal accumulation of ROS in the two mammary gland cell lines. The results showed more intrinsic ROS accumulation, potentially leading to more oxidative damage in the ANXA1−/−, suggesting that ANXA1 potentially plays a protective role in mammary gland cells. This protective role of ANXA1 or lack of responses to DNA damage phenotype in the ANXA1−/− as compared with ANXA1+/− cells was further confirmed by the second functional study using WST assay whereby the cells were subjected to another form of DNA damage (oxidative stress). The WST reagent, which is used for the spectrophotometric quantification of cell proliferation, growth, viability, and chemo-sensitivity in the assay, measures the overall activity of the mitochondrial dehydrogenases and hence indicates the metabolically active cells. Although the overexpression of Yap1 in the ANXA1−/− cells (the down-regulated protein in ANXA1−/− cells with the highest fold change) failed to reverse its intrinsic “damage-accumulated” phenotype, Yap1 was able to rescue its cellular response toward externally introduced oxidative damage significantly, suggesting the direct relationship between these regulated proteins and DNA damage response when ANXA1 is depleted in the mammary gland cells. All the above observations reveal how ANXA1 could be protective via the regulation of proteins involved in the DNA damage response, apoptosis, and caspase cascade pathways in normal mammary gland cells.
The cluster of up-regulated proteins in ANXA−/− cells is mainly associated with tissue remodeling/wound repair process as well as cell adhesion and motility. Wound healing has long been established to be an essential physiological process characterized by four phases including immediate response, inflammatory response, proliferation, migration and contraction phase, and resolution phase (41). Being an anti-inflammatory mediator, ANXA1 contributes to overall resolution by counteracting the process of leukocyte extravasation (42), as well as mediating functions of glucocorticoids, in an attempt to dampen inflammation. It is interesting to note that ANXA1 has been proposed to hold anti-migratory effects because its elevated level induced by the anti-inflammatory property of glucocorticoids inhibits leukocytes migration during inflammation as well as by other mechanisms (43). However, these were in conjunction with the observation that the presence of increased ANXA1 led to inhibition of adhesion of the inflammatory cells to the endothelial (44, 45). One of the mechanisms proposed in this adhesion inhibition is the down-regulation and shedding of a cell adhesion molecule l-selectin on the leukocytes (46, 47).
Down-regulation of ANXA1 has also been shown in carcinogenic models to inhibit cell migration/invasion (15, 48), suggesting that ANXA1 could possibly be pro-migratory in carcinogenesis. Recent studies from our lab and others have demonstrated that ANXA1 is indeed pro-migratory in cancer cells, whereas silencing ANXA1 reduces metastatic capability (15). Altogether, cell motility plays a central part in coordinating normal physiological and pathological processes including wound healing and metastasis. Cell motility/migration summarizes the dynamic integrated system of the “on” and “off” movement of a cell by changes in the expression of proteins involved, particularly those involved in the formation and disassembly of cell adhesion sites (49). Our data confirm that ANXA1 does make a difference in cell motility even in normal cells. In the absence of ANXA1, a number of proteins involved in cellular adhesion are highly up-regulated, making the “on” movement of a cell more difficult. For example, talin, one of the up-regulated proteins identified in our ANXA1−/− mammary gland cells, is especially important in sustaining cell spreading and adhesion (50, 51). β-Parvin, a protein that directly interacts with integrin-linked kinase and is important for stabilizing focal adhesions (52–54), has been shown to be expressed in normal mammary gland but down-regulated in advanced breast cancer and breast cancer cell lines (55). It is the most up-regulated protein with the highest SILAC ratio in our analysis among the proteins involved in cell adhesion/migration. However, we noticed that proteins such as EGFR and MMP14 that promote cell migration (under overexpression conditions) (56–58) are also up-regulated. It has been documented that EGFR activation induces focal adhesion disassembly in mouse fibroblasts (59). Up-regulation of MMP14, a member of MMP family involved in the breakdown of extracellular matrix (60), has been known to promote tumor cell invasions (61–63). Nevertheless, our proteomics data, which provided an overview of the systemic changes in protein expressions, are indicative of the collaborative effect of the overall protein expressions in the context of ANXA1 depletion. Hence, despite the up-regulation of EGFR and MMP14, up-regulation of other proteins involved in cell adhesion, especially those with high SILAC ratios, could possibly override the pro-migratory property of these two proteins. We showed this effect by knocking down the most up-regulated protein β-parvin in ANXA1−/− cells and had observed a reversal of its “sticky” phenotype. These observations suggest that β-parvin, possibly together with other up-regulated proteins involved in focal adhesions, is able to override the pro-migratory capability of EGFR and MMP14, deriving the characteristic of adhesiveness for ANXA1−/− cells. All the above supporting data explain the low migratory index in ANXA1−/− as compared with ANXA1+/−, because these cells are more sticky (Fig. 4A).
In summary, we have shown how the use of SILAC on normal mammary gland epithelial cells from ANXA1-deficient and heterozygous mice has allowed us to further understand the role of ANXA1 in the development of mammary glands. While the combination of SILAC, quantitative mass spectrometry, and bioinformatics analysis provides global levels of protein expression, further pathway analysis of dysregulated proteins elucidates the decisive role of ANXA1 in cellular processes. This information not only adds to the list of known functions of ANXA1 but also helps us understand how perturbation in ANXA1 expression in normal mammary glands may contribute to initiation and progression in breast carcinoma. Finally, these findings lead us to a better understanding of deregulated hubs in human cancers that may eventually aid the development of therapeutic targets for restoring normal cellular function.
Acknowledgments
We thank Ler Siok Ghee, Eric Tan, and Rachel Li (mass spectrometry); Yeh-Shiu Chu (video camera microscope); and Prakash Hande (Comet Imager software). pCDNA-Yap (mouse) construct was a kind gift from RUNX group (Cancer Science Institute Singapore).
Footnotes
* This work was supported by the Agency for Science, Technology and Research (A*STAR). The costs of publication of this article were defrayed in part by the payment of page charges.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- EGFR
- epidermal growth factor receptor
- FBS
- fetal bovine serum
- MMP
- matrix metalloproteinase
- PAI1
- plasminogen activator inhibitor 1
- ROS
- reactive oxygen species
- SILAC
- stable isotope labeling of amino acids in cell culture
- STAT
- signal transducer and activator of transcription
- TIMP
- tissue inhibitor of metalloproteinase
- DMEM
- Dulbecco's modified Eagle's medium
- si-β-parvin
- β-parvin small interfering RNA
- WST
- water soluble Tetrazolium salts.
REFERENCES
- 1. Crompton M. R., Moss S. E., Crumpton M. J. (1988) Diversity in the lipocortin/calpactin family. Cell 55, 1–3 [DOI] [PubMed] [Google Scholar]
- 2. Perretti M., D'Acquisto F. (2009) Annexin A1 and glucocorticoids as effectors of the resolution of inflammation. Nat. Rev. Immunol. 9, 62–70 [DOI] [PubMed] [Google Scholar]
- 3. Wallner B. P., Mattaliano R. J., Hession C., Cate R. L., Tizard R., Sinclair L. K., Foeller C., Chow E. P., Browing J. L., Ramachandran K. L., Pepinsky B. R. (1986) Cloning and expression of human lipocortin, a phospholipase A2 inhibitor with potential anti-inflammatory activity. Nature 320, 77–81 [DOI] [PubMed] [Google Scholar]
- 4. De B. K., Misono K. S., Lukas T. J., Mroczkowski B., Cohen S. (1986) A calcium-dependent 35-kilodalton substrate for epidermal growth factor receptor/kinase isolated from normal tissue. J. Biol. Chem. 261, 13784–13792 [PubMed] [Google Scholar]
- 5. Alldridge L. C., Harris H. J., Plevin R., Hannon R., Bryant C. E. (1999) The annexin protein lipocortin 1 regulates the MAPK/ERK pathway. J. Biol. Chem. 274, 37620–37628 [DOI] [PubMed] [Google Scholar]
- 6. Arur S., Uche U. E., Rezaul K., Fong M., Scranton V., Cowan A. E., Mohler W., Han D. K. (2003) Annexin I is an endogenous ligand that mediates apoptotic cell engulfment. Dev. Cell 4, 587–598 [DOI] [PubMed] [Google Scholar]
- 7. Lim L. H., Pervaiz S. (2007) Annexin 1: The new face of an old molecule. FASEB J. 21, 968–975 [DOI] [PubMed] [Google Scholar]
- 8. Hu N., Flaig M. J., Su H., Shou J. Z., Roth M. J., Li W. J., Wang C., Goldstein A. M., Li G., Emmert-Buck M. R., Taylor P. R. (2004) Comprehensive characterization of annexin I alterations in esophageal squamous cell carcinoma. Clin. Cancer Res. 10, 6013–6022 [DOI] [PubMed] [Google Scholar]
- 9. Xin W., Rhodes D. R., Ingold C., Chinnaiyan A. M., Rubin M. A. (2003) Dysregulation of the annexin family protein family is associated with prostate cancer progression. Am. J. Pathol. 162, 255–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Garcia Pedrero J. M., Fernandez M. P., Morgan R. O., Herrero Zapatero A., Gonzalez M. V., Suarez Nieto C., Rodrigo J. P. (2004) Annexin A1 down-regulation in head and neck cancer is associated with epithelial differentiation status. Am. J. Pathol. 164, 73–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. de Coupade C., Gillet R., Bennoun M., Briand P., Russo-Marie F., Solito E. (2000) Annexin 1 expression and phosphorylation are upregulated during liver regeneration and transformation in antithrombin III SV40 T large antigen transgenic mice. Hepatology 31, 371–380 [DOI] [PubMed] [Google Scholar]
- 12. Bai X. F., Ni X. G., Zhao P., Liu S. M., Wang H. X., Guo B., Zhou L. P., Liu F., Zhang J. S., Wang K., Xie Y. Q., Shao Y. F., Zhao X. H. (2004) Overexpression of annexin 1 in pancreatic cancer and its clinical significance. World J. Gastroenterol. 10, 1466–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yi M., Schnitzer J. E. (2009) Impaired tumor growth, metastasis, angiogenesis and wound healing in annexin A1-null mice. Proc. Natl. Acad. Sci. U.S.A. 106, 17886–17891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. de Graauw M., van Miltenburg M. H., Schmidt M. K., Pont C., Lalai R., Kartopawiro J., Pardali E., Le Dévédec S. E., Smit V. T., van der Wal A., Van't Veer L. J., Cleton-Jansen A. M., ten Dijke P., van de Water B. (2010) Annexin A1 regulates TGF-β signaling and promotes metastasis formation of basal-like breast cancer cells. Proc. Natl. Acad. Sci. U.S.A. 107, 6340–6345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bist P., Leow S. C., Phua Q. H., Shu S., Zhuang Q., Loh W. T., Nguyen T. H., Zhou J. B., Hooi S. C., Lim L. H. (2011) Annexin-1 interacts with NEMO and RIP1 to constitutively activate IKK complex and NF-κB: Implication in breast cancer metastasis. Oncogene 30, 3174–3185 [DOI] [PubMed] [Google Scholar]
- 16. Rhee D. K., Park S. H., Jang Y. K. (2008) Molecular signatures associated with transformation and progression to breast cancer in the isogenic MCF10 model. Genomics 92, 419–428 [DOI] [PubMed] [Google Scholar]
- 17. Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 18. Shevchenko A., Tomas H., Havlis J., Olsen J. V., Mann M. (2006) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1, 2856–2860 [DOI] [PubMed] [Google Scholar]
- 19. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 20. Cox J., Matic I., Hilger M., Nagaraj N., Selbach M., Olsen J. V., Mann M. (2009) A practical guide to the MaxQuant computational platform for SILAC-based quantitative proteomics. Nat. Protoc. 4, 698–705 [DOI] [PubMed] [Google Scholar]
- 21. Beere H. M., Wolf B. B., Cain K., Mosser D. D., Mahboubi A., Kuwana T., Tailor P., Morimoto R. I., Cohen G. M., Green D. R. (2000) Heat-shock protein 70 inhibits apoptosis by preventing recruitment of procaspase-9 to the Apaf-1 apoptosome. Nat. Cell Biol. 2, 469–475 [DOI] [PubMed] [Google Scholar]
- 22. Takahashi A., Matsumoto H., Nagayama K., Kitano M., Hirose S., Tanaka H., Mori E., Yamakawa N., Yasumoto J., Yuki K., Ohnishi K., Ohnishi T. (2004) Evidence for the involvement of double-strand breaks in heat-induced cell killing. Cancer Res. 64, 8839–8845 [DOI] [PubMed] [Google Scholar]
- 23. Greenbaum D., Colangelo C., Williams K., Gerstein M. (2003) Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biol. 4, 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Strano S., Monti O., Pediconi N., Baccarini A., Fontemaggi G., Lapi E., Mantovani F., Damalas A., Citro G., Sacchi A., Del Sal G., Levrero M., Blandino G. (2005) The transcriptional coactivator Yes-associated protein drives p73 gene-target specificity in response to DNA Damage. Mol. Cell 18, 447–459 [DOI] [PubMed] [Google Scholar]
- 25. Levy D., Adamovich Y., Reuven N., Shaul Y. (2007) The Yes-associated protein 1 stabilizes p73 by preventing Itch-mediated ubiquitination of p73. Cell Death Differ. 14, 743–751 [DOI] [PubMed] [Google Scholar]
- 26. Kastan M. B., Canman C. E., Leonard C. J. (1995) p53, cell cycle control and apoptosis: Implications for cancer. Cancer Metastasis Rev. 14, 3–15 [DOI] [PubMed] [Google Scholar]
- 27. Wei C. L., Wu Q., Vega V. B., Chiu K. P., Ng P., Zhang T., Shahab A., Yong H. C., Fu Y., Weng Z., Liu J., Zhao X. D., Chew J. L., Lee Y. L., Kuznetsov V. A., Sung W. K., Miller L. D., Lim B., Liu E. T., Yu Q., Ng H. H., Ruan Y. (2006) A global map of p53 transcription-factor binding sites in the human genome. Cell 124, 207–219 [DOI] [PubMed] [Google Scholar]
- 28. Kastan M. B., Onyekwere O., Sidransky D., Vogelstein B., Craig R. W. (1991) Participation of p53 protein in the cellular response to DNA damage. Cancer Res. 51, 6304–6311 [PubMed] [Google Scholar]
- 29. García-Cao I., García-Cao M., Martín-Caballero J., Criado L. M., Klatt P., Flores J. M., Weill J. C., Blasco M. A., Serrano M. (2002) “Super p53” mice exhibit enhanced DNA damage response, are tumor resistant and age normally. EMBO J. 21, 6225–6235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sharpless N. E., DePinho R. A. (1999) The INK4A/ARF locus and its two gene products. Curr. Opin. Genet. Dev. 9, 22–30 [DOI] [PubMed] [Google Scholar]
- 31. Sherr C. J. (1998) Tumor surveillance via the ARF-p53 pathway. Genes Dev. 12, 2984–2991 [DOI] [PubMed] [Google Scholar]
- 32. Frosina G., Fortini P., Rossi O., Carrozzino F., Raspaglio G., Cox L. S., Lane D. P., Abbondandolo A., Dogliotti E. (1996) Two pathways for base excision repair in mammalian cells. J. Biol. Chem. 271, 9573–9578 [DOI] [PubMed] [Google Scholar]
- 33. Nair S., Hande M. P., Lim L. H. (2010) Annexin-1 protects MCF7 breast cancer cells against heat-induced growth arrest and DNA damage. Cancer Lett. 294, 111–117 [DOI] [PubMed] [Google Scholar]
- 34. Fadeel B., Ottosson A., Pervaiz S. (2008) Big wheel keeps on turning: Apoptosome regulation and its role in chemoresistance. Cell Death Differ. 15, 443–452 [DOI] [PubMed] [Google Scholar]
- 35. Zha J., Weiler S., Oh K. J., Wei M. C., Korsmeyer S. J. (2000) Posttranslational N-myristoylation of BID as a molecular switch for targeting mitochondria and apoptosis. Science 290, 1761–1765 [DOI] [PubMed] [Google Scholar]
- 36. Luo X., Budihardjo I., Zou H., Slaughter C., Wang X. (1998) Bid, a Bcl2 interacting protein, mediates cytochrome c release from mitochondria in response to activation of cell surface death receptors. Cell 94, 481–490 [DOI] [PubMed] [Google Scholar]
- 37. Li H., Zhu H., Xu C. J., Yuan J. (1998) Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell 94, 491–501 [DOI] [PubMed] [Google Scholar]
- 38. Petrella A., Festa M., Ercolino S. F., Zerilli M., Stassi G., Solito E., Parente L. (2005) Induction of annexin-1 during TRAIL-induced apoptosis in thyroid carcinoma cells. Cell Death Differ. 12, 1358–1360 [DOI] [PubMed] [Google Scholar]
- 39. Solito E., Kamal A., Russo-Marie F., Buckingham J. C., Marullo S., Perretti M. (2003) A novel calcium-dependent proapoptotic effect of annexin 1 on human neutrophils. FASEB J. 17, 1544–1546 [DOI] [PubMed] [Google Scholar]
- 40. Wiseman H., Halliwell B. (1996) Damage to DNA by reactive oxygen and nitrogen species: Role in inflammatory disease and progression to cancer. Biochem. J. 313, 17–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shaw T. J., Martin P. (2009) Wound repair at a glance. J. Cell Sci. 122, 3209–3213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Perretti M., Gavins F. N. (2003) Annexin 1: An endogenous anti-inflammatory protein. News Physiol. Sci. 18, 60–64 [DOI] [PubMed] [Google Scholar]
- 43. Parente L., Solito E. (2004) Annexin 1: More than an anti-phospholipase protein. Inflamm. Res. 53, 125–132 [DOI] [PubMed] [Google Scholar]
- 44. Lim L. H., Solito E., Russo-Marie F., Flower R. J., Perretti M. (1998) Promoting detachment of neutrophils adherent to murine postcapillary venules to control inflammation: Effect of lipocortin 1. Proc. Natl. Acad. Sci. U.S.A. 95, 14535–14539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mancuso F., Flower R. J., Perretti M. (1995) Leukocyte transmigration, but not rolling or adhesion, is selectively inhibited by dexamethasone in the hamster post-capillary venule. Involvement of endogenous lipocortin 1. J. Immunol. 155, 377–386 [PubMed] [Google Scholar]
- 46. Strausbaugh H. J., Rosen S. D. (2001) A potential role for annexin 1 as a physiologic mediator of glucocorticoid-induced l-selectin shedding from myeloid cells. J. Immunol. 166, 6294–6300 [DOI] [PubMed] [Google Scholar]
- 47. de Coupade C., Solito E., Levine J. D. (2003) Dexamethasone enhances interaction of endogenous annexin 1 with l-selectin and triggers shedding of l-selectin in the monocytic cell line U-937. Br. J. Pharmacol. 140, 133–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Babbin B. A., Lee W. Y., Parkos C. A., Winfree L. M., Akyildiz A., Perretti M., Nusrat A. (2006) Annexin I regulates SKCO-15 cell invasion by signaling through formyl peptide receptors. J. Biol. Chem. 281, 19588–19599 [DOI] [PubMed] [Google Scholar]
- 49. Lauffenburger D. A., Horwitz A. F. (1996) Cell migration: A physically integrated molecular process. Cell 84, 359–369 [DOI] [PubMed] [Google Scholar]
- 50. Nieves B., Jones C. W., Ward R., Ohta Y., Reverte C. G., LaFlamme S. E. (2010) The NPIY motif in the integrin β1 tail dictates the requirement for talin-1 in outside-in signaling. J. Cell Sci. 123, 1216–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhang X., Jiang G., Cai Y., Monkley S. J., Critchley D. R., Sheetz M. P. (2008) Talin depletion reveals independence of initial cell spreading from integrin activation and traction. Nat. Cell Biol. 10, 1062–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yamaji S., Suzuki A., Kanamori H., Mishima W., Yoshimi R., Takasaki H., Takabayashi M., Fujimaki K., Fujisawa S., Ohno S., Ishigatsubo Y. (2004) Affixin interacts with α-actinin and mediates integrin signaling for reorganization of F-actin induced by initial cell-substrate interaction. J. Cell Biol. 165, 539–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Matsuda C., Kameyama K., Suzuki A., Mishima W., Yamaji S., Okamoto H., Nishino I., Hayashi Y. K. (2008) Affixin activates Rac1 via βPIX in C2C12 myoblast. FEBS Lett. 582, 1189–1196 [DOI] [PubMed] [Google Scholar]
- 54. Sepulveda J. L., Wu C. (2006) The parvins. Cell Mol Life Sci 63, 25–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mongroo P. S., Johnstone C. N., Naruszewicz I., Leung-Hagesteijn C., Sung R. K., Carnio L., Rustgi A. K., Hannigan G. E. (2004) β-Parvin inhibits integrin-linked kinase signaling and is downregulated in breast cancer. Oncogene 23, 8959–8970 [DOI] [PubMed] [Google Scholar]
- 56. Zaman M. H., Trapani L. M., Sieminski A. L., Mackellar D., Gong H., Kamm R. D., Wells A., Lauffenburger D. A., Matsudaira P. (2006) Migration of tumor cells in 3D matrices is governed by matrix stiffness along with cell-matrix adhesion and proteolysis. Proc. Natl. Acad. Sci. U.S.A. 103, 10889–10894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Andl C. D., Mizushima T., Oyama K., Bowser M., Nakagawa H., Rustgi A. K. (2004) EGFR-induced cell migration is mediated predominantly by the JAK-STAT pathway in primary esophageal keratinocytes. Am. J. Physiol. Gastrointest. Liver Physiol. 287, G1227–G1237 [DOI] [PubMed] [Google Scholar]
- 58. Verbeek B. S., Adriaansen-Slot S. S., Vroom T. M., Beckers T., Rijksen G. (1998) Overexpression of EGFR and c-erbB2 causes enhanced cell migration in human breast cancer cells and NIH3T3 fibroblasts. FEBS Lett. 425, 145–150 [DOI] [PubMed] [Google Scholar]
- 59. Xie H., Pallero M. A., Gupta K., Chang P., Ware M. F., Witke W., Kwiatkowski D. J., Lauffenburger D. A., Murphy-Ullrich J. E., Wells A. (1998) EGF receptor regulation of cell motility: EGF induces disassembly of focal adhesions independently of the motility-associated PLCγ signaling pathway. J. Cell Sci. 111, 615–624 [DOI] [PubMed] [Google Scholar]
- 60. Nagase H., Woessner J. F., Jr. (1999) Matrix metalloproteinases. J. Biol. Chem. 274, 21491–21494 [DOI] [PubMed] [Google Scholar]
- 61. Nagakawa O., Murakami K., Yamaura T., Fujiuchi Y., Murata J., Fuse H., Saiki I. (2000) Expression of membrane-type 1 matrix metalloproteinase (MT1-MMP) on prostate cancer cell lines. Cancer Lett. 155, 173–179 [DOI] [PubMed] [Google Scholar]
- 62. Nomura H., Sato H., Seiki M., Mai M., Okada Y. (1995) Expression of membrane-type matrix metalloproteinase in human gastric carcinomas. Cancer Res. 55, 3263–3266 [PubMed] [Google Scholar]
- 63. Singh S., Singh U. P., Grizzle W. E., Lillard J. W., Jr. (2004) CXCL12-CXCR4 interactions modulate prostate cancer cell migration, metalloproteinase expression and invasion. Lab. Invest. 84, 1666–1676 [DOI] [PubMed] [Google Scholar]