Abstract
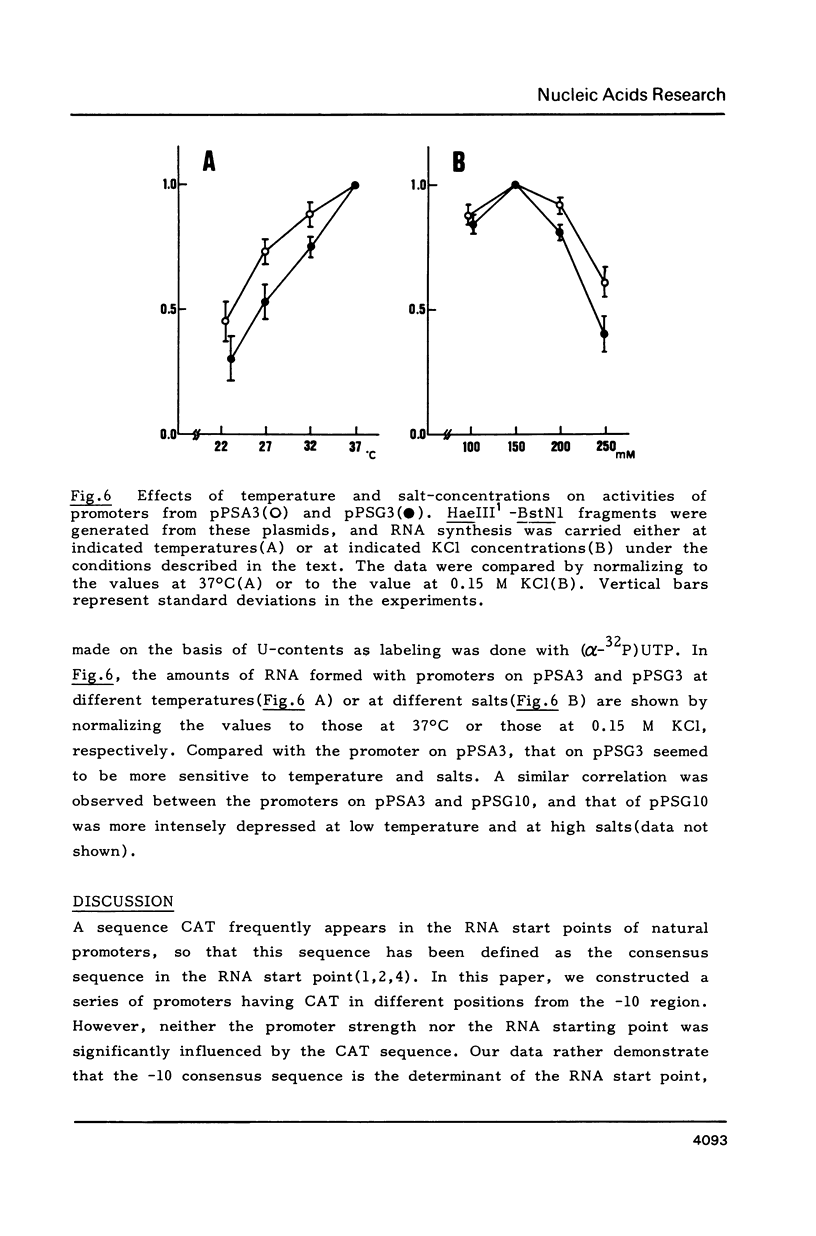
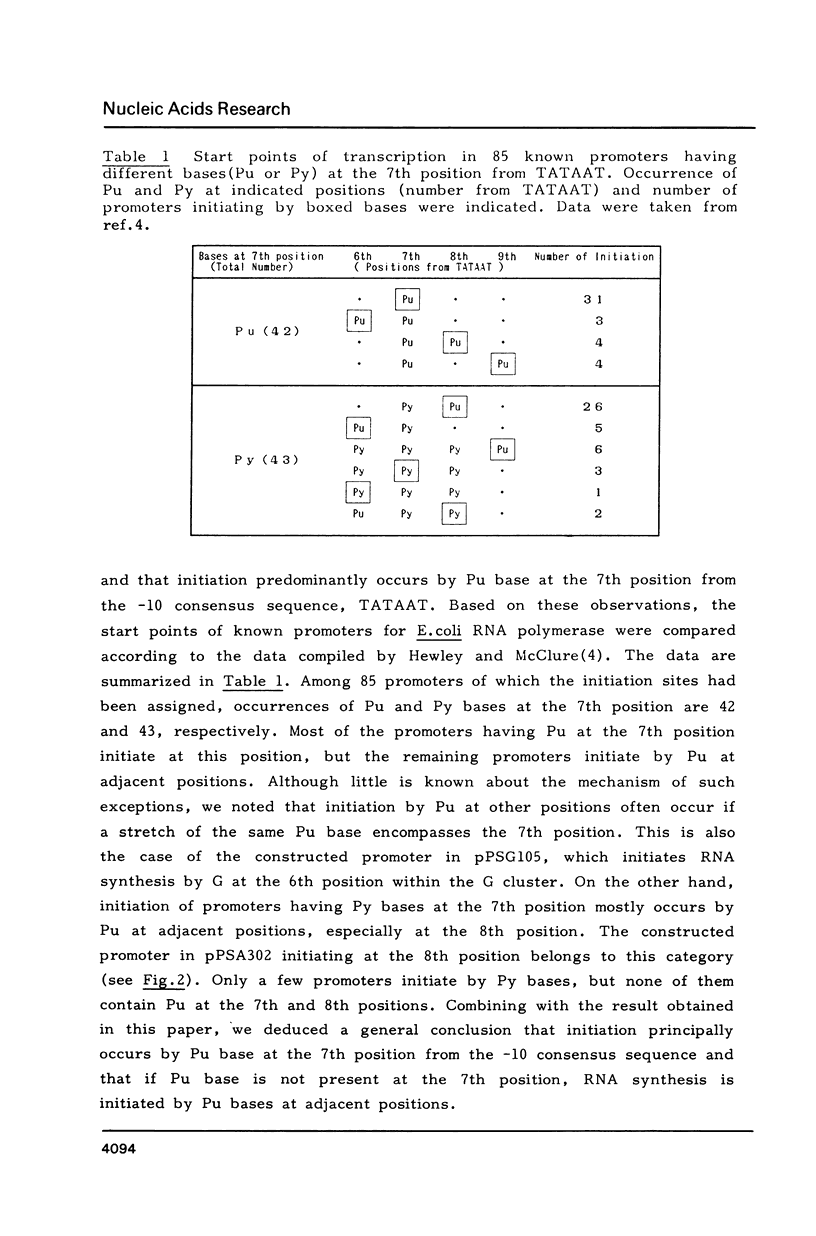
Starting from a synthetic E. coli promoter with the consensus sequences at -35 and -10 regions, a sequence CAT frequently occurred in the RNA start points of natural promoters was introduced in the downstream of the consensus sequences, and the sequences around the RNA start points as well as the relative positions of CAT from the consensus sequences were altered. Analysis of the RNA start points and strength of these synthetic promoters in an in vitro transcription system provided evidence that the RNA start point was principally fixed by distance from the -10 consensus sequence (TATAAT). Neither the promoter strength nor the RNA start point was significantly influenced by the CAT sequence. The sequences around the RNA start points rather seemed to exert influence on the response of promoter to temperature and salts.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aoyama T., Takanami M., Ohtsuka E., Taniyama Y., Marumoto R., Sato H., Ikehara M. Essential structure of E. coli promoter: effect of spacer length between the two consensus sequences on promoter function. Nucleic Acids Res. 1983 Sep 10;11(17):5855–5864. doi: 10.1093/nar/11.17.5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn G. T., Wells R. D. The leftward promoter of bacteriophage lambda. Structure, biological activity, and influence by adjacent regions. J Biol Chem. 1981 Feb 25;256(4):2003–2009. [PubMed] [Google Scholar]
- Klein R. D., Wells R. D. Effects of neighboring DNA homopolymers on the biochemical and physical properties of the Escherichia coli lactose promoter. I. Cloning and characterization studies. J Biol Chem. 1982 Nov 10;257(21):12954–12961. [PubMed] [Google Scholar]
- Melnikova A. F., Beabealashvilli R., Mirzabekov A. D. A study of unwinding of DNA and shielding of the DNA grooves by RNA polymerase by using methylation with dimethylsulphate. Eur J Biochem. 1978 Mar;84(1):301–309. doi: 10.1111/j.1432-1033.1978.tb12169.x. [DOI] [PubMed] [Google Scholar]
- Munson L. M., Mandecki W., Caruthers M. H., Reznikoff W. S. Oligonucleotide mutagenesis of the lacPUV5 promoter. Nucleic Acids Res. 1984 May 11;12(9):4011–4017. doi: 10.1093/nar/12.9.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Siebenlist U. RNA polymerase unwinds an 11-base pair segment of a phage T7 promoter. Nature. 1979 Jun 14;279(5714):651–652. doi: 10.1038/279651a0. [DOI] [PubMed] [Google Scholar]
- Siebenlist U., Simpson R. B., Gilbert W. E. coli RNA polymerase interacts homologously with two different promoters. Cell. 1980 Jun;20(2):269–281. doi: 10.1016/0092-8674(80)90613-3. [DOI] [PubMed] [Google Scholar]
- Sugimoto K., Sugisaki H., Okamoto T., Takanami M. Studies on bacteriophage fd DNA. IV. The sequence of messenger RNA for the major coat protein gene. J Mol Biol. 1977 Apr 25;111(4):487–507. doi: 10.1016/s0022-2836(77)80065-x. [DOI] [PubMed] [Google Scholar]
- Travers A. A. Conserved features of coordinately regulated E. coli promoters. Nucleic Acids Res. 1984 Mar 26;12(6):2605–2618. doi: 10.1093/nar/12.6.2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace R. B., Johnson M. J., Suggs S. V., Miyoshi K., Bhatt R., Itakura K. A set of synthetic oligodeoxyribonucleotide primers for DNA sequencing in the plasmid vector pBR322. Gene. 1981 Dec;16(1-3):21–26. doi: 10.1016/0378-1119(81)90057-3. [DOI] [PubMed] [Google Scholar]
- Wang J. C., Jacobsen J. H., Saucier J. M. Physiochemical studies on interactions between DNA and RNA polymerase. Unwinding of the DNA helix by Escherichia coli RNA polymerase. Nucleic Acids Res. 1977;4(5):1225–1241. doi: 10.1093/nar/4.5.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youderian P., Bouvier S., Susskind M. M. Sequence determinants of promoter activity. Cell. 1982 Oct;30(3):843–853. doi: 10.1016/0092-8674(82)90289-6. [DOI] [PubMed] [Google Scholar]
- von Hippel P. H., Bear D. G., Morgan W. D., McSwiggen J. A. Protein-nucleic acid interactions in transcription: a molecular analysis. Annu Rev Biochem. 1984;53:389–446. doi: 10.1146/annurev.bi.53.070184.002133. [DOI] [PubMed] [Google Scholar]