Abstract
There is a pressing and continued need for improved predictive power in preclinical pharmaceutical toxicology assessment as substantial numbers of drugs are still removed from the market, or from late-stage development, because of unanticipated issues of toxicity. In recent years a number of consortia have been formed with a view to integrating -omics molecular profiling strategies to increase the sensitivity and predictive power of preclinical toxicology evaluation. In this study we report on the LC-MS based proteomic analysis of the effects of the hepatotoxic compound EMD 335823 on liver from rats using an integrated discovery to targeted proteomics approach. This compound was one of a larger panel studied by a variety of molecular profiling techniques as part of the InnoMed PredTox Consortium. Label-free LC-MS analysis of hepatotoxicant EMD 335823 treated animals revealed only moderate correlation of individual protein expression with changes in mRNA expression observed by transcriptomic analysis of the same liver samples. Significantly however, analysis of the protein and transcript changes at the pathway level revealed they were in good agreement. This higher level analysis was also consistent with the previously suspected PPARα activity of the compound. Subsequently, a panel of potential biomarkers of liver toxicity was assembled from the label-free LC-MS proteomics discovery data, the previously acquired transcriptomics data and selected candidates identified from the literature. We developed and then deployed optimized selected reaction monitoring assays to undertake multiplexed measurement of 48 putative toxicity biomarkers in liver tissue. The development of the selected reaction monitoring assays was facilitated by the construction of a peptide MS/MS spectral library from pooled control and treated rat liver lysate using peptide fractionation by strong cation exchange and off-gel electrophoresis coupled to LC-MS/MS. After iterative optimization and quality control of the selected reaction monitoring assay panel, quantitative measurements of 48 putative biomarkers in the liver of EMD 335823 treated rats were carried out and this revealed that the panel is highly enriched for proteins modulated significantly on drug treatment/hepatotoxic insult. This proof-of-principle study provides a roadmap for future large scale pre-clinical toxicology biomarker verification studies whereby putative toxicity biomarkers assembled from multiple disparate sources can be evaluated at medium-high throughput by targeted MS.
The inability of current preclinical toxicology evaluation methods to predict early, and with good accuracy, that a drug candidate will have to be removed from development (or from the market) because of toxicicity/safety issues is a serious bottleneck in the drug development pipeline (1). Novel omics profiling technologies have the potential to provide more effective preclinical predictive models for toxicity (2). By performing detailed and comprehensive molecular profiling of animal or cell-based models that have been exposed to known toxic insults, it should be possible to catalog the spectrum of molecular changes that cause or accompany a particular mechanism of toxicity. It is reasonable to assume that molecular changes underlying, or induced by, toxicologic mechanisms will be manifested at earlier time points and at lower dose levels than are required for classical toxicology evaluation endpoints. Hence, the basic premise of preclinical predictive systems toxicology is to perform molecular profiling experiments for a range of compounds, potentially hundreds, displaying various toxicities and deriving biomarker signatures related to given toxicological mechanisms or endpoints. This approach has recently begun to enjoy some success with a panel of seven urinary protein biomarkers for nephrotoxicity having been deemed acceptable in the context of nonclinical drug development for the detection of acute drug-induced kidney toxicity in a joint evaluation by the Food and Drug Administration (FDA) and European Medicines Agency (EMEA) (3). Consortia-based efforts have been established to systematically investigate drug-induced liver, and other toxicities, however, progress has been less apparent (4, 5). The integrated EU Framework 6 Project InnoMed PredTox (6) was such a consortium, consisting of 14 pharmaceutical companies, three universities, and two technology providers, focused on assessing the potential of combining the molecular profiling techniques of transcriptomics, proteomics, and metabolomics with conventional toxicology measurements, to provide improved decision making in preclinical safety evaluation. The general conclusion from the PredTox consortium was that to some extent the “omics” technologies can help toxicologists to make better informed decisions during exploratory toxicological studies, however, integrating data sets from different molecular profiling technologies proved problematic (6). In particular, the proteomics studies performed in the context of the PredTox project were restricted to the use of SELDI (7) and two-dimensional electrophoresis (8) approaches which, although undertaken rigorously, displayed limited proteome coverage and identified relatively small numbers of modulated proteins. This limitation significantly hampered efforts aimed at integrated analysis of protein expression changes with the transcriptomics or metabolomics data sets. In order to address this issue with a view to increasing proteome coverage and then subsequently providing methods for robust and sensitive protein quantification in future studies of this type, we undertook a reanalysis of livers from rats treated with one of the PredTox compounds, EMD 335823, using LC-MS-based proteomics methods. For more specific information on the outcome of previous detailed studies, in particular with regard to gene expression profiling and mechanistic analyses, we refer the reader to a summary of the InnoMed PredTox Consortium activities surround EMD 335823 (9). The study described herein is a re-analysis of archived liver samples from the same in vivo dosing experiments. First, a discovery proteomics screen of liver from hepatotoxicant treated rats by MS1 intensity-based label-free liquid chromatography (LC)-MS (10) was performed. Second, a panel of putative biomarkers assembled from the label-free LC-MS study, a previous transcriptomics study (9), and literature sources (11), were measured simultaneously in a larger cohort of hepatotoxicant treated rat livers by targeted MS using selected reaction monitoring (12).
The goals of this study were to determine the feasibility of using LC-MS-based proteomics to augment and facilitate large-scale efforts in the direction of preclinical toxicology evaluation and systems toxicology. There are essentially two ways which advanced proteomics methodologies could contribute to this field. The first concerns the elucidation of biochemical and mechanistic aspects of toxicological phenotypes. The second is in the determination of biomarkers associated with the prediction of a given toxicological event. Although tissue-based analysis of the target organ of toxicity (as was carried out in this study) is directly appropriate for the first goal, the second goal remains a more complex prospect. In the ideal case a biomarker would be directly measureable in an accessible body fluid to facilitate longitudinal measurements, in addition to the possibility of transferring such a biomarker from preclinical to clinical utility, however, the difficulties associated with plasma/urine biomarker discovery and validation are well described (13). The utility of tissue-based biomarkers is less clear as histological evaluation is routinely applied and, as such, a clear sensitivity benefit for novel tissue-based biomarkers over histopathology would have to be demonstrated. A more likely route may be the transfer of promising biomarkers candidates from tissue to plasma-based assays in the medium-long term.
MS1 intensity-based label-free LC-MS (10) (as opposed to spectral counting-based label-free LC-MS (14)) has emerged as an attractive alternative to isotope labeling-based strategies for preclinical or clinical studies where relatively large numbers of samples need to be analyzed and integrating metabolic or chemical labeling into the sample preparation may be problematic. Although the achievable proteome coverage is not as high with label-free approaches as can be realized with isotope labeling which routinely incorporate extensive fractionation, substantial numbers of proteins can be quantified and identified by additionally employing a directed MS/MS approach incorporating re-injection of samples with inclusion lists to supplement the peptide identifications acquired in data dependent analyses (15, 16). The data analysis associated with the MS1 label-free approach, in particular the alignment of MS1 feature maps, remains a challenge. In addition, the success of this approach rests on maintaining stability and reproducibility in the chromatography, as well as mass accuracy and intensity measurements. Although recent advances in software and instrument robustness have made the label-free LC-MS approach feasible for small-medium scale discovery proteomics study with preclinical or clinical samples, the use of this approach in very large-scale studies is likely to be complex and currently difficult to achieve. Verification of biomarker panels for preclinical toxicology evaluation will most likely require the analysis of large numbers of compounds with well characterized toxicological properties and, as such, a technology platform that can reproducibly and sensitively measure proteins in a targeted fashion is required.
The use of selected reaction monitoring (SRM)1 for proteomics studies has emerged in recent years as a powerful method for sensitive, robust, and increasingly routine targeted quantification of proteins in complex biological samples (12, 17). These characteristics have led to wide adoption and development of the technique for the targeted quantification of discrete sets of proteins for studies in both model systems (18–20) and clinical samples (21–23), as well as studies specifically focused on drug toxicology (24). Initially the development of reliable SRM assays was very time consuming and manual, however, in recent years methods and software have been developed that have substantially decreased the time required for the development of robust and sensitive SRM assays (25–28). This means that SRM assays targeting tens to hundreds of proteins can be developed in a matter of weeks and deployed indefinitely in large-scale targeted proteomics studies. The SRM method is also inherently flexible and can be easily refined to integrate additional new proteins into an assay panel that arise, for example, from ongoing discovery studies, or to remove proteins with low discriminatory power that are no longer required.
In this study SRM was utilized as a means to undertake the measurement of a panel of putative biomarker candidates assembled from disparate sources, namely a discovery proteomics screen by label-free LC-MS, a previous transcriptomics study where the same liver samples were analyzed (9), and hepatotoxicity biomarker candidates taken from literature (11), into a single assay panel of 48 rat liver proteins. The primary set of proteins included in the SRM panel was derived from an MS1 intensity-based label-free LC-MS discovery proteomics study of a subset of the hepatotoxicant treated rat livers also conducted in the context of this study. These studies provide a significant proof-of-principle demonstration for future preclinical toxicology studies whereby (1) label-free LC-MS can provide putative biomarkers and mechanistic information on the toxicological insult, and, (2) putative biomarkers from multiple sources can be integrated into an SRM assay panel that can be deployed at medium-high throughput for large scale verification studies involving substantial numbers of well-characterized toxicants, and later for more sensitive toxicology evaluation for drugs under early development.
EXPERIMENTAL PROCEDURES
In Vivo Drug Treatment Study Design
Details of the compounds, animal studies, and classical toxicology evaluation procedures as well as further details of the InnoMed PredTox Consortium methods have been published previously (7, 29–31). The experiments described here focused on a single compound study in the InnoMed PredTox Consortium designated FP005ME. In this study Wistar rats were treated daily for 3 or 14 days with vehicle, a nontoxic dose (15 mg/kg), or a high dose (350 mg/kg) of the known hepatotoxic compound (EMD 335823, see supplemental Fig. S1 for chemical structure) the latter dose chosen to induce significant hepatotoxicity (liver necrosis, fibrosis, and bile duct necrosis/hyperplasia). Each drug treatment group contained five rats with a total of 30 rats across the six treatment groups analyzed. Animal experimentation plans underwent an ethical review within the project and were carried out according to the local regulations and permissions of each participant company and in accordance with the guidelines of the European Council on Experimental Animal Care. Additional details on the in vivo study design are given the supplementary information and supplemental Tables S1, S2, S3, and S4.
Protein Extraction From Liver and Digestion With Trypsin
Protein extraction from liver samples was as previously described (7). Briefly, the frozen left lateral lobe of the liver (100 ± 50 mg) was ground with a mortar and pestle under liquid nitrogen. Powdered tissue was transferred into a tube containing 125 μl lysis buffer I (10 mm Tris/pH 7.0.5, 1 mm EDTA, 0.2 m sucrose, Benzonase 25 U/μl (Calbiochem, San Diego, CA), protease inhibitor mixture (Set III - 100 mm AEBSF hydrochloride, 80 μm aprotinin (bovine lung), 5 mm bestatin, 1.5 mm E-64, 2 mm leupeptin hemisulfate, 1 mm pepstatin A, Calbiochem) and completely suspended. Afterward, 875 μl lysis buffer II (7 m urea, 2 m thiourea, 4% (w/v) 3-[(3-cholamidopropyl)dimethylammonio]propanesulfonate, 40 mm dithiothreitol, 20 mm spermine) was added and pipeted 30 times to aid suspension. The protein extracts were mixed at room temperature on a rotary shaker at 500 rpm for 1 h to ensure complete cell lysis and solubilization of protein. To separate membranous components and other insoluble debris, samples were ultracentrifuged for 30 min at 10 °C and 74,000 × g. The supernatant was aliquoted and stored at −80 °C.
Two hundred micrograms of the liver protein extracts were reduced (5 mm TCEP, 20 min, room temperature) and alkylated (10 mm iodoacetamide, 30 min room temperature in the dark) before precipitation with six volumes of acetone for 2 h at −20 °C. The samples were centrifuged for 10 min at 5000 × g and 4 °C, and the pellets were resolubilized in Rapigest 1.3% (w/v), 50 mm NH3HCO3 or 5% (w/v) trifluoroethanol, 50 mm NH3HCO3 and incubated with 2 μg sequencing grade modified porcine trypsin, (overnight, 37 °C). Rapigest containing samples were acidified with 2% (v/v) formic acid (4 h, RT) and centrifuged (13,000 × g, 10 min) to remove the Rapigest hydrolysis products. The samples were transferred to clean tubes, evaporated to dryness in a centrifugal evaporator (60 °C, 150 × g), and resuspended in 100 μl of 0.1% (v/v) formic acid, 3% (v/v) acetonitrile prior to injection.
Label-Free LC-MS
Samples were analyzed using a 6520 quadrupole-time of flight (Q-TOF) mass spectrometer connected online to a 1200 Series nanoflow high performance liquid chromatograph (HPLC) via an orthogonal spray HPLC-Chip/MS interface (Agilent Technologies). Digested protein (3 μg) was chromatographed with a 90 min gradient from 3% (v/v) acetonitrile, 0.1% (v/v) formic acid to 40% (v/v) acetonitrile, 0.1% formic acid using a HPLC-Chip equipped with a 75 μm × 150 mm, 5 μm C-18 300SB-Zorbax analytical column and a 160 nl Zorbax 300SB-C18 5 μm enrichment column. The mass spectrometer was operated using two duty cycles. Each of the biological replicates (designated A31-A35 in the vehicle control group and A41-A45 in the treatment group), as well as six replicates of a pool of these samples to measure technical variance, were injected using a duty cycle intended to maximize the information content of the MS1 scans. This duty cycle included 1 MS scan for 333 milliseconds, and two MS/MS scans for 333 milliseconds each. Setting the MS1 scan to 333 milliseconds allowed for summing of 3212 TOF transients. Reducing the number of MS/MS spectra acquired to two resulted in a duty cycle lasting only ∼1 s ensuring a large number of data points across the chromatographic elution of each peptide. The pooled sample was injected a further two times using a duty cycle intended to maximize the number of peptide MS/MS spectra acquired, and hence the number of peptide/protein identifications. This standard “shotgun” duty cycle included 1 MS scan for 333 milliseconds, and 8 MS/MS scans for 333 milliseconds each. All spectra were collected in profile mode. After the initial round of data analysis, the pooled sample was injected a further 3 times using targeted inclusion lists in an attempt to identify the most statistically discriminating peptides that were not identified in the initial injections (32).
Analysis of Label-Free LC-MS Data Set
Data files from the label-free LC-MS analysis were converted from the Agilent proprietary .d format to .mzXML using the Trapper converter distributed as part of the Trans Proteomic Pipeline (40). The mzXML files were imported into Progenesis LC-MS (v2.6 - Nonlinear Dynamics, UK) at which point features were automatically detected. A representative reference run was selected from the pooled sample injections and all other runs were aligned to the reference run in a pair wise fashion. Alignment was achieved by the manual placement of 15–30 alignment vectors followed by use of the automated alignment algorithm within Progenesis LC-MS. The alignment was inspected for accuracy and additional manual vectors were added if required. Features were filtered for charge state (charge state 2–7 accepted), and retention time in the peptide elution part of the gradient (mobile phase composition ∼8–35% acetonitrile). Univariate (ANOVA) and multivariate (principal components analysis) statistical analyses were performed using Progenesis LC-MS. To integrate MS features with protein identities, MS/MS spectra from all runs in the analysis were centroided and exported as a single .mgf file and searched against the International Protein Index (IPI) Rat database (version 3.53 concatenated with common laboratory contaminant proteins and reverse entries for false identification rate estimation) using Mascot (v2.2 - Matrix Sciences) (33). The results in Mascot .xml format were imported back into Progenesis LC-MS where the peptide identifications were mapped back onto the quantitative peptide data. Peptide level data were rolled up to protein level using the Protein View in Progenesis LC-MS and summary statistics (fold change, ANOVA p value, q-value, and power) were calculated. Proteins with a fold change >1.5 and ANOVA p value < 0.05 were considered significant. False identification rates at the peptide and protein levels were calculated with respect to decoy database hits (58). Pathway analyses for label-free LC-MS as well as the previously acquired gene expression microarray data (9) were performed using Ingenuity Pathway Analysis (Ingenuity Systems, Redwood City, CA).
Acquisition and Construction of MS/MS Spectral Libraries to Aid SRM Development
Multidimensional separations in the form of OFFGEL electrophoresis or strong cation exchange chromatography prior to reversed phase LC-MS/MS analysis were performed using a pooled rat liver lysate sample for the purpose of constructing an MS/MS spectral library to aid SRM development. For OFFGEL electrophoresis rat liver extract was trypsin-digested and peptides (from 200 μg protein digest) were resuspended in 3.6 ml of 2% (v/v) carrier ampholytes, pH 3–10 (GE HealthCare) and loaded onto a prehydrated immobilized pH gradient (IPG) gel strip using a 24-well partition frame on an Agilent 3100 OFFGEL fractionator. The peptide sample was electrophoresed according to manufacturer's instructions for 50 kVh at a maximum of 50 μA or 4500 V. For strong cation exchange chromatography a Pepsil strong cation exchange (SCX) column (Column Technology Inc., CA), 0.32 mm × 10 cm, 5 μm particle size, 300 Å pore size, was fitted between the autosampler and the injection valve of a 1200 Series nanoflow HPLC allowing the SCX column to be in line for peptide loading and elution steps, and out of line for the reversed phase gradient. Peptide fractions were eluted from the SCX column with increasing steps of sodium acetate in 0.1% (v/v) formic acid, 3% (v/v) ACN (10 steps of sodium acetate at the following concentrations: 0, 12.5, 25, 37.5, 50, 75, 100, 150, 250, and 500 mm). Each OFFGEL fraction (analyzed offline) or SCX fraction (analyzed online) was loaded on the reversed phase enrichment column and subjected to reversed phase LC-MS/MS essentially as described above using the shotgun duty cycle.
The data files from the LC-MS/MS analysis of rat liver extract SCX and OFFGEL fractions were converted from the Agilent proprietary .d format to .mzXML using Trapper (v4.3.1), and peak lists in the .mgf format were extracted using mzXML2Search (v4.3.1). The peak lists were searched against the IPI Rat database (version 3.53 concatenated with common laboratory contaminant proteins and reverse entries for false identification rate estimation —80,062 entries including decoys) using Mascot v2.1 (33) and X!Tandem (k-score) v2 (34, 35) with the following parameters: semitryptic cleavage allowed, two missed cleavages permitted, methionine oxidation variable modification, carbamidomethylated cysteine fixed modification, precursor mass tolerance 20 ppm, fragment mass tolerance 50 ppm. These search engines have previously been shown to provide complimentary results (36). Results files from the search engines were converted to the .pepXML format and analyzed using PeptideProphet (37) to estimate the false discovery rate using the decoy hits to establish the negative distribution in the model (38). InterProphet was then used to combine the results from the two search engine used, and rollup to a minimal protein list was achieved with ProteinProphet (39). Trapper, mzXML2Search, PeptideProphet, InterProphet, and ProteinProphet are all components of the Trans Proteomic Pipeline (40, 41). A spectral library was constructed from the InterProphet pepXML results files and centroided peak list mzXML files via Skyline v0.6 (25), which utilizes Bibliospec (42) for spectral library construction.
Target Selection, Design, and Performance of SRM Assays
The selection of proteins to include in SRM assay development was based on (1) differential regulation in the label-free LC-MS study, (2) differential regulation in the gene expression microarray study previously conducted (9), (3) literature sources including a recent literature review of hepatotoxicity biomarker candidates (11) in addition to previously published results from the InnoMed PredTox Consortium (7, 30), and, (4) a selection of housekeeping genes/proteins that were not differentially regulated in the label-free LC-MS or gene expression microarray studies. A complete list of the genes/proteins selected for the panel is given in Table I. Mapping to UniProt accessions, where required (e.g. from Affymetrix accessions), was performed with BioMart v0.7 (43). UniProt accessions were imported into a Skyline document (25) and only proteins for which at least one high quality library peptide MS/MS spectrum was available were retained. Peptides were filtered based on a unique mapping to the protein of interest (“proteotypic”) against the UniProt Complete Proteome for Rattus novegicus (release 2010_07) using the Skyline Unique Peptides tool. As the UniProt Complete Proteome database for Rattus norvegicus is currently only partially curated (∼7500 SwissProt entries and 23,500 TrEmbl entries), proteins for which SwissProt and TrEmbl entries were present that had greater than 90% identity as determined by BLAST (44) were considered redundant for the purpose of choosing proteotypic peptides. The remaining peptides were additionally filtered according to the following criteria: length between 7 and 25 amino acid residues; methionine containing peptides excluded; ragged ends excluded (i.e. adjacent cleavage sites such as KK, KR, RK, or RR). Carbamidomethylation was set as a fixed modification on cysteine residues. The 10 most abundant singly or doubly charged b or y product ions excluding b1-b3 and y1-y3 per peptide were automatically selected for the transition list which was then manually inspected and edited with reference to the library MS/MS spectra. The transition list was split into methods containing no more than 130 transitions with the following parameters: dwell time set to 20 ms; collision energy calculated based on the equation ((m/z × 0.32) – 4.8) V; MS1 resolution set to “wide” and MS2 resolution set to “unit”; fragmentor voltage set to 130 V. A representative sample consisting of a pool of 30 rat liver digests, including drug treated and vehicle controls, was introduced into a 6460 triple quadrupole (QqQ) mass spectrometer connected online to a 1200 Series nanoflow HPLC via an orthogonal spray HPLC-Chip/MS interface (Agilent Technologies). Trypsin-digested protein (4 μg) was chromatographed with a 25 min peptide elution gradient from 3% (v/v) acetonitrile, 0.1% (v/v) formic acid to 40% (v/v) acetonitrile, 0.1% formic acid using a HPLC-Chip equipped with a 75 μm × 150 mm, 5 μm C-18 300SB-Zorbax analytical column and a 160 nl Zorbax 300SB-C18 5 μm enrichment column. The resulting data files were imported into Skyline and manually inspected for quality, and agreement between the relative intensity of product ions from the QqQ SRM data and the QqTOF MS/MS library data was assessed using the Skyline dot product calculation (45). Peptides with high quality measureable signals were retained and a single dynamic SRM instrument method (or scheduled method) was constructed. The collision energy for each transition was optimized by testing five steps of 3 V either side of the estimated collision energy voltage and the collision energy with the highest response that was automatically selected by Skyline. The analytical reproducibility of the optimized SRM assay was determined by 10 serial injections of the pooled liver digest with alternating blank injections. Subsequently, each of the 30 digested liver protein extracts from drug treated or control animals was analyzed using the optimized SRM method in randomized order with alternating blank injections. Raw data files were imported into Skyline and the SRM chromatography was manually inspected for co-elution of transitions for the same peptide and appropriate peak integration. Peak area values were exported and summary statistics were calculated. Protein level statistics across drug treatment groups (fold change and t test p value) were calculated by summing peptide abundances where multiple peptides per protein were measured. In a secondary step, in order to determine concordance between quantitative values for multiple peptides mapping to a protein, protein level statistics were also considered by calculating the median and standard deviation of peptide level fold change ratios (included in supplemental material).
Table I. SRM determined protein fold change versus time matched vehicle control after drug treatment. Protein fold change is reported as the ratio among drug treatment groups of median summed peptide abundances; HK, housekeeping protein (control); PX, proteomics label-free LC-MS; GX, gene expression (transcriptomics); LIT, literature source; Veh/Veh indicates the fold change of the day 15 vehicle over the day 4 vehicle animals where no (drug induced) effect is expected; bold font indicates 0.5 > fold change > 2.0; T-test p values are indicated by *<0.05, **<0.01, ***<0.001.
| Uniprot acc. | Protein | Source | Peptides | Low dose |
High dose |
Veh/Veh | ||
|---|---|---|---|---|---|---|---|---|
| Day 4 | Day 15 | Day 4 | Day 15 | |||||
| O35077 | Glycerol-3-phosphate dehydrogenase [NAD+], cytoplasmic | HK | 1 | 1.15 | 1.05 | 1.17 | 1.26 | 0.98 |
| P04642 | l-lactate dehydrogenase A chain | HK | 3 | 0.92 | 0.9 | 1.18 | 1.01 | 0.92 |
| P10719 | ATP synthase subunit beta, mitochondrial | HK | 5 | 1.02 | 1.08 | 1.01 | 1.02 | 0.89 |
| P15999 | ATP synthase subunit alpha, mitochondrial | HK | 5 | 1.08 | 1.08 | 1 | 0.82 | 0.76 |
| P85108 | Tubulin beta-2A chain | HK | 1 | 1.15 | 0.97 | 0.88 | 0.9 | 0.86 |
| P05178 | Cytochrome P450 2C6 | PX | 3 | 1.06 | 1.59** | 3.43*** | 6.28*** | 0.65* |
| P23965 | 3,2-trans-enoyl-CoA isomerase, mitochondrial | PX | 1 | 1.18 | 1.45 | 6.37*** | 10.46*** | 1 |
| Q4V8F9 | Hydroxysteroid dehydrogenase-like protein 2 | PX | 1 | 0.85 | 1.02 | 4.11*** | 2.93*** | 1.44 |
| Q64591 | 2,4-dienoyl-CoA reductase, mitochondrial | PX | 2 | 1.07 | 1.33* | 2.95*** | 6.21** | 1.01 |
| O88267 | Acyl-coenzyme A thioesterase 1 | PX + GX | 1 | 0.71 | 0.31 | 1188.27*** | 690.64** | 1.18 |
| P04903 | Glutathione S-transferase alpha-2 | PX + GX | 1 | 0.99 | 1.18 | 2.81*** | 5.96*** | 0.89 |
| P07687 | Epoxide hydrolase 1 | PX + GX | 3 | 0.98 | 1.16 | 2.86*** | 6.07*** | 0.72 |
| P07896 | Peroxisomal bifunctional enzyme | PX + GX | 4 | 1.19 | 1.36 | 17.23*** | 10.13*** | 0.72 |
| P08516 | Cytochrome P450 4A10 | PX + GX | 2 | 1.01 | 1.3 | 19.49*** | 13.89*** | 0.85 |
| P09875 | UDP-glucuronyltransferase 2B1 | PX + GX | 1 | 1.08 | 0.82 | 1.23* | 1.64** | 1.19 |
| P14141 | Carbonic anhydrase 3 | PX + GX | 5 | 1.01 | 1.09 | 0.66* | 0.5*** | 0.73 |
| P21775 | 3-ketoacyl-CoA thiolase A, peroxisomal | PX + GX | 4 | 1.03 | 1.07 | 7.88*** | 7.22*** | 0.85 |
| P38918 | Aflatoxin B1 aldehyde reductase member 3 | PX + GX | 1 | 1.24 | 0.86 | 3.64*** | 6.77** | 0.79 |
| P51647 | Retinal dehydrogenase 1 | PX + GX | 2 | 1.66 | 2.06 | 13.19*** | 34.41*** | 0.7 |
| Q6I7R1 | Dehydrogenase/reductase (SDR family) member 7 | PX + GX | 2 | 1.08 | 0.82 | 0.81 | 0.36** | 0.98 |
| O55171 | Acyl-coenzyme A thioesterase 2, mitochondrial | GX | 1 | 1.1 | 1.02 | 4.87*** | 4.91** | 0.94 |
| P00176 | Cytochrome P450 2B1 | GX | 1 | 1.35 | 1.22* | 54.5*** | 26.37** | 0.95 |
| P04182 | Ornithine aminotransferase, mitochondrial | GX | 2 | 0.91 | 1.29 | 0.21*** | 0.42** | 0.87 |
| P04906 | Glutathione S-transferase P | GX | 1 | 1.01 | 1.11 | 0.93 | 1.81 | 0.98 |
| P05369 | Farnesyl pyrophphate synthase | GX | 1 | 0.84 | 1.18 | 0.63 | 1.97* | 0.93 |
| P06757 | Alcohol dehydrogenase 1 | GX | 3 | 1.21 | 1.21 | 1.21 | 1.94** | 0.71 |
| P13697 | NADP-dependent malic enzyme | GX | 2 | 1.38 | 0.88 | 1.92** | 6.62** | 1.15 |
| P36365 | Dimethylaniline monooxygenase [N-oxide-forming] 1 | GX | 1 | 0.96 | 0.94 | 0.63 | 0.47 | 0.53 |
| Q5XI60 | Receptor expression-enhancing protein 6 | GX | 1 | 0.87 | 0.9 | 1.72*** | 2.7*** | 0.86 |
| Q64654 | Lanterol 14-alpha demethylase | GX | 1 | 0.87 | 0.94 | 0.84 | 1.53* | 0.78 |
| Q6TEK3 | Vitamin K epoxide reductase complex subunit 1-like protein 1 | GX | 1 | 1.06 | 1.14 | 1.28 | 1.43* | 0.93 |
| Q9EQ76 | Dimethylaniline monooxygenase [N-oxide-forming] 3 | GX | 5 | 1.06 | 1.12 | 0.81 | 0.39*** | 1.15 |
| B4F7D0 | Prodh protein (Fragment) | GX | 1 | 0.75 | 1.13 | 0.59 | 0.76 | 0.89 |
| Q5XFW5 | FK506 binding protein 11 | GX | 1 | 0.99 | 0.97 | 1.29* | 1.93* | 0.8* |
| P01048 | T-kininogen 1 | LIT [30] | 1 | 1.14 | 0.93 | 1.33 | 1.21 | 1.08 |
| P55159 | Serum paraoxonase/arylesterase 1 | LIT [30] | 2 | 1.13 | 1.11 | 1.55** | 1.38 | 1.09 |
| O89049 | Thioredoxin reductase 1, cytoplasmic | LIT [11] | 2 | 1.02 | 1.04 | 1.61** | 1.96*** | 0.94 |
| P04041 | Glutathione peroxidase 1 | LIT [11] | 4 | 1.2 | 0.94 | 1.14 | 0.66* | 1.02 |
| P04762 | Catalase | LIT [11] | 5 | 1.08 | 1.17 | 1.12 | 1.25 | 0.81* |
| P04785 | Protein disulfide-isomerase | LIT [11] | 3 | 0.98 | 1.01 | 0.92 | 1.42** | 0.89 |
| P06761 | 78 kDa glucose-regulated protein | LIT [11] | 3 | 0.98 | 0.93* | 1.08 | 1.54*** | 0.85 |
| P07632 | Superoxide dismutase [Cu-Zn] | LIT [11] | 3 | 0.98 | 1.01 | 0.96 | 1.03 | 0.88 |
| P07895 | Superoxide dismutase [Mn], mitochondrial | LIT [11] | 2 | 0.97 | 1 | 0.9 | 1.12 | 0.97 |
| P14480 | Fibrinogen beta chain | LIT [11] | 3 | 1.03 | 0.78 | 0.89 | 1.19 | 1.16 |
| P18418 | Calreticulin | LIT [11] | 1 | 1.01 | 0.99 | 0.99 | 1.36** | 0.88 |
| P35565 | Calnexin | LIT [11] | 2 | 0.99 | 1.04 | 0.98 | 1.13 | 0.94* |
| P48199 | C-reactive protein | LIT [11] | 2 | 0.81 | 0.68 | 1.21 | 1.03 | 1.05 |
| P31044 | Phphatidylethanolamine-binding protein 1 | LIT [7] | 2 | 1.05 | 0.96 | 0.9* | 0.89 | 0.87* |
RESULTS
Classical Toxicology Evaluation
No toxicological findings were apparent in animals receiving the vehicle (0 mg/kg/day) or low (15 mg/kg/day) doses at either time point. Minimal hypertrophy of the hepatocytes was recorded in most animals receiving the high dose (350 mg/kg/day) at day 3. Hypertrophy is generally not considered as a toxic effect but rather as an adaptive response to the xenobiotic insult, which can be because of (1) an increase in smooth endoplasmic reticulum (mainly related to the increased production of drug metabolizing enzymes), (2) peroxisomal proliferation, or, (3) mitochondrial swelling (46). Clear drug-induced histopathological findings, in addition to hypertrophy of hepatocytes, were observed in animals receiving the high dose at day 14. The five animals of this group were affected to a somewhat varying degree with lesions including marked bile duct inflammation or hyperplasia and bile duct and liver cell necrosis. Further details of the classical toxicology findings and evaluation methods are given in supplemental Table S1, as well as additional publications from the InnoMed PredTox Consortium (7, 29, 30).
Label-Free LC-MS Analysis
To determine the differential modulation of liver proteins in hepatotoxicant EMD 335823 treated rats a label-free LC-MS study was undertaken using nLC-MS/MS analysis. The MS data set, consisting of 23 LC-MS runs, was imported into Progenesis LC-MS v2.6 and subjected to alignment with additional user inspection and manual placement of additional alignment vectors until a visually adequate alignment was obtained (for an example of alignment see supplemental Fig. S2). The initial round of MS feature detection returned 144,415 features and this was then filtered by (1) retention time in the peptide elution portion of the gradient, (2) 2 ≤ charge state ≤ 7, and (3) number of isotopologs in the peptide isotopic envelope ≥ 3 (see supplemental Figs. S3 and S4 for details of feature detection and filtering). This resulted in a feature set containing 40,444 features which was assumed to consist predominantly of peptides (note: this feature set is not deconvoluted at the level of charge state or PTMs and so does not necessarily contain 40,444 unique peptide features by sequence).
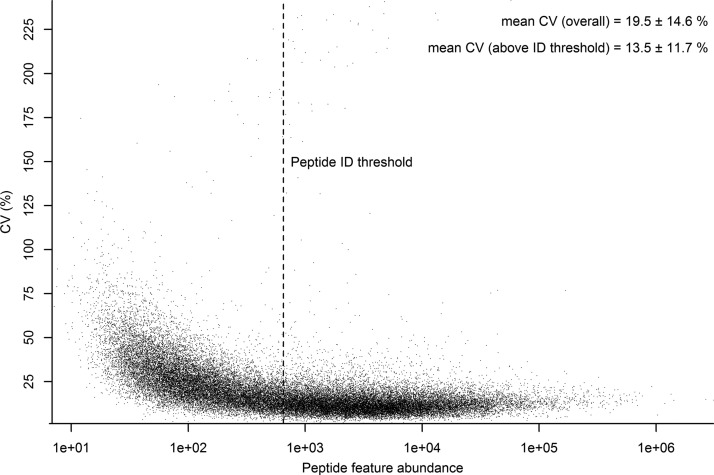
In order to determine the technical variance of the label-free method six consecutive injections of the same sample (a sample pool consisting of all five vehicle and five drug treated liver samples) were undertaken and the data analyzed as detailed in the Experimental Procedures. Fig. 1 shows a plot of the % coefficient of variation (% CV) versus peptide feature abundance for the six measured technical replicates. The plot shows a clear trend whereby, as may be expected, peptide features with higher abundance show greater reproducibility compared with features at the lower end of the abundance. The mean % CV for the 40,444 features detected was 19.5%. Although quantitative information for all detected peptide features can be obtained from the MS1 level mass spectra, only a small subset of these features have associated informative MS2 spectra from which peptide/protein identifications can be assigned. The set of peptide features that can be assigned identifications is also strongly biased toward higher abundance. In Fig. 1 the lowest abundance at which a peptide identification was confidently assigned is indicated by a line designated “Peptide ID threshold.” The set above this threshold consists of 22,186 features for which the mean % CV is 13.6%. It is worth noting, however, that even above this threshold the fraction of peptide features confidently identified remains small at ∼20%, with a strong bias toward identification of higher abundance features within this subset.
Fig. 1.
Technical variance of peptide feature abundance by label-free LC-MS. The technical variance, as measured by % CV ± standard deviation across six technical replicates of a representative sample, is plotted against their average abundance on the log 10 scale for all 40,444 detected features. The peptide threshold indicates the lowest abundance at which a peptide identity was confidently assigned to an associated MS/MS spectrum.
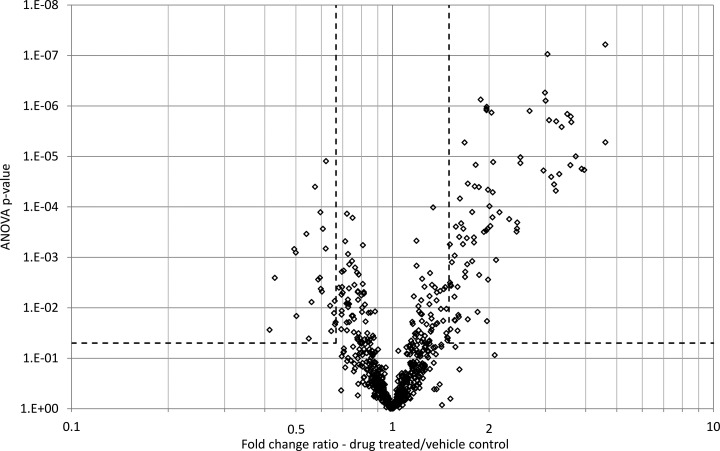
The 96,193 MS/MS spectra acquired from all of the samples were exported from Progenesis LC-MS as a single .mgf file and searched using Mascot. A total of 2,797 peptides (FDR = 2.5%) mapping to 809 proteins (FDR = 3.1%) were identified, with 717 of these proteins being identified by more than one peptide. Each protein was identified by an average of 4.7 peptides. The peptide/protein identifications were mapped back onto the peptide quantification data and rolled up to the protein level using the Progenesis LC-MS protein view. Fig. 2 shows a volcano plot relating the fold change and ANOVA p value calculated for each identified protein. One hundred twenty-two proteins were determined to be modulated more than 1.5-fold with ANOVA p value less than 0.05 (corresponding M/A plots are shown in supplemental Fig. S5). The identities and summary statistics for all of the proteins quantified and identified in this label-free LC-MS study are shown in supplemental Table S2. In general terms, a permissive approach was adopted for the statistical cutoff's as the purpose of the label-free LC-MS study was as a discovery screen to identify candidates for the targeted SRM study and, as such, it was deemed that some false positives could be tolerated for subsequent potential exclusion by targeted protein verification.
Fig. 2.
Protein modulation in liver on hepatotoxicant treatment as determined by label-free LC-MS. The volcano plot summarizes the protein modulation on drug treatment showing the t test p value plotted against protein fold change. Data points in lower center area of the plot have a fold change close to 1 and a p value approaching 1 and indicate no significant change, whereas points in the upper left and upper right quadrants indicate significant negative and positive changes in protein abundance respectively. Corresponding M/A plots are given in supplemental Fig. S5.
Comparison of Discovery Proteomics Data With Gene Expression Data
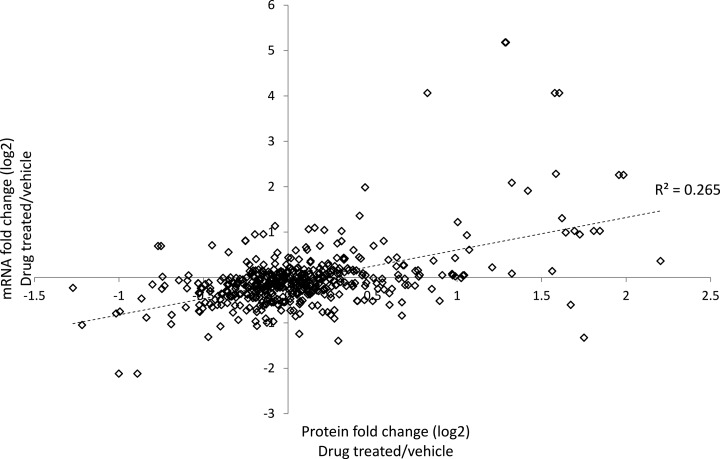
The list of differentially modulated proteins determined by the label-free LC-MS analysis was compared with the results of a previously acquired gene expression data set (using the Affymetrix Whole Genome Array platform) arising from separate liver sections from the same drug treated animals analyzed in this study (9). Fig. 3 shows the correlation between corresponding protein and transcript changes for which accessions could be unambiguously mapped (530 protein to transcript accession mappings). There was only a very modest correlation between protein and transcript modulation (R2 = 0.265). Significantly, there were a substantial number of proteins determined to be differentially modulated (0.67 > fold change ratio > 1.5) that did not show a change in abundance at the transcript level (0.9 < fold change ratio < 1.1) (see supplemental Table S3). Proteins in this category include multiple toxicologically relevant phase I and phase II drug metabolising enzymes (i.e. cytochrome P450s, UDP glycosyltransferases, glutathione S-transferases).
Fig. 3.
Correlation between hepatotoxicant induced changes in protein and corresponding transcript levels. Transcript fold change (log 2) as determined by microarray analysis is shown plotted against the protein fold change (log 2) as determined by label-free LC-MS where accessions could be unambiguously mapped (530 proteins).
A pathway over-representation analysis demonstrated that despite the modest correlation in expression changes at the individual protein/transcript level, there was a higher degree of agreement between protein and transcript changes at the pathway level with an overlap of 12 pathways between the top 20 over-represented pathways determined from the proteomics analysis with the top 20 from the transcriptomics analysis (see supplemental Fig. S6). The overlapping pathways include xenobiotic metabolism, fatty acid metabolism, LPS/IL-1 mediated inhibition of RXR function, CAR/RXR function, PXR/RXR function, aryl hydrocarbon receptor signaling, cytochrome P450 panel - substrate is a fatty acid (rat), cytochrome P450 panel - substrate is a xenobiotic (human), cytochrome P450 panel - substrate is a fatty acid (mouse), cytochrome P450 panel - substrate is a fatty acid (human), FXR/RXR activation, and cholesterol biosynthesis. A detailed discussion of the pathway analysis with respect to possible toxicological mechanisms is included in a publication summarizing studies on EMD 335823 in the context of the InnoMed PredTox Consortium (9). As the pathway analysis from the previous transcriptomics studies and the proteomics data described in this article have very similar regulations, we refer the reader to the discussion of toxicological mechanisms therein.
Construction of a Rat Liver MS/MS Spectral Library to Aid SRM Development
One of the most substantial benefits of the SRM approach over other targeted protein measurement strategies (such as Western blot or ELISA) is the ability to multiplex the assay to include potentially tens to hundreds of proteins in a single analytical assay. The development of robust and reliable SRM assays is greatly facilitated by the availability of appropriate MS/MS data for the peptides for which SRM measurements are being developed. Although comprehensive peptide MS/MS data sets exist for proteins of a number of organisms we quickly realized that a similar resource for rat peptides was not readily (publicly) available. In order to facilitate the rapid development of SRM assays of rat for use in, for example, future preclinical toxicology studies, we reasoned that a reasonably comprehensive tandem MS database for the peptides/proteins of interest was required (12, 27, 47). To this end, a tandem MS data set was obtained from peptides resulting from the tryptic digestion of a sample pool of all drug treated and vehicle control rats from which liver protein extracts were prepared. The complex whole lysate peptide sample was fractionated by OFFGEL electrophoresis or strong cation exchange chromatography before reversed phase separation and analysis by QqTOF tandem MS. The processing and database searching of the 53 LC-MS/MS raw data files containing 652,868 MS/MS spectra resulted in the assignment of 43,150 MS/MS spectra to 7515 unique peptide sequences (0.7% FDR estimated by PeptideProphet). The identified peptide sequences mapped to 1687 unique proteins in the IPI Rat database (v3.53) (1.4% FDR estimated by ProteinProphet). There was a significant increase in peptide identifications (25–37%) because of the combination of results via InterProphet from two search engines, namely Mascot (33) and X!Tandem (k-score) (34, 35), over either search engine alone (see supplemental Fig. S7). The raw data files associated with this spectral library have been deposited at the PeptideAtlas repository (28) and are available for download under the sample accession “PAe001466”. Additionally, this spectral library is provided in the form of a Bibliospec library (42) with the Skyline share archive file in the supplemental Material such that the entire spectral library can be manually inspected using the spectral library viewer in the open source and freely available software Skyline (25).
In order to provide further confidence in this data set the tissue expression of the identified proteins as determined by previous knowledge from the UniGene EST database (48) was examined. This analysis shows a corresponding annotation for liver expression in 496 of the proteins identified which is substantially more than matching to any other tissue (see supplemental Fig. S8). In addition, an analysis of the overrepresented pathways as determined from the KEGG (Kyoto Encyclopedia of Genes and Genomes) database (49) are consistent with known liver function (see supplemental Fig. S9).
Development and Optimization of a Hepatotoxicity SRM Assay Panel From Multiple Disparate Discovery Sources
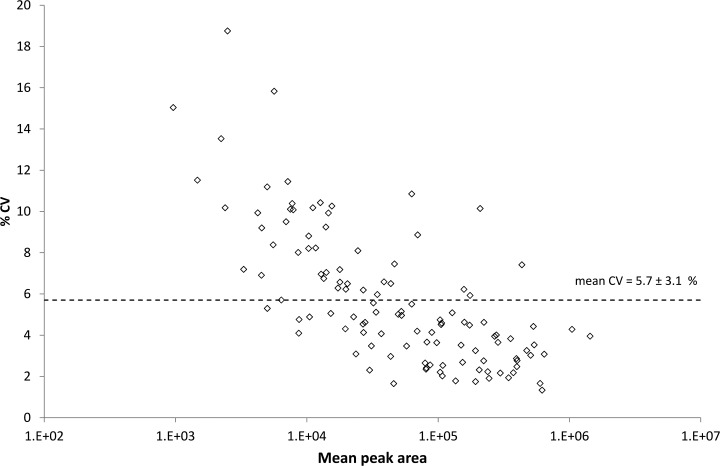
We sought to combine the proteins shown to change in liver in response to the hepatotoxicant EMD 335823 with other potential markers of hepatoxicity to generate a panel of proteins for measurement by SRM with a view to generating a “hepatotoxicity marker protein panel.” The development of the SRM assay panel consisted of, (1) assembly a list of putative biomarker protein candidates from multiple sources, (2) construction of SRM transition lists for proteotypic peptides determined with respect to the rat liver MS/MS library, (3) establishing peptide suitabililty and detectability with SRM via multiple unscheduled injections of a representative (pooled) sample, (4) quality control of peptide SRM signals with reference to the rat liver library MS/MS spectra, and (5) optimization of the SRM assay panel including choice of transitions, retention time scheduling and collision energy optimization. supplemental Fig. S10 outlines in detail the steps taken to assemble the initial biomarker candidate list. Briefly, the list of putative biomarker candidates was drawn from significantly modulated proteins in the label-free LC-MS dataset (53 proteins with 0.5 > fold change > 2.0 and ANOVA p value < 0.01), significantly modulated transcripts from the gene expression data set (200 transcripts with 0.55 > fold change > 1.9 and ANOVA p value < 0.01), and a recent literature review of potential hepatotoxicity biomarkers both in liver tissue and plasma (27 proteins). In addition to the putative hepatotoxicity biomarkers, a selection of housekeeping proteins, shown to be invariant on drug treatment in both the label-free LC-MS and transcriptomics studies, were chosen to act as controls. After mapping to Uniprot accessions, the list was reduced to 245 proteins, which were then cross-referenced with the set of proteins that had been confidently identified in the rat liver MS/MS data set. This reduced the list to 108 biomarker candidates, which had been previously detected by QqTOF MS. The bulk of this reduction is accounted for by candidates arising from the transcriptomics study, with only 51 of the 168 candidate proteins having been previously detected by MS, which is in contrast to the proteins arising from the label-free LC-MS study for which, by definition, all proteins had available MS/MS spectra (see supplemental Fig. S10). This list of 108 proteins was further filtered to 57 proteins because of the availability of suitable tryptic peptides on which to base the SRM assay (i.e. unique peptide to protein mapping, no methionine, peptide size 7–25 amino acid residues, no directly adjacent tryptic cleavage sites, manual quality inspection of the library MS/MS spectrum). The detectability, or not, of peptides mapping to this set of proteins in a representative sample derived from a pool of all samples by SRM was then established (see supplemental Fig. S11 for details). A list of transitions consisting of 1681 entries derived from 8to 10 SRM transitions for each of the 174 peptides (1–5 peptides per protein) for all 57 proteins was assembled. This transition list was subdivided across 15 instrument methods each with not more than 130 SRM transitions per sample run. The instrument was set with a dwell time of 20 milliseconds facilitating a duty cycle of ∼2.5–3.0 s which resulted in the acquisition of at least 8 to 10 measured data points across the chromatographic peptide elution. The SRM signals for each of the peptides were evaluated manually for quality including signal to noise ratio, co-elution of the SRM chromatograms the Skyline dot product (which measures the correlation between SRM transition peak areas and ion intensity in the corresponding library MS/MS spectrum) (45), and the predicted retention time. Retention times were calibrated using peptides derived from a set of six standard proteins chromatographed with the same solvent gradient. After manual quality evaluation the SRM method for each peptide was reduced to the five most intense transitions without interference and a single method using retention time scheduling and hence combining all peptide transitions was developed. Supplemental Fig. S12 shows a histogram of the dot product calculation for the measurement of 109 peptides (from 48 proteins) with the majority of peptides demonstrating a dot product > 0.975 and all peptides > 0.925. This data provides a very high level of confidence that the SRM assays are indeed measuring the peptides of interest and not interfering peptide or other analytes. Finally, each of the 545 transitions (five per peptide) in the refined and time-scheduled method were optimized for collision energy using five steps of 3 V either side of the formula predicted optimization energy with the optimal collision energy value was being automatically selected by Skyline and incorporated into the finalized SRM method. Overlaid representative SRM chromatography for the optimized SRM assay containing all 545 transitions is shown in supplemental Fig. S13. Finally, the technical reproducibility of the optimized method was determined by 10 serial injections of a representative sample which resulted in a mean CV of 5.7% (see Fig. 4). Furthermore, the data revealed that peptides were quantified across more than three orders of SRM signal abundance and, in general, although the reproducibility decreased marginally with decreasing SRM signal intensity but no individual peptides displayed a CV of greater than 20%.
Fig. 4.
Technical reproducibility for peptide measurement by SRM. The technical variance, as measured by % CV ± standard deviation across 10 technical replicate injections of a representative sample, is plotted against mean peak area on the log 10 scale for all 109 peptides monitored by SRM.
Targeted Measurement of a Putative Hepatotoxicity Biomarker Panel in Drug Treated Rat Liver
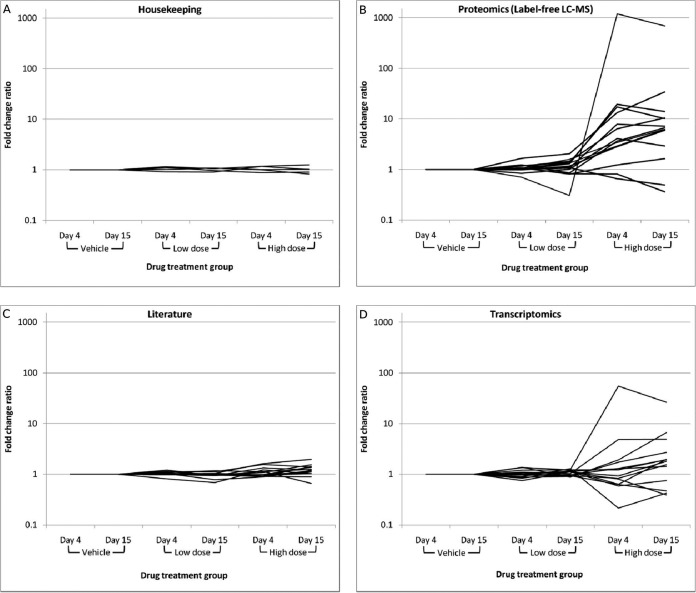
The optimized SRM method to simultaneously measure 48 proteins using 109 proteotypic peptides, and 545 transitions that constitutes a putative hepatotoxicity biomarker panel was applied to 30 individual liver samples from rats treated daily with EMD 335823 at vehicle, low, or high dose for 3 or 14 days (5 rats per treatment group). Table I shows the proteins targeted in the SRM assay and the associated fold change ratios versus time matched vehicle control, calculated by summing the peptide abundances for a given protein, taking the median value across the five animals in the treatment group, and ratioing the values between treatment groups. supplemental Table S4 is analogous to Table I except the protein fold change ratio is given as the median of the peptide fold change ratios ± standard deviation and is intended to provide a measure of the concordance of multiple peptides mapping to the same protein, which is in general high for this dataset. A representative example of peptide to protein concordance is given in supplemental Figs. S14 and S15, which show the SRM peptide abundances for all 30 animals plotted for the three measured peptides mapping to Cytochrome P450 2C6 and representative chromatography for the same three peptides from three animals respectively. The bulk of significantly regulated proteins in the SRM analysis originate from the label-free LC-MS proteomics and transcriptomics data sets, and appear in the high dose groups (day 4 and day 15). The fold change ratio between the vehicle treatment groups at day 4 and day 15 are also shown as a control and, in general, these ratios are close to 1. Fig. 5 shows the fold change ratio versus time matched vehicle control for each treatment group, separated by the source of the putative biomarker (housekeeping, proteomics - label-free MS, literature, or transcriptomics). This plot emphasizes the enrichment of differentially modulated proteins in high dose treatment rats for proteins in the proteomics and transcriptomics groups (Figs. 5B and 5D), while also highlighting essentially no change in the proteins selected as housekeeping controls (Fig. 5A; see also supplemental Table S7 for raw peak areas of housekeeping controls). Interestingly, only a few proteins in the “literature” group (Fig. 5C) which were selected largely from putative biomarkers in a recent review (albeit with some suggested as plasma, and not tissue biomarkers) (11), displayed differential modulation, however, fold changes in this case were in the range 1.5–2.0 and, as such, relatively modest.
Fig. 5.
Protein fold change ratio versus time matched vehicle control as measured by SRM. The mean protein fold change ratio versus the time matched vehicle control as determined SRM measurement is plotted with respect to the drug treatment group (dose and time point). The plots are separated by the source of the measured protein. (A) Housekeeping control protein profiles, (B) profiles for protein selected from label-free LC-MS studies, (C) profiles for proteins selected as literature biomarker candidates, (D) profiles for proteins selected based on transcriptomics data.
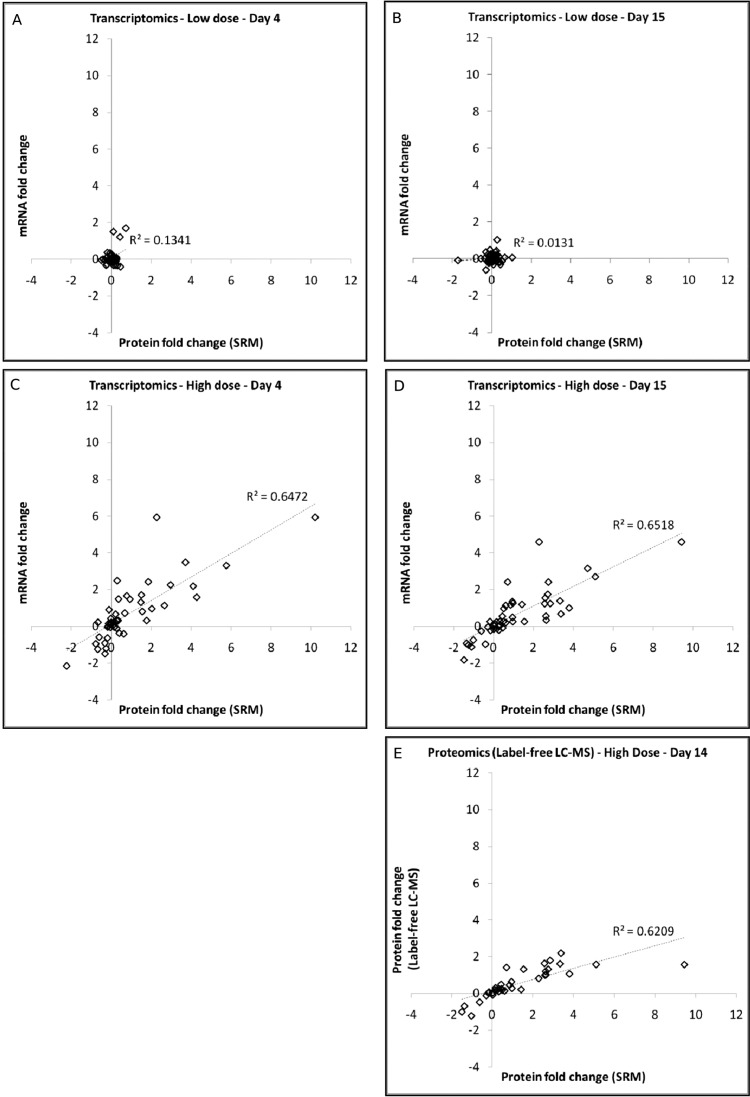
To determine the agreement on drug treatment in changes at the individual protein/transcript level, the results from the SRM-based protein measurements and microarray-based mRNA measurements, and the SRM-based protein measurements and the label-free LC-MS-based protein measurements were directly compared. Fig. 6 shows a comparison of the median log2 fold change ratios from the SRM data plotted against the median log2 fold changes ratios determined in either the transcriptomics (Figs. 6A–6D), or label-free LC-MS proteomics (Fig. 6E) from which the putative biomarker proteins were selected. In the low dose groups (Figs. 6A and 6B), where there are essentially no significant changes and no correlation is observed. This in contrast to the high dose groups at days 4 and 15 (Figs. 6C and 6D) where there are a large number of differentially modulated transcripts and these show a substantial correlation with the corresponding SRM-based protein measurements (R2 = 0.65 for high dose, day 4 and R2 = 0.65 for high dose, day 15). Similarly, the label-free LC-MS based protein measurements (panel e), which were only carried out for the high dose, day 15 treatment group, are well correlated with the SRM-based protein measurements (R2 = 0.62). Interestingly, the correlation between the label-free LC-MS and SRM measurements are skewed by a single data point which displays a very high fold change in the SRM measurement. The inflated fold change ratio for this protein is due to the absence of signal in the corresponding SRM measurement for the vehicle control group (i.e. the value for the high dose treatment group is ratioed to a baseline value from the vehicle control creating an artificially high fold change ratio). If this single data point is removed from the plot, the R2 value increases to 0.78 indicating a high level of correlation.
Fig. 6.
Correlation of SRM-based protein fold change with the corresponding transcript or label-free LC-MS-based protein fold change. The protein modulation (given as log twofold change versus time-matched vehicle control) as measured by SRM is compared with the transcript or protein fold change as measured by microarray analysis (panels A–D) or by label-free LC-MS (panel E). The comparison of SRM to transcriptomics data is given for 4 treatment groups (panel A, low dose treatment, day 4; panel B, low dose treatment day 15; panel C, high dose treatment, day 15; panel D, high dose treatment, day 15), whereas the comparison of SRM to label-free LC-MS is only given for a single treatment group (panel E, high dose treatment, day 15) as this was the only treatment group analyzed by label-free LC-MS (in addition to the time-matched vehicle control).
There was variation in the toxicological phenotype of individual animals in response to EMD 335823 as determined by classical toxicological evaluation (see supplemental Table S1). It was of interest therefore to interpret changes in the abundance of putative biomarker proteins at the level of the individual animal, as opposed to drug treatment groups. When this analysis was undertaken some interesting correlations between protein biomarkers and toxicological phenotype emerged. For example, supplemental Figs. S16–S19 show the SRM abundance and representative SRM chromatography for each animal for peptides mapping to T-Kininogen and Fibrinogen beta chain. These proteins show substantial elevation in animals 42 and 45 following high dose after 15 days of treatment. These were the same two animals that showed the severest lesions in the histopathology evaluation, including marked bile duct inflammation with bile duct and liver cell necrosis and were determined to be outliers from their treatment group in the original transcriptomics data set (9).
DISCUSSION
There is a clear and pressing need for better tools to predict the potential for toxicity of new drugs—specifically with higher accuracy and at an earlier stage of drug development (1). Furthermore, there remains substantial interest in pursuing the use of omics technologies as a route to more sensitive preclinical toxicology evaluation (2), and this is evidenced by the large number of consortia focused on this topic (5). The InnoMed PredTox consortium represented one such effort comprising pharmaceutical companies, academic groups, and technology providers in Europe (6). In addition to the desire for more effective early toxicology evaluation, the field of pharmaceutical toxicology also represents a very useful test case scenario for the development of biomarkers in general. The reason for this is that toxicology evaluation could make use of both tissue-based, and peripheral fluid-based biomarkers. Notably, in current preclinical toxicology evaluation the tissues of drug treated animals are routinely analyzed by histopathology, which means de facto that tissue samples are available for omics analysis at the level of DNA, RNA, protein, or metabolites. The likelihood of discovering biomarker sets with predictive value from the tissue in which the toxicological phenotype is observed seems initially more probable than approaches which focus solely on blood plasma or urine. With this said, there is also substantial interest in deriving toxicity biomarkers measurable in peripheral fluids for the purpose of more routine monitoring in animal toxicology studies and or for translation into humans as drug safety biomarkers. This “stepping-stone” approach of first establishing biomarker sets in the tissue of interest, and later attempting translation into peripheral fluid measurements represents an attractive strategy but one in which there is likely to be “attrition” of many candidates. The opportunity to measure in a targeted and multianalyte approach (i.e. via SRM) candidate protein biomarkers represents a significant advantage to support this “stepping-stone” approach.
In this study we provide a roadmap for biomarker discovery and verification in preclinical toxicology evaluation by demonstrating the integration of data reporting potential biomarker candidates from multiple disparate sources into a targeted MS-based assay utilizing selected reaction monitoring on a triple quadrupole platform which can easily be deployed in large sample cohorts. Clearly, the approach is customizable to other candidate protein biomarker panels and the overall SRM development strategy as undertaken here potentially represents a valuable workflow tool for the generation of such assays. Here, the biomarker candidate list was assembled from three sources. First, a discovery label-free LC-MS study (utilizing MS1 intensities for quantification) was performed in liver samples from a subset of rats treated with a known hepatotoxicant EMD 335823. Second, differentially expressed genes from an earlier transcriptomic study of the same rat livers (EMD 335823 treated) were integrated as potential biomarker candidates. Third, we incorporated a set of literature biomarker candidates, primarily from a recent review of biomarkers related to drug-induced liver injury (11). To facilitate SRM assay development we built a substantial library of MS/MS spectra assigned with high confidence to tryptic peptides arising from a large pool of rat liver samples. The MS/MS spectral library data set has been deposited in PeptideAtlas and has additionally already been exploited by additional studies aimed at elucidating unanticipated post translational modifications relating to drug toxicology (50). Specific SRM assays for 48 biomarker candidates were developed via iterative rounds of optimization and quality control. The combined optimized SRM assays were then applied to the full set of 30 liver samples from the PredTox EMD 335823 drug treatment study.
The initial source of biomarker candidates included in the optimized SRM assay panel was a label-free LC-MS study of livers from rats treated for 15 days with a high dose of EMD 335823 or vehicle control. The results from the label-free LC-MS analysis demonstrated that relative quantification could be achieved with high technical reproducibility (mean CV = 13.6% for identified peptides), and that of the 809 proteins quantified, 111 were significantly modulated in their abundance in the high dose, day 15 treatment group. The enrichment of differentially modulated proteins in pathways such as xenobiotic metabolism, cytochrome P450s, PPARα, PPARα/RXR activation, fatty acid metabolism, oxidative stress, and oxidative stress response mediated by Nrf2 indicate that the detected proteins are indeed related directly to drug response. These results are relevant in the context of toxicological response to drug treatment, and in the context of the suspected PPARα activity of the compound in question (9). There was also significant overlap with pathways over-represented in the transcriptomics studies of the same samples. This pathway correlation is in contrast to direct comparison of individual protein and corresponding transcript changes which showed only weak correlation, similar to findings that been previously reported for studies in which corresponding protein and transcript changes have been compared (51, 52). In fact, the added value of performing the proteomic analysis is demonstrated by the measurement of a significant group of 21 toxicologically relevant modulated proteins (i.e. mainly phase I + II enzymes - see supplemental Table S3) that showed no change in abundance in the transcriptomic studies.
Although MS1 intensity-based label-free LC-MS provides a very useful strategy for discrete discovery proteomics studies, the overhead associated with instrument time and complex data analysis makes this approach currently difficult to implement in the type of large-scale and/or multisite studies that are required for the verification of biomarker sets. By comparison, targeted MS using SRM provides a platform for reproducible and sensitive measurement of panels of proteins at medium-high throughput (12). Additionally, SRM is inherently flexible, with the ability to easily adapt the panel of proteins to be measured based on emerging information from ongoing studies. The time for development of reliable SRM assays is rapidly being reduced due to novel software tools and data resources (26). In particular, the availability of high quality peptide MS/MS spectra for the proteins of interest enables rapid selection of appropriate peptides and transitions, as well as providing confidence in the final SRM assay by correlation of fragment ion intensities between the library MS/MS spectrum and SRM measurements (27, 47). In many instances such libraries are readily available in the public domain however we found this not to be the case for rat liver proteins and so for this reason, we assembled a high quality library of peptide MS/MS spectra derived from a pool of rat liver samples (hepatotoxicant treated and vehicle controls) before undertaking SRM assay development. This spectral library containing more than 7500 peptide assigned QqTOF mass spectra has been provided as a public resource for future SRM development related to pre-clinical toxicology studies. It is, however, worth noting that during the SRM development process some proteins were not included in the final SRM assay due to a lack of peptide MS/MS spectra. This was particularly the case for biomarker candidates arising from the transcriptomics study, where of the 168 proteins initially shortlisted for assay development, only 51 had associated peptide MS/MS spectra and could be progressed toward SRM assays (see supplemental Fig. S10). Acquisition of more comprehensive MS/MS spectral libraries will certainly facilitate SRM development, and recently MS/MS analysis of synthetic peptide libraries created specifically for this purpose has been undertaken in yeast (27), with similar efforts in human ongoing. The acquisition of such spectral libraries in key pre-clinical toxicology model systems such as rat would greatly accelerate efforts to develop SRM-based biomarker panels.
Optimization and quality control of the SRM assays of the final panel of 48 proteins was achieved in a relatively short but intensive period. This included 41 injections on the QqQ mass spectrometer totalling ∼37 h of instrument time. All assay development, optimization, and quality control were done in an iterative fashion (see supplemental Fig. S11, a summary of the optimization workflow) via the Skyline Targeted Proteomics Environment. The use of Skyline greatly accelerated the assay development cycle by automating/accelerating previously manual or semi-automated tasks relating to: peptide and transition selection with respect library MS/MS spectra and background proteome databases; filtering of peptides and transitions; collision energy optimization; retention time scheduling; comparison of SRM data to library MS/MS spectra; peak integration; and data review. In particular, the dot product calculation in Skyline (45), which scores the similarity between SRM intensities for measured transitions against the library MS/MS spectrum, provides a very useful filter to determine authentic peptide signals from interferences (see supplemental Fig. S12 for a summary of dot product values for the optimized SRM assays in this study). Despite the substantial time-saving in informatics tasks facilitated by Skyline, there is still a considerable manual inspection and quality control effort required to verify that the chosen SRM assay coordinates are in fact representative of the targeted peptide, and not interfering species. Currently, methods to determine and control the false discovery rate for SRM experiments in an automated and objective manner via decoy transition sets are under development (53) and this should help to reduce the burden of manual inspection in SRM studies. The SRM development cycle undertaken here also included a measurement of the technical variance of the SRM assays as determined by the coefficient of variation across 10 technical replicate injections of a representative pooled sample (see Fig. 4). A significant benefit of the SRM strategy is the excellent technical precision, demonstrated in this study by a mean CV of 5.7%, which is a substantial improvement over what is generally available with other proteomics methodologies. It is this technical precision, combined with the ability to consistently measure every analyte of interest (i.e. no missing values), that makes the SRM strategy very attractive for the type of large scale analyses that will be required for biomarker verification in the pre-clinical, and potentially, clinical toxicology evaluation arena.
The results of the SRM analysis of liver from drug-treated and vehicle control rats demonstrate that by selecting biomarker candidates from multiple relevant sources we have enriched for a subset of proteins which are significantly modulated on treatment with the hepatotoxic agent EMD 335823. In particular, the proteins derived from the label-free LC-MS and transcriptomics studies also demonstrated modulation in the high dose treated animals at both day 4 and day 15 (see Table I and Fig. 5). Interestingly, proteins selected from literature sources (11) showed less marked modulation on treatment, although some proteins were significantly regulated. Proteins included as housekeeping controls showed no significant modulation between drug-treated and time-matched vehicle control rats and comparisons between the vehicle treated rats at day 4 and day 15 also showed no significant changes. This indicates that the statistical cut-offs applied in the discovery experiments were appropriate and that changes observed in the drug-treated animals are highly like to represent true positive observations.
Additionally, a reasonable correlation was noted between protein fold change as determined by SRM-based compared with label-free LC-MS-based measurements, and between protein and transcript fold change as determined by SRM-based compared with microarray-based measurements (see Fig. 6). This result validates the premise of using lower-throughput discovery proteomic and transcriptomic approaches to gather biomarker candidates for subsequent medium-high throughput SRM based protein measurements in large-scale verification studies.
The statistical evaluation in this study used a staged approach. A relatively nonstrict set of fold change and t test cut-offs were employed in the label-free LC-MS discovery data analysis with the rationale to include as many viable biomarker candidates as possible and to potentially remove false positives later using the more precise and higher throughput SRM measurements. This strategy will continue to become more feasible as the number of peptides/proteins measureable by SRM, currently limited by duty cycle considerations, increases with new instrumentation and acquisition paradigms. Data from the SRM analyses were also ultimately evaluated by fold change and t test statistics, which while apparently effective for ranking candidate markers, are essentially ad hoc filters. Further effort is required from the proteomics community to evaluate the use more sophisticated methods largely developed for microarray analysis, for example, modified t-tests (54) and t test relative to a threshold (TREAT) (55), in protein biomarker studies. Additionally, empirical Bayes methods have already been implemented in proteomics studies and could provide a more rigorous path to significance analysis for biomarker research (56).
Although label-free SRM quantification can be performed in relatively smaller studies such as this one assuming the sample processing is well controlled and the samples are closely related in background and overall protein composition, the eventual goal is to extend this type of study to larger-scale consortium type efforts. In this case there are a range of considerations affecting quantitative performance that must be accounted for. This study employed a set of housekeeping proteins selected as controls to detect any systematic biases that might have adversely effected the detection of true drug-treatment related changes in relative protein concentrations. This strategy, however, will not detect matrix effects for individual analytes, and is not likely to prove successful on a larger scale. Substantial efforts have been made toward evaluating and controlling biases that might arise in large scale SRM studies, in particular by the National Cancer Institute's Clinical Proteomic Technologies for Cancer (CPTC) initiative (21). In particular, the use of stable isotope dilution standards provides a method to control for errors due, for example, to matrix effects or intersite operator biases. As such, due care should be taken with respect to previous knowledge on potential sources of error in the planning future large-scale studies in the preclinical toxicology arena.
The classical toxicology evaluation in this study (see supplemental Table S1 for histopathology results) showed the most marked findings at day 15 in the high dose treated rats and this included necrosis, fibrosis and bile duct necrosis/hyperplasia. By comparison, at day 4 in the high dose group only hepatocyte hypertrophy was reported in 3 of 5 animals and this is considered essentially an adaptive response and not a toxic insult (46). In the SRM-based proteomics analysis, however, the majority of proteins modulated at day 14 in the high dose group also showed a significant change in the day 4 group at high dose. This significant finding suggests that using multiplex SRM-based protein measurements has important potential for increased sensitivity in predicting toxicity over the classical toxicology evaluation methods. Increased sensitivity for prediction of toxicity is, in essence, one of the primary goals of systems toxicology approaches (2). It should be noted, however, that the protein changes observed at the early time point may not necessarily reflect more sensitive markers of overt toxicity but could rather indicate adaptive changes on drug treatment. As such, for a more complete characterization of potential toxicity biomarkers, studies using finer time and dosing intervals will certainly be required. Essentially no changes were observed by transcriptomics, classical toxicology evaluation, or SRM-based protein measurements for animals in the low dose group at either time point indicating that the dosage point was too low, also suggesting that future studies using finer dosing intervals would be extremely useful to determine dose dependent effects using SRM-based multiplex protein measurements.
This study represents a key proof-of-principle demonstration in which hepatotoxicity biomarker candidates from diverse sources including discovery proteomics, transcriptomics, and literature mining, are integrated into a panel enriched for putative biomarkers which following SRM assay development can be robustly measured in a medium-high throughput fashion via SRM. In this case only a single hepatotoxic compound was studied, however, given the substantial diversity of toxic insults which occur in liver (and indeed other toxicologically relevant tissues) large numbers of studies will need to be performed with a wide variety of model toxicant compounds with well defined toxicological characteristics to establish signatures of toxicity. Multiple consortia have begun to address this problem (4, 5) and substantial datasets arising from these consortia will continue to be published going forward. The flexibility and scalability of SRM, as illustrated in this proof of principle study, makes it an ideal candidate for integrating putative biomarkers from these diverse data types into panels for testing in large-scale verification studies, and as a tool for toxicology evaluation in drug development.
Data Availability
Raw data associated with the label-free LC-MS and SRM studies in the project are available in the mzML format from Tranche (www.proteomecommons.org) (57) using the hash CE7M45cJHCYhqvlL7CLvR/VpB0bZIdfH9Rck0fgX1ahymwr3NFdELucR50iZpregHUZ3MfMUHHevvWgbuNI8M3UAELsAAAAAAAApIA== and raw data associated with the rat liver MS/MS spectral library are available from PeptideAtlas (www.peptideatlas.org) (28) using the accession “PAe001466.” All information regarding the SRM assays and the related spectral library is available in the Skyline share archive file in the supplemental material and can be inspected using the freely available software Skyline (25). Additionally, a spreadsheet containing key information regarding the configured SRM assays (transitions, etc.) is included in the supplementary material.
Footnotes
* We gratefully acknowledge funding for BC from the Agilent Foundation under a UCD Newman Fellowship. Equipment funding and support for technical assistance is acknowledged from Science Foundation Ireland and UCD. The UCD Conway Institute Mass Spectrometry Resource is acknowledged for instrument support and technical assistance. The UCD Conway Institute and the Proteome Research Centre is funded by the Programme for Research in Third Level Institutions, as administered by the Higher Education Authority of Ireland. We would like to thank all other members of the InnoMed PredTox Consortium. Funding of the Integrated Project, InnoMed, is acknowledged under the European Union's Sixth Framework Programme.
 This article contains supplemental Methods, Figs. S1 to S19, Tables S1 to S8.
This article contains supplemental Methods, Figs. S1 to S19, Tables S1 to S8.
1 The abbreviations used are:
- SRM
- selected reaction monitoring
- ACN
- acetonitrile
- EST
- expressed sequence tag
- FDR
- false discovery rate
- IAA
- iodoacetamide
- nLC-MS/MS
- nanoflow liquid chromatography – tandem mass spectrometry
- PTM
- post translational modification
- QqTOF
- quadrupole time-of-flight mass spectrometer
- QqQ
- triple quadrupole mass spectrometer
- TCEP
- tris(2-carboxyethyl)phosphine.
REFERENCES
- 1. Kola I., Landis J. (2004) Can the pharmaceutical industry reduce attrition rates? Nat. Rev. Drug Discov. 3, 711–715 [DOI] [PubMed] [Google Scholar]
- 2. Waters M. D., Fostel J. M. (2004) Toxicogenomics and systems toxicology: aims and prospects. Nat. Rev. Genet. 5, 936–948 [DOI] [PubMed] [Google Scholar]
- 3. Dieterle F., Sistare F., Goodsaid F., Papaluca M., Ozer J. S., Webb C. P., Baer W., Senagore A., Schipper M. J., Vonderscher J., Sultana S., Gerhold D. L., Phillips J. A., Maurer G., Carl K., Laurie D., Harpur E., Sonee M., Ennulat D., Holder D., Andrews-Cleavenger D., Gu Y. Z., Thompson K. L., Goering P. L., Vidal J. M., Abadie E., Maciulaitis R., Jacobson-Kram D., Defelice A. F., Hausner E. A., Blank M., Thompson A., Harlow P., Throckmorton D., Xiao S., Xu N., Taylor W., Vamvakas S., Flamion B., Lima B. S., Kasper P., Pasanen M., Prasad K., Troth S., Bounous D., Robinson-Gravatt D., Betton G., Davis M. A., Akunda J., McDuffie J. E., Suter L., Obert L., Guffroy M., Pinches M., Jayadev S., Blomme E. A., Beushausen S. A., Barlow V. G., Collins N., Waring J., Honor D., Snook S., Lee J., Rossi P., Walker E., Mattes W. (2010) Renal biomarker qualification submission: a dialog between the FDA-EMEA and Predictive Safety Testing Consortium. Nat. Biotechnol. 28, 455–462 [DOI] [PubMed] [Google Scholar]
- 4. Mattes W. B. (2008) Public consortium efforts in toxicogenomics. Methods Mol. Biol. 460, 221–238 [DOI] [PubMed] [Google Scholar]
- 5. Gallagher W. M., Tweats D., Koenig J. (2009) Omic profiling for drug safety assessment: current trends and public-private partnerships. Drug Discov. Today 14, 337–342 [DOI] [PubMed] [Google Scholar]
- 6. Suter L., Schroeder S., Meyer K., Gautier J. C., Amberg A., Wendt M., Gmuender H., Mally A., Boitier E., Ellinger-Ziegelbauer H., Matheis K., Pfannkuch F. (2011) EU Framework 6 Project: Predictive Toxicology (PredTox)-overview and outcome. Toxicol. Appl. Pharmacol. 252, 73–84 [DOI] [PubMed] [Google Scholar]
- 7. Collins B. C., Sposny A., McCarthy D., Brandenburg A., Woodbury R., Pennington S. R., Gautier J. C., Hewitt P., Gallagher W. M. (2010) Use of SELDI MS to discover and identify potential biomarkers of toxicity in InnoMed PredTox: a multi-site, multi-compound study. Proteomics 10, 1592–1608 [DOI] [PubMed] [Google Scholar]
- 8. Com E., Gruhler A., Courcol M., Gautier J. C. (2011) Protocols of two-dimensional difference gel electrophoresis to investigate mechanisms of toxicity. Methods Mol. Biol. 691, 187–203 [DOI] [PubMed] [Google Scholar]
- 9. Sposny A., Schmitt C. S., Hewitt P. (2011) Mechanistic investigation of EMD 335 823s hepatotoxicity using multiple omics profiling technologies, in Handbook of Systems Toxicology, Casciano D. A., Saura S. C., Editors., John Wiley & Sons Ltd. pp. 389–346 [Google Scholar]
- 10. America A. H., Cordewener J. H. (2008) Comparative LC-MS: a landscape of peaks and valleys. Proteomics 8, 731–749 [DOI] [PubMed] [Google Scholar]
- 11. Amacher D. E. (2010) The discovery and development of proteomic safety biomarkers for the detection of drug-induced liver toxicity. Toxicol. Appl. Pharmacol. 245, 134–142 [DOI] [PubMed] [Google Scholar]
- 12. Lange V., Picotti P., Domon B., Aebersold R. (2008) Selected reaction monitoring for quantitative proteomics: a tutorial. Mol. Syst. Biol. 4, 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Anderson N. L., Anderson N. G. (2002) The human plasma proteome: history, character, and diagnostic prospects. Mol. Cell. Proteomics 1, 845–867 [DOI] [PubMed] [Google Scholar]
- 14. Lundgren D. H., Hwang S. I., Wu L., Han D. K. (2010) Role of spectral counting in quantitative proteomics. Expert Rev. Proteomics 7, 39–53 [DOI] [PubMed] [Google Scholar]
- 15. Schmidt A., Claassen M., Aebersold R. (2009) Directed mass spectrometry: towards hypothesis-driven proteomics. Curr. Opin. Chem. Biol. 13, 510–517 [DOI] [PubMed] [Google Scholar]
- 16. Domon B., Aebersold R. (2010) Options and considerations when selecting a quantitative proteomics strategy. Nat. Biotechnol. 28, 710–721 [DOI] [PubMed] [Google Scholar]
- 17. Kitteringham N. R., Jenkins R. E., Lane C. S., Elliott V. L., Park B. K. (2009) Multiple reaction monitoring for quantitative biomarker analysis in proteomics and metabolomics. J. Chromatogr. B 877, 1229–1239 [DOI] [PubMed] [Google Scholar]
- 18. Picotti P., Bodenmiller B., Mueller L. N., Domon B., Aebersold R. (2009) Full dynamic range proteome analysis of S. cerevisiae by targeted proteomics. Cell 138, 795–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jovanovic M., Reiter L., Picotti P., Lange V., Bogan E., Hurschler B. A., Blenkiron C., Lehrbach N. J., Ding X. C., Weiss M., Schrimpf S. P., Miska E. A., Grosshans H., Aebersold R., Hengartner M. O. (2010) A quantitative targeted proteomics approach to validate predicted microRNA targets in C. elegans. Nat. Methods 7, 837–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lange V., Malmström J. A., Didion J., King N. L., Johansson B. P., Schäfer J., Rameseder J., Wong C. H., Deutsch E. W., Brusniak M. Y., Buhlmann P., Björck L., Domon B., Aebersold R. (2008) Targeted quantitative analysis of Streptococcus pyogenes virulence factors by multiple reaction monitoring. Mol. Cell. Proteomics 7, 1489–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Addona T. A., Abbatiello S. E., Schilling B., Skates S. J., Mani D. R., Bunk D. M., Spiegelman C. H., Zimmerman L. J., Ham A. J., Keshishian H., Hall S. C., Allen S., Blackman R. K., Borchers C. H., Buck C., Cardasis H. L., Cusack M. P., Dodder N. G., Gibson B. W., Held J. M., Hiltke T., Jackson A., Johansen E. B., Kinsinger C. R., Li J., Mesri M., Neubert T. A., Niles R. K., Pulsipher T. C., Ransohoff D., Rodriguez H., Rudnick P. A., Smith D., Tabb D. L., Tegeler T. J., Variyath A. M., Vega-Montoto L. J., Wahlander A., Waldemarson S., Wang M., Whiteaker J. R., Zhao L., Anderson N. L., Fisher S. J., Liebler D. C., Paulovich A. G., Regnier F. E., Tempst P., Carr S. A. (2009) Multi-site assessment of the precision and reproducibility of multiple reaction monitoring-based measurements of proteins in plasma. Nat. Biotechnol. 27, 633–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schiess R., Wollscheid B., Aebersold R. (2009) Targeted proteomic strategy for clinical biomarker discovery. Mol. Oncol. 3, 33–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Whiteaker J. R., Zhang H., Zhao L., Wang P., Kelly-Spratt K. S., Ivey R. G., Piening B. D., Feng L. C., Kasarda E., Gurley K. E., Eng J. K., Chodosh L. A., Kemp C. J., McIntosh M. W., Paulovich A. G. (2007) Integrated pipeline for mass spectrometry-based discovery and confirmation of biomarkers demonstrated in a mouse model of breast cancer. J. Proteome Res. 6, 3962–3975 [DOI] [PubMed] [Google Scholar]
- 24. Jenkins R. E., Kitteringham N. R., Hunter C. L., Webb S., Hunt T. J., Elsby R., Watson R. B., Williams D., Pennington S. R., Park B. K. (2006) Relative and absolute quantitative expression profiling of cytochromes P450 using isotope-coded affinity tags. Proteomics 6, 1934–1947 [DOI] [PubMed] [Google Scholar]
- 25. Maclean B., Tomazela D. M., Shulman N., Chambers M., Finney G. L., Frewen B., Kern R., Tabb D. L., Liebler D. C., Maccoss M. J. (2010) Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cham Mead J. A., Bianco L., Bessant C. (2010) Free computational resources for designing selected reaction monitoring transitions. Proteomics 10, 1106–1126 [DOI] [PubMed] [Google Scholar]
- 27. Picotti P., Rinner O., Stallmach R., Dautel F., Farrah T., Domon B., Wenschuh H., Aebersold R. (2010) High-throughput generation of selected reaction-monitoring assays for proteins and proteomes. Nat. Methods 7, 43–46 [DOI] [PubMed] [Google Scholar]
- 28. Deutsch E. W., Lam H., Aebersold R. (2008) PeptideAtlas: a resource for target selection for emerging targeted proteomics workflows. EMBO Rep. 9, 429–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mulrane L., Rexhepaj E., Smart V., Callanan J. J., Orhan D., Eldem T., Mally A., Schroeder S., Meyer K., Wendt M., O'Shea D., Gallagher W. M. (2008) Creation of a digital slide and tissue microarray resource from a multi-institutional predictive toxicology study in the rat: an initial report from the PredTox group. Exp. Toxicol. Pathol. 60, 235–245 [DOI] [PubMed] [Google Scholar]
- 30. Adler M., Hoffmann D., Ellinger-Ziegelbauer H., Hewitt P., Matheis K., Mulrane L., Gallagher W. M., Callanan J. J., Suter L., Fountoulakis M. M., Dekant W., Mally A. (2010) Assessment of candidate biomarkers of drug-induced hepatobiliary injury in preclinical toxicity studies. Toxicol. Lett. 196, 1–11 [DOI] [PubMed] [Google Scholar]
- 31. Hoffmann D., Adler M., Vaidya V. S., Rached E., Mulrane L., Gallagher W. M., Callanan J. J., Gautier J. C., Matheis K., Staedtler F., Dieterle F., Brandenburg A., Sposny A., Hewitt P., Ellinger-Ziegelbauer H., Bonventre J. V., Dekant W., Mally A. (2010) Performance of novel kidney biomarkers in preclinical toxicity studies. Toxicol. Sci. 116, 8–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schmidt A., Gehlenborg N., Bodenmiller B., Mueller L. N., Campbell D., Mueller M., Aebersold R., Domon B. (2008) An integrated, directed mass spectrometric approach for in-depth characterization of complex peptide mixtures. Mol. Cell. Proteomics 7, 2138–2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Perkins D. N., Pappin D. J., Creasy D. M., Cottrell J. S. (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- 34. Craig R., Beavis R. C. (2004) TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20, 1466–1467 [DOI] [PubMed] [Google Scholar]
- 35. MacLean B., Eng J. K., Beavis R. C., McIntosh M. (2006) General framework for developing and evaluating database scoring algorithms using the TANDEM search engine. Bioinformatics 22, 2830–2832 [DOI] [PubMed] [Google Scholar]
- 36. Searle B. C., Turner M., Nesvizhskii A. I. (2008) Improving sensitivity by probabilistically combining results from multiple MS/MS search methodologies. J. Proteome Res. 7, 245–253 [DOI] [PubMed] [Google Scholar]
- 37. Keller A., Nesvizhskii A. I., Kolker E., Aebersold R. (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- 38. Choi H., Nesvizhskii A. I. (2008) Semisupervised model-based validation of peptide identifications in mass spectrometry-based proteomics. J. Proteome Res. 7, 254–265 [DOI] [PubMed] [Google Scholar]
- 39. Nesvizhskii A. I., Keller A., Kolker E., Aebersold R. (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 [DOI] [PubMed] [Google Scholar]
- 40. Keller A., Eng J., Zhang N., Li X. J., Aebersold R. (2005) A uniform proteomics MS/MS analysis platform utilizing open XML file formats. Mol. Syst. Biol. 1, 2005.0017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Deutsch E. W., Mendoza L., Shteynberg D., Farrah T., Lam H., Tasman N., Sun Z., Nilsson E., Pratt B., Prazen B., Eng J. K., Martin D. B., Nesvizhskii A. I., Aebersold R. (2010) A guided tour of the Trans-Proteomic Pipeline. Proteomics 10, 1150–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Frewen B. E., Merrihew G. E., Wu C. C., Noble W. S., MacCoss M. J. (2006) Analysis of peptide MS/MS spectra from large-scale proteomics experiments using spectrum libraries. Anal. Chem. 78, 5678–5684 [DOI] [PubMed] [Google Scholar]
- 43. Smedley D., Haider S., Ballester B., Holland R., London D., Thorisson G., Kasprzyk A. (2009) BioMart–biological queries made easy. BMC Genomics 10, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410 [DOI] [PubMed] [Google Scholar]
- 45. Sherwood C. A., Eastham A., Lee L. W., Risler J., Vitek O., Martin D. B. (2009) Correlation between y-type ions observed in ion trap and triple quadrupole mass spectrometers. J. Proteome Res. 8, 4243–4251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cattley R. C., Popp J. A. (2002) Liver, in Handbook of Toxicologic Pathology, 2nd Edition, Haschek W. M., Rousseaux C. G., Wallig M. A., Editors., Academic Press [Google Scholar]
- 47. Prakash A., Tomazela D. M., Frewen B., Maclean B., Merrihew G., Peterman S., Maccoss M. J. (2009) Expediting the development of targeted SRM assays: using data from shotgun proteomics to automate method development. J. Proteome Res. 8, 2733–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wheeler D. L., Barrett T., Benson D. A., Bryant S. H., Canese K., Chetvernin V., Church D. M., Dicuccio M., Edgar R., Federhen S., Feolo M., Geer L. Y., Helmberg W., Kapustin Y., Khovayko O., Landsman D., Lipman D. J., Madden T. L., Maglott D. R., Miller V., Ostell J., Pruitt K. D., Schuler G. D., Shumway M., Sequeira E., Sherry S. T., Sirotkin K., Souvorov A., Starchenko G., Tatusov R. L., Tatusova T. A., Wagner L., Yaschenko E. (2008) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 36, D13–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kanehisa M., Goto S. (2000) KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dasari S., Chambers M. C., Codreanu S. G., Liebler D. C., Collins B. C., Pennington S. R., Gallagher W. M., Tabb D. L. (2011) Sequence tagging reveals unexpected modifications in toxicoproteomics. Chem. Res. Toxicol. 24, 204–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. de Godoy L. M., Olsen J. V., Cox J., Nielsen M. L., Hubner N. C., Fröhlich F., Walther T. C., Mann M. (2008) Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature 455, 1251–1254 [DOI] [PubMed] [Google Scholar]
- 52. Xia D., Sanderson S. J., Jones A. R., Prieto J. H., Yates J. R., Bromley E., Tomley F. M., Lal K., Sinden R. E., Brunk B. P., Roos D. S., Wastling J. M. (2008) The proteome of Toxoplasma gondii: integration with the genome provides novel insights into gene expression and annotation. Genome Biol. 9, R116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Reiter L., Rinner O., Picotti P., Huttenhain R., Beck M., Brusniak M. Y., Hengartner M. O., Aebersold R. (2011) mProphet: automated data processing and statistical validation for large-scale SRM experiments. Nat Methods 8, 430–435 [DOI] [PubMed] [Google Scholar]
- 54. Witten D., Tibshirani A. (2007) A comparison of fold change and the t-statistic for microarray data analysis. Technical Report, Stanford University. [Google Scholar]
- 55. McCarthy D. J., Smyth G. K. (2009) Testing significance relative to a fold-change threshold is a TREAT. Bioinformatics 25, 765–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Margolin A. A., Ong S. E., Schenone M., Gould R., Schreiber S. L., Carr S. A., Golub T. R. (2009) Empirical Bayes analysis of quantitative proteomics experiments. PLoS One 4, e7454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hill J. A., Smith B. E., Papoulias P. G., Andrews P. C. (2010) ProteomeCommons.org collaborative annotation and project management resource integrated with the Tranche repository. J. Proteome Res. 9, 2809–2811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Elias J. E., Gygi S. P. (2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods 7, 207–214 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Raw data associated with the label-free LC-MS and SRM studies in the project are available in the mzML format from Tranche (www.proteomecommons.org) (57) using the hash CE7M45cJHCYhqvlL7CLvR/VpB0bZIdfH9Rck0fgX1ahymwr3NFdELucR50iZpregHUZ3MfMUHHevvWgbuNI8M3UAELsAAAAAAAApIA== and raw data associated with the rat liver MS/MS spectral library are available from PeptideAtlas (www.peptideatlas.org) (28) using the accession “PAe001466.” All information regarding the SRM assays and the related spectral library is available in the Skyline share archive file in the supplemental material and can be inspected using the freely available software Skyline (25). Additionally, a spreadsheet containing key information regarding the configured SRM assays (transitions, etc.) is included in the supplementary material.