Abstract
The Ku heterodimer plays an essential role in non-homologous end-joining and other cellular processes including transcription, telomere maintenance and apoptosis. While the function of Ku is regulated through its association with other proteins and nucleic acids, the specific composition of these macromolecular complexes and their dynamic response to endogenous and exogenous cellular stimuli are not well understood. Here we use quantitative proteomics to define the composition of Ku multicomponent complexes and demonstrate that they are dramatically altered in response to UV radiation. Subsequent biochemical assays revealed that the presence of DNA ends leads to the substitution of RNA-binding proteins with DNA and chromatin associated factors to create a macromolecular complex poised for DNA repair. We observed that dynamic remodeling of the Ku complex coincided with exit of Ku and other DNA repair proteins from the nucleolus. Microinjection of sheared DNA into live cells as a mimetic for double strand breaks confirmed these findings in vivo.
The Ku heterodimer (hereafter referred to as “Ku”) is composed of 70 kDa (Ku70) and 86 kDa (Ku86) subunits that associate in stoichiometric quantities and bind DNA ends with high affinity as a first step in the nonhomologous end-joining DNA repair pathway. Ku binding to double strand breaks results in recruitment of the catalytic subunit of DNA-dependent protein kinase DNA-PKcs to form the holo-enzyme DNA-PK, followed by the exonuclease Artemis and the XRCC4-like factor XLF (1). The final stage of repair requires polymerase activity and the ligation of the DNA ends by a complex composed of the x-ray repair cross-complementing protein XRCC4 and DNA-ligase-IV (2–4). In addition to its DNA-directed activities, Ku and DNA-PKcs are known to regulate other cellular functions such as RNA-transcription (5–11), processing (12, 13) and telomere maintenance (12, 14).
The functional diversity of Ku suggests that its cellular activities are regulated by distinct sets of molecular interactions. To date, Ku protein partners include the RNA helicase RHA and several heterogeneous ribonucleoproteins (13), the Werner Syndrome associated protein WRN (15–17), poly(ADP-ribose) polymerase I PARP-1 (17, 18), the yin-yang transcription factor YY1 (19), histone H2AX (20), and telomerase (12, 14). Binary protein-protein interactions with Ku70 were also delineated by two-hybrid screen (21), whereas mass spectrometry-based approaches have been used to identify multicomponent protein complexes that copurify with Ku (22, 23). These data provide a useful inventory of Ku protein partners, although the dynamics of specific interactions in response to exogenous or endogenous stimuli remain unknown. In addition, the mechanisms by which Ku selectively engages with DNA versus RNA and whether these nucleic acid interactions are dependent on Ku protein associations, is largely unexplored. Finally, although Ku resides in distinct nuclear compartments (24, 25), the relationship between Ku localization and the molecular composition of its associated macromolecular complex is unresolved.
In this study, we used biochemical and proteomics approaches to delineate Ku molecular interactions, localization, and dynamic response to DNA damage. Ultraviolet (UV) irradiation resulted in the recruitment of DNA-binding proteins to the multi-component Ku complex and a concomitant loss of RNA-binding proteins. Addition of DNA ends to the Ku complex in vitro induced the loss of RNA-mediated protein interactions. Moreover, we observed a striking translocation of Ku from the nucleolus to the nucleoplasm upon microinjection of DNA ends into living cells. Our data support a model in which DNA ends act as a molecular switch, mediating the transition of Ku from a ribonucleoprotein complex into a DNA-bound protein complex poised to initiate DNA repair.
EXPERIMENTAL PROCEDURES
Cell Culture
MDA-MB-231 human breast adenocarcinoma cells (American Type Culture Collection) were cultured in RPMI medium containing 10% (v/v) fetal bovine serum, 100 units/ml penicillin, and 100 mg/ml streptomycin (Invitrogen). HeLa-S3 cells (American Type Culture Collection) where cultured in DMEM containing 10% (v/v) newborn calf serum (Invitrogen, Carlsbad, CA) and antibiotics.
Immunofluorescence
Cells grown on 12 mm glass cover-slips were fixed with 4% paraformaldehyde for 10 min, extracted with methanol for 15 min and washed with phosphate-buffered saline (PBS). To optimize nucleolar visualization, cells were first permeabilized with 0.1% Triton X-100 for 30 s on ice, and subsequently fixed with 4% paraformaldehyde for 10 min (T/PFA1). The cells were incubated with the following primary antibodies: mouse anti-Ku86 (111, NeoMarkers), mouse anti Ku-70 (N3H10, NeoMarkers), mouse anti-SSRP1 (Biolegend, San Diego, CA), mouse anti-WRN (Sigma), mouse anti-RPL19 (Sigma), mouse anti-B23/NPM1 (Santa Cruz Biotechnology, Santa Cruz, CA), rabbit anti-NOL1/NOP2 (Calbiochem, San Diego, CA), all at a 1:100 dilution with the exception of 1:400 dilution of NOL1, for 1 h at 37 °C. Goat anti-rabbit and goat anti-mouse IgG conjugated to Alexa Fluor-488 and Alexa Fluor-594 secondary antibodies (Molecular Probes, Eugene, OR) were diluted 1:300 in PBS. DNA was visualized by DAPI staining (Sigma). Cover slips were mounted with ClearMount (Zymed Laboratories Inc. South San Francisco, CA) for microinjection experiments, or mounted with VectaShield hard set mounting medium.
Immunoblotting
Protein samples were heated at 70 °C for 10 min in reducing buffer, separated by NuPAGE electrophoresis (Invitrogen) and transferred to nitrocellulose membranes (Bio-Rad, Hercules, CA) before immunoblotting. We used mouse antibodies directed against Ku70, Ku86 (see above), DNA-PKcs (18–2, Neomarkers), SSRP1 and SPT16 (Biolegends) and FLAG (Sigma). Rabbit antibodies were used against WRN (Santa Cruz Biotechnology), RHA (a kind gift from Dr. Jeff Parvin, The Ohio State University), YB-1, and Histone H2A (Cell Signaling Technologies, Danvers, MA). Anti-mouse and anti-rabbit secondary antibodies were from Amersham Biosciences, Pierce, or Cell Signaling. Immunoreactive bands were visualized with Supersignal West Pico chemiluminescent reagent (Pierce) and a Fujifilm luminescent image analyzer LAS (Fuji, Tokyo Japan).
Nuclear Extract Preparation and Tandem Affinity Purification (TAP)
Parental HeLa-S3 cells or HeLa-S3 cells expressing a doubly tagged FLAG-HA Ku86 (supplemental Fig. S1) were mechanically harvested and washed with PBS. Nuclear extracts were prepared as previously described (26). Briefly, HeLa-S3 cells were resuspended in hypotonic buffer (10 mm Tris pH 8, 10 mm KCl, 1.5 mm MgCl2, 10 mm 2-mercapto-ethanol, and a 1 × protease inhibitor mix [Roche]) and incubated on ice for 10 min. The cell membranes were then disrupted using a dounce tissue grinder (Wheaton) and nuclei were collected by centrifugation at 1000 × g for 10 min at 4 °C, resuspended in 1 ml of hypotonic buffer and centrifuged as above. Nuclei were flash frozen in liquid nitrogen and stored at −80 °C until use. The resulting nuclear pellet was extracted with four volumes of FLAG-IP/lysis buffer (50 mm Tris pH 8, 150 mm NaCl, 10% glycerol, 2 mm EDTA, 0.5% Nonidet P-40 and a 1 × protease inhibitor mix (Roche) for 45 min at 4 °C. The insoluble material was pelleted by centrifugation at 15,000 × g for 10 min at 4 °C. The resulting supernatant was called “nuclear extract.” The Ku86 complex was purified through tandem affinity purification using FLAG and HA antibody agarose conjugates (Sigma and Santa-Cruz, respectively) (27). Purified complexes were separated on 4–12% NuPAGE BisTris gels (Invitrogen), and visualized by SilverQuest silver staining (Invitrogen).
RNA Analysis
Protein complexes were purified by TAP except that 20 units/ml of RNaseOUT (Invitrogen) was added during the procedure. Coprecipitating RNAs were resolved on 1.2% agarose gels for the analysis of the 18 and 5.8S ribosomal RNA or on a 6% acrylamide/urea gel for U3snoRNA and transferred to nylon membranes. The 18S and 5.8S probes were amplified from genomic DNA using the following primers: 18S-Fwd: GTA GTG ACG AAA AAT AAC; 18S-Rev AAG GGC AGG GAC TTA ATC; 5.8S-Fwd TCG TGC GTC GAT GAA GAA; 5.8S-Rev CGC TCA GAC AGG CGT AGC. PCR products were gel purified and uniformly labeled with α-p32 dCTP. U3snoRNA was detected using the single-stranded oligonucleotide: ACC ACT CAG ACC GCG TTC TCT CCC TCT CAC labeled at the 5′ end. Quantitative measurement of 18S ribosomal RNA was performed after purification of co-precipitating RNA using the RNeasy micro kit (Qiagen, Valencia, CA), reverse transcription (Superscript III, Invitrogen) and real-time qPCR using TaqMan assay Hs03928985_g1 (Applied Biosystems). The abundance of the RNA was calculated using the 2−ΔCt method and normalized to the abundance of Ku-FLAG-HA in each purified complex (28). The average abundance and standard deviation across three technical replicates was calculated and plotted relative to the normalized abundance of 18S present in the complex at t = 0. RNA from the t = 0 condition that were not submitted to reverse transcription were used as negative control. This −RT control did not yield any detectable signal in a parallel assay.
Nuclease Treatment
Ku86 protein complexes were purified by FLAG affinity purification as described above and kept on beads. Anti-FLAG agarose beads were washed sequentially in lysis buffer and in nuclease digestion buffer (50 mm NaCl, 10 mm Tris pH 7.5, 10% Glycerol, 2.5 mm MgCl2 and 0.5 mm CaCl2). The beads were resuspended in nuclease digestion buffer and equally distributed between three tubes containing either: 10 μl of RNase-free DNase (Qiagen) and 1 μl RNase-OUT (Invitrogen); 2 μl of DNase-free RNase (Roche) and 1 mm EDTA or buffer only. Reactions were carried for 15 min at room temperature (RT) and stopped by two washes in lysis buffer. The protein complexes were eluted by excess FLAG peptide and were either directly analyzed by silver staining and immunoblotting using indicated antibodies or further purified by HA IP before analysis.
Chromatin and Plasmid DNA Preparation and Competition Experiments
Recombinant pcs2 plasmid was prepared using a maxi-preparation kit (Qiagen) and resuspended at a concentration of 0.5 μg/μl in water. To obtain chromatin, HeLa-S3 cell nuclei were extracted with lysis buffer as described above. Insoluble material (chromatin) was pelleted by centrifugation at 15,000 × g for 30 min at 4 °C and gently re-suspended in one volume of lysis buffer. Purified chromatin or plasmid DNA were UV irradiated (1000 J/m2) on small drops of DNA solution spread on Parafilm (Pechiney). For chromatin competition, 50 μl of intact, sonicated or UV irradiated chromatin was added to the nuclear extract at the start of the purification and protein complexes were purified by tandem FLAG-HA purification as described above. For plasmid competition, 20 μl of either intact, sonicated (as a mimic for double strand breaks) or UV irradiated plasmid was added to Ku protein complexes immobilized on anti-FLAG agarose beads and incubated at 4 °C for 30 min. Beads were subsequently pelleted and supernatants collected. Ku86 complexes were eluted from the beads with excess FLAG peptide as described above. Supernatants and complexes were analyzed by silver staining and immunoblotting.
UV Irradiation and Biological Replicates
Six 15 cm plates of parental or HeLa-S3 FLAG-HA Ku86 were processed for each time point. Media was removed from the plates just prior to irradiation and centrifuged at 500 × g for 5 min to remove floating cells. UV-C light (300 J/m2) was administered to cells using a Stratalinker 2400 (Stratagene). The old media, supplemented with 5 ml of fresh media was added back to the plates after irradiation. Cells were collected and fractionated 30 min, 2.5 or 5 h after UV irradiation. Control cells for the reference time point (t = 0) were processed as above but not irradiated. Biological replicates consisted of two independent experiments (irradiation, extract preparation and TAP) performed several weeks apart.
Protein Digestion, iTRAQ Labeling, and LC-MS/MS Data Acquisition
Proteins were denatured and reduced by adding an equal volume of a solution containing 0.2% RapiGest (Waters), and 10 mm dithiotreitol, and incubating for 30 min at 56 °C. Reduced proteins were digested with 500 ng of trypsin overnight at 37 °C. The resulting tryptic peptides were desalted on 96-well plate embedded with reversed phase resin (C18-Microelution plate, Waters, Milford, MA): each well was equilibrated by a succession of washes with 0.1% trifluoroacetic acid (TFA), 80% methanol, and two final washes with 0.1% TFA (400 μl each). Tryptic peptides were diluted with an equal volume of 0.1% TFA and applied to equilibrated wells. Bound peptides were washed twice with 0.1% TFA (450 μl). After the final wash, peptides were eluted with 80% methanol (150 μl) and dried by vacuum centrifugation. Excess HA peptide was removed as previously described (29). For isobaric tags for relative and absolute quantification (iTRAQ) labeling, each of the four samples before (t = 0) or 0.5, 2.5, and 5 h after UV irradiation was re-suspended in 30 μl of 0.5 m TEAB, and labeled for 1 h at RT with 70 μl of the appropriate isobaric tag reagents solution (Applied Biosystems): 114 (t = 0); 115 (t = 0.5); 116 (t = 2.5); 117 (t = 5). Each sample was dried by vacuum centrifugation, re-suspended in 0.1% TFA (35 μl) and the four samples were pooled. The combined, labeled peptide mixture was again desalted on a C18-Microelution plate as described above, dried by vacuum centrifugation and re-suspended in a 25% acetonitrile and 0.1% formic acid solution. Peptides were fractionated by strong cation exchange chromatography (SCX) in batch format (Poros HS 20, Applied Biosystems), captured on SCX media previously equilibrated with loading buffer (25% acetonitrile, 0.1% formic acid) and washed with loading buffer (50 μl). Bound peptides were sequentially eluted with 20 μl of the loading buffer containing increasing concentrations of KCl (50, 100, 150, 200, and 300 mm). The eluates, along with the column flow-through collected during sample loading, were dried to 5 μl by vacuum centrifugation, and diluted to 30 μl with 0.1% TFA. SCX eluates and the flow-through samples were loaded onto a precolumn using a NanoAcquity Sample Manager and UPLC pump. Peptides were gradient eluted (1–30% B in 120 min; A = 0.2 m aqueous acetic acid, B = 0.2 m acetic acid in acetonitrile) at a flow rate of < 50 nL/min to an analytical column as described previously (30, 31). Mass spectrometry data acquisition was performed in information dependent mode on the QSTAR Elite equipped with a Digital Picoview ESI positioning platform (New Objective, Woburn, MA) using the following settings: MS scan 300 ≤ m/z ≤ 1500; top eight most abundant MS/MS scans using low resolution for precursor isolation; 2.0 s. accumulation with enhance all mode; 1.8 kV ESI voltage.
LC-MS/MS Data Analysis
LC-MS/MS data were submitted in native format to ProteinPilot (version 3.0, Revision Number 114732, Applied Biosystems) using the default parameters for the precursor and fragment ion mass tolerance. The Paragon algorithm (version 3.0.0.0, 113442) was used to search the Homo sapiens subset of the RefSeq database (downloaded September 2010) containing 38939 entries. The search parameters included trypsin as the enzyme specificity, iTRAQ as a fixed modification on Lysine residues and peptide amino termini. An unused ProtScore threshold > 1.30 was specified, corresponding to a protein confidence score of 95%. The RefSeq accession number for the representative member (“winner”) of each protein group was mapped to the corresponding Entrez Gene ID and official HGNC Gene Symbol. This Gene ID was used to match proteins across the two biological replicates. The peptide and protein summary data files were exported in comma-separated format for further analysis in spreadsheet format. Final peptide and protein data, along with annotated MS/MS spectra were exported to mzResults files (32) and are provided in Supplementary Materials. The iTRAQ115/iTRAQ114, iTRAQ116/iTRAQ114 and iTRAQ117/iTRAQ114 ratio of Ku86 was used to normalize the corresponding ratio for all proteins detected. The logarithm of normalized ratios was calculated for the subset of proteins common between the two biological replicates for which quantitative data was available. An unweighted pair group method with arithmetic mean (UPGMA) approach was used for unsupervised hierarchical clustering (Spotfire, Tibco, Palo Alto, CA). All mzResults (32) files which include annotated MS/MS spectra are available as supplementary files. Annotated MS/MS spectra for peptides reported in supplemental Table S3 are also reported in spreadsheet format (supplemental Table S4 Annotated Spectra for Replicate Analyses of Ku Complexes). These along with the associated native mass spectrometry data files are also available for download at: http://blaispathways.dfci.harvard.edu/mz/?id=doi:10.1074/mcp.M111.013581.
Gene Annotation Co-occurrence Analysis
Gene IDs corresponding to proteins from each cluster were individually submitted to GeneCodis (version 2.0, http://genecodis.dacya.ucm.es/analysis/), selecting Homo sapiens as the source for the annotations. To accurately calculate statistical significance of gene annotation co-occurrence in our dataset, a subset of the human proteome corresponding to 2230 nuclear proteins with GO term #0031981 (“nuclear lumen”) and its children was assembled and used as background in the enrichment analysis.
Microinjection
Microinjections were performed under a Fluovert inverted microscope (Leitz) attached to a micromanipulator (Leitz) using femtotips pipettes (Eppendorf). Rabbit control marker IgG (Jackson Immunoresearch Laboratories, West Grove, PA) was microinjected at 0.5 mg/ml in PBS with or without plasmid DNA (0.1 mg/ml) as indicated. Microinjections were done into the nuclei of MDA-MB-231 cells at room temperature. The cells were returned to normal culture conditions immediately after microinjection and fixed for immunofluorescence 30 min later.
RESULTS
Ku Switches From a Ribonucleo- to a Deoxyribonucleo-Protein Complex in Response to UV-Induced DNA Damage
Although DNA-PK is best known for its role in response to double strand breaks (DSB) resulting from ionizing radiation, recent evidence has shown that its catalytic subunit (DNA-PKcs) is phosphorylated by ATR in response to UV-induced replication stress, suggesting a novel role in this particular DNA damage response pathway (33). We recently reported that the Ku heterodimer, which is essential for the recruitment of DNA-PKcs to sites of DNA damage, re-localizes from the nucleolus to the nucleoplasm after UV irradiation (34). This observation was corroborated in a recent study that broadly characterized the structural and proteomic changes in the nucleolus in response to UV radiation (35).
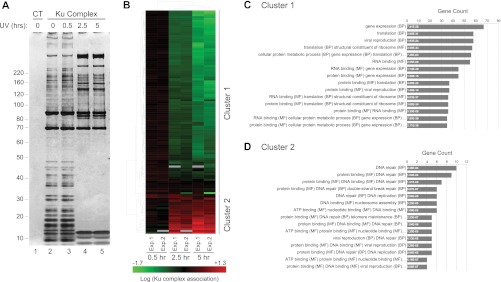
To determine the dynamics of Ku protein interactions, we isolated Ku86 by tandem affinity purification (TAP) from cells across a time course after UV irradiation. We observed the recruitment of several high molecular weight proteins at latter time points and the concomitant loss of low molecular weight proteins from the complex (Fig. 1A). To identify members of the Ku protein complex and quantify changes in their association in response to UV radiation, we utilized a gel-free, quantitative proteomics approach. Unsupervised hierarchical clustering of these data revealed two protein groups that readily segregated based on their relative abundance in the Ku complex after UV irradiation (Fig. 1B and supplemental Table S1 Summary of quantitative LC-MS data). Functional enrichment analysis indicated that Cluster 1 was enriched for cellular processes related to RNA metabolism (gene expression, translation, RNA binding, and ribosome structure) (Fig. 1C and supplemental Table S2 Functional enrichment analysis). By contrast, Cluster 2 was enriched for GO terms related to DNA repair, replication, and DNA binding (Fig. 1D). These data suggest that the complex had transitioned from an RNA-binding to a DNA-binding macromolecular assembly in response to UV radiation.
Fig. 1.
UV irradiation alters the composition of Ku86 macromolecular complexes. A, Protein complexes were purified from parental HeLa-S3 cells or HeLa-S3 cells expressing doubly tagged FLAG-HA Ku86 (supplemental Fig. S1). Representative silver stain analysis of nuclear Ku86 complexes purified before (t = 0, lane 2) or after (t = 0.5 h, 2.5 h, 5 h, lanes 3, 4, and 5, respectively) UV irradiation (300 J/m2), along with a “mock” complex purified from parental HeLa-S3 cells (lane 1). B, Hierarchical clustering of quantitative proteomic data obtained after iTRAQ LC-MS/MS analysis of biological replicates: 118 Ku86 associated proteins detected in two independent experiments (Exp.1 and Exp.2) primarily segregate between two clusters based on their response to UV irradiation. Green and red, respectively, indicate decreased and increased association after UV irradiation (see also supplemental Table S1 Summary of quantitative LC-MS data). (C and D) Functional enrichment analysis of proteins from Cluster 1 and 2. The number of genes represented in (B) for each category is provided on the horizontal axis. The corresponding adjusted p value is indicated within each bar. For clarity, only the first three co-occurrence terms are listed. See supplemental Table S2 Functional enrichment analysis, for the full list of annotations.
UV Radiation Induces Changes in the Localization of a Subset of Ku Interactors
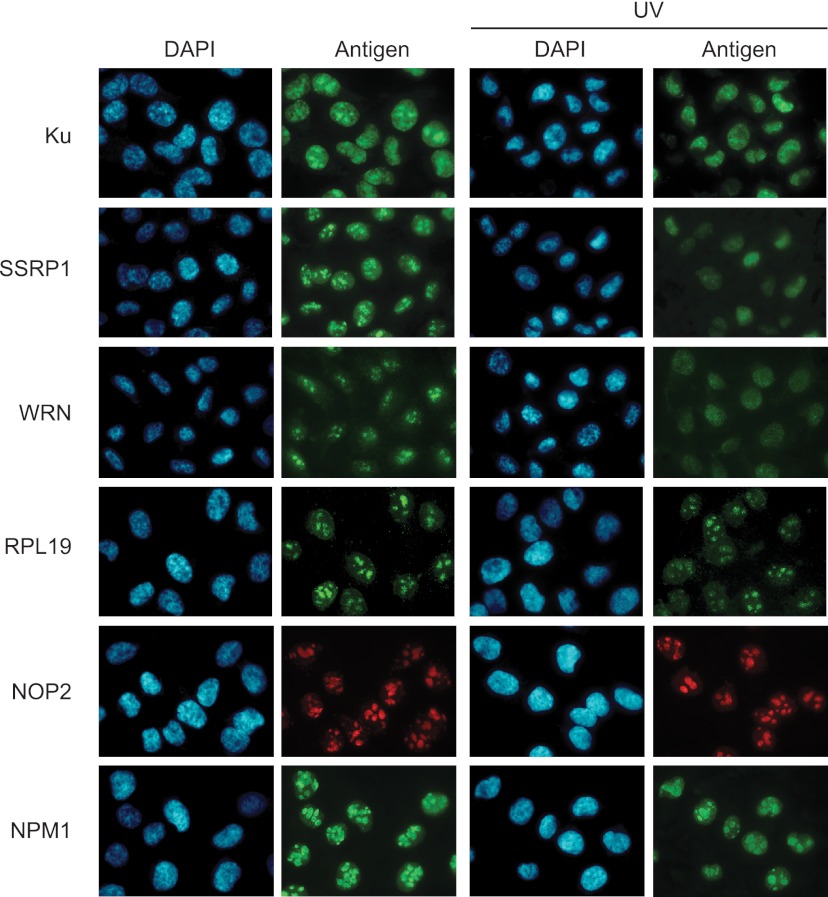
Because the nucleolar localization of Ku is lost upon genotoxic stress (34), we investigated UV-induced changes in subcellular localization of Ku-associated proteins (Fig. 1) in MDA-MB-231 breast cancer cells. Unlike HeLa-S3, these adherent cells are amenable to immunofluorescence staining and importantly, exhibit a DNA damage-dependent nucleolar fraction of Ku (34). Similar to the localization pattern observed for Ku, both SSRP1 and WRN were found in the nucleolus prior to UV irradiation but exhibited diffuse nuclear staining thereafter. In contrast, the 60S ribosomal protein RPL19 and the nucleolar protein NOP2 remained in the nucleolus after UV treatment, as did nucleophosmin NPM1, a nucleolar marker (Fig. 2). These observations, taken together with recent findings (35), suggest that Ku protein complexes exit the nucleolus in response to UV-induced DNA damage, and that this change in localization is accompanied by remodeling of Ku protein associations from largely RNA-binding to primarily DNA-binding proteins.
Fig. 2.
UV irradiation mediates nuclear re-localization of Ku complex components. Immunofluorescence analysis of Ku complex components in response to UV radiation. MDA-MB-231 cells were exposed to UV radiation and processed 2.5 h later for immunofluorescence following a triton extraction and paraformaldehyde fixation protocol (T/PFA). Ku, WRN and the DNA binding protein SSRP1 re-localize from the nucleolus to the nucleoplasm after irradiation with UV. The nucleolar and ribosomal proteins RPL19 and NOP2, which interact primarily with Ku in undamaged cells remain localized in the nucleolus after UV irradiation. The stability of staining for the nucleolar marker NPM1 indicates that nucleolar integrity is preserved after UV radiation.
Ku Interacts with Ribonucleoproteins in an RNA Dependent Manner
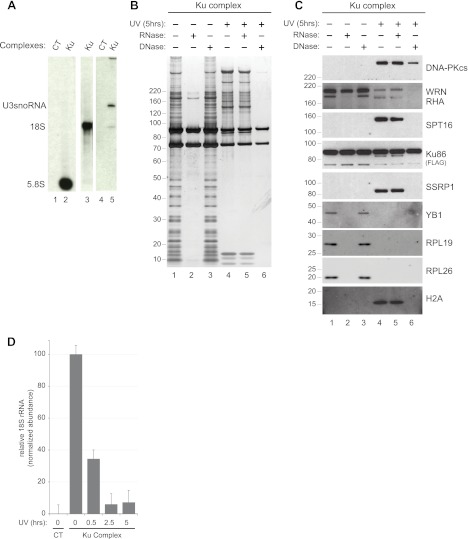
Because Ku resides at least partially in the nucleolus and interacts with various ribosomal proteins (Supplemental Table S1 Summary of quantitative LC-MS data), we asked whether ribosomal RNAs were also present in the Ku complex. Indeed, 5.8S and 18S ribosomal RNAs were specifically detected in purified Ku complexes (Fig. 3A). Interestingly, we also detected U3snoRNA, a small nucleolar RNA implicated in early pre-rRNA cleavage and 40S subunit assembly (36), consistent with the observation that Ku interacts with proteins involved in ribosomal RNA processing (the guanine nucleotide binding protein-like 3, GNL3, and NOP2) (37, 38) and ribosomal biogenesis (the biogenesis of ribosomes in Xenopus protein BRIX and the glutamate-rich WD repeat containing protein1, GRWD1) (supplemental Table S3 Peptide Data for Replicate Analyses of Ku Complex) (39, 40). These results demonstrate that both ribosomal RNAs and ribosomal proteins specifically associate with Ku in intact cells.
Fig. 3.
Ku exists as distinct ribonucleo- and deoxyribonucleo-protein complexes. A, Northern blot analysis of RNA co-precipitating with Ku. Complexes were purified from nuclear extracts of parental HeLa-S3 (CT, lanes 1 and 4) or HeLa-S3 expressing Ku86-FLAG-HA cells (Ku) and Northern blot was used to probe for ribosomal RNA (5.8S and 18S, lanes 2 and 3) and U3snoRNA (lane 5). B, Silver stain analysis of nuclear Ku86 complexes purified before (lanes 1–3) or 5 h after UV irradiation (lanes 4–6). The complexes were incubated with buffer alone (lanes 1 and 4) or with RNase (lanes 2 and 5) or DNase (lanes 3 and 6) prior to tandem affinity purification. C, Western blot analysis of the Ku complexes shown in (B). Complexes were probed for DNA-PKcs, SPT16, SSRP1, WRN, RHA, Ku86-FLAG-HA, YB-1, RPL26, RPL19 and H2A. D, Quantitative RT-PCR analysis of 18S rRNA in nuclear Ku86 complexes after UV irradiation. Nuclear Ku complexes along with a “mock” complex were purified from parental HeLa-S3 cells as described in Fig. 1A. Copurifying 18S RNA was quantified after reverse transcription by real-time RT-PCR. Average abundance and standard deviation across three technical replicates is plotted relative to the normalized abundance of 18S present in the complex at t = 0.
To determine if protein interactions within the Ku86 complex were indirectly mediated by nucleic acids, we treated the semipurified complex from control and UV irradiated cells with RNase or DNase. In complexes purified from control cells, RNase treatment resulted in the loss of nearly all protein associations, whereas DNase had no detectable effect (Fig. 3B). In contrast, RNase treatment had no effect on the composition of the Ku complex purified from cells exposed to UV irradiation, while DNase treatment subsequent to UV-induced damage led to the complete loss of interaction between the Ku heterodimer and its associated proteins (Fig. 3B). Western blot analysis (Fig. 3C) revealed that the interactions between Ku86 and several RNA associated proteins (the RNA helicase RHA, the Y-box protein YB1, and ribosomal proteins RPL19 and RPL26) were eliminated when the complex was purified from control cells and subsequently treated with ribonuclease. By contrast, the interaction between Ku and WRN was only marginally disrupted by RNase treatment alone, but was substantially reduced by DNase when complexes were purified from cells subjected to UV radiation. As anticipated, DNA-PKcs was strongly recruited to the Ku complex purified from irradiated cells. We also observed the recruitment of both components of the FACT (facilitator of transcription on chromatin templates) complex: suppressor of Ty protein SPT16 and the structure specific recognition protein SSRP1, as well as the core histone H2A, in response to UV radiation (Fig. 3C). Interestingly some interactions were lost (SPT16, WRN, SSRP1, H2A), or significantly reduced (DNA-PKcs) when the complex was treated with DNase, suggesting that DNA at least partially mediates these interactions.
Our results clearly demonstrate that ribosomal RNAs are integral components of the Ku complex and likely mediate the association between Ku and RNA-binding proteins. The loss of this particular protein class after UV irradiation suggests that the interaction between Ku and RNAs themselves might be regulated in response to UV exposure. In fact, we observed that the abundance of the 18S rRNA in Ku complexes did indeed decrease in a time dependent manner after UV irradiation (Fig. 3D and supplemental Fig. S2). Together, these findings reveal the existence of at least two very different Ku complexes: In control cells, Ku exists as a large ribonucleoprotein assembled around an RNA scaffold from which it dissociates upon DNA damage and re-assembles as an RNA-independent complex enriched for DNA-binding and repair factors.
DNA Ends are Sufficient to Induce Remodeling of the Ku Complex
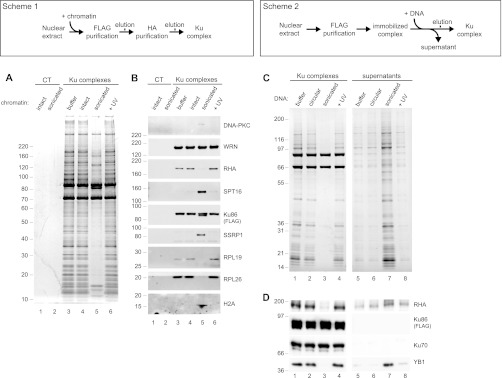
We wondered if the dramatic remodeling observed for the Ku complex resulted from its binding to sites of damaged chromatin. Prior to tandem affinity purification of Ku, we prepared nuclear extracts from intact cells and added intact, UV-irradiated or sonicated (to mimic DSB) chromatin (Fig. 4, Scheme 1). Only incubation with sonicated chromatin led to significant changes in complex composition (Fig. 4A). In fact, the complex purified under these conditions resembled the complex purified from cells exposed to UV radiation (Fig. 1). We confirmed the specific dissociation of the RNA-binding proteins RHA, RPL19, and RPL26, and the concomitant recruitment of the FACT DNA-binding complex (SPT16 and SSRP1) in addition to H2A when the complex was purified in the presence of sonicated chromatin (Fig. 4B). Interestingly, we observed a stable interaction between WRN and Ku, and only a marginal recruitment of DNA-PKcs under these experimental conditions, suggesting that other events in addition to chromatin binding may regulate these interactions. Nonetheless, these results suggest that binding to damaged chromatin is sufficient to induce profound changes in the composition of the Ku complex.
Fig. 4.
DNA ends induce coordinate substitution of RNA associated proteins with DNA binding factors in Ku multicomponent complexes. (A and B: Scheme 1) (C and D: Scheme 2) Biochemical manipulation of nuclear extracts to monitor assembly of Ku complexes in the presence of intact or sonicated chromatin. A, Silver stain analysis of control (lanes 1 and 2) or nuclear Ku (lanes 3–6) complexes purified in the presence of buffer (lane 3) or intact (lanes 1 and 4), sonicated (lane 2 and 5) or UV irradiated (lane 6) chromatin. B, Western blot analysis of the protein complexes shown in (A). Complexes were probed for DNA-PKcs, SPT16, SSRP1, WRN, RHA, Ku86-FLAG-HA, RPL26, RPL19, and H2A. (C and D) Biochemical manipulation of immobilized Ku complexes to identify factors stably bound or released in response to incubation with DNA mimicking double-strand breaks. C, Silver stain analysis of complexes and supernatants resulting from incubation of immobilized complexes with buffer alone (lanes 1 and 5) or with circular (lanes 2 and 6), sonicated (lanes 3 and 7), or UV irradiated (lanes 4 and 8) plasmid DNA. D, Western blot analysis of the protein complexes and supernatants shown in (C).
To further investigate this remodeling event in the absence of factors present in the cell extract, we incubated semi-purified Ku complexes that were immobilized on beads with circular, UV-irradiated or sonicated plasmid DNA (Fig. 4, Scheme 2). Again, we observed that only sonicated DNA efficiently disrupted the interactions between Ku and its binding partners (Fig. 4C). We confirmed these observations by Western blot analysis showing that both RHA and YB1, but not Ku70, dissociated from bead-immobilized Ku and appeared in the supernatant (Fig. 4D). These results indicate that DNA ends, independent of other cellular factors, are sufficient to disrupt interactions between Ku and its RNA binding partners. Collectively our results provide compelling evidence that DNA ends induce a multi-step remodeling of Ku associated proteins that ultimately results in a complex bound to chromatin and poised for DNA repair.
Microinjection of DNA Ends Induces Loss of Nucleolar Ku In Vivo
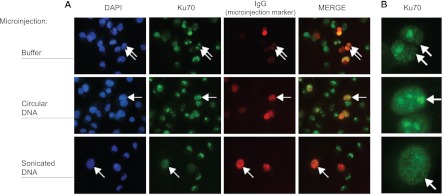
The nucleolus is the site of ribosomal RNA transcription, processing and subunit assembly (41). Ku interacts with many of the RNA binding and processing factors that participate in these steps and importantly relocates from the nucleolus to the nucleoplasm upon treatment with genotoxic agents (34, 35). These observations, along with data herein showing that RNA-binding proteins are displaced from the Ku complex by fragmented chromatin in vitro (Fig. 4), led us to speculate that DNA ends would be sufficient to induce exit of Ku from nucleoli in vivo. To test this hypothesis we microinjected circular or sonicated plasmids into the live MDA-MB-231 breast cancer cells and monitored the localization of Ku70. We observed stable nucleolar localization of Ku70 in cells microinjected with IgG alone or in combination with circular plasmid DNA (Fig. 5A). However, microinjection of IgG plus sonicated plasmid DNA resulted in a diffuse staining pattern for Ku70 throughout the nucleus (Figs. 5A and 5B). Similar to the re-localization of Ku observed in response to UV radiation (Fig. 2), the introduction of DNA ends in the nucleus was sufficient to induce exit of Ku from the nucleolus in vivo.
Fig. 5.
DNA ends are sufficient to induce exit of Ku from the nucleolus in vivo. A, MDA-MB-231 cells were microinjected with rabbit immunoglobulin (IgG) as an injection marker, alone (top row) or in combination with circular (second row) or sonicated (third row) plasmid DNA and processed for immunofluorescence following a triton extraction and paraformaldehyde fixation protocol (T/PFA). Cells were labeled with an anti-rabbit IgG antibody to identify microinjected cells (red channel) and with an anti-mouse IgG antibody to detect endogenous Ku70 (green channel). Nuclei were stained with DAPI (blue channel). B, Cropped views of microinjected cells from (A) showing highly magnified nuclei. Arrows indicate representative microinjected cells.
Taken together our data suggest that the biochemical switch from a ribonucleoprotein complex to one dominated by DNA binding and repair factors coincides with the migration of Ku from the nucleolus to sites of DSBs.
DISCUSSION
Ku is best known for its role in response to the appearance of double strand breaks, primarily generated by DNA damage or as a consequence of immune gene rearrangement. In both situations, Ku binds the newly formed DNA ends and, through the recruitment of multiple factors, promotes repair of the lesion by non-homologous end-joining. Interestingly, Ku is also implicated in the proper function of telomeres, the naturally occurring DNA ends at the extremities of chromosomes (42). Ku is also known to bind with nano-molar affinity to a variety of RNAs (43): In yeast and higher eukaryotes, Ku binds to the stem loop structure of TLC1, the RNA component of telomerase, and promotes telomere addition (14, 44). In addition, Ku was shown to interact with RHA in an RNA dependent manner (13), although the identity of RNAs mediating this interaction was not investigated. These studies raise the possibility that distinct Ku complexes, functionally directed to various aspects of RNA metabolism or to the maintenance of DNA structural integrity, exist in eukaryotic cells.
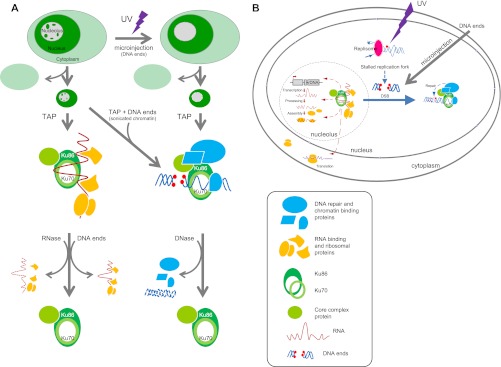
Herein we show that in the absence of DNA damage, Ku forms a large ribonucleoprotein complex containing ribosomal RNAs and multiple RNA-binding proteins. Using a combination of TAP and quantitative mass spectrometry, we show that UV radiation leads to a time-dependent dissociation of this functional class of proteins and a concomitant recruitment of DNA-binding proteins (Fig. 1). We further demonstrate that in undamaged cells, Ku exists as an RNase-dependent ribonucleoprotein but becomes RNase resistant and DNase sensitive after exposure to UV radiation (Fig. 3B). This transition is accompanied by a time-dependent dissociation between Ku and the 18S ribosomal RNA (Fig. 3D) and the relocalization of Ku from the nucleolus to the nucleoplasm (Fig. 2). Using purified complexes in in vitro assays, we demonstrate that remodeling of the Ku complex is initiated by its binding to DNA ends (Fig. 4C). Finally, microinjection of DNA ends into non-damaged cells induces the exit of Ku from the nucleolar compartment (Fig. 5). Taken together our results converge in support of the hypothesis that the appearance of DNA ends triggers a concomitant change in the molecular composition and sub-nuclear localization of Ku (Fig. 6).
Fig. 6.
Schematic summary of (A) experiments and observations in support of (B) the hypothesis that DNA ends alter the molecular composition and nuclear localization of Ku multicomponent complexes.
Although our data detail the effect of UV radiation on the composition of Ku complexes, these results must be considered in the context of broader, proteome-wide responses. In fact Moore et al. recently reported that the proteomic content and structure of the nucleolus is dramatically altered in response to UV radiation (35). Interestingly, these effects occurred over a time period of several hours, similar to what we observed for remodeling of the Ku complex. In both cases, the kinetics suggest that the responses are not directly mediated by typical UV-induced DNA damage products (cyclopyrimidine dimers and 6–4 photoproducts). This hypothesis is supported by the fact that only DNA ends, and not UV-irradiated DNA, altered the composition of Ku complexes in vitro and localization in vivo. Instead, we speculate that remodeling occurs in response to double strand breaks that appear subsequent to UV-induced replication fork stall and collapse (Fig. 6B). The recruitment of TONSL, and RPA1 (supplemental Table S1 Summary of quantitative LC-MS data), two factors involved in the response to replication stress (45, 46), further supports our hypothesis. This idea is also consistent with recent studies demonstrating a coordinate activation of ATR and DNA-PK in response to UV-induced replication stress (33, 47).
Although the majority of studies on Ku and DNA-PKcs are related to DNA repair, several reports have also demonstrated that these proteins regulate gene transcription, translation, and other RNA-directed activities. For example, DNA-PK has been shown to regulate the activity of RNA-Polymerase I, the enzyme responsible for rRNA precursor transcription in the nucleolus (7, 9). This finding is consistent with our observation that Ku interacts with factors involved in rRNA processing (NOP2 and GNL3), and suggests that DNA-PK may also regulate rRNA synthesis at the post-transcriptional level. Interestingly, Ku, in a complex with RPL26 and nucleolin, was shown to bind to the 5′UTR of the p53 mRNA and regulate its translation after DNA damage (48). Together, these results suggest that Ku may control gene expression at both the global level, by controlling ribosomal subunits synthesis and at the gene-specific level by regulating the translational efficiency of specific mRNAs. This hypothesis is corroborated by a recent study showing that the catalytic subunit of DNA-PK plays an important role in translational reprogramming in response to UV irradiation (49). Our findings suggest that regulating the sub-nuclear localization of Ku may allow cells to fine tune protein expression in response to DNA damage and raise the interesting possibility that DNA-PK may play a role in coordinating the nucleolar response to other types of cellular stress (50).
Importantly, our results suggest that the canonical function of Ku in DNA repair and its more recently recognized role in gene expression rely on two mutually exclusive but interrelated complexes: one composed of ribosomal proteins and RNA-binding factors and the other composed of proteins involved in DNA repair and metabolism (Fig. 6A). The fact that DNA ends trigger both a molecular transition between these two complexes along with relocation of Ku from the nucleolus to the nucleoplasm (Fig. 6B) may constitute a simple mechanism by which cells couple the repair of DNA after genotoxic insult to gene expression and protein translation.
Acknowledgments
We thank James Webber and Scott Ficarro for bioinformatics support and Vincent Cheng for proteomics sample preparation.
Footnotes
* This work was supported by the National Institutes of Health (NIH) (P50HG004233 and P01NS047572), the Susan Smith Center for Women's Cancers, and the Strategic Research Initiative at the Dana-Farber Cancer Institute (to J. A. M.), in addition to NIH (R37GM36373-24) to P. A. S., along with support from the Marsha Rivkin Foundation, The Catt Family Foundation, and The James and Kimberly Pallotta Fund for Ovarian Cancer Research (to J. D. I.).
 This article contains supplemental Figs. S1 and S2, Tables S1 to S4 and Results S1 and S2.
This article contains supplemental Figs. S1 and S2, Tables S1 to S4 and Results S1 and S2.
1 The abbreviations used are:
- PFA
- paraformaldehyde
- DSB
- double strand break
- iTRAQ
- isobaric tags for relative and absolute quantification
- NHEJ
- non-homologous end-joining
- rDNA
- ribosomal DNA
- RP
- reversed phase
- rRNA
- ribosomal RNA
- SCX
- strong cation exchange
- TAP
- tandem-affinity purification
- UTR
- untranslated region
- UV
- ultraviolet.
Gene names abbreviations;
- ATR
- Ataxia telangiectasia mutated and Rad3 related
- BRIX
- Biogenesis of ribosomes in Xenopus
- DNA-PK
- DNA-dependent protein kinase
- DNA-PKcs
- DNA-dependent protein kinase catalytic subunit
- GNL3
- guanine binding protein-like 3
- GRWD1
- glutamate-rich WD repeat containing protein1
- hnRNP
- heterogeneous nuclear ribonucleoprotein
- NOP2
- nucleolar protein 2
- NPM1
- nucleophosmin 1
- RHA
- RNA helicase A
- RPS and RPL
- ribosomal protein (small and large subunits, respectively)
- SPT16
- suppressor of Ty 16
- SSRP1
- Structure specific recognition protein 1
- U3 snoRNA
- U3 small nucleolar RNA
- XLF
- XRCC4 like factor
- XRCC4
- X-ray repair cross-complementing protein 4
- YB1
- Y-box-binding protein 1
- YY1
- yin-yang 1.
REFERENCES
- 1. Yano K., Morotomi-Yano K., Wang S. Y., Uematsu N., Lee K. J., Asaithamby A., Weterings E., Chen D. J. (2008) Ku recruits XLF to DNA double-strand breaks. EMBO Rep. 9, 91–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Calsou P., Delteil C., Frit P., Drouet J., Salles B. (2003) Coordinated assembly of Ku and p460 subunits of the DNA-dependent protein kinase on DNA ends is necessary for XRCC4-ligase IV recruitment. J. Mol. Biol. 326, 93–103 [DOI] [PubMed] [Google Scholar]
- 3. Ma Y., Lu H., Tippin B., Goodman M. F., Shimazaki N., Koiwai O., Hsieh C. L., Schwarz K., Lieber M. R. (2004) A biochemically defined system for mammalian nonhomologous DNA end joining. Mol. Cell 16, 701–713 [DOI] [PubMed] [Google Scholar]
- 4. Meek K., Gupta S., Ramsden D. A., Lees-Miller S. P. (2004) The DNA-dependent protein kinase: the director at the end. Immunol. Rev. 200, 132–141 [DOI] [PubMed] [Google Scholar]
- 5. Chibazakura T., Watanabe F., Kitajima S., Tsukada K., Yasukochi Y., Teraoka H. (1997) Phosphorylation of human general transcription factors TATA-binding protein and transcription factor IIB by DNA-dependent protein kinase–synergistic stimulation of RNA polymerase II basal transcription in vitro. Eur. J. Biochem. 247, 1166–1173 [DOI] [PubMed] [Google Scholar]
- 6. Dvir A., Peterson S. R., Knuth M. W., Lu H., Dynan W. S. (1992) Ku autoantigen is the regulatory component of a template-associated protein kinase that phosphorylates RNA polymerase II. Proc. Natl. Acad. Sci. U.S.A. 89, 11920–11924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kuhn A., Gottlieb T. M., Jackson S. P., Grummt I. (1995) DNA-dependent protein kinase: a potent inhibitor of transcription by RNA polymerase I. Genes Dev. 9, 193–203 [DOI] [PubMed] [Google Scholar]
- 8. Kuhn A., Stefanovsky V., Grummt I. (1993) The nucleolar transcription activator UBF relieves Ku antigen-mediated repression of mouse ribosomal gene transcription. Nucleic Acids Res. 21, 2057–2063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Labhart P. (1995) DNA-dependent protein kinase specifically represses promoter-directed transcription initiation by RNA polymerase I. Proc. Natl. Acad. Sci. U.S.A. 92, 2934–2938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Michaelidis T. M., Grummt I. (2002) Mechanism of inhibition of RNA polymerase I transcription by DNA-dependent protein kinase. Biol. Chem. 383, 1683–1690 [DOI] [PubMed] [Google Scholar]
- 11. Woodard R. L., Anderson M. G., Dynan W. S. (1999) Nuclear extracts lacking DNA-dependent protein kinase are deficient in multiple round transcription. J. Biol. Chem. 274, 478–485 [DOI] [PubMed] [Google Scholar]
- 12. Ting N. S., Pohorelic B., Yu Y., Lees-Miller S. P., Beattie T. L. (2009) The human telomerase RNA component, hTR, activates the DNA-dependent protein kinase to phosphorylate heterogeneous nuclear ribonucleoprotein A1. Nucleic Acids Res. 37, 6105–6115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang S., Schlott B., Görlach M., Grosse F. (2004) DNA-dependent protein kinase (DNA-PK) phosphorylates nuclear DNA helicase II/RNA helicase A and hnRNP proteins in an RNA-dependent manner. Nucleic Acids Res. 32, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ting N. S., Yu Y., Pohorelic B., Lees-Miller S. P., Beattie T. L. (2005) Human Ku70/80 interacts directly with hTR, the RNA component of human telomerase. Nucleic Acids Res. 33, 2090–2098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cooper M. P., Machwe A., Orren D. K., Brosh R. M., Ramsden D., Bohr V. A. (2000) Ku complex interacts with and stimulates the Werner protein. Genes Dev. 14, 907–912 [PMC free article] [PubMed] [Google Scholar]
- 16. Karmakar P., Piotrowski J., Brosh R. M., Jr., Sommers J. A., Miller S. P., Cheng W. H., Snowden C. M., Ramsden D. A., Bohr V. A. (2002) Werner protein is a target of DNA-dependent protein kinase in vivo and in vitro, and its catalytic activities are regulated by phosphorylation. J. Biol. Chem. 277, 18291–18302 [DOI] [PubMed] [Google Scholar]
- 17. Li B., Comai L. (2000) Functional interaction between Ku and the werner syndrome protein in DNA end processing. J. Biol. Chem. 275, 39800. [DOI] [PubMed] [Google Scholar]
- 18. Galande S., Kohwi-Shigematsu T. (1999) Poly(ADP-ribose) polymerase and Ku autoantigen form a complex and synergistically bind to matrix attachment sequences. J. Biol. Chem. 274, 20521–20528 [DOI] [PubMed] [Google Scholar]
- 19. Sucharov C. C., Helmke S. M., Langer S. J., Perryman M. B., Bristow M., Leinwand L. (2004) The Ku protein complex interacts with YY1, is up-regulated in human heart failure, and represses alpha myosin heavy-chain gene expression. Mol. Cell. Biol. 24, 8705–8715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Heo K., Kim H., Choi S. H., Choi J., Kim K., Gu J., Lieber M. R., Yang A. S., An W. (2008) FACT-mediated exchange of histone variant H2AX regulated by phosphorylation of H2AX and ADP-ribosylation of Spt16. Mol. Cell 30, 86–97 [DOI] [PubMed] [Google Scholar]
- 21. Yang C. R., Yeh S., Leskov K., Odegaard E., Hsu H. L., Chang C., Kinsella T. J., Chen D. J., Boothman D. A. (1999) Isolation of Ku70-binding proteins (KUBs). Nucleic Acids Res. 27, 2165–2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bürckstummer T., Bennett K. L., Preradovic A., Schutze G., Hantschel O., Superti-Furga G., Bauch A. (2006) An efficient tandem affinity purification procedure for interaction proteomics in mammalian cells. Nat. Methods 3, 1013–1019 [DOI] [PubMed] [Google Scholar]
- 23. Zhou F., Cardoza J. D., Ficarro S. B., Adelmant G. O., Lazaro J. B., Marto J. A. (2010) Online nanoflow RP-RP-MS reveals dynamics of multicomponent Ku complex in response to DNA damage. J Proteome Res 9, 6242–6255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Higashiura M., Shimizu Y., Tanimoto M., Morita T., Yagura T. (1992) Immunolocalization of Ku-proteins (p80/p70): localization of p70 to nucleoli and periphery of both interphase nuclei and metaphase chromosomes. Exp. Cell Res. 201, 444–451 [DOI] [PubMed] [Google Scholar]
- 25. Reeves W. H. (1985) Use of monoclonal antibodies for the characterization of novel DNA-binding proteins recognized by human autoimmune sera. J. Exp. Med. 161, 18–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dignam J. D., Lebovitz R. M., Roeder R. G. (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11, 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nakatani Y., Ogryzko V. (2003) Immunoaffinity purification of mammalian protein complexes. Methods Enzymol. 370, 430–444 [DOI] [PubMed] [Google Scholar]
- 28. Livak K. J., Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 29. Ivanov A. R. L. A. V. (2011) Sample preparation in biological mass spectrometry, Springer, Dordrecht [Google Scholar]
- 30. Ficarro S. B., Adelmant G., Tomar M. N., Zhang Y., Cheng V. J., Marto J. A. (2009) Magnetic bead processor for rapid evaluation and optimization of parameters for phosphopeptide enrichment. Anal. Chem. 81, 4566–4575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ficarro S. B., Zhang Y., Lu Y., Moghimi A. R., Askenazi M., Hyatt E., Smith E. D., Boyer L., Schlaeger T. M., Luckey C. J., Marto J. A. (2009) Improved electrospray ionization efficiency compensates for diminished chromatographic resolution and enables proteomics analysis of tyrosine signaling in embryonic stem cells. Anal. Chem. 81, 3440–3447 [DOI] [PubMed] [Google Scholar]
- 32. Webber J. T., Askenazi M., Marto J. A. (2011) mzResults: an interactive viewer for interrogation and distribution of proteomics results. Mol. Cell. Proteomics 10, M110.003970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yajima H., Lee K. J., Zhang S., Kobayashi J., Chen B. P. (2009) DNA double-strand break formation upon UV-induced replication stress activates ATM and DNA-PKcs kinases. J. Mol. Biol. 385, 800–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dejmek J., Iglehart J. D., Lazaro J. B. (2009) DNA-dependent protein kinase (DNA-PK)-dependent cisplatin-induced loss of nucleolar facilitator of chromatin transcription (FACT) and regulation of cisplatin sensitivity by DNA-PK and FACT. Mol. Cancer Res. 7, 581–591 [DOI] [PubMed] [Google Scholar]
- 35. Moore H. M., Bai B., Boisvert F. M., Latonen L., Rantanen V., Simpson J. C., Pepperkok R., Lamond A. I., Laiho M. (2011) Quantitative proteomics and dynamic imaging of the nucleolus reveals distinct responses to UV and ionizing radiation. Mol. Cell. Proteomics 10, M111.009241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fatica A., Tollervey D. (2002) Making ribosomes. Curr. Opin. Cell Biol. 14, 313–318 [DOI] [PubMed] [Google Scholar]
- 37. Hong B., Brockenbrough J. S., Wu P., Aris J. P. (1997) Nop2p is required for pre-rRNA processing and 60S ribosome subunit synthesis in yeast. Mol. Cell. Biol. 17, 378–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Romanova L., Grand A., Zhang L., Rayner S., Katoku-Kikyo N., Kellner S., Kikyo N. (2009) Critical role of nucleostemin in pre-rRNA processing. J. Biol. Chem. 284, 4968–4977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kaser A., Bogengruber E., Hallegger M., Doppler E., Lepperdinger G., Jantsch M., Breitenbach M., Kreil G. (2001) Brix from xenopus laevis and brx1p from yeast define a new family of proteins involved in the biogenesis of large ribosomal subunits. Biol. Chem. 382, 1637–1647 [DOI] [PubMed] [Google Scholar]
- 40. Gratenstein K., Heggestad A. D., Fortun J., Notterpek L., Pestov D. G., Fletcher B. S. (2005) The WD-repeat protein GRWD1: potential roles in myeloid differentiation and ribosome biogenesis. Genomics 85, 762–773 [DOI] [PubMed] [Google Scholar]
- 41. Kressler D., Hurt E., Bassler J. (2010) Driving ribosome assembly. Biochim. Biophys. Acta 1803, 673–683 [DOI] [PubMed] [Google Scholar]
- 42. Downs J. A., Jackson S. P. (2004) A means to a DNA end: the many roles of Ku. Nat. Rev. Mol. Cell Biol. 5, 367–378 [DOI] [PubMed] [Google Scholar]
- 43. Dynan W. S., Yoo S. (1998) Interaction of Ku protein and DNA-dependent protein kinase catalytic subunit with nucleic acids. Nucleic Acids Res. 26, 1551–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Stellwagen A. E., Haimberger Z. W., Veatch J. R., Gottschling D. E. (2003) Ku interacts with telomerase RNA to promote telomere addition at native and broken chromosome ends. Genes Dev. 17, 2384–2395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Robison J. G., Elliott J., Dixon K., Oakley G. G. (2004) Replication protein A and the Mre11.Rad50.Nbs1 complex co-localize and interact at sites of stalled replication forks. J. Biol. Chem. 279, 34802–34810 [DOI] [PubMed] [Google Scholar]
- 46. O'Donnell L., Panier S., Wildenhain J., Tkach J. M., Al-Hakim A., Landry M. C., Escribano-Diaz C., Szilard R. K., Young J. T., Munro M., Canny M. D., Kolas N. K., Zhang W., Harding S. M., Ylanko J., Mendez M., Mullin M., Sun T., Habermann B., Datti A., Bristow R. G., Gingras A. C., Tyers M. D., Brown G. W., Durocher D. (2010) The MMS22L-TONSL complex mediates recovery from replication stress and homologous recombination. Mol. Cell 40, 619–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yajima H., Lee K. J., Chen B. P. (2006) ATR-dependent phosphorylation of DNA-dependent protein kinase catalytic subunit in response to UV-induced replication stress. Mol. Cell. Biol. 26, 7520–7528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Takagi M., Absalon M. J., McLure K. G., Kastan M. B. (2005) Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell 123, 49–63 [DOI] [PubMed] [Google Scholar]
- 49. Powley I. R., Kondrashov A., Young L. A., Dobbyn H. C., Hill K., Cannell I. G., Stoneley M., Kong Y. W., Cotes J. A., Smith G. C., Wek R., Hayes C., Gant T. W., Spriggs K. A., Bushell M., Willis A. E. (2009) Translational reprogramming following UVB irradiation is mediated by DNA-PKcs and allows selective recruitment to the polysomes of mRNAs encoding DNA repair enzymes. Genes Dev. 23, 1207–1220 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 50. Boulon S., Westman B. J., Hutten S., Boisvert F. M., Lamond A. I. (2010) The nucleolus under stress. Mol. Cell 40, 216–227 [DOI] [PMC free article] [PubMed] [Google Scholar]