Abstract
To investigate the quantitative response of energy metabolic pathways in human MCF-7 breast cancer cells to hypoxia, glucose deprivation, and estradiol stimulation, we developed a targeted proteomics assay for accurate quantification of protein expression in glycolysis/gluconeogenesis, TCA cycle, and pentose phosphate pathways. Cell growth conditions were selected to roughly mimic the exposure of cells in the cancer tissue to the intermittent hypoxia, glucose deprivation, and hormonal stimulation. Targeted proteomics assay allowed for reproducible quantification of 76 proteins in four different growth conditions after 24 and 48 h of perturbation. Differential expression of a number of control and metabolic pathway proteins in response to the change of growth conditions was found. Elevated expression of the majority of glycolytic enzymes was observed in hypoxia. Cancer cells, as opposed to near-normal MCF-10A cells, exhibited significantly increased expression of key energy metabolic pathway enzymes (FBP1, IDH2, and G6PD) that are known to redirect cellular metabolism and increase carbon flux through the pentose phosphate pathway. Our quantitative proteomic protocol is based on a mass spectrometry-compatible acid-labile detergent and is described in detail. Optimized parameters of a multiplex selected reaction monitoring (SRM) assay for 76 proteins, 134 proteotypic peptides, and 401 transitions are included and can be downloaded and used with any SRM-compatible mass spectrometer. The presented workflow is an integrated tool for hypothesis-driven studies of mammalian cells as well as functional studies of proteins, and can greatly complement experimental methods in systems biology, metabolic engineering, and metabolic transformation of cancer cells.
Adaptation of cancer cells to hypoxia, lack of nutrients, and abnormal hormonal stimulation is known to alter their metabolism (1, 2). “Aerobic glycolysis,” also referred as the Warburg effect, is a unique characteristic of rapidly proliferating cancer cells (3). Deregulating cellular energetics and reprogramming of metabolism are the emerging hallmarks of cancer (4). There is also an increasing number of epidemiologic evidence that link cancer risk with metabolic disorders such as diabetes and obesity (5).
Until recently, the metabolic transformation of cancer cells was studied primarily at the level of genome (6), transcriptome (7), and metabolome (8). These studies discovered new mutations (9, 10), cancer-related alternative splicing isoforms (11), and altered enzyme activities in human cancers (2). Subsequent clinical applications included diagnostic imaging (12), prognosis (13), and identification of compounds targeting tumor metabolism (14).
To fully understand the events and outcomes of metabolic transformation of cancer cells, quantitative proteomic approaches are required to complement existing genomic, transcriptomic, and metabolomic approaches. Proteomic methods provide additional levels of information, such as protein abundances, post-translational modifications, dynamics of protein turn-over, which cannot be accurately predicted using other -omic approaches.
Typically, expression of enzymes in cellular metabolic pathways is measured by ELISA or immunoblot assays, which provide information for a very limited number of enzymes. Multiplex proteomic assays, on the contrary, could reveal simultaneous rearrangement of protein expression in entire metabolic pathways. Mass spectrometry-based proteomics, complemented with chemical and metabolic labeling approaches, is a powerful tool for global analysis of protein expression. However, these approaches have very limited throughput and require extensive sample preparation, complex data analysis, and verification of their results by independent assays. Besides, metabolic labeling is applicable to actively dividing cell lines, but not to the primary cells.
Targeted proteomic assays present an attractive complementary tool devoid of the abovementioned limitations. Selected reaction monitoring (SRM)1 assays can simultaneously monitor hundreds of proteins in hundreds of different biological samples. Targeted proteomics was previously used to measure protein abundances in metabolic pathways in bacteria and yeast (15–17). Analysis of proteins in mammalian cells poses additional challenges such as detection of multiple enzyme isoforms and almost 10-fold lower protein abundance per cell. To our knowledge, analysis of multiple energy metabolic pathways in mammalian cells by SRM has not been realized to date.
Here, we present an integrated SRM-based label-free proteomic workflow for measurement of the relative expression of proteins in entire metabolic pathways of mammalian cells (Fig. 1). Our SRM-compatible sample preparation protocol allows for accurate measurement of changes in protein expression between two biological conditions in less than a day. In our protocol, protein abundances are normalized to an array of high-abundance housekeeping proteins. Such normalization allows for a label-free analysis, reduces variability, and facilitates accurate measurement of relatively small changes in protein expression under different growth conditions.
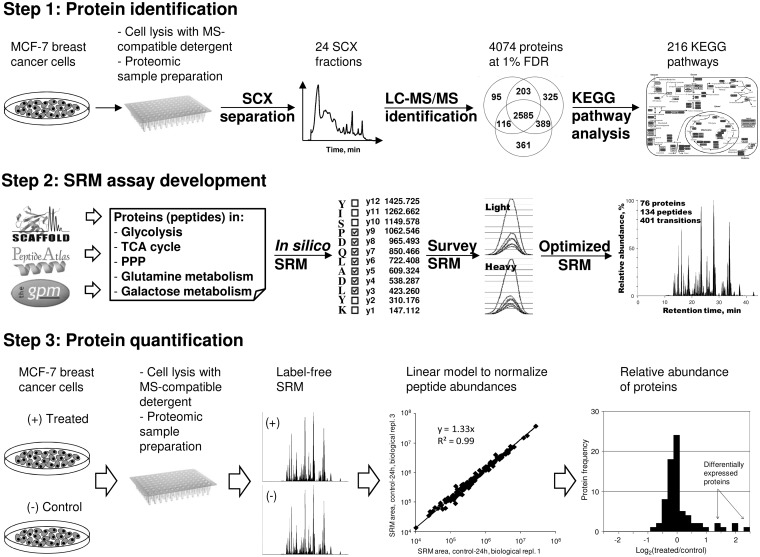
Fig. 1.
Schematic presentation of a workflow for SRM measurement of the relative expression of proteins in mammalian cells. In Step 1, MCF-7 cells were lysed, and proteins were subjected to 2D-LC-MS/MS identification. Data analysis revealed 4074 unique proteins in 216 KEGG pathways. In Step 2, a multiplex SRM assay was developed for proteins in energy metabolic pathways and included 76 proteins, 134 peptides, and 401 transitions. In Step 3, MCF-7cells were grown in different conditions, lysed, and subjected to the proteomic sample preparation. The peak area for each peptide was measured with a multiplex SRM assay and normalized using a statistical linear model. Protein areas were used to calculate relative abundances and identify differentially expressed proteins in the entire metabolic pathways in mammalian cells grown under different conditions. PPP, the pentose phosphate pathway.
As a proof-of-principle experiment, MCF-7 breast cancer cells were exposed to different growth conditions such as normoxia, hypoxia, galactose-rich-glucose-free media, and estradiol stimulation. These conditions roughly mimic the exposure of cells in the breast cancer tissue to the intermittent hypoxia, starvation, and hormonal stimulation. Using a multiplex SRM assay, we measured relative abundances of nearly all proteins in major energy metabolic pathways, such as glycolysis/gluconeogenesis, TCA cycle, and the pentose phosphate pathway (Fig. 2). A set of control proteins was used to validate cellular response to the given growth conditions. Using our SRM assay, we also compared cancer (MCF-7) and near-normal (MCF-10A) breast cells.
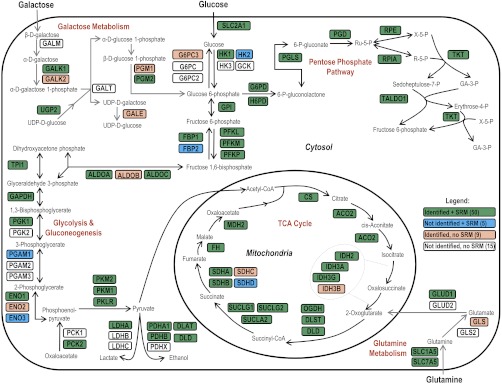
Fig. 2.
Proteins and protein isoforms in glycolysis/gluconeogenesis, TCA cycle, pentose phosphate pathway, galactose metabolism, and glutamine metabolism pathways. In total, out of 79 proteins, 50 proteins were identified by LC-MS/MS and quantified by SRM (highlighted in green), nine proteins were identified, but not quantified (highlighted in pink), five proteins were not identified, but quantified (highlighted in blue), and 15 proteins were neither identified, nor quantified (not highlighted). Enzymes in the presented energy metabolic pathways catalyze: (1) conversion of glucose or galactose through glycolysis into pyruvate, which is used in the TCA cycle to produce NADH; (2) conversion of glutamine to 2-oxoglutarate, which is used in the TCA cycle (3) generation of NADPH and conversion of glucose or galactose through the pentose phosphate pathway into ribulose 5-phosphate or erythrose-4-phosphate, used in the synthesis of nucleotides and aromatic aminoacids, respectively.
MATERIALS AND METHODS
Cell Lines
The breast cancer MCF-7 and breast epithelial MCF-10A cell lines were purchased from the American Type Culture Collection, Manassas, VA. MCF-7 cells were maintained in Dulbecco's Modified Eagle Medium (DMEM)—high glucose (25 mm) culture medium (Invitrogen) supplemented with 10% fetal bovine serum. MCF-10A cells were maintained in Dulbecco's modified Eagle's medium and F-12 medium (DMEM/F-12) supplemented with 10% fetal bovine serum, epidermal growth factor (20 ng/ml), hydrocortisone (0.5 μg/ml), and insulin (10 μg/ml). All cells were cultured in a humidified incubator at 37 °C and 5% CO2.
MCF-7 Proteome Identification by 2D-LC-MS/MS
Three biological replicates of ∼3 × 106 MCF-7 cells (500 μg of total protein) were used. Before cell lysis, culture media were discarded; cells were washed three times with phosphate-buffered saline (PBS) buffer to remove traces of fetal bovine serum, trypsinized, and centrifuged at 9000 rpm. The cell pellet was resuspended with 200 μl of 0.1% acid-labile detergent RapiGest SF (sodium-3-[(2-methyl-2-undecyl-1,3-dioxolan-4-yl)-methoxyl]-1-propanesulfonate, Waters, Milford, MA) in 25 mm ammonium bicarbonate, vortexed, and sonicated three times for 30 s. All lysates were centrifuged for 20 min at 15,000 rpm at 4 °C, even though no debris was typically observed after lysis. Total protein concentration was measured using a Coomassie (Bradford) protein assay reagent (Pierce). Proteins in detergent solution were denatured at 60 °C, and the disulfide bonds were reduced with 10 mm dithiothreitol. Following reduction, the samples were alkylated with 20 mm iodoacetamide. Samples were then digested overnight at 37 °C with sequencing grade modified trypsin (Promega, Madison, WI). Trypsin/total protein ratio of 1:30 was used. After digestion, RapiGest SF detergent was cleaved with trifluoroacetic acid, 1% final concentration, and samples were centrifuged at 1500 rpm. Upon removal of Rapigest, tryptic peptides were diluted to 500 μl with strong cation exchange (SCX) mobile phase A (0.26 m formic acid in 5% acetonitrile; pH 2–3) and loaded directly onto a 500 μl loop connected to a PolySULFOETHYL A™ column (2.1 mm ID × 200 mm, 5 μm, 200 Å, The Nest Group Inc., MA). The SCX chromatography and fractionation were performed on an HPLC system (Agilent 1100) using a 60-min two-step gradient, which was optimized to provide a uniform elution of peptides based on the absorption at 280 nm. An elution buffer that contained all components of mobile phase A with the addition of 1 m ammonium formate was introduced at 10 min and increased to 20% at 30 min and then to 100% at 45 min. Fractions were collected every 2 min from the 10 min time point onwards. This resulted in the collection of 24 fractions (400 μl each). Peptides in each fraction were identified by liquid chromatography-tandem MS (LC-MS/MS) as previously described (18, 19). Briefly, peptides were extracted with 10 μl OMIX C18 tips, eluted with 64.5% acetonitrile, diluted to 40 μl with 0.1% formic acid, and loaded onto a 3 cm C18 trap column (with an inner diameter of 150 μm; New Objective), packed in-house with 5 μm Pursuit C18 (Varian, Lake Forest, CA). Eluted peptides from the trap column were subsequently loaded onto a resolving analytical PicoTip Emitter column, 5 cm in length (with an inner diameter of 75 μm and 8 μm tip, New Objective) and packed in-house with 3 μm Pursuit C18 (Varian, Lake Forest, CA). The trap and analytical columns were operated on the EASY-nLC system (Proxeon Biosystems, Odense, Denmark), and this liquid chromatography setup was coupled online to an LTQ-Orbitrap XL hybrid mass spectrometer (Thermo Fisher Scientific, San Jose, CA) using a nano-ESI source (Proxeon Biosystems, Odense, Denmark). Peptides were separated using a 60-min gradient and analyzed in data dependent mode in which a full MS1 scan acquisition from 450–1450 m/z in the Orbitrap mass analyzer (resolution 60,000) was followed by MS2 scan acquisition of the top six parent ions in the linear ion trap mass analyzer. The following parameters were enabled: monoisotopic precursor selection, charge state screening, and dynamic exclusion. In addition, charge states of +1, >4 and unassigned charge states were not subjected to MS2 fragmentation. For protein identification and data analysis, XCalibur software (v. 2.0.5; Thermo Fisher) was used to generate RAW files of each MS run. The RAW files were subsequently used to generate Mascot Generic Files (MGF) through extract_msn on Mascot Daemon (version 2.2.2). Once generated, MGFs were searched with two search engines, Mascot (Matrix Science, London, UK; version 2.2) and X!Tandem (Global Proteome Machine Manager; version 2006.06.01). Searches were conducted against the nonredundant Human IPI database (v. 3.62; 167,894 forward and reverse protein sequences) using the following parameters: fully tryptic cleavages, 7 ppm precursor ion mass tolerance, 0.4 Da fragment ion mass tolerance, allowance of one missed cleavage and fixed modifications of carbamidomethylation of cysteines. Variable modifications included oxidation of methionine, pyro-Glu from glutamine of the n-terminuss-carbamoylmethylcystein cyclization at N terminus, deamidation of glutamine, oxidation of tryptophan, and acetylation of the n-terminus. The files generated from MASCOT (DAT files) and X!Tandem (XML files) for the three replicates were then integrated through Scaffold 2 software (version 2.06; Proteome Software Inc., Portland, Oregon) resulting in a nonredundant list of identified proteins per sample. Results were filtered separately using the X!Tandem LogE filter and Mascot ion-score filters on Scaffold to achieve a protein false discovery rate (FDR) of 1.0%. FDR was calculated as [2×FP/(TP+FP)]×100, where FP (false positive) is the number of proteins that were identified based on sequences in the reverse database component and TP (true positive) is the number of proteins that were identified based on sequences in the forward database component. Scaffold protein report was generated and uploaded onto Protein Center Professional Edition (v. 3.5.2.1; Proxeon Bioinformatics, Odense, Denmark) to facilitate KEGG pathway analysis and visualization. Peptide and protein identifications reports are included into supplemental Tables S1 and S2. Scaffold file with all spectra, peptide and protein identifications including annotated spectra for single-peptide identifications was deposited at Tranche Proteome Commons Database (http://proteomecommons.org/tranche) with the hash code 0HdNVoQPyPNfeYjcaZXwvDUKxfV926AVv4ljTK47qaRZ4YAzg3IrvH497s/XzsIK08yXLYuKNz932jSuh0muSF1BK/8AAAAABOTsTA== Peptide spectra were deposited with the hash code
4olJ/dS6OT5hxnz22JSKfPvBRt4yBVSi6w1awKEKYe/5xoOFfmAyI7+6oKZIUgVmTWAzQEpSu2FigIhZiXEKikycEH4AAAAAANRUEw==.
Cell Culture and Growth Conditions for SRM Quantification
For SRM-based experiments, three biological replicates for each growth condition were used. Approximately 0.5 × 106 cells were seeded individually into six-well tissue culture plates and left for 1 day for cell attachment. For the galactose-based experiment, cell medium was changed to glucose-free DMEM supplemented with 25 mm galactose and 10% fetal bovine serum. For the hypoxia-based experiment, cells were cultured under hypoxic conditions (0.1% O2). Control cells for both experiments were maintained in regular media with no galactose, 25 mm glucose, and cultured at 18% O2. For the estradiol stimulation experiment, MCF-7 cells were transferred to a phenol red-free Roswell Park Memorial Institute 1640 culture medium supplemented with 10% the charcoal-dextran-stripped fetal bovine serum and grown for a day. Following this, cells were stimulated with either estradiol (10 nm final concentration) or 0.1% ethanol (control cells) and were grown for 24 and 48 h, trypsinized and lysed. We ensured that cells still grew as a monolayer in 48 h after change of growth conditions or estradiol stimulation.
Cell Lysis and Protein Digestion for SRM Quantification
The cell pellet was resuspended with 100 μl of 0.1% acid-labile detergent RapiGest SF (sodium-3-[(2-methyl-2-undecyl-1,3-dioxolan-4-yl)-methoxyl]-1-propanesulfonate, Waters) in 25 mm ammonium bicarbonate, vortexed, and sonicated three times for 30 s. All lysates were centrifuged for 20 min at 15,000 rpm at 4 °C, even though no debris was typically observed after lysis. Total protein concentration was measured using a Coomassie (Bradford) protein assay reagent (Pierce). Proteins (60 μg) in detergent solution were transferred to the 96-well plate, denatured at 60 °C, and the disulfide bonds were reduced with 10 mm dithiothreitol. PCR thermocycler (MasterCycler 5332, Eppendorf, Germany) was used to incubate all samples on a 96-well plate at the corresponding temperatures. Following reduction, the samples were alkylated with 20 mm iodoacetamide. Samples were then digested overnight at 37 °C with sequencing grade modified trypsin (Promega). Trypsin:total protein ratio of 1:30 was used. After digestion, RapiGest SF detergent was cleaved with trifluoroacetic acid, 1% final concentration, and samples in the 96-well plate were centrifuged at 1500 rpm. Peptides were extracted with 10 μl OMIX C18 tips (Varian) using 12-channel pipettes and eluted with 10 μl 64.5% acetonitrile. Recovery from Omix C18 microextraction tips was measured by SRM using several peptides of BSA digest and was estimated as 82%. In this work, a heavy isotope-labeled internal standard peptide LSEPAELTDAVK* peptide was spiked into each digest and used as a quality control for C18 microextraction. Following microextraction, peptides were diluted to 130 μl to provide three, 40 μl injections from a 96-well plate. The following precautions were taken to minimize deterioration of peptides, such as oxidation of methionines and deamidation of asparagines and glutamines, during storage and analysis: (1) peptides in 96-well plates were stored at −20 °C until the use and then analyzed within a day; (2) plates were sealed with silicone rubber sealing mats and kept at 6 °C during analysis.
Development of SRM Assays
To facilitate protein quantification, we intended to identify for each protein two peptides with the most intense and reproducible SRM signal. Initially, GPM proteomics database (http://mrm.thegpm.org) was used to select top 5–8 peptides for each protein based on the occurrence of +2 ions. Peptides were then confirmed in SRM atlas (http://www.srmatlas.org) or in our 2D-LC-MS/MS identification data. Fully tryptic and doubly charged peptides with 7–20 aminoacids were chosen. Peptides with a significant occurrence of +3 ion (>10% of +2 ion) according to the GPM proteomics database or to our identification data were not considered. Peptides with methionine, tryptophan, and N-terminal cysteine residues were avoided, if possible. A list of nearly 600 peptides was uploaded to Pinpoint and was used to design in silico survey SRM methods. An equimolar mixture of regular MCF-7 cells and SILAC-labeled MCF-7 cells (13C6, 15N2 l-Lysine, +8 Da, and 13C6 l-Arginine, +6 Da) was used to experimentally test nearly 10,000 transitions. In the first step of method development, 7–8 peptides and about 125 transitions were included into each of 80 survey SRM methods and run in a nonscheduled mode with 12 ms scan times per transition. In the second step, 4–5 peptides and around 60 transitions were included into each of 40 survey SRM methods and run in a nonscheduled mode with 25 ms scans per transition. Retention times, relative intensities of peptides, three most intense and selective transitions per peptide were recorded at that step. Transitions with fragment m/z higher than precursor m/z were preferable, but transitions with lower m/z were not excluded if had high intensity (especially at proline residue), and low noise. As a reference to exclude possible interferences, we used SRM signal of SILAC cells. In the third step, around 25 light and heavy peptides and near 150 transitions (three most intense transitions per peptide) were scheduled in five survey SRM methods and analyzed with 10–60 ms scans. At that step, the following parameters were verified, recorded or tuned, if required: (1) retention time of light and heavy peptides and scheduling intervals; (2) heavy-to-light ratios of transitions; (3) selectivity of transitions and possible interferences; (4) scan times. If multiple peptides per protein were detected, two peptides with the highest SRM area and significantly different retention times were chosen. In general, there was a clear correlation between protein abundance in the cell lysate and the number of peptides suitable for SRM quantification. All peptides were also analyzed with the Basic Local Alignment Search Tool (BLAST) at http://blast.ncbi.nlm.nih.gov/Blast.cgi to ensure that peptides were unique to each protein isoform. At the fourth step, 76 proteins, 134 peptides, and 401 most intense and reproducible transitions were scheduled in a single multiplex scheduled SRM method within 3.5-min (±1.75 min) intervals during a 60 min LC gradient. Scan times were optimized for each peptide in the final SRM method to ensure the measurement of 15–20 points per LC peak per transition. Optimized regression to calculate collision energies CE = 0.0316 × m/z + 1.7802 (r2 = 0.95) was used.
Protein Quantification by SRM
Peptides were separated by 60-min C18 reversed-phase liquid chromatography (EASY-nLC, Proxeon, Odense, Denmark) and analyzed by a triple-quadrupole mass spectrometer (TSQ Vantage, Thermo Fisher Scientific Inc., San Jose, CA) using a nanoelectrospray ionization source, as previously described (18–20). Analytical nanoLC column performed well several days before and after analysis, so stability of SRM signal was not compromised. Reproducibility of SRM signal was ensured by running a QC solution of 0.25 fmol/μl BSA every nine runs. The upper level of signal instability of our system could be estimated by the technical variation of analysis (median CV 4%). Carry-over was evaluated with a blank injection and was estimated in the range 0.05–0.2%. To minimize the carry-over effect, analysis of control cells was followed by the analysis of treated cells for each biological condition. We also assumed that the effect of ionization suppression was reproducible in all runs because of the identical cell lysate matrix, low variation of LC retention times (1.8% median CV), and the fixed position of the nanoESI tip to ensure the constant electric field. The maximum level of signal instability because of the intermittent ionization suppression can be estimated with biological variation of analysis.
Data Analysis
Raw files recorded for each sample were analyzed using Pinpoint software, and CSV files with peptide areas were extracted (supplemental table S3). It should be mentioned that 73 peaks in the set of 13,132 peaks (134 peptides measured in 98 runs) were not detected due to their drift outside the scheduled window (∼0.5% of all measurements). For peptides lacking a single technical replicate, or a single biological replicate, or two or three biological replicates, the average of two technical replicates, or two peptides in the remaining two biological replicates, or only the second peptide in three biological replicates were used for the analysis, respectively. The statistics software package and programming environment R was used for data normalization and analysis. Normalization of peptide areas was performed on the log2-transformed peak areas. The first injection of replicate 1 of Control-24 h cells was used as the reference, and the subsequent replicates were normalized to that reference by the means of a normalization constant estimated by a linear model (supplemental information). Normalization constants and normalized areas are presented in the supplemental Tables S3 and S4. After normalization, we first computed r2 (square of Pearson correlation coefficient) and RMS (Root Mean Square) errors between all pairs of samples (supplemental Table S5). One biological replicate of hypoxia-48h sample had significantly lower correlation with other replicates (0.75 and 0.76 versus 0.99) and thus was excluded from analysis. All other pairs of biological replicates for each condition showed consistent pattern of correlation. The median r2 was 0.995 across LC-SRM injections and 0.978 across biological replicates. Second, we performed quality control to remove peptides that were the consistent outliers. We computed ratios of 28 pairs of MCF-7 biological replicates (except the third replicate of hypoxia-48), found the median ratio and the standard deviation, and flagged outlier peptides with p < 0.05 based on the normal approximation of the distribution of the ratios. Then we computed how many times (out of 28) each peptide was flagged as an outlier and then removed peptides that were outliers 10 or more times (FPR of not being an outlier was 6 × 10−7). As a result, we removed 6 peptides (supplemental Table S3). Poor reproducibility of these peptides may be because of the poorly controlled or irreproducible biological, digestion or LC separation effects. For example, four of six peptides had relatively short length (7–9 aminoacids) and eluted at the beginning of LC gradient (12–16 min). Peptide LSEPAELTDAVK*, a quality control for microextraction, was not used in data analysis. Normalized peptide areas for each protein with two peptides were summed to obtain the protein area (supplemental Table S6). Biological reproducibility of protein areas (median CV 7%) was estimated with three replicates (supplemental Table S7 and Fig. S4).
To calculate relative abundance of proteins in control and treatment groups, we used a linear model to fit protein areas and growth times (supplemental information), and assess the significance of treatment. The Benjamini and Hochberg approach was used to obtain FDR corrected p values (q-values) (21). We used q-value < 0.05 as a cutoff to select differentially expressed proteins (supplemental Table S8). Because we were interested in proteins with high differential ratios, we presented in Table I only proteins with ratios outside the range of two standard deviations from the mean (0.72–1.30). The mean ratio (1.01) and its standard deviation (0.14) were calculated based on ratios of 12 high-abundance house-keeping proteins in galactose-grown, hypoxia, and estradiol stimulation samples.
Table I. Differentially expressed proteins and their relative abundance in 48 hours after treatment (in parentheses). FDR-corrected p values (q-values) <0.05 were used to select differentially expressed proteins. Presented proteins have treated/conrol ratios outside the range of two standard deviations from the mean (0.72–1.30).
| Positive control proteins |
Metabolic proteins |
|||
|---|---|---|---|---|
| Up-regulated | Down-regulated | Up-regulated | Down-regulated | |
| High-galactose-no-glucose media | SLC2A1 (2.0) | ALDOC (1.7) | ||
| HSPA5 (1.7) | ||||
| HYOU1 (1.4) | ||||
| Hypoxia | SLC2A1 (3.0)a | PGR (0.5) | ALDOC (3.1)a | FBP2 (0.7) |
| GREB1 (0.6) | GPI (1.9) | |||
| LDHA (1.9)a | ||||
| PGK1 (1.8)a | ||||
| PFKP (1.7)a | ||||
| SLC7A5 (1.7) | ||||
| ALDOA (1.5)a | ||||
| SDHA (1.5) | ||||
| PKM1/M2 (1.4)a | ||||
| GAPDH (1.4)a | ||||
| ENO1 (1.3)a | ||||
| PGAM1 (1.3)a | ||||
| Estradiol stimulation | TFF1 (5.7)b | SLC1A5 (1.4) | FBP1 (0.6)b | |
| PGR (4.1)b | FBP2 (0.6)b | |||
| GREB1 (4.1)b | ||||
| TFRC (3.1) | ||||
| TPD52L1 (2.8)b | ||||
| MCF-10A | GLUD1 (5.4) | TFF1 (0.01) | ||
| LDHA (4.9) | FBP1 (0.01) | |||
| SDHB (3.2) | FBP2 (0.1) | |||
| UGP2 (2.8) | IDH2 (0.1) | |||
| FH (2.4) | G6PD (0.1) | |||
| PGM2 (2.2) | GALK1(0.1) | |||
| HK1 (2.2) | GREB1 (0.2) | |||
| TUBB2C (1.9) | TPD52L1 (0.2) | |||
| IDH3G (1.9) | SLC1A5 (0.2) | |||
| PDHA1 (1.9) | PES1 (0.2) | |||
| PDHB (1.8) | TFRC (0.3) | |||
| TUBB (1.7) | PCK2 (0.3) | |||
| ACO2 (1.7) | PFKL (0.3) | |||
| SUCLA2 (1.7) | RPIA (0.3) | |||
| DLAT (1.6) | ALDOC (0.4) | |||
| PGD (0.4) | ||||
| HSP90AB1 (0.5) | ||||
| HYOU1 (0.5) | ||||
| PGR (0.5) | ||||
| GAPDH (0.6) | ||||
| RPL6 (0.6) | ||||
| SUCLG2 (0.6) | ||||
| ALDOA (0.7) | ||||
| RPL27A (0.7) | ||||
a Promoters or enhancers of these genes have hypoxia-responsive elements (HRE).
b Promoters or enhancers of these genes have estrogen-responsive elements (ERE).
RESULTS AND DISCUSSION
Proteomic Protocol
Accuracy and reproducibility of quantitative proteomics measurements rely to a great extent on the robustness of the sample preparation protocol. Our goal here was to develop a straightforward protocol that integrated mammalian cell culture and accurate SRM quantification. We used a mass spectrometry-compatible acid-cleavable detergent Rapigest that was originally proposed as an acid-labile detergent for protein denaturation (22). In this work, we optimized the use of Rapigest for cell lysis. We also tested SDS- and CHAPS-based protocols and found that all three protocols provided similar number of protein identifications using 2D-LC-MS/MS (unpublished data). SDS and CHAPS detergents, however, are removed with additional steps of dialysis and lyophilization, may cause sample loss and make the whole proteomic protocol more tedious and less robust. Rapigest-based protocol facilitated quick and easy cell lysis, protein denaturation, digestion, and preparation of peptides for mass spectrometry analysis. This protocol was successfully used in our laboratory for the proteomic analysis of breast and prostate cancer cells, human amniocytes, and human proximal tubule cells. Rapigest-based protocol was also found as efficient as SDS-based protocol in terms of identification of membrane proteins. Recently, similar protocol was used to quantify by LC-MS/MS nearly 7300 proteins in the human osteosarcoma cells (23). In this work, we report that Rapigest-based protocol is efficient, straightforward, and may be widely used for protein identification by LC-MS/MS and for functional studies of proteins in mammalian cells by SRM.
Protein Identification
We used a described cell lysis protocol followed by strong cation-exchange chromatography and reversed-phase-nanoLC-ESI-MS/MS for protein identification (Fig. 1). We identified in MCF-7 cell lysate 4074 proteins (at false discovery rate of 1.0%), which represents one of the largest proteome of MCF-7 cells ever reported (supplemental Tables S1 and S2). A total of 2738 proteins were identified with at least two peptides. Identified proteins were annotated to 216 distinct protein pathways using the KEGG pathway database (supplemental Table S9). Rapigest-based sample preparation protocol was efficient with identification of membrane proteins as GO Cellular Component analysis revealed (supplemental Fig. S1). Interestingly, nearly all enzymes in the major cellular metabolic pathways were identified. This allowed us to select distinct enzymes and enzyme isoforms in glycolysis/gluconeogenesis, TCA cycle, pentose phosphate pathway, galactose metabolism, and glutamine metabolism pathways (Fig. 2). In addition, there were five proteins (seven peptides) in the metabolic pathways that were not identified, but for which we were able to develop SRM assays.
Development of SRM Assay
Sets of survey SRM methods were designed for five to eight peptides per protein, for light- and heavy-isotope labeled peptides. An equimolar mixture of digests of normal and SILAC-labeled heavy MCF-7 cells was used to survey five to eight transitions per peptide (supplemental Fig. S2). In total, nearly 10,000 transitions, 600 unique peptides, and 90 proteins were surveyed.
To ensure high selectivity and the correct identity of each peak, we applied the following set of criteria for each peptide:
Peptide sequence uniqueness: all peptide sequences were analyzed with BLAST to ensure sequence uniqueness. The only exception was made for some high-abundant proteins selected for normalization, such as beta-tubulins, for which peptides could represent several members of the protein family;
Correspondence of LC retention times of light- and heavy-peptide forms;
Correlation of SRM retention time of a peptide to its discovery retention time (supplemental Fig. S3); this test should eliminate spurious peptides with multiple isobaric transitions, but different hydrophobicities;
Superposition of transitions; light- and heavy-peptide forms should have a minimum of six to eight overlaid y-ion transitions. This number of transitions is sufficient to ensure correct peptide identity in complex mixtures (24);
Order of transitions: same order of y-ion transition intensities for light- and heavy-peptide forms, e.g. y1>y2>y3;
Integrated area of all transitions; area of light and heavy peptides should be equal in the equimolar mixture of lysates.
To ensure high selectivity, all transitions were tested in the same cell lysate matrix that was used in the final SRM analysis. Finally, we verified, recorded or tuned the following parameters: (1) retention times and scheduling intervals; (2) selectivity of transitions and possible interferences; (3) scan times; (4) collision energies. Following that, one or two peptides per protein and the three most intense, selective and reproducible transitions per peptide were chosen; 134 peptides representing 76 proteins were multiplexed in a single SRM assay (supplemental Fig. S3).
In the future, the proposed assay can be upgraded with four to five peptides per protein, as well as synthetic or concatenated peptide standards, or with SILAP, super-SILAC, and whole protein standards (25–28). Such assay should provide very accurate estimation of protein abundance. It should also resolve some uncertainties and avoid inconsistencies because of the post-translational modifications and proteolytic cleavage of proteins, incomplete trypsin digestion, acquired peptide modifications, ionization suppression, nonlinear response, and poor LC reproducibility. One of the limitations of our present work is an assumption that all these parameters remain the same in MCF-7 cells under three different biological conditions and in MCF-10A cells.
SRM Analysis of Breast Cancer Cells
Three biological replicates of MCF-7 breast cancer cells were cultured under different growth conditions for 24 and 48 h, then were collected and lysed. All lysates were simultaneously subjected to the sample preparation protocol, and the peak area for each peptide was measured with a multiplex SRM assay. Because the unfractionated digest of the whole cell lysate was used, only medium-to-high-abundance proteins were quantified. Additional fractionation may increase sensitivity of analysis to a hundred or so protein copies per cell (15) but this extra step will significantly decrease the throughput.
The median CV of retention times of peptides in all conditions was 1.8%, so 3.5 min scheduling interval was sufficient to multiplex all peptides in a 60-min LC gradient. Reproducibility of retention times of selected peptides is presented at Fig. 3. The median variation of SRM area in technical replicates was 4% (supplemental Fig. S4), whereas the median biological variation before normalization was 15% (Fig. 4). Dynamic range of SRM analysis was 3.5 orders of magnitude, with the lowest range for progesterone receptor and the highest range for beta-actin (supplemental Fig. S5). This range correlated well with the previously reported levels of progesterone receptor, ∼50,000 copies per MCF-7 cell (29), and beta-actin, ∼108 copies per cell (30).
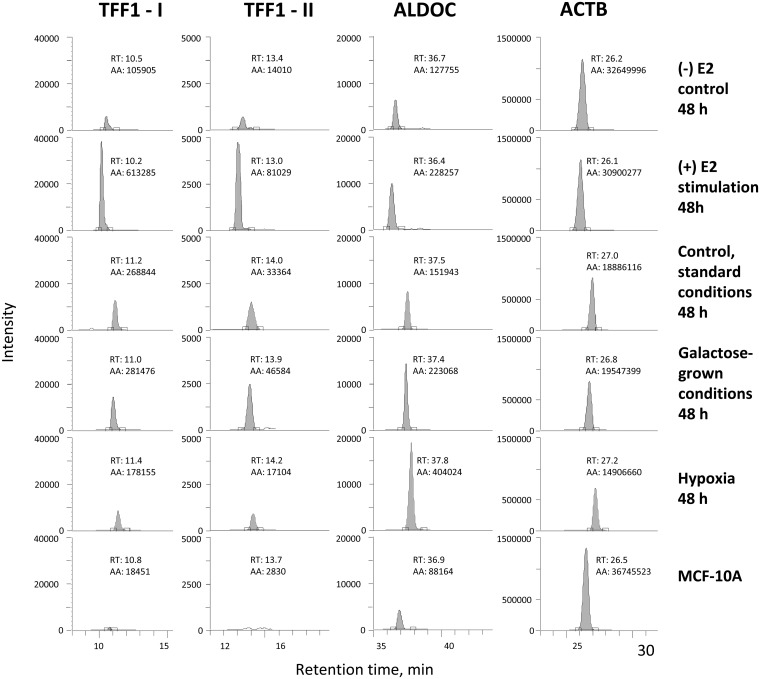
Fig. 3.
Reproducibility of SRM signal and retention times of four selected peptides in different biological conditions, and in MCF-10A cells. TFF1-I and TFF1-II are peptides GCCFDDTVR and QNCGFPGVTPSQCANK of trefoil factor 1, respectively.
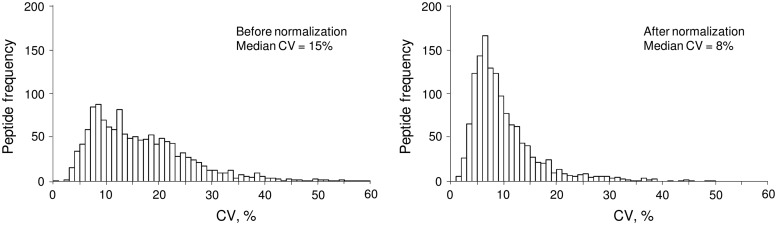
Fig. 4.
Biological variation of SRM areas of peptides in all growth conditions before and after normalization. Normalization with a linear model reduced median biological variation from 15 to 8%.
Data Analysis
Correct interpretation of multiplex SRM assay data requires a robust statistical analysis. In this work, we first used a statistical linear model to calculate normalized peptide areas. Normalization was required to correct total protein measurements and essentially normalize peptide abundances to the same number of cells in each biological condition. Normalization decreased variability (Fig. 4) and facilitated more accurate label-free analysis of relative abundances of proteins.
Protein abundances were computed using the sum of normalized peptide areas. Such method provided the most accurate results in case proteotypic peptides had significantly different SRM areas (31). This method assigned different weights to peptides, so peptides with significantly lower SRM areas (and often significantly higher CVs) did not have any considerable impact.
Another statistical linear model was used to calculate the change of relative abundances of proteins as a function of growth time and corresponding q-values (FDR corrected p values) to correct for multiple testing. Such correction is required when more than one parameter is measured simultaneously, and, thus, is required to analyze data generated with any multiplex SRM assay. This model also allowed discriminating early- and late-response proteins through the comparison of ratio increase in 24 and 48 h (supplemental Table S8).
Growth in Galactose-Rich-Glucose-Free Media
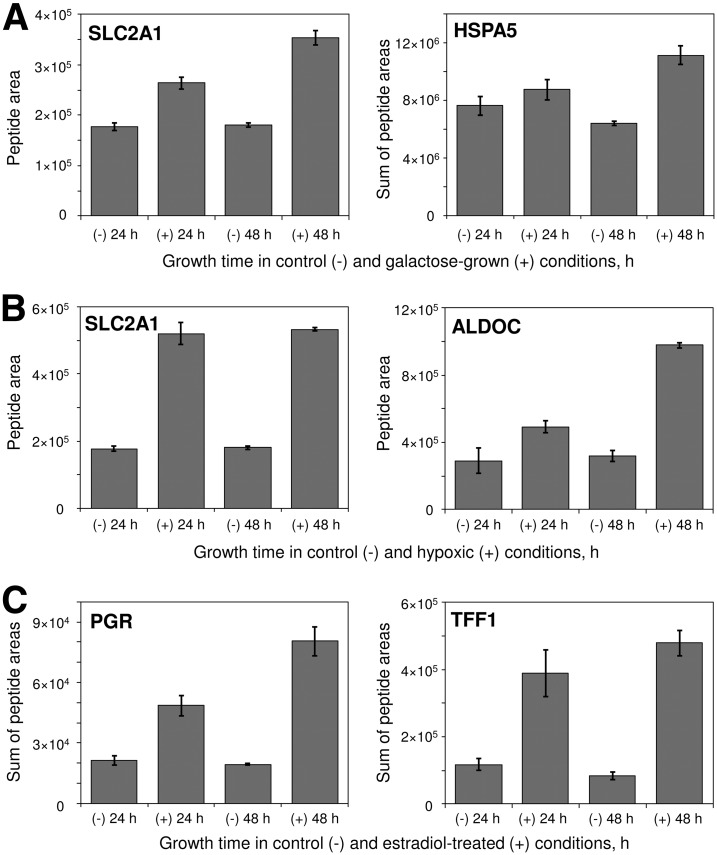
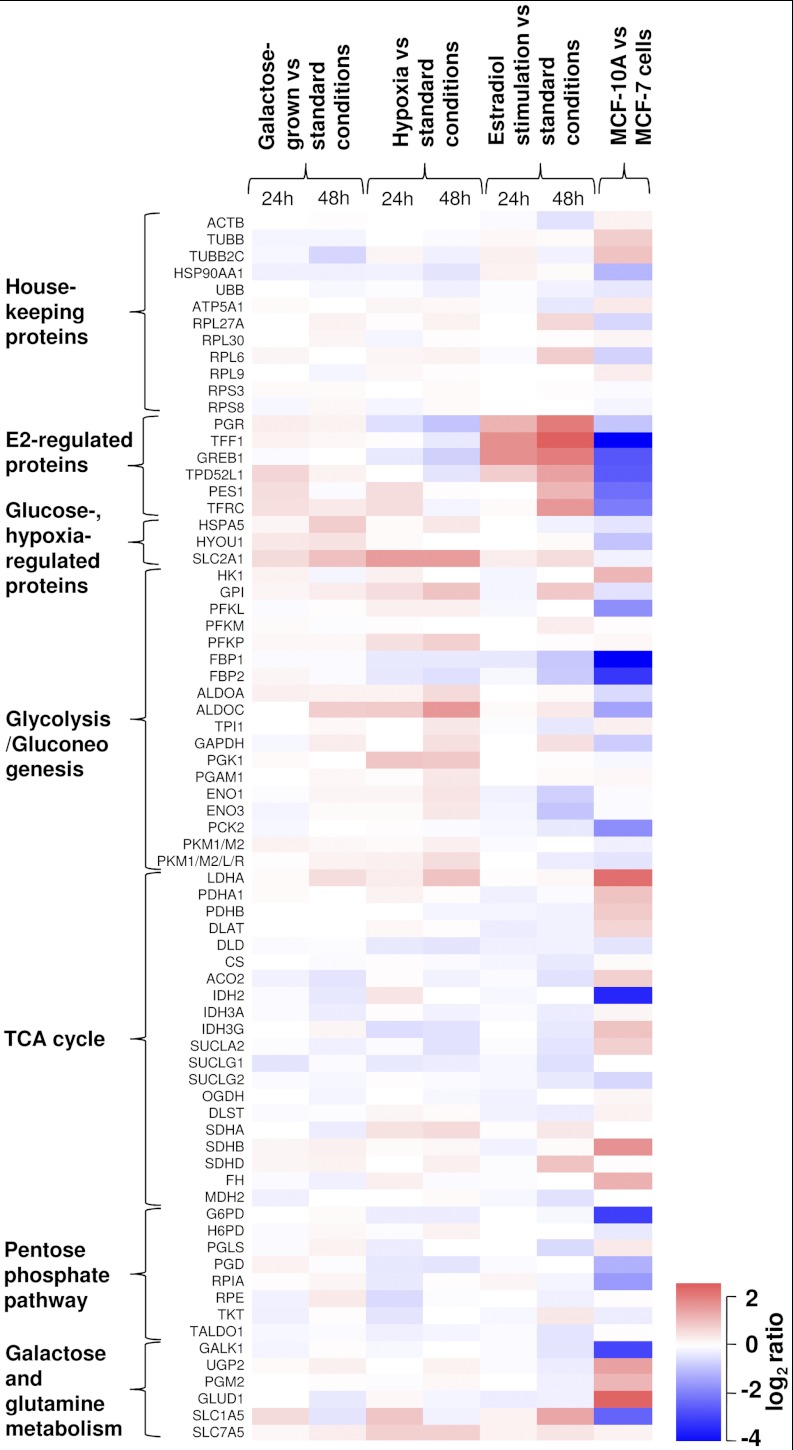
Growth in galactose-rich-glucose-free media resulted in the expression of several glucose-regulated proteins (Table I, Figs. 5–6). There was a twofold increased expression of SLC2A1 (glucose transporter protein 1), 1.7-fold increased expression of HSPA5 (glucose-regulated protein 78kDa), and 1.4-fold increased expression of HYOU1 (glucose-regulated and hypoxia up-regulated protein), which validated our biological model. Changes in the metabolic pathways included increased expression of glycolytic pathway aldolase ALDOC. Interestingly, expression of galactokinase, GALK1, which is a protein highly up-regulated by galactose in yeast, did not change in MCF-7 cells grown in the galactose-rich media. This different response to galactose between yeast and mammalian cells was previously described for hepatocytes (32).
Fig. 5.
Time-dependent expression of selected proteins in control and galactose-grown (A), hypoxic (B), and estradiol-treated (C) conditions. Columns represent normalized peptide area or the sum of normalized peptide areas, whereas error bars show variation of area in three biological replicates.
Fig. 6.
Relative abundance of housekeeping, control and metabolic pathway proteins in MCF-7 cells under different growth conditions, and in near-normal MCF-10A cells as compared with MCF-7 cells. In galactose-grown conditions, differentially expressed proteins included SLC2A1 (glucose transporter protein 1), HSPA5 (glucose-regulated protein 78kDa), HYOU1 (glucose-regulated and hypoxia up-regulated protein 1) and ALDOC (aldolase C). Hypoxic conditions resulted in differential expression of SLC2A1, estrogen-regulated proteins and a number of proteins in glycolysis. Estradiol stimulation changed expression of estrogen-regulated proteins, but did not significantly affect energy metabolic pathways. Significant difference in protein expression levels was found between MCF-7 (cancer) and MCF-10A (near-normal) breast cells.
Hypoxia
Growth in hypoxic conditions (0.1% oxygen) resulted in the elevated expression of a considerable number of hypoxia-regulated and metabolic proteins (Table I, Figs. 5 and 6). This included SLC2A1 as well as ten glycolytic enzymes. For example, there was a 3.1-fold increase in expression of aldolase C, which is known to be directly regulated in epithelial cells exposed to hypoxia through the hypoxia-responsive element (33). It total, increased expression of 11 of 12 proteins in Table I was previously reported in hypoxic conditions (34–36). Expression of nine of 12 proteins was regulated directly through the hypoxia-inducible factor (HIF) because the corresponding genes had hypoxia-responsive elements (HREs) in their promoter regions (37, 38). As determined by the comparison of relative rates of protein expression in 24 and 48 h (supplemental Table S8), expression of glucose transporter protein SLC2A1 sharply increased in hypoxia, while slowly increasing in the galactose-grown conditions. Thus, SLC2A1 is presumably an early-response gene in hypoxia.
In total, we observed an elevated expression of enzymes involved in 9 out of 11 enzymatic reactions that convert glucose into lactate. On the contrary, fructose-1,6-bisphosphatase 2, an enzyme reverting glycolysis to gluconeogenesis, was found down-regulated. It was fascinating to observe such simultaneous effect of hypoxia on breast cancer cells.
Interestingly, hypoxia also resulted in a consistent decrease of amounts of estrogen-regulated proteins PGR and GREB1. Such effect was mediated through the degradation of estrogen receptor by proteasomal degradation pathway and repression of estrogen receptor mRNA expression (39). Better understanding of estrogen regulation in hypoxia may unveil the mechanism of transformation of breast cancers to the estrogen-independent forms.
Estradiol Stimulation
Estradiol stimulation resulted in the strong over-expression of known estrogen-regulated proteins (PGR, TFF1, GREB1, TPD52L1, and TFRC). A gradual increase of protein expression was observed after 24 and 48 h of growth (Fig. 5). Differentially increased metabolic proteins included glycolytic fructose-1,6-bisphosphatases FBP1 and FBP2. Interestingly, promoter regions of these genes had estrogen-responsive elements (EREs) (40).
Increased expression of PGR, TFF1, and GREB1 genes upon estradiol stimulation is well-documented in the literature, and was intensively studied at mRNA level (41–43). GREB1, TPD52L1, and TFF1 genes are currently investigated as prognostic and predictive markers of breast cancer (44–46), so presented SRM assay can be used to investigate expression of corresponding proteins in breast cancer cells and tissues. Estradiol-mediated expression of TPD52L1 was previously reported, but its cellular function is still not known (47). Using our SRM-based workflow, multiple transcriptomics-based data sets of estrogen-regulated genes can be validated, and functional roles of many estrogen-regulated proteins can be elucidated (40, 47). In addition, measurement of estrogen-regulated proteins in the presence of increasing concentrations of drug candidates can facilitate cell-based screening for agonists and inhibitors of estrogen receptor and accurate determination of their effective and inhibitory concentrations.
Cancer Versus Near-normal Breast Cancer Cells
Impressive difference in protein expression was found between MCF-7 (cancer) and MCF-10A (near-normal) breast cells (Fig. 6). Cancer cells had significantly elevated levels of FBP1, FBP2 (gluconeogenesis), IDH2 (TCA cycle), and G6PD (the pentose phosphate pathway).
Elevated expression of FBP1, FBP2, and G6PD in cancer cells increases glucose flux from glycolysis into the pentose phosphate pathway and redirects cellular energy metabolism toward increased biosynthesis (48). Interestingly, the IDH2 enzyme has recently captured a lot of attention in relation to metabolic transformation of cancer cells (9, 10). Cancer-specific mutations of IDH2 discovered in gliomas and leukemias lead to accumulation of the uncommon metabolite 2-hydroxyglutarate and to profound changes in the cellular metabolome (49). Here, we searched our MCF-7 MS/MS identification data and found no known IDH2 mutations (R172K, R172M, R172W) (50). However, IDH2 was considerably elevated in cancer cells. Recently, IDH2 expression was found significantly elevated in three breast cancer cell lines that represented late stages of tumor (51). Levels of estrogen-regulated proteins, especially TFF1, were significantly lower in near-normal MCF-10A cells that were known not to express an estrogen receptor (52). Similar trend for protein abundance in MCF-10A versus MCF-7 cells has recently been found based on spectral counting comparison of shotgun proteomics data (53). However, because of the limited dynamic range of spectral counting, such comparison provided reliable results only for high-abundance proteins.
Concluding Remarks
To summarize, we presented here an SRM-based proteomic workflow to monitor relative expression of proteins in the entire energy metabolic pathway in mammalian cells exposed to different growth conditions.
We want to emphasize the advancements made in the present work. In general, in this work we optimized multiple proteomic and cell biology techniques and integrated them into a single workflow with a minimal number of sample preparation steps to facilitate quantitative comparison of protein expression in mammalian cells. First, we optimized the use of acid-labile mass spectrometry compatible detergent (Rapigest) for lysis of mammalian cells. Followed by Rapigest-assisted protein denaturation, our proteomic protocol was carried out in a single well of a 96-well plate and was devoid of common steps of detergent removal and lyophilization that might lead to sample loss and low reproducibility of analysis. Second, this work is one of the first reports on SRM-based quantitative proteomic workflows for mammalian cells. Our workflow allows monitoring changes in protein expression in the whole metabolic pathway, in multiple growth conditions and time points, and with multiple biological and technical replicates. Present SRM workflow and the data analysis model can be used to validate the results of global discovery proteomics experiments, such as SILAC. In addition, our statistical model facilitates identification of early- and late-response proteins and may possibly differentiate between directly and indirectly regulated proteins. Third, we present quantitative protein assays that are currently lacking in the field of metabolic transformation of cancer cells. Alterations in cellular metabolism in hypoxia and upon abnormal hormone stimulation are widely investigated in breast cancer studies and often rely on measurement of mRNA expression, but not proteins (54). Fourth, we confirmed by SRM, an alternative protein quantification method, a well-known expression of a number of proteins (such as SLC2A1, HSPA5, LDHA, PGR, and TFF1) in mammalian cells exposed to glucose deprivation, hypoxia, and estradiol stimulation. Hypoxia- or estrogen-responsive elements in the promoters of corresponding genes were found for the majority of differentially expressed metabolic and control proteins. Fifth, we measured some estrogen-regulated proteins (GREB1 and TPD52L1), for which quantitative protein assays, such as ELISA, are not available.
Our quantitative proteomic protocol based on an acid-labile mass spectrometry-compatible detergent is described in detail. Presented SRM assay includes 76 proteins, 134 proteotypic peptides, the three most intense and selective SRM transitions, and optimal instrumental parameters (supplemental Table S10). This large set of parameters can be easily shared between SRM-compatible mass spectrometers and used as a foundation for developing comprehensive assays to measure protein expression in all metabolic pathways in the cell.
It should be mentioned that the present assay allows quantifying changes in protein expression, but not in enzyme activity. Enzyme activity in the living cell is often regulated by multiple post-translational mechanisms, regulatory proteins, and cofactors and cannot always be correlated with enzyme concentration. To fully comprehend the complexity of the cellular metabolism, the targeted proteomics assays should be complemented with metabolomic, transcriptomic, and activity-based profilings of cellular pathways (55, 56). In addition, presented workflow cannot be applied to low-abundance proteins expressed in mammalian cells at low copy numbers (<10,000 copies per cell). Quantification of low-abundance proteins would require an additional step of protein or peptide separation and may decrease the throughput and reproducibility of the assay. In our experience, however, a large fraction of proteins amenable to 2D-LC-MS/MS identification can be quantified by SRM in the unfractionated digest of cell lysate or biological fluid.
In summary, the presented workflow is an integrated tool for hypothesis-driven studies of mammalian cells as well as functional studies of proteins, and can greatly complement experimental methods in systems biology, metabolic engineering, and metabolic transformation of cancer cells.
Acknowledgments
We thank members of our laboratory for helpful discussions and Joyce Lai for assistance with preparation of figures.
Footnotes
* This work was supported by the Canadian Institutes of Health Research (CIHR) post-doctoral fellowship to A. P. D. and Ontario Graduate Scholarship (OGS) to M. P. P.
 This article contains supplemental Tables S1 to S10, Figs. S1 to S5 and supplemental information.
This article contains supplemental Tables S1 to S10, Figs. S1 to S5 and supplemental information.
1 The abbreviations used are:
- SRM
- selected reaction monitoring
- 2D-LC-MS/MS
- two-dimensional liquid chromatography-tandem mass spectrometry
- CE
- collision energy
- CV
- coefficient of variation
- Da
- Daltons
- ELISA
- enzyme-linked immunosorbent assay
- FDR
- false discovery rate
- LC
- liquid chromatography
- MCF-7
- human breast adenocarcinoma cell line
- MCF-10A
- near normal human mammary epithelial cell line
- MS/MS
- tandem mass spectrometry
- PPP
- pentose phosphate pathway
- SCX
- strong cation exchange chromatography
- TCA cycle
- tricarboxylic acid cycle.
REFERENCES
- 1. Gatenby R. A., Gillies R. J. (2004) Why do cancers have high aerobic glycolysis? Nat. Rev. Cancer 4, 891–899 [DOI] [PubMed] [Google Scholar]
- 2. DeBerardinis R. J., Lum J. J., Hatzivassiliou G., Thompson C. B. (2008) The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 7, 11–20 [DOI] [PubMed] [Google Scholar]
- 3. Warburg O. (1956) On respiratory impairment in cancer cells. Science 124, 269–270 [PubMed] [Google Scholar]
- 4. Hanahan D., Weinberg R. A. (2011) Hallmarks of cancer: the next generation. Cell 144, 646–674 [DOI] [PubMed] [Google Scholar]
- 5. Giovannucci E., Harlan D. M., Archer M. C., Bergenstal R. M., Gapstur S. M., Habel L. A., Pollak M., Regensteiner J. G., Yee D. (2010) Diabetes and cancer: a consensus report. CA Cancer J. Clin. 60, 207–221 [DOI] [PubMed] [Google Scholar]
- 6. Gasparre G., Porcelli A. M., Bonora E., Pennisi L. F., Toller M., Iommarini L., Ghelli A., Moretti M., Betts C. M., Martinelli G. N., Ceroni A. R., Curcio F., Carelli V., Rugolo M., Tallini G., Romeo G. (2007) Disruptive mitochondrial DNA mutations in complex I subunits are markers of oncocytic phenotype in thyroid tumors. Proc. Natl. Acad. Sci. U.S.A. 104, 9001–9006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Duarte N. C., Becker S. A., Jamshidi N., Thiele I., Mo M. L., Vo T. D., Srivas R., Palsson B. Ø. (2007) Global reconstruction of the human metabolic network based on genomic and bibliomic data. Proc. Natl. Acad. Sci. U.S.A. 104, 1777–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ramanathan A., Wang C., Schreiber S. L. (2005) Perturbational profiling of a cell-line model of tumorigenesis by using metabolic measurements. Proc. Natl. Acad. Sci. U.S.A. 102, 5992–5997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yan H., Parsons D. W., Jin G., McLendon R., Rasheed B. A., Yuan W., Kos I., Batinic-Haberle I., Jones S., Riggins G. J., Friedman H., Friedman A., Reardon D., Herndon J., Kinzler K. W., Velculescu V. E., Vogelstein B., Bigner D. D. (2009) IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 360, 765–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mardis E. R., Ding L., Dooling D. J., Larson D. E., McLellan M. D., Chen K., Koboldt D. C., Fulton R. S., Delehaunty K. D., McGrath S. D., Fulton L. A., Locke D. P., Magrini V. J., Abbott R. M., Vickery T. L., Reed J. S., Robinson J. S., Wylie T., Smith S. M., Carmichael L., Eldred J. M., Harris C. C., Walker J., Peck J. B., Du F., Dukes A. F., Sanderson G. E., Brummett A. M., Clark E., McMichael J. F., Meyer R. J., Schindler J. K., Pohl C. S., Wallis J. W., Shi X., Lin L., Schmidt H., Tang Y., Haipek C., Wiechert M. E., Ivy J. V., Kalicki J., Elliott G., Ries R. E., Payton J. E., Westervelt P., Tomasson M. H., Watson M. A., Baty J., Heath S., Shannon W. D., Nagarajan R., Link D. C., Walter M. J., Graubert T. A., DiPersio J. F., Wilson R. K., Ley T. J. (2009) Recurring mutations found by sequencing an acute myeloid leukemia genome. N. Engl. J. Med. 361, 1058–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Christofk H. R., Vander Heiden M. G., Harris M. H., Ramanathan A., Gerszten R. E., Wei R., Fleming M. D., Schreiber S. L., Cantley L. C. (2008) The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature 452, 230–233 [DOI] [PubMed] [Google Scholar]
- 12. Gambhir S. S. (2002) Molecular imaging of cancer with positron emission tomography. Nat. Rev. Cancer 2, 683–693 [DOI] [PubMed] [Google Scholar]
- 13. van den Bent M. J., Dubbink H. J., Marie Y., Brandes A. A., Taphoorn M. J., Wesseling P., Frenay M., Tijssen C. C., Lacombe D., Idbaih A., van Marion R., Kros J. M., Dinjens W. N., Gorlia T., Sanson M. (2010) IDH1 and IDH2 mutations are prognostic but not predictive for outcome in anaplastic oligodendroglial tumors: a report of the European Organization for Research and Treatment of Cancer Brain Tumor Group. Clin. Cancer Res. 16, 1597–1604 [DOI] [PubMed] [Google Scholar]
- 14. Tennant D. A., Durán R. V., Gottlieb E. (2010) Targeting metabolic transformation for cancer therapy. Nat. Rev. Cancer 10, 267–277 [DOI] [PubMed] [Google Scholar]
- 15. Picotti P., Bodenmiller B., Mueller L. N., Domon B., Aebersold R. (2009) Full dynamic range proteome analysis of S. cerevisiae by targeted proteomics. Cell 138, 795–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Costenoble R., Picotti P., Reiter L., Stallmach R., Heinemann M., Sauer U., Aebersold R. (2011) Comprehensive quantitative analysis of central carbon and amino-acid metabolism in Saccharomyces cerevisiae under multiple conditions by targeted proteomics. Mol. Syst. Biol. 7, 464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schmidt A., Beck M., Malmström J., Lam H., Claassen M., Campbell D., Aebersold R. (2011) Absolute quantification of microbial proteomes at different states by directed mass spectrometry. Mol. Syst. Biol. 7, 510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Drabovich A. P., Diamandis E. P. (2010) Combinatorial peptide libraries facilitate development of multiple reaction monitoring assays for low-abundance proteins. J. Proteome Res. 9, 1236–1245 [DOI] [PubMed] [Google Scholar]
- 19. Makawita S., Smith C., Batruch I., Zheng Y., Ruckert F., Grutzmann R., Pilarsky C., Gallinger S., Diamandis E. P. (2011) Integrated proteomic profiling of cell line conditioned media and pancreatic juice for the identification of pancreatic cancer biomarkers. Mol. Cell. Proteomics 10, 10.1074/mcp.M111.008599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Drabovich A. P., Jarvi K., Diamandis E. P. (2011) Verification of male infertility biomarkers in seminal plasma by multiplex selected reaction monitoring assay. Mol. Cell. Proteomics 10, 10.1074/mcp.M110.004127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Benjamini Y., Hochberg Y. (1995) Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. Roy. Statist. Soc. 57, 289–300 [Google Scholar]
- 22. Yu Y. Q., Gilar M., Lee P. J., Bouvier E. S., Gebler J. C. (2003) Enzyme-friendly, mass spectrometry-compatible surfactant for in-solution enzymatic digestion of proteins. Anal. Chem. 75, 6023–6028 [DOI] [PubMed] [Google Scholar]
- 23. Beck M., Schmidt A., Malmstroem J., Claassen M., Ori A., Szymborska A., Herzog F., Rinner O., Ellenberg J., Aebersold R. (2011) The quantitative proteome of a human cell line. Mol. Syst. Biol. 7, 549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Prakash A., Tomazela D. M., Frewen B., Maclean B., Merrihew G., Peterman S., Maccoss M. J. (2009) Expediting the development of targeted SRM assays: using data from shotgun proteomics to automate method development. J. Proteome Res. 8, 2733–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Beynon R. J., Doherty M. K., Pratt J. M., Gaskell S. J. (2005) Multiplexed absolute quantification in proteomics using artificial QCAT proteins of concatenated signature peptides. Nat. Methods 2, 587–589 [DOI] [PubMed] [Google Scholar]
- 26. Yu K. H., Barry C. G., Austin D., Busch C. M., Sangar V., Rustgi A. K., Blair I. A. (2009) Stable isotope dilution multidimensional liquid chromatography-tandem mass spectrometry for pancreatic cancer serum biomarker discovery. J. Proteome Res. 8, 1565–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Geiger T., Cox J., Ostasiewicz P., Wisniewski J. R., Mann M. (2010) Super-SILAC mix for quantitative proteomics of human tumor tissue. Nat. Methods 7, 383–385 [DOI] [PubMed] [Google Scholar]
- 28. Stergachis A. B., MacLean B., Lee K., Stamatoyannopoulos J. A., MacCoss M. J. (2011) Rapid empirical discovery of optimal peptides for targeted proteomics. Nat. Methods 8, 1041–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nogueira C. R., Brentani M. M. (1996) Triiodothyronine mimics the effects of estrogen in breast cancer cell lines. J. Steroid Biochem. Mol. Biol. 59, 271–279 [DOI] [PubMed] [Google Scholar]
- 30. Zuo X., Lee K., Speicher DW. (2004) Electrophoretic prefractionation for comprehensive analysis of proteomes. In: Speicher D. W., ed. Proteome Analysis: Interpreting the Genome, pp. 93–115, Elsevier Science, New York [Google Scholar]
- 31. Carrillo B., Yanofsky C., Laboissiere S., Nadon R., Kearney R. E. (2010) Methods for combining peptide intensities to estimate relative protein abundance. Bioinformatics 26, 98–103 [DOI] [PubMed] [Google Scholar]
- 32. Zaret K. S., Stevens K. A. (1990) Selection and analysis of galactose metabolic pathway variants of a mouse liver cell line. Mol. Cell. Biol. 10, 4582–4589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jean J. C., Rich C. B., Joyce-Brady M. (2006) Hypoxia results in an HIF-1-dependent induction of brain-specific aldolase C in lung epithelial cells. Am. J. Physiol. Lung Cell Mol. Physiol. 291, L950–956 [DOI] [PubMed] [Google Scholar]
- 34. Semenza G. L. (2009) Regulation of cancer cell metabolism by hypoxia-inducible factor 1. Semin. Cancer Biol. 19, 12–16 [DOI] [PubMed] [Google Scholar]
- 35. Mense S. M., Sengupta A., Zhou M., Lan C., Bentsman G., Volsky D. J., Zhang L. (2006) Gene expression profiling reveals the profound upregulation of hypoxia-responsive genes in primary human astrocytes. Physiol. Genomics 25, 435–449 [DOI] [PubMed] [Google Scholar]
- 36. Lu S., Gu X., Hoestje S., Epner D. E. (2002) Identification of an additional hypoxia responsive element in the glyceraldehyde-3-phosphate dehydrogenase gene promoter. Biochim. Biophys. Acta 1574, 152–156 [DOI] [PubMed] [Google Scholar]
- 37. Ortiz-Barahona A., Villar D., Pescador N., Amigo J., del Peso L. (2010) Genome-wide identification of hypoxia-inducible factor binding sites and target genes by a probabilistic model integrating transcription-profiling data and in silico binding site prediction. Nucleic Acids Res. 38, 2332–2345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Benita Y., Kikuchi H., Smith A. D., Zhang M. Q., Chung D. C., Xavier R. J. (2009) An integrative genomics approach identifies Hypoxia Inducible Factor-1 (HIF-1)-target genes that form the core response to hypoxia. Nucleic Acids Res. 37, 4587–4602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ryu K., Park C., Lee Y. (2011) Hypoxia-inducible factor 1 alpha represses the transcription of the estrogen receptor alpha gene in human breast cancer cells. Biochem. Biophys. Res. Commun. 407, 831–836 [DOI] [PubMed] [Google Scholar]
- 40. Bourdeau V., Deschênes J., Métivier R., Nagai Y., Nguyen D., Bretschneider N., Gannon F., White J. H., Mader S. (2004) Genome-wide identification of high-affinity estrogen response elements in human and mouse. Mol. Endocrinol. 18, 1411–1427 [DOI] [PubMed] [Google Scholar]
- 41. Kocanova S., Kerr E. A., Rafique S., Boyle S., Katz E., Caze-Subra S., Bickmore W. A., Bystricky K. (2010) Activation of estrogen-responsive genes does not require their nuclear co-localization. PLoS Genet. 6, e1000922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hnatyszyn H. J., Liu M., Hilger A., Herbert L., Gomez-Fernandez C. R., Jorda M., Thomas D., Rae J. M., El-Ashry D., Lippman M. E. (2010) Correlation of GREB1 mRNA with protein expression in breast cancer: validation of a novel GREB1 monoclonal antibody. Breast Cancer Res. Treat. 122, 371–380 [DOI] [PubMed] [Google Scholar]
- 43. Zheng Z. Y., Bay B. H., Aw S. E., Lin V. C. (2005) A novel antiestrogenic mechanism in progesterone receptor-transfected breast cancer cells. J. Biol. Chem. 280, 17480–17487 [DOI] [PubMed] [Google Scholar]
- 44. Abba M. C., Sun H., Hawkins K. A., Drake J. A., Hu Y., Nunez M. I., Gaddis S., Shi T., Horvath S., Sahin A., Aldaz C. M. (2007) Breast cancer molecular signatures as determined by SAGE: correlation with lymph node status. Mol. Cancer Res. 5, 881–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Taylor K. J., Sims A. H., Liang L., Faratian D., Muir M., Walker G., Kuske B., Dixon J. M., Cameron D. A., Harrison D. J., Langdon S. P. (2010) Dynamic changes in gene expression in vivo predict prognosis of tamoxifen-treated patients with breast cancer. Breast Cancer Res. 12, R39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dunbier A. K., Anderson H., Ghazoui Z., Folkerd E. J., A'Hern R., Crowder R. J., Hoog J., Smith I. E., Osin P., Nerurkar A., Parker J. S., Perou C. M., Ellis M. J., Dowsett M. (2010) Relationship between plasma estradiol levels and estrogen-responsive gene expression in estrogen receptor-positive breast cancer in postmenopausal women. J. Clin. Oncol. 28, 1161–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Charpentier A. H., Bednarek A. K., Daniel R. L., Hawkins K. A., Laflin K. J., Gaddis S., MacLeod M. C., Aldaz C. M. (2000) Effects of estrogen on global gene expression: identification of novel targets of estrogen action. Cancer Res. 60, 5977–5983 [PubMed] [Google Scholar]
- 48. Jiang P., Du W., Wang X., Mancuso A., Gao X., Wu M., Yang X. (2011) p53 regulates biosynthesis through direct inactivation of glucose-6-phosphate dehydrogenase. Nat. Cell Biol. 13, 310–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gross S., Cairns R. A., Minden M. D., Driggers E. M., Bittinger M. A., Jang H. G., Sasaki M., Jin S., Schenkein D. P., Su S. M., Dang L., Fantin V. R., Mak T. W. (2010) Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate dehydrogenase 1 and 2 mutations. J. Exp. Med. 207, 339–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Reitman Z. J., Yan H. (2010) Isocitrate dehydrogenase 1 and 2 mutations in cancer: alterations at a crossroads of cellular metabolism. J. Natl. Cancer Inst. 102, 932–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Geiger T., Madden S. F., Gallagher W. M., Cox J., Mann M. (2012) Proteomic portrait of human breast cancer progression identifies novel prognostic markers. Cancer Res. 72, 2428–2439 [DOI] [PubMed] [Google Scholar]
- 52. Soule H. D., Maloney T. M., Wolman S. R., Peterson W. D., Jr., Brenz R., McGrath C. M., Russo J., Pauley R. J., Jones R. F., Brooks S. C. (1990) Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res. 50, 6075–6086 [PubMed] [Google Scholar]
- 53. Bateman N. W., Sun M., Hood B. L., Flint M. S., Conrads T. P. (2010) Defining central themes in breast cancer biology by differential proteomics: conserved regulation of cell spreading and focal adhesion kinase. J. Proteome Res. 9, 5311–5324 [DOI] [PubMed] [Google Scholar]
- 54. Favaro E., Lord S., Harris A. L., Buffa F. M. (2011) Gene expression and hypoxia in breast cancer. Genome Med. 3, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Nomura D. K., Dix M. M., Cravatt B. F. (2010) Activity-based protein profiling for biochemical pathway discovery in cancer. Nat. Rev. Cancer 10, 630–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Schwanhäusser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., Selbach M. (2011) Global quantification of mammalian gene expression control. Nature 473, 337–342 [DOI] [PubMed] [Google Scholar]