Abstract
Protein kinase signaling is fundamental to cell homeostasis and is deregulated in all cancers but varies between patients. Understanding the mechanisms underlying this heterogeneity is critical for personalized targeted therapies. Here, we used a recently established LC-MS/MS platform to profile protein phosphorylation in acute myeloid leukemia cell lines with different sensitivities to kinase inhibitors. The compounds used in this study were originally developed to target Janus kinase, phosphatidylinositol 3-kinase, and MEK. After further validation of the technique, we identified several phosphorylation sites that were inhibited by these compounds but whose intensities did not always correlate with growth inhibition sensitivity. In contrast, several hundred phosphorylation sites that correlated with sensitivity/resistance were not in general inhibited by the compounds. These results indicate that markers of pathway activity may not always be reliable indicators of sensitivity of cancer cells to inhibitors that target such pathways, because the activity of parallel kinases can contribute to resistance. By mining our data we identified protein kinase C isoforms as one of such parallel pathways being more active in resistant cells. Consistent with the view that several parallel kinase pathways were contributing to resistance, inhibitors that target protein kinase C, MEK, and Janus kinase potentiated each other in arresting the proliferation of multidrug-resistant cells. Untargeted/unbiased approaches, such as the one described here, to quantify the activity of the intended target kinase pathway in concert with the activities of parallel kinase pathways will be invaluable to personalize therapies based on kinase inhibitors.
Protein kinase signaling networks control cell proliferation, survival, motility, and metabolism and are deregulated in diseases such as cancer (1, 2). Inhibitors that primarily target the HER2, vascular endothelial growth factor receptor, epidermal growth factor receptor, and BCR-Abl kinases have been approved for the treatment of specific cancers (3, 4), whereas others, including compounds targeting the PI3K/Akt/mTOR,1 MEK/ERK, and JAK/STAT kinase networks, are being advanced toward the clinic (5–12). However, cancers are biologically heterogeneous, and it has become clear that the wiring of the proliferative kinase networks vary profoundly between cells (1, 13) and that cancer cells developing in different contexts vary in their kinase requirements for proliferation (14, 15). The practical consequence of this heterogeneity is that cancers respond to kinase inhibitors to different extents. Targeted therapies will therefore require a systematic identification and monitoring of deregulated kinase pathways in a given cell population. Members of these downstream pathways may then serve both as useful additional drug targets as well as biomarkers for personalized medicine (16).
Systematic gene sequencing efforts have uncovered thousands of mutations in kinases in essentially all cancer types (17–20). Although cancer may ultimately result from the collective set of genetic mutations, establishing a functional connection between specific mutations and kinase signaling pathway activation is difficult with our current knowledge of the network components and how these are wired (21, 22). Methods for direct and systematic quantification of kinase activation should therefore complement genetic studies. Correlating these activities with biological end points (e.g., with the sensitivity to inhibitors or to RNA interference probes) or with genetic mutations could then be used to investigate kinase pathways at the system level, within their given molecular, biological, and physiological contexts.
Advances in MS and separation technology have resulted in the development of several techniques that can be used for global and systematic analysis of signaling pathways. For example, previous studies have shown the potential of MS to quantify the activities of selected kinases in a multiplex manner (23–25). Another approach to systematically quantify overall kinase pathways is based on quantitative phosphoproteomic techniques, which measure global physiological phosphorylation. Powerful and robust MS-based phosphoproteomics methods have been described (26–30) and are being used to provide insights into how kinase signaling pathways contribute to biology (27, 31–33).
In this study, we have applied quantitative phosphoproteomics to analyze global phosphorylation in acute myeloid leukemia (AML) cell lines with different sensitivities to inhibitors originally developed to target PI3K, MEK, and JAK pathways. Our aim was to investigate two alternative hypotheses to account for the resistance of these cells to kinase inhibitors: namely that (i) activation of the inhibitor target(s) may be the main determinant of conferring sensitivity to the compounds (as stated by the theory of oncogene addiction (34)) or (ii) resistance to kinase inhibitors can be due to the activity of parallel pathways in the kinase network, as sometimes (but not always) observed in other cancer types (35, 36). For this, we first identified (by LC-MS/MS) phosphorylation sites inhibited by three different kinase inhibitors in three different pairs of resistant/sensitive cell lines. We then asked whether those phosphorylation sites (which are markers of the kinase activities inhibited by the compounds) would correlate with sensitivity to the inhibition of proliferation by the compounds. We found that cells that differed in their relative sensitivity to kinase inhibitors had markedly different patterns in basal global phosphorylation in the absence of inhibitors, with some phosphorylation patterns correlating with sensitivity/resistance. The intensities of activity markers did not correlate with inhibition of cell proliferation, arguing that readouts of pathway activity are not necessarily reliable indicators of cellular sensitivities to inhibitors targeting such pathways. These findings also indicate that some cancer cells are not addicted to a single overactive kinase pathway and are consistent with the emerging view that susceptibility to an inhibitor may be not only due to activation of the target kinase but also to activation of parallel pathways (37). By mining our data, we found that several PKC isoforms were more phosphorylated at sites known to correlate with their activity in AML resistant to kinase inhibitors, suggesting a role for PKCs in resistance to kinase inhibitors. A pharmacological approach was then used to test whether combining different kinase inhibitors affected the proliferation of sensitive and resistant AML cells. These results were consistent with the phosphoproteomics data and showed that multidrug-resistant AML cells proliferated using different kinase pathways parallel to the ones targeted by the drugs.
EXPERIMENTAL PROCEDURES
Cell Culture
AML cell lines included in the COSMIC cancer genome project (19) (AML-193, CMK, CTS, HEL, Kasumi-1, KG-1, MV4-11, and P31/FUJ) and murine NIH 3T3 fibroblasts were routinely cultured in Dulbecco's modified Eagle's medium at 37 °C in a humidified atmosphere of 5% CO2 in medium supplemented with 10% bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin. AML cell lines were maintained at ∼5–10 × 105 cell/ml in RPMI supplemented with 50 μm β-mercaptoethanol.
When indicated, the effect of kinase inhibitors on phosphorylation was determined by incubating cells with the respective inhibitor (1 μm PI-103, 500 nm MEK I inhibitor, or 500 nm JAK I inhibitor (catalog numbers 528100, 444937, and 420099 from Calbiochem, respectively)) for 1 h before cell lysis. AML cells were seeded at 5 × 105 cells/ml in fresh medium the day before the experiment. Each culture contained 5 × 106 cells in 10 ml, and three independent cell cultures were used per cell line and condition. The cells were harvested by centrifugation at 300 × g for 10 min and washed twice with ice-cold PBS containing 1 mm pV and 1 mm sodium fluoride. Lysis was performed using a denaturing buffer (20 mm HEPES, pH 8.0, 8 m urea, 1 mm pV, 1 mm sodium fluoride, 2.5 mm sodium pyrophosphate, 1 mm β-glycerolphosphate) at a concentration of 10 × 106 cells/ml. Further protein solubilization was achieved by sonication. Lysate debris was cleared by centrifugation at 20,000 × g for 10 min, and protein concentration of the supernatants was determined by Bradford assay. The samples were then kept frozen at −80 °C until further analysis.
Validation of label-free quantitative phosphoproteomic was carried out in NIH 3T3 cells using an approach for assessing quantification accuracy (38). Briefly, the cells were seeded at ∼35% confluency and cultured for 24 h, when cells reached ∼70% confluency. After preincubation at 37 °C for 30 min with the phosphatase inhibitor pV at a final concentration of 1 mm, the cells were then washed twice and trypsinized off the flask following the harvesting protocol described above. Extracts of pV-treated cells were mixed with decreasing amounts of extracts of nontreated cells (100, 90, 70, 50, 30, 10, and 0%) so that each experimental point contained 0.5 mg of protein to obtain a complex phosphopeptide titration curve to test matrix contribution to the performance of the LC-MS/MS quantitative method.
Sensitivity of AML Cell Lines to Drug Treatment
Eight cell lines (AML-193, CMK, CTS, HEL, Kasumi-1, KG-1, MV4-11, and P31/FUJ) were seeded in 96-well plates at 105 cell/ml in triplicate for each condition. After a recovery period of 2 h, the cells were treated with increasing concentrations (1 nm, 10 nm, 100 nm, 1 μm, and 10 μm) of MEK I inhibitor (Calbiochem), JAK I inhibitor (Calbiochem), PI-103 (Calbiochem), and GF 109203X (Sigma). As controls, the cells were both treated with the vehicle (DMSO) and left untreated. After 48 or 72 h treatment, cell viability was assessed by MTS assay (CellTiter 96® AQueous One Solution cell proliferation assay; Promega Corporation, Madison, WI) and standard curves used to calculate cell numbers. Cell proliferation was calculated as the number of cells after inhibitor treatment minus number of cells seeded. According to the suppliers, the IC50 values of these inhibitors are 12 nm for MEK (MEK I inhibitor), 15 nm for JAK1 (JAK I inhibitor), 2 nm and 5–200 nm for PI3K isoforms and mTORC1 (PI-103), respectively, and 8–20 nm for PKC isoforms (GF 109203X).
Protein Digestion and Solid Phase Extraction
Sample reduction and alkylation were performed with 4.1 mm DTT and 8.3 mm iodoacetamide in the dark and at room temperature for 15 min each. After diluting the samples to 2 m urea with HEPES, pH 8.0, trypsinization was performed using 10 TAME (p-toluene-sulfonyl-L-arginine methyl ester) units of immobilized 1-chloro-3-tosylamido-7-amino-2-heptanone-trypsin/5 × 106 cells for 16 h at 37 °C. Digestion was stopped by adding TFA at a final concentration of 1%. The resultant peptide solutions were desalted using Sep-Pak C18 columns (Waters UK Ltd., Manchester, UK) according to the manufacturer's guidelines. Peptide elution was carried out with 5 ml of 50% ACN, 0.1% TFA.
Immobilized Metal Ion Affinity Chromatography
Phosphopeptide separation was achieved using an adapted immobilized metal ion affinity chromatography enrichment protocol (38). In short, each sample was incubated for 30 min at room temperature with 300 μl of Fe(III)-coated-Sepharose high performance beads used as a 50% slurry in 50% ACN, 0.1% TFA. Unbound peptides were discarded, and beads were washed with 300 μl of 50% ACN, 0.1% TFA twice. The enriched phosphopeptide fraction was eluted with 300 μl of 1.5% ammonia water, pH 11. A second elution using 50 μl of 1.5% ammonia water, pH 11, containing 50% ACN allowed further enrichment. Eluted peptides were acidified by adding 10% formic acid and finally dried in a SpeedVac and stored at −80 °C.
Nanoflow-Liquid Chromatography Tandem Mass Spectrometry (LC-MS/MS)
For phosphoproteomic experiments, dried phosphopeptides enriched samples were dissolved in 10 μl of 0.1% TFA and analyzed in a LC-MS/MS system. The latter consisted of an LTQ-Orbitrap XL mass spectrometer (Thermo Fisher Scientific) connected online to a nanoflow ultrahigh pressure liquid chromatography (nanoAcquity, Waters/Micromass) that delivered a flow rate of 5 μl/min (loading) and 400 nl/min (elution) with an operating back pressure of ∼3,000 psi. Separations were performed in a BEH 100-μm × 100-mm column (Waters/Micromass). The mobile phases were solution A, 0.1% formic acid in LC-MS grade water and solution B, 0.1% FA in LC-MS grade ACN. Gradient runs were from 1% B to 35% B in 100 min followed by a 5-min wash at 85% B and a 7-min equilibration step at 1% B. Full scan survey spectra (m/z 350–1600) were acquired in the LTQ-Orbitrap XL with a resolution of 60000 at m/z 400. A data-dependent analysis in multistage acquisition mode was employed in which the five most abundant multiply charged ions present in the survey spectrum were automatically mass-selected and fragmented by collision-induced dissociation (normalized collision energy 35%). MS scans were followed by five MS/MS scans (m/z 50–2000), which allow the acquisition of at least 15 data points/chromatographic peak. Dynamic exclusion was enabled with the exclusion list restricted to 500 entries, exclusion duration of 40 s, and mass window of 10 ppm.
Data Analysis
LTQ-Orbitrap MS/MS data were smoothed and centroided using Mascot Distiller. The processed files were searched against the mouse sequence library in the international protein index (IPI Mouse v3.49, 165169 sequences) or SwissProt (version 09-2010 containing 20,286 human entries) using the Mascot search engine. Searches were automated with Mascot Daemon (v2.2.2; Matrix Science, London, UK). The parameters included, choosing trypsin as digestion enzyme with one missed cleavage allowed, carbamidomethyl (C) was set as fixed modification, and pyro-glu (N-terminal), oxidation (M), and phospho (STY) were variable modifications. The data sets were searched with a mass tolerance of ± 7 ppm and a fragment mass tolerance of ± 800 mmu. The hits were considered significant when the Mascot expectation value was <0.05. The false positive rate as estimated by searches against a decoy database was less than 2%. Sites of modification are reported when they had delta scores <20. Delta scores were calculated as described (39). Otherwise the site of modification was deemed to be ambiguous; in such cases phosphopeptides are reported as the start-end residues within the protein sequence.
Phosphopeptides identified by Mascot with a statistical significant threshold were placed in a database of peptides quantifiable by LC-MS and quantified by PESCAL. This program, written in-house for the automation of label-free LC-MS data analysis (40), quantifies the intensities of the peptides present in the database across all the samples one wishes to compare. PESCAL uses the m/z, z, and retention time of the selected peptides to construct extracted ion chromatograms (XICs) for the first three isotopes of each ion. This applies restrictions on the molecular mass, retention time, charge, and isotope distributions, which permits confident identification of the LC-MS elution profiles corresponding to the studied phosphopeptides. Windows for XIC construction were 5 ppm and 2.5 min for m/z and retention time, respectively. Chromatograms were aligned using the retention times of peaks common to all samples by the application of an algorithm based on linear regression. The intensity values could then be calculated by determining the peak height of each individual XIC. The resulting quantitative data were parsed into Excel files for further normalized and statistical analysis. Peptide intensities were normalized to the total chromatogram intensity and to the mean intensities across the samples being compared.
LC-Multiple Reaction Monitoring
To validate our label-free quantitative strategy, selected reaction monitoring was performed in a linear ion trap of an LTQ-Orbitrap XL using the same LC conditions as above. Twenty-five of the identified phosphopeptides were monitored in samples containing increasing amounts of extracts of pV-treated cells. Typical chromatographic peak widths were 20–40 s at the base, which ensured collection of >10 points per chromatographic peak. Quantification was performed by adding the peak areas of XICs of the three most intense MS/MS fragment ions with an m/z greater than that of the parent ion. Xcalibur was used to automate these analyzes. The integration of each chromatographic peak was corrected manually to ensure accurate quantification.
Statistical Analysis
The averaged data are expressed as the means ± S.D. (for phosphoproteomics data) or means ± S.E. (for cell viability and proliferation data). Statistical differences in phosphopeptide intensities were assessed by Student's t test and adjusted for multiple testing using the Benjamini Hochberg false discovery rate method. The results were considered statistically significant at the p < 0.05 level after correction for multiple testing. R was used for principal component analysis.
Western Blot
25 μg of protein were mixed with loading buffer (30 mm Tris-HCl, pH 6.5, 1% SDS (w/v), 5% glycerol (v/v), 0.0005% bromphenol blue (w/v)). The membranes were blocked with TBS-Tween 20 supplemented with 5% skimmed milk (w/v), incubated with primary antibody overnight at 4 °C, with secondary antibody for 1 h at room temperature and developed with enhanced chemiluminiscence reagents. Anti-ERK1/2 phosphorylated at Thr-202 and Tyr-204 (Cell Signaling; catalog number 9106), anti-ERK1/2 (Cell Signaling; catalog number 9102), anti-mouse peroxidase (GE Healthcare; catalog number NXA901), and anti-rabbit peroxidase (GE Healthcare; catalog number NA934V) were used at 1:2000, 1:2000, 1:5000, and 1:5000 dilution, respectively. Band intensities were quantified with a GS800 calibrated densitometer (Bio-Rad).
RESULTS
Analytical Strategy
Our aim was to profile and quantify global phosphorylation across several cell lines, either untreated or treated with different inhibitors, and to perform biological replicates for statistical analysis. We therefore developed a label-free MS strategy for quantitative phosphoproteomics because this would allow comparison of an, in principle, unlimited number of samples and replicates. Label-free quantification of phosphorylation by LC-MS using peak intensities (areas or heights) of XICs has been described (41–44). The analytical strategy proposed here is based on this approach and uses peak heights of XICs as the quantitative readout of phosphopeptides detected by LC-MS/MS. This is akin to selected ion monitoring (45) but exploits the resolution of the Orbitrap analyzer and chromatographic reproducibility and alignment to construct XICs with very narrow windows of mass and relative retention times. We have previously reported that >90% of phosphopeptides analyzed using this strategy can be quantified with good precision and with accuracy deviations <50% (38).
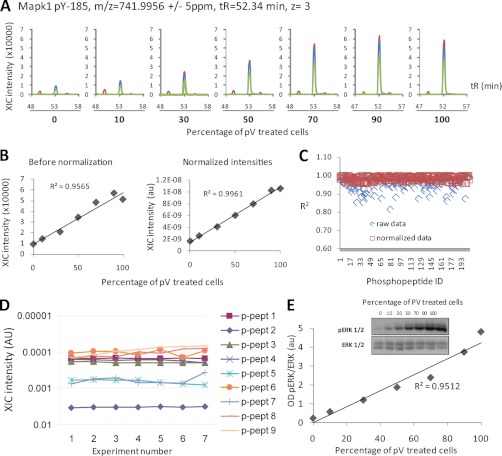
To further assess the accuracy of quantification of our techniques, and the potential of matrix effects contributing to variability, we performed a series of experiments in which cells treated with pervanadate (pV), a protein-tyrosine phosphatase inhibitor, were mixed in different proportions with untreated cells. As expected, pV treatment augmented protein phosphorylation not only on Tyr but also on Ser and Thr, given that although pV is tyrosine phosphatase inhibitor, this compound also increases the phosphorylation on Ser and Thr residues as a knock-on effect (41). This increase in phosphorylation was proportional to the percentage of pV-treated cell extract added to the mixture (Fig. 1A), indicating that the linearity of quantification was not affected by changes in sample composition or matrix effects. An example of this is the quantification of the mouse ERK1 Tyr(P)-185 phosphopeptide that, after normalization using total ion chromatogram intensity, showed linearity of signal as a function of pV-treated cells added to the mixture with R2 > 0.99 (Fig. 1B). In addition to Tyr(P)-185 ERK, the intensities of all 200 phosphopeptides that showed enhanced intensities upon pV treatment also correlated to the amount of pV-treated cells in the sample, with R2 >0.95 after normalization (Fig. 1C; illustrative peptides are shown in supplemental Fig. 1). In addition to operating in a linear dynamic range, the technique was also found to be highly reproducible and comparable with selected reaction monitoring (Fig. 1D and supplemental Fig. 2). There was also a quantitative correlation between label-free LC-MS and Western blot analysis (Fig. 1E), although quantitative LC-MS data, as returned by this approach, showed a better linear correlation and was thus more accurate than quantification by immunoblotting.
Fig. 1.
Accuracy and precision of phosphopeptide quantification by label-free LC-MS/MS. Extracts of pV-treated cells were mixed with extracts of untreated cells at the proportions shown in A, followed by enrichment of phosphopeptides and quantification by LC-MS. A, XICs of a phosphopeptide matching MAPK Tyr(P)-185 in samples containing increasing amounts of pV-treated cells. XICs of the first, second, and third isotopes are shown in blue, red, and green, respectively. B, plot of data, as returned by Pescal, demonstrating the linearity of quantification before and after normalization. C, the linearity of the quantitative data was confirmed by the analysis of 200 additional phosphopeptides induced upon pV treatment. After normalization, R2 > 0.95 for each of the analyzed phosphopeptides was achieved. D, plot of XIC intensities of nine phosphopeptides in seven replicates demonstrating the precision (reproducibility) of the analysis. E, aliquots of lysates from the experiment shown in A–C were analyzed by immunoblotting using an antibody against MAPK Tyr(P)-185 (ERK1/2). The plot shows that phosphosite quantification by immunoblotting is not as linear as quantification by LC-MS (compare E with B). OD, optical density.
Basal Phosphoproteomic Analysis of Sensitive versus Resistant Leukemia Cells
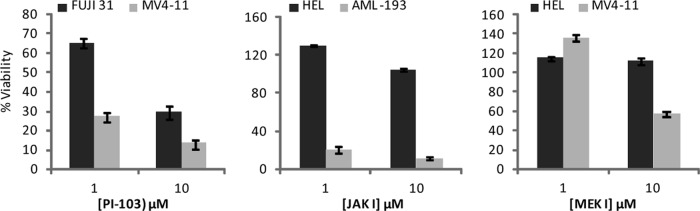
We next treated a panel of AML cell lines with increasing concentrations of inhibitors originally developed to target PI3K (PI-103 (9)), MEK (MEK I; Calbiochem (46)), or JAK (JAK I; Calbiochem (47)), revealing a range of sensitivities for growth inhibition (supplemental Fig. 3). These inhibitors were chosen because the pathways being targeted are known to have a role in AML (11). Cell line pairs with different sensitivity to the inhibitors used were chosen for further analysis (Fig. 2), namely P31/FUJ versus MV4-11 (relatively resistant versus relatively sensitive to PI-103), HEL versus AML-193 (relatively resistant versus relatively sensitive to JAK I), and HEL versus MV4-11 (relatively resistant versus relatively sensitive to MEK I).
Fig. 2.
AML cell lines with different sensitivity to signaling inhibitors. Survival of the selected cell lines used for phosphoproteomic analysis was measured by MTS assay after incubation for 72 h with the indicated doses of inhibitors. The data points shown are the means ± S.E. (n = 3). Viability was expressed relative to cells treated with vehicle only.
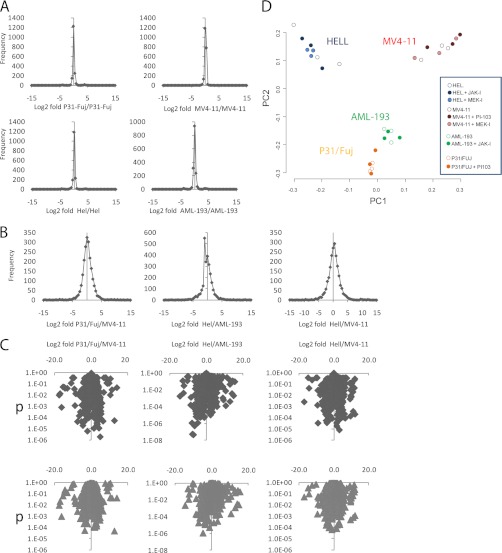
Basal phosphorylation was measured in six replicates (individual cell preparations) to assess the reproducibility of our MS approach and the potential for significant differences to arise by chance during analysis. The means of three arbitrarily chosen replicates from each cell line were compared with the means of the other three. As expected, the frequencies of Log2 fold changes within a cell line were centered on zero (no change), and >95% of phosphopeptide measurements were within −0.5 and 0.5 log2 fold change (Fig. 3A). After correction for multiple testing, only one measurement of ∼16,000 had a p < 0.05 (supplemental Fig. 4), indicating that the contribution of the quantification step to false discovery rates was negligible.
Fig. 3.
Overview of phosphoproteomic data in AML cell lines with different sensitivities to kinase inhibitors. A, phosphorylation was measured by LC-MS in six cell preparations. Graphs show frequency of phosphopeptide fold change of three randomly selected replicates compared with the other three. B and C, frequency distributions (B) and Volcano plots (C) of phosphopeptide fold changes versus p values in cell line pairs resistant/sensitive to PI-103, JAK-I, or MEK-I. D, principal component analysis of AML cell lines based on global phosphorylation data.
We next compared basal phosphorylation between each of the three sensitive/resistant cell line pairs shown in Fig. 2. The Log2 fold differences in basal phosphorylation between cell line pairs also centered around zero (Fig. 3B), but the spread of fold change differences was much greater than when same cell replicates were analyzed (compare frequency distributions in Fig. 3, A and B). To identify differences in basal phosphorylation of cell lines, we chose threshold cutoff values of p < 0.05 (by t test and Benjamini-Hochberg multiple testing correction) and Log2 fold between −1.5 and 1.5 because, as discussed above, these values resulted in no positive identification simply by chance upon comparing replicates (supplemental Fig. 4). These cutoff criteria are the same as described elsewhere (44). As shown in the volcano plots in Fig. 3C, hundreds of phosphopeptides showed significant and robust differences between sensitive and resistant cell lines. The quality of XICs derived from phosphopeptides with significant differences between cell lines was validated manually.
Correlation of Known Pathway Markers with Drug Sensitivity of AML Cells
We also compared global phosphorylation in cell lines before and after treatment with inhibitors. This analysis was performed in triplicate (three individual cell preparations per experimental condition) and quantified ∼2,100 phosphopeptides across these conditions. Principal component analysis revealed that replicates of individual cell lines grouped together and that cell lines could not be separated by principal component analysis based on inhibitor treatment only (Fig. 3D), indicating that there were not obvious outliers in the experiment and that the differences in phosphorylation between cell lines was larger than those caused by the inhibitors.
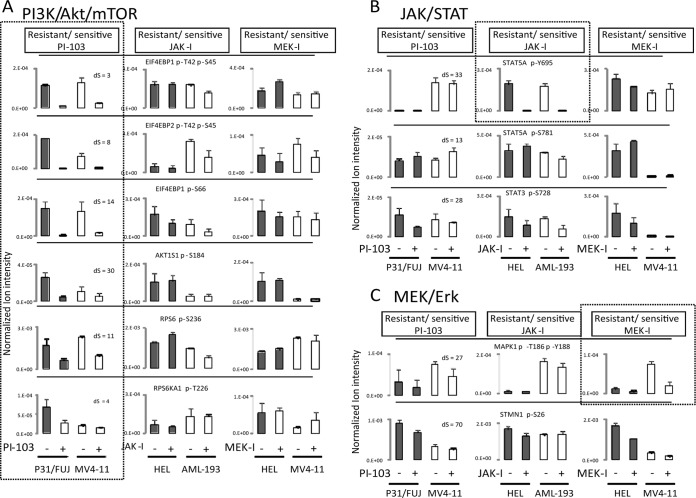
Susceptibility to kinase inhibitors is often rationalized by the theory of oncogene addiction, which predicts that cells with overactive kinases may respond to inhibitors that target such kinases to a greater extent than cells in which the inhibitor targets are less active (34). Therefore, we examined the phosphorylation of sites known to be downstream of kinases targeted by the compounds, i.e., markers of pathway activation. As illustrated in Fig. 4, several sites known to be downstream of PI3K, MEK, and JAK were detected in our analysis and decreased upon treatment of the cells with the respective inhibitors. For example, sites on EIF4BP1 (also known as 4EBP1), AKTS1 (also known as PRAS40), and RPS6 (also known as ribosomal S6), previously demonstrated to be downstream of PI3K (48), were inhibited by PI-103 but not by JAK-I or MEK-I (Fig. 4A). Similarly, phosphorylation on STAT5A Tyr(P)-694, a downstream effector of JAK (49), was specifically affected by JAK inhibition (Fig. 4B). As for MEK I, the MAPK1 (also known as ERK2) sites downstream of MEK were inhibited in MV4-11, although this site was undetectable in HEL (Fig. 4C). Supplemental Fig. 5 shows the chromatograms used to quantify the phosphopeptides in Fig. 4. Overall, these data provide further validation of our approach as a means to quantify signaling downstream of kinase inhibitors.
Fig. 4.
Phosphorylation levels of MEK/MAPK, PI3K/AKT/mTOR, and JAK/STAT pathway components in AML cell lines with different sensitivity to kinase inhibitors. Quantitation of phosphorylation on sites known to be downstream of PI3K/Akt/mTOR (A), JAK/STAT (B), or MEK (C) as a function of cell treatment with PI-103, JAK-I, or MEK-I. Markers of known inhibitor targets are shown in dashed boxes for cells treated with the respective compound. The data are the means ± S.D. (n = 3 biological replicates). Confidence in phosphorylation site location was determined by calculating Delta scores (see “Experimental Procedures”), which are shown in the first column for each phosphopeptide. The chromatograms used to assess expression of these pathway markers for PI3K/mTOR, JAK/STAT, and MEK/ERK were inspected visually and shown in supplemental Fig. 4, A–C for the cell pairs resistant/sensitive to PI-103, JAK-I, and MEK-I, respectively.
There was often no correlation between basal phosphorylation of specific pathway activity markers and drug sensitivity, as measured by growth inhibition, in that cells that were sensitive to drug inhibition did not necessarily have high basal phosphorylation of the pathway marker. Indeed, the basal intensities of phosphorylation on EIF4BP1, AKTS1, and RPS6 did not correlate with biological sensitivity to PI-103 (Fig. 4A). Similarly, the level of basal STAT5A Tyr(P)-694 did not correlate with sensitivity to JAK inhibition (Fig. 4iB). As for MEK I, MAPK1 sites downstream of MEK, did correlate with the sensitivity of MV4-11 to the MEK inhibitor in that the resistant cell line had a low basal phosphorylation on this site (Fig. 4C). Taken together, these data show that pathway inhibitors invariably inhibited the phosphorylation of known pathway activity markers, independent of their basal levels of phosphorylation, both in sensitive and resistant cell lines. However, the basal intensities of these pathway markers did not always correlate with resistance/sensitive to the same compounds.
Identification of New Phosphorylation Site Markers of Signaling Inhibitor Efficacy and Their Correlation to Sensitivity
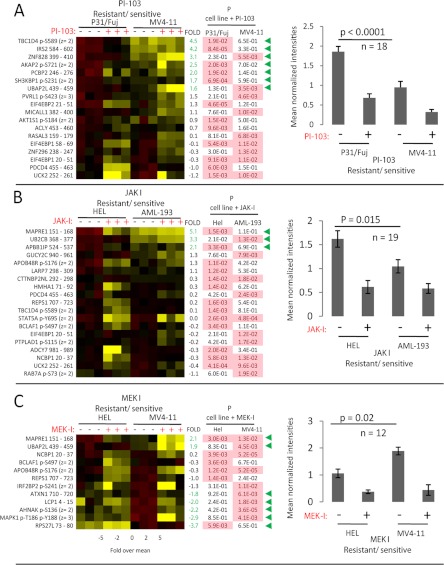
Investigative compounds, such as the ones used in this study, in addition to effects on the intended target, often have off-target effects on other kinases. To investigate whether the activity of these other kinases were the determinants in coffering sensitivity to the inhibitors, we mined our phosphoproteomics data to identify additional phosphorylation sites inhibited by the compounds used in this study; these phosphorylation sites can thus serve as markers of the activity of the kinase(s) that are actually being inhibited by the compounds, in addition to those for which the inhibitors were developed. Unbiased mining of our phosphoproteomics data with the stringent criteria outlined above revealed a total of 18, 19, and 12 phosphopeptides that were significantly inhibited by PI-103, JAK-I, and MEK-I, respectively (Fig. 5 and supplemental Fig. 6, A–C show the chromatograms used to quantify these phosphopeptides). The phosphorylation sites in these peptides could be genuine activity markers of the intended target kinase and/or of off-target kinase(s) inhibited by the compounds. To assess whether the differences pathway markers being affected by the inhibitors were statistically significant, the data were normalized to the mean across observations, so that a mean of the expression of all phosphopeptides in a sample could be obtained (Fig. 5, A–C, right panels).
Fig. 5.
Identification of additional markers of compound activities and their correlation with sensitivity. The intensities of phosphorylation sites observed to be inhibited in resistant and/or sensitive cells as a result of inhibitor treatment were measured and shown for PI-103 (A), JAK-I (B), and MEK-I (C). The left panels show the data visualized in heat maps, with bar charts in the far right panels showing the means of the averaged data across all peptides shown in the heat map. The column headed with “FOLD basal” shows the Log2 fold change difference for each named phosphopeptide between untreated resistant and sensitive cells. The p values (n = 3, Student's t test) derive from the comparison of changes in phosphorylation of each peptide in the named cell line upon treatment with the inhibitor, with significant reductions (p < 0.05) of phosphorylation highlighted in pink. Green arrowheads indicate phosphopeptides that significantly differ in basal phosphorylation between untreated cell lines (determined as in Fig. 3). The chromatograms used to quantitate these pathway markers are shown in supplemental Fig. 5, A–C.
Phosphorylation sites that were inhibited by the compounds include known pathway markers such as EIF4EBP1 Ser(P)-66, STAT5A Tyr(P)-595, and MAPK1 Thr(P)-195 Tyr(P)-199 (also shown in Fig. 4) and additional, novel ones. As illustrated in Fig. 5 (right panels), this analysis showed that phosphorylation of the newly identified activity markers of PI-103 targets were on average more intense in the resistant cell line P31/FUJ compared with the sensitive MV4-11 (Fig. 5A). Similarly, novel activity markers of JAK-I targets were more intense in HEL than in AML-193 (the JAK-I-resistant/sensitive cell pair; Fig. 5B). Illustrative examples of sites not previously associated (to the best of our knowledge) with signaling targeted by these inhibitors include AKAP2 Ser(P)-721 for PI-103 (Fig. 5A), NCBP1 p20/37 for JAK-I (Fig. 5B), and APOB48R Ser(P)-176 for MEK-I (Fig. 5C). When analyzing specific peptides, seven and three sites (labeled in Fig. 5, A and B, with green arrows) downstream of PI-103 and JAK-I, respectively, were more phosphorylated in the untreated basal state in the resistant cells relative to the sensitive ones, with a Log2 fold of >1.5, whereas none of these was more phosphorylated in sensitive cells at this fold change threshold. In contrast, previously uncharacterized markers of kinases targeted by MEK-I were more prominent in MV4-11 than in HEL (sensitive and resistant to MEK-I, respectively; Fig. 5C). Five and two sites were more phosphorylated at basal level in cells sensitive and resistant to MEK-I, respectively (marked with a green arrow in Fig. 5C). The partial overlap in the sites that were regulated by the different compounds in each cell line pair (Fig. 5) might reflect differences in substrate expression and/or kinase activity between cells.
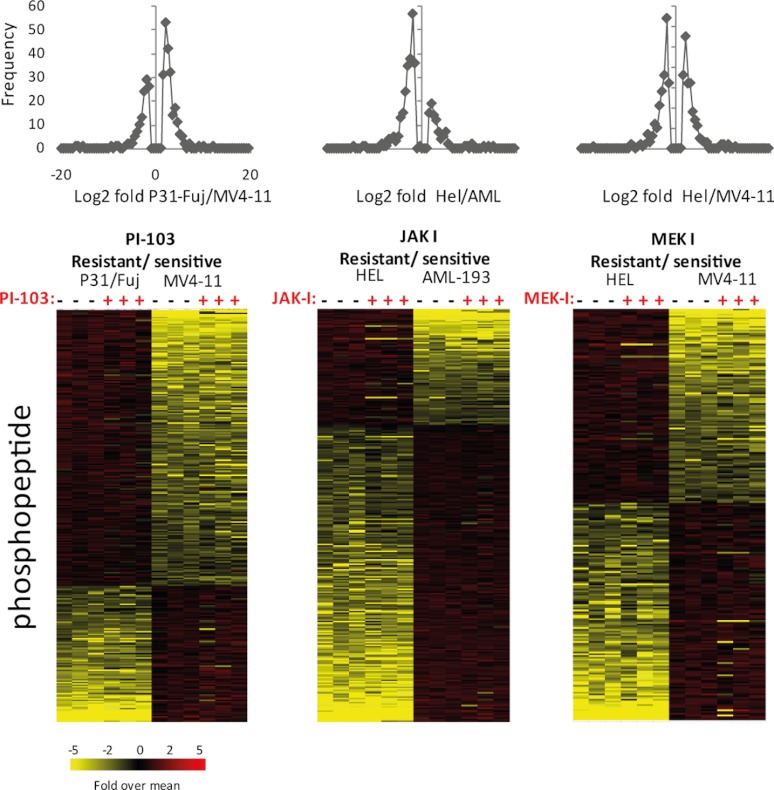
Basal State Phosphorylation Sites That Correlate with Resistance/Sensitivity to Signaling Inhibitors
The activity of pathways parallel to those being targeted can contribute to the resistance of cancer cells to kinase inhibitors (37). We therefore next examined the basal phosphorylation on sites found to be quantitatively different among cell line pairs, regardless of whether or not these were affected by the compounds. Using the stringent criteria of 1.5-fold difference and p < 0.05 (after correction for multiple testing), the intensities of 300, 295, and 235 phosphopeptides ions were found to be significantly different in the untreated PI-103, JAK-I, and MEK-I cell pairs, respectively (Fig. 6). The quality of this data set was further assessed by examining the intensities of phosphopeptides identified with more than one charge showing CV <20% on average (supplemental Fig. 7). We next explored the impact of drug treatment on the phosphorylation of these sites and found that sites that correlated with sensitivity to the compounds were in general not inhibited, indicating that kinase pathways that correlated with sensitivity were not kinases targeted by the inhibitors (Fig. 6). As discussed above, the exception was for sites that correlated with MEK-I sensitivity, whose intensities on MAPK1, AHNAK, Plastin-2, and 40 S ribosomal protein 27 did correlate with sensitivity (Fig. 5C), being >3-fold greater in sensitive than in resistant cells.
Fig. 6.
Phosphopeptides that are robustly and significantly different in cell lines with different sensitivities to signaling inhibitors. Phosphopeptides in exponentially growing, non-drug-treated cells that show Log2 fold changes more than 1.5 or less than −1.5 and p < 0.05 (after multiple testing correction as described in the main text) were selected and represented as frequency of fold change (top panels) or in heat maps clustered by fold change (bottom panels). Quantitative data of three biological replicates are shown separately.
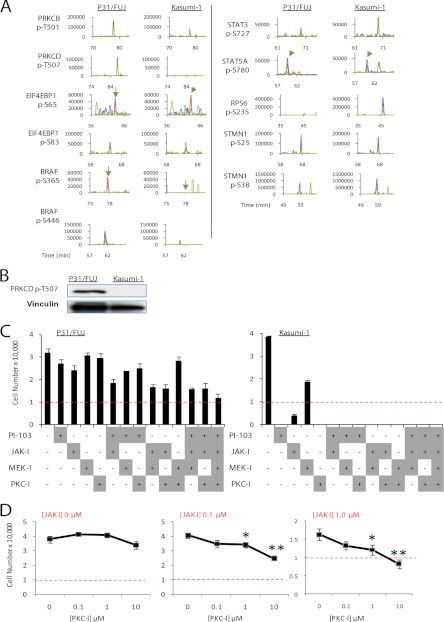
To test whether these results would hold in a different cell line pair, we compared phosphorylation in the P31/FUJ cells relative to the Kasumi-1 cell line, which showed marked differences in sensitivity to several kinase inhibitors (Fig. 7 and Ref. 38). Consistent with the data above, markers of pathway activities, such as phosphorylation sites on 4EBP1 and STAT isoforms, were not increased in the basal state in the sensitive cell line (Fig. 7A), although phosphorylation on RPS6 was more prominent in this line relative to the resistant one. Interestingly, phosphorylation of PKC isoforms on sites known to correlate with their kinase activity (Thr-501 on PRKCB and Thr-507 on PRCKD (50)), were found to be more prominent in the resistant P31/FUJ relative to the sensitive Kasumi-1 by LC-MS (Fig. 7A) and by immunoblotting (Fig. 7B). These results suggest that PKC activities may be elevated in P31/FUJ (relatively resistant to a battery of kinase inhibitors) compared with Kasumi-1 (sensitive to all kinase inhibitors tested).
Fig. 7.
Kinase pathways cooperate to drive the proliferation of multidrug-resistant cells. A, XICs of peptides in P31/FUJ and Kasumi-1 bearing phosphorylation sites markers of pathway activation. Signals for the first, second, and third isotopes are depicted in blue, red, and green, respectively. Vertical arrows indicate the position of the named phosphopeptide in the chromatogram. B, Western blot of Thr(P)-507 PRKCD. C, proliferation of cells exposed to different combinations of kinase inhibitors. Inhibitor treatment was for 48 h at 100 nm (PI-103), 1 μm (JAK-I), 10 μm (MEK-I), or 10 μm (PKC-I). The initial number of cells seeded was 10,000 (indicated with a red dotted line). The data points are the means ± S.E. (n = 5). D, proliferation of P31/FUJ cells exposed to different combinations of JAK-I and PKC-I showing that these inhibitors potentiate each other in inhibiting cell viability. Note that combination of 1 μm JAK-I with 10 μm PKC-I resulted in a lower number of cells compared with input, indicative of cell death. The data points are the means ± S.E. (n = 5). *, p < 0.05; **, p < 0.01.
Anti-proliferative Effects of Combined Treatment with Kinase Inhibitors
Collectively, our data indicated that activity markers of a given pathway were not always indicators of sensitivity/resistance to inhibitors of such pathways, which implies that susceptibility of cancer cells to kinase inhibitors may not only be due to the activity of the pathway being targeted. Because pathways that correlated with sensitivity/resistance were, in most cases, not targets of the inhibitors tested, we hypothesized that the resistant cancer cells used in this study may be proliferating via pathways parallel to those being targeted, so that the combination of active kinase pathways may determine whether these cells respond to the inhibitors.
To test whether the P31/FUJ cell line was resistant to several kinase inhibitors because they activated several kinase pathways “parallel” to those being inhibited by the compounds, we measured the proliferation of cells exposed to different combinations of the kinase inhibitors used. The compounds tested were PI3K, JAK, and MEK, as well as BIM-1, an inhibitor that was originally developed to target PKC isoforms (51). Interestingly, several inhibitor combinations potentiated each other in inhibiting P31/FUJ proliferation, with the combinations of JAK+MEK and JAK+BIM-1 inhibitors being most effective. Indeed, BIM-1, by itself, did not reduce P31/FUJ proliferation (Fig. 7C, left panel), but its combination with JAK-I reduced proliferation in a concentration-dependent manner (Fig. 7D, middle and right panels), supporting the notion that BIM-1 and JAK-I potentiate each other in inhibition of cell proliferation. We also measured inhibition to cell proliferation induced by Go9676, another inhibitor developed to target several PCK isoforms (52). As for the cell proliferation data of BIM-1, Go9676 efficiently reduced the proliferation of the P31/Fuj multidrug-resistant cell line (supplemental Fig. 8), a reduction that was enhanced by a combination of inhibitors. These results are consistent with the view that P31/FUJ proliferate using the combination of kinases inhibited by the compounds. The cell proliferation data are also in line with our quantitative phosphoproteomics data showing increased phosphorylation in activity markers of several proliferative pathways in resistant cancer cells relative to sensitive ones and reinforce the notion that phosphoproteomics may be used to identify and inform which combination of active kinase pathways contributes to the proliferation of cancer cells.
DISCUSSION
A common feature of virtually all cancers is that they activate kinase pathways that allow them to acquire many of the traits of the malignant phenotype. Kinases are therefore suitable targets for the development of anti-cancer drugs. Identifying the patient population that may respond to specific kinase inhibitors is critical for the success of therapies targeting these enzymes (53). There consequently is great interest in characterizing markers of pathway activation (16, 54) with the view that these can serve as markers of drug sensitivity. However, an emerging concept is that cancer cells can activate several components of the kinase network, allowing proliferation to be maintained in the wake of treatment with agents targeting a single kinase (37).
Our data indicate that although the sensitivity of cancer cells to kinase inhibitors could on occasion be predicted by examining the activation of the target, as observed in other systems (22, 34), this may not always be the case. Indeed, in this study we first identified several novel markers of the activities of kinases inhibited by compounds developed to target MEK and PI3K/mTOR and JAK (Fig. 5, note that these activity markers are readouts of the actual kinase activities that are being inhibited irrespective of whether or not these are the intended target). We then asked whether these activity markers would correlate with the sensitivity of AML cells to such compounds and could thus be used to predict susceptibility to treatment with these inhibitors; in other words, whether markers of kinase activity would also be markers of sensitivity to the compounds. We found that although markers of kinase activities being targeted by MEK-I were on average more intense in the cell line sensitive to this inhibitor (Fig. 5C), activity markers of kinases inhibited by PI-103 and JAK-I were more intense in resistant cells (Figs. 4 and 5, A and B). Therefore, pathway activation did not seem to be the main determinant in making cells susceptible to inhibitors of such pathways in the cell models used in this study.
By mining our data we found that the signals of phosphorylated PKCs were more intense in resistant than in sensitive cells (Fig. 7). PKCs have been implicated in the biology of AML (55) and can contribute to the resistance to chemotherapeutic drugs by interfering with the cell response triggered by these drugs or by increasing their clearance as a result of activating P-glycoprotein (56, 57). Here we provide evidence that PKCs may also be implicated in AML resistance to different kinase inhibitors for the first time. The compounds tested inhibited their target to the same extent in resistant and sensitive cells (Figs. 4 and 5), thus indicating that differences in resistance were not due to differences in how these cells metabolize the inhibitors. The main determinant in conferring resistance to the inhibitors tested in this study appeared to be the activation of pathways parallel to those being targeted by the compounds. Thus, resistant cells seem to have wired the kinase network differently than sensitive cells. This is reflected by both the markedly different patterns of phosphorylation between resistant and sensitive cell lines (Figs. 3 and 6) and also by the fact that the proliferation of sensitive cell lines, such as Kasumi-1, was inhibited by most kinase inhibitors tested, whereas resistant cells (P31/FUJ) were only inhibited by a combination of them (Fig. 7). Indeed, kinase inhibitors having PKC, MEK, and JAK kinases as intended targets potentiated each other in inhibiting the viability of P31/FUJ cells. Although the kinase inhibitors may inhibit other kinases, in addition to those for which they were originally developed, the pharmacological data shown in Fig. 7 and supplemental Fig. 8 demonstrate that cells resistant to the inhibition by single compounds use several different (and seemingly redundant) routes to proliferate.
These conclusions imply that knowledge of the activation status of the pathway being targeted may not always be sufficient to predict sensitivity to an inhibitor of such pathway. Our data is consistent with (i) the emerging notion that cancer cells are not always addicted to single oncogenes (58), (ii) recent studies showing that markers of PI3K and MEK pathway activities are poor predictors of response to PI3K and MEK inhibitors, respectively (9, 59), (iii) the notion that kinase pathways can cooperate to drive cancer cells proliferation in some systems (60), and iv) the view that a combination of inhibitors may be more effective in arresting cancer cell growth than agents that target single kinases (37). Techniques for systematic and deep quantification of signaling in cancer cells, such as the approach reported here, may be used in the future to profile deregulated kinase networks in the tumor of each patient, allowing the oncologist to choose the best kinase therapy based on the activation and circuitry of the different kinase pathways within the network.
Here we have used MS to identify activity markers of kinases inhibited by compounds having PI3K, MEK, and JAK as intended targets and to profile kinase signaling in cancer cells of different sensitivity to these inhibitors. The advantage of MS is that these analyses were done without a preconception of the kinases that may be inhibited by a particular compound or the pathways that may be involved. The MS-based quantitative approach used in this study was found to provide accurate and reproducible data (Figs. 1 and 2). Analysis of replicates showed that the potential for random identification was minimal (Fig. 2 and supplemental Fig. 3) and analysis of peptides with different charges confirmed that the quantification data was reproducible and accurate (supplemental Fig. 7). The observation that kinase inhibitors reduced phosphorylation of known compound targets (Fig. 4) provided further confidence of the data. Our findings that several kinase pathways cooperate in the resistance of leukemia cell lines to kinase inhibitors and the potential implication of PKC isoforms in this process exemplifies the power of the approach described here for cell signaling studies in cancer cells.
Acknowledgments
We thank members of the Centre for Cell Signaling, Mike Waterfield and Subham Basu for feedback on the manuscript. We thank Peter Parker for advice on PKC inhibitors and signaling.
Footnotes
* This work was supported by Biotechnology and Biological Sciences Research Council Grant BB/G015023/1, funds from Atlantic Philanthropies, Barts and the London Charity Grant 297/298 and 297/997, and Medical Research Council Grant G0800914.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- PI3K
- phosphatidylinositol 3-kinase
- AML
- acute myeloid leukemia
- JAK
- Janus kinase
- ERK
- extracellular signal-regulated kinase
- MAPK
- mitogen-activated protein kinase
- MEK
- MAPK/ERK kinase
- PKC
- protein kinase C
- mTOR
- mammalian target of rapamycin
- STAT
- signal transducers and activators of transcription
- XIC
- extracted ion chromatogram
- pV
- pervanadate.
REFERENCES
- 1. Manning B. D. (2009) Challenges and opportunities in defining the essential cancer kinome. Sci. Signal. 2, pe15. [DOI] [PubMed] [Google Scholar]
- 2. Hanahan D., Weinberg R. A. (2011) Hallmarks of cancer: The next generation. Cell 144, 646–674 [DOI] [PubMed] [Google Scholar]
- 3. Baselga J. (2006) Targeting tyrosine kinases in cancer: The second wave. Science 312, 1175–1178 [DOI] [PubMed] [Google Scholar]
- 4. Baselga J., Albanell J. (2001) Mechanism of action of anti-HER2 monoclonal antibodies. Ann. Oncol. 12, (Suppl. 1) S35–S41 [DOI] [PubMed] [Google Scholar]
- 5. Benekli M., Baumann H., Wetzler M. (2009) Targeting signal transducer and activator of transcription signaling pathway in leukemias. J. Clin. Oncol. 27, 4422–4432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Di Cosimo S., Baselga J. (2008) Targeted therapies in breast cancer: Where are we now? Eur. J. Cancer 44, 2781–2790 [DOI] [PubMed] [Google Scholar]
- 7. Druker B. J. (2002) STI571 (Gleevec) as a paradigm for cancer therapy. Trends Mol. Med. 8, S14–S18 [DOI] [PubMed] [Google Scholar]
- 8. Liu P., Cheng H., Roberts T. M., Zhao J. J. (2009) Targeting the phosphoinositide 3-kinase pathway in cancer. Nat. Rev. Drug Discov. 8, 627–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Raynaud F. I., Eccles S., Clarke P. A., Hayes A., Nutley B., Alix S., Henley A., Di-Stefano F., Ahmad Z., Guillard S., Bjerke L. M., Kelland L., Valenti M., Patterson L., Gowan S., de Haven Brandon A., Hayakawa M., Kaizawa H., Koizumi T., Ohishi T., Patel S., Saghir N., Parker P., Waterfield M., Workman P. (2007) Pharmacologic characterization of a potent inhibitor of class I phosphatidylinositide 3-kinases. Cancer Res. 67, 5840–5850 [DOI] [PubMed] [Google Scholar]
- 10. Rinehart J., Adjei A. A., Lorusso P. M., Waterhouse D., Hecht J. R., Natale R. B., Hamid O., Varterasian M., Asbury P., Kaldjian E. P., Gulyas S., Mitchell D. Y., Herrera R., Sebolt-Leopold J. S., Meyer M. B. (2004) Multicenter phase II study of the oral MEK inhibitor, CI-1040, in patients with advanced non-small-cell lung, breast, colon, and pancreatic cancer. J. Clin. Oncol. 22, 4456–4462 [DOI] [PubMed] [Google Scholar]
- 11. Scholl C., Gilliland D. G., Fröhling S. (2008) Deregulation of signaling pathways in acute myeloid leukemia. Semin. Oncol. 35, 336–345 [DOI] [PubMed] [Google Scholar]
- 12. Serra V., Markman B., Scaltriti M., Eichhorn P. J., Valero V., Guzman M., Botero M. L., Llonch E., Atzori F., Di Cosimo S., Maira M., Garcia-Echeverria C., Parra J. L., Arribas J., Baselga J. (2008) NVP-BEZ235, a dual PI3K/mTOR inhibitor, prevents PI3K signaling and inhibits the growth of cancer cells with activating PI3K mutations. Cancer Res. 68, 8022–8030 [DOI] [PubMed] [Google Scholar]
- 13. Schlabach M. R., Luo J., Solimini N. L., Hu G., Xu Q., Li M. Z., Zhao Z., Smogorzewska A., Sowa M. E., Ang X. L., Westbrook T. F., Liang A. C., Chang K., Hackett J. A., Harper J. W., Hannon G. J., Elledge S. J. (2008) Cancer proliferation gene discovery through functional genomics. Science 319, 620–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Grueneberg D. A., Degot S., Pearlberg J., Li W., Davies J. E., Baldwin A., Endege W., Doench J., Sawyer J., Hu Y., Boyce F., Xian J., Munger K., Harlow E. (2008) Kinase requirements in human cells: I. Comparing kinase requirements across various cell types. Proc. Natl. Acad. Sci. U.S.A. 105, 16472–16477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Grueneberg D. A., Li W., Davies J. E., Sawyer J., Pearlberg J., Harlow E. (2008) Kinase requirements in human cells: IV. Differential kinase requirements in cervical and renal human tumor cell lines. Proc. Natl. Acad. Sci. U.S.A. 105, 16490–16495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sawyers C. L. (2008) The cancer biomarker problem. Nature 452, 548–552 [DOI] [PubMed] [Google Scholar]
- 17. Wood L. D., Parsons D. W., Jones S., Lin J., Sjöblom T., Leary R. J., Shen D., Boca S. M., Barber T., Ptak J., Silliman N., Szabo S., Dezso Z., Ustyanksky V., Nikolskaya T., Nikolsky Y., Karchin R., Wilson P. A., Kaminker J. S., Zhang Z., Croshaw R., Willis J., Dawson D., Shipitsin M., Willson J. K., Sukumar S., Polyak K., Park B. H., Pethiyagoda C. L., Pant P. V., Ballinger D. G., Sparks A. B., Hartigan J., Smith D. R., Suh E., Papadopoulos N., Buckhaults P., Markowitz S. D., Parmigiani G., Kinzler K. W., Velculescu V. E., Vogelstein B. (2007) The genomic landscapes of human breast and colorectal cancers. Science 318, 1108–1113 [DOI] [PubMed] [Google Scholar]
- 18. Jones S., Zhang X., Parsons D. W., Lin J. C., Leary R. J., Angenendt P., Mankoo P., Carter H., Kamiyama H., Jimeno A., Hong S. M., Fu B., Lin M. T., Calhoun E. S., Kamiyama M., Walter K., Nikolskaya T., Nikolsky Y., Hartigan J., Smith D. R., Hidalgo M., Leach S. D., Klein A. P., Jaffee E. M., Goggins M., Maitra A., Iacobuzio-Donahue C., Eshleman J. R., Kern S. E., Hruban R. H., Karchin R., Papadopoulos N., Parmigiani G., Vogelstein B., Velculescu V. E., Kinzler K. W. (2008) Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 321, 1801–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Forbes S. A., Tang G., Bindal N., Bamford S., Dawson E., Cole C., Kok C. Y., Jia M., Ewing R., Menzies A., Teague J. W., Stratton M. R., Futreal P. A. (2010) COSMIC (the Catalogue of Somatic Mutations in Cancer): A resource to investigate acquired mutations in human cancer. Nucleic Acids Res. 38, D652–D657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Greenman C., Stephens P., Smith R., Dalgliesh G. L., Hunter C., Bignell G., Davies H., Teague J., Butler A., Stevens C., Edkins S., O'Meara S., Vastrik I., Schmidt E. E., Avis T., Barthorpe S., Bhamra G., Buck G., Choudhury B., Clements J., Cole J., Dicks E., Forbes S., Gray K., Halliday K., Harrison R., Hills K., Hinton J., Jenkinson A., Jones D., Menzies A., Mironenko T., Perry J., Raine K., Richardson D., Shepherd R., Small A., Tofts C., Varian J., Webb T., West S., Widaa S., Yates A., Cahill D. P., Louis D. N., Goldstraw P., Nicholson A. G., Brasseur F., Looijenga L., Weber B. L., Chiew Y. E., DeFazio A., Greaves M. F., Green A. R., Campbell P., Birney E., Easton D. F., Chenevix-Trench G., Tan M. H., Khoo S. K., Teh B. T., Yuen S. T., Leung S. Y., Wooster R., Futreal P. A., Stratton M. R. (2007) Patterns of somatic mutation in human cancer genomes. Nature 446, 153–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Haber D. A., Settleman J. (2007) Cancer: Drivers and passengers. Nature 446, 145–146 [DOI] [PubMed] [Google Scholar]
- 22. Weinstein I. B. (2002) Cancer. Addiction to oncogenes: The Achilles heal of cancer. Science 297, 63–64 [DOI] [PubMed] [Google Scholar]
- 23. Cutillas P. R., Khwaja A., Graupera M., Pearce W., Gharbi S., Waterfield M., Vanhaesebroeck B. (2006) Ultrasensitive and absolute quantification of the phosphoinositide 3-kinase/Akt signal transduction pathway by mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 103, 8959–8964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cutilllas P. R. (2006) Activitomics: Mulitiplex analysis of kinase activities by mass spectrometry, in Proceedings of the 54th American Society for Mass Spectrometry Conference of Mass Spectrometry and Allied Topics, Seattle, WA, May 28–June 1, 2006 [Google Scholar]
- 25. Kubota K., Anjum R., Yu Y., Kunz R. C., Andersen J. N., Kraus M., Keilhack H., Nagashima K., Krauss S., Paweletz C., Hendrickson R. C., Feldman A. S., Wu C. L., Rush J., Villén J., Gygi S. P. (2009) Sensitive multiplexed analysis of kinase activities and activity-based kinase identification. Nat. Biotechnol. 27, 933–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Thingholm T. E., Jensen O. N., Larsen M. R. (2009) Analytical strategies for phosphoproteomics. Proteomics 9, 1451–1468 [DOI] [PubMed] [Google Scholar]
- 27. Huang P. H., Mukasa A., Bonavia R., Flynn R. A., Brewer Z. E., Cavenee W. K., Furnari F. B., White F. M. (2007) Quantitative analysis of EGFRvIII cellular signaling networks reveals a combinatorial therapeutic strategy for glioblastoma. Proc. Natl. Acad. Sci. U.S.A. 104, 12867–12872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., Mann M. (2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648 [DOI] [PubMed] [Google Scholar]
- 29. Wolf-Yadlin A., Hautaniemi S., Lauffenburger D. A., White F. M. (2007) Multiple reaction monitoring for robust quantitative proteomic analysis of cellular signaling networks. Proc. Natl. Acad. Sci. U.S.A. 104, 5860–5865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ballif B. A., Roux P. P., Gerber S. A., MacKeigan J. P., Blenis J., Gygi S. P. (2005) Quantitative phosphorylation profiling of the ERK/p90 ribosomal S6 kinase-signaling cassette and its targets, the tuberous sclerosis tumor suppressors. Proc. Natl. Acad. Sci. U.S.A. 102, 667–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Carretero J., Shimamura T., Rikova K., Jackson A. L., Wilkerson M. D., Borgman C. L., Buttarazzi M. S., Sanofsky B. A., McNamara K. L., Brandstetter K. A., Walton Z. E., Gu T. L., Silva J. C., Crosby K., Shapiro G. I., Maira S. M., Ji H., Castrillon D. H., Kim C. F., García-Echeverría C., Bardeesy N., Sharpless N. E., Hayes N. D., Kim W. Y., Engelman J. A., Wong K. K. (2010) Integrative genomic and proteomic analyses identify targets for Lkb1-deficient metastatic lung tumors. Cancer Cell 17, 547–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hoffert J. D., Pisitkun T., Knepper M. A. (2011) Phosphoproteomics of vasopressin signaling in the kidney. Expert Rev. Proteomics 8, 157–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rikova K., Guo A., Zeng Q., Possemato A., Yu J., Haack H., Nardone J., Lee K., Reeves C., Li Y., Hu Y., Tan Z., Stokes M., Sullivan L., Mitchell J., Wetzel R., Macneill J., Ren J. M., Yuan J., Bakalarski C. E., Villen J., Kornhauser J. M., Smith B., Li D., Zhou X., Gygi S. P., Gu T. L., Polakiewicz R. D., Rush J., Comb M. J. (2007) Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 131, 1190–1203 [DOI] [PubMed] [Google Scholar]
- 34. Sawyers C. L. (2009) Shifting paradigms: The seeds of oncogene addiction. Nat. Med. 15, 1158–1161 [DOI] [PubMed] [Google Scholar]
- 35. Liu P., Cheng H., Santiago S., Raeder M., Zhang F., Isabella A., Yang J., Semaan D. J., Chen C., Fox E. A., Gray N. S., Monahan J., Schlegel R., Beroukhim R., Mills G. B., Zhao J. J. (2011) Oncogenic PIK3CA-driven mammary tumors frequently recur via PI3K pathway-dependent and PI3K pathway-independent mechanisms. Nat. Med. 17, 1116–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Muellner M. K., Uras I. Z., Gapp B. V., Kerzendorfer C., Smida M., Lechtermann H., Craig-Mueller N., Colinge J., Duernberger G., Nijman S. M. (2011) A chemical-genetic screen reveals a mechanism of resistance to PI3K inhibitors in cancer. Nat. Chem. Biol. 7, 787–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Knight Z. A., Lin H., Shokat K. M. (2010) Targeting the cancer kinome through polypharmacology. Nat. Rev. Cancer 10, 130–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Casado P., Cutillas P. R. (2011) A self-validating quantitative mass spectrometry method for assessing the accuracy of high-content phosphoproteomic experiments. Mol. Cell. Proteomics 10.1074/mcp.M110.003079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Savitski M. M., Lemeer S., Boesche M., Lang M., Mathieson T., Bantscheff M., Kuster B. (2011) Confident phosphorylation site localization using the Mascot Delta Score. Mol. Cell. Proteomics 10.1074/mcp.M110.003830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cutillas P. R., Vanhaesebroeck B. (2007) Quantitative profile of five murine core proteomes using label-free functional proteomics. Mol. Cell. Proteomics 6, 1560–1573 [DOI] [PubMed] [Google Scholar]
- 41. Cutillas P. R., Geering B., Waterfield M. D., Vanhaesebroeck B. (2005) Quantification of gel-separated proteins and their phosphorylation sites by LC-MS using unlabeled internal standards: Analysis of phosphoprotein dynamics in a B cell lymphoma cell line. Mol. Cell. Proteomics 4, 1038–1051 [DOI] [PubMed] [Google Scholar]
- 42. Nguyen V., Cao L., Lin J. T., Hung N., Ritz A., Yu K., Jianu R., Ulin S. P., Raphael B. J., Laidlaw D. H., Brossay L., Salomon A. R. (2009) A new approach for quantitative phosphoproteomic dissection of signaling pathways applied to T cell receptor activation. Mol. Cell. Proteomics 8, 2418–2431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Stulemeijer I. J., Joosten M. H., Jensen O. N. (2009) Quantitative phosphoproteomics of tomato mounting a hypersensitive response reveals a swift suppression of photosynthetic activity and a differential role for hsp90 isoforms. J. Proteome Res. 8, 1168–1182 [DOI] [PubMed] [Google Scholar]
- 44. Bodenmiller B., Wanka S., Kraft C., Urban J., Campbell D., Pedrioli P. G., Gerrits B., Picotti P., Lam H., Vitek O., Brusniak M. Y., Roschitzki B., Zhang C., Shokat K. M., Schlapbach R., Colman-Lerner A., Nolan G. P., Nesvizhskii A. I., Peter M., Loewith R., von Mering C., Aebersold R. (2010) Phosphoproteomic analysis reveals interconnected system-wide responses to perturbations of kinases and phosphatases in yeast. Sci. Signal. 3, rs4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fierens C., Stöckl D., Thienpont L. M., De Leenheer A. P. (2001) Strategies for determination of insulin with tandem electrospray mass spectrometry: Implications for other analyte proteins? Rapid Commun. Mass Spectrom. 15, 1433–1441 [DOI] [PubMed] [Google Scholar]
- 46. Wityak J., Hobbs F. W., Gardner D. S., Santella J. B., 3rd, Petraitis J. J., Sun J. H., Favata M. F., Daulerio A. J., Horiuchi K. Y., Copeland R. A., Scherle P. A., Jaffe B. D., Trzaskos J. M., Magolda R. L., Trainor G. L., Duncia J. V. (2004) Beyond U0126: Dianion chemistry leading to the rapid synthesis of a series of potent MEK inhibitors. Bioorg. Med. Chem. Lett. 14, 1483–1486 [DOI] [PubMed] [Google Scholar]
- 47. Pedranzini L., Dechow T., Berishaj M., Comenzo R., Zhou P., Azare J., Bornmann W., Bromberg J. (2006) Pyridone 6, a pan-Janus-activated kinase inhibitor, induces growth inhibition of multiple myeloma cells. Cancer Res. 66, 9714–9721 [DOI] [PubMed] [Google Scholar]
- 48. Nojima H., Tokunaga C., Eguchi S., Oshiro N., Hidayat S., Yoshino K., Hara K., Tanaka N., Avruch J., Yonezawa K. (2003) The mammalian target of rapamycin (mTOR) partner, raptor, binds the mTOR substrates p70 S6 kinase and 4E-BP1 through their TOR signaling (TOS) motif. J. Biol. Chem. 278, 15461–15464 [DOI] [PubMed] [Google Scholar]
- 49. Buitenhuis M., Coffer P. J., Koenderman L. (2004) Signal transducer and activator of transcription 5 (STAT5). Int. J. Biochem. Cell Biol. 36, 2120–2124 [DOI] [PubMed] [Google Scholar]
- 50. Liu Y., Belkina N. V., Graham C., Shaw S. (2006) Independence of protein kinase C-delta activity from activation loop phosphorylation: Structural basis and altered functions in cells. J. Biol. Chem. 281, 12102–12111 [DOI] [PubMed] [Google Scholar]
- 51. Toullec D., Pianetti P., Coste H., Bellevergue P., Grand-Perret T., Ajakane M., Baudet V., Boissin P., Boursier E., Loriolle F., et al. (1991) The bisindolylmaleimide GF 109203X is a potent and selective inhibitor of protein kinase C. J. Biol. Chem. 266, 15771–15781 [PubMed] [Google Scholar]
- 52. Martiny-Baron G., Kazanietz M. G., Mischak H., Blumberg P. M., Kochs G., Hug H., Marmé D., Schächtele C. (1993) Selective inhibition of protein kinase C isozymes by the indolocarbazole Go 6976. J. Biol. Chem. 268, 9194–9197 [PubMed] [Google Scholar]
- 53. Duffy M. J., O'Donovan N., Crown J. (2011) Use of molecular markers for predicting therapy response in cancer patients. Cancer Treat. Rev. 37, 151–159 [DOI] [PubMed] [Google Scholar]
- 54. Pajic M., Scarlett C. J., Chang D. K., Sutherland R. L., Biankin A. V. (2011) Preclinical strategies to define predictive biomarkers for therapeutically relevant cancer subtypes. Hum. Genet. 130, 93–101 [DOI] [PubMed] [Google Scholar]
- 55. Ruvolo P. P., Zhou L., Watt J. C., Ruvolo V. R., Burks J. K., Jiffar T., Kornblau S., Konopleva M., Andreeff M. (2011) Targeting PKC-mediated signal transduction pathways using enzastaurin to promote apoptosis in acute myeloid leukemia-derived cell lines and blast cells. J. Cell. Biochem. 112, 1696–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hu M., Liu Y., Deng C., Han R., Jia Y., Liu S., Jiang Z., Cao X., He L., Zhang Q. (2011) Enhanced invasiveness in multidrug resistant leukemic cells is associated with overexpression of P-glycoprotein and cellular inhibitor of apoptosis protein. Leuk. Lymphoma 52, 1302–1311 [DOI] [PubMed] [Google Scholar]
- 57. Rumsby M. G., Drew L., Warr J. R. (1998) Protein kinases and multidrug resistance. Cytotechnology 27, 203–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Luo J., Solimini N. L., Elledge S. J. (2009) Principles of cancer therapy: Oncogene and non-oncogene addiction. Cell 136, 823–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Dry J. R., Pavey S., Pratilas C. A., Harbron C., Runswick S., Hodgson D., Chresta C., McCormack R., Byrne N., Cockerill M., Graham A., Beran G., Cassidy A., Haggerty C., Brown H., Ellison G., Dering J., Taylor B. S., Stark M., Bonazzi V., Ravishankar S., Packer L., Xing F., Solit D. B., Finn R. S., Rosen N., Hayward N. K., French T., Smith P. D. (2010) Transcriptional pathway signatures predict MEK addiction and response to selumetinib (AZD6244). Cancer Res. 70, 2264–2273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Morgan-Lappe S., Woods K. W., Li Q., Anderson M. G., Schurdak M. E., Luo Y., Giranda V. L., Fesik S. W., Leverson J. D. (2006) RNAi-based screening of the human kinome identifies Akt-cooperating kinases: A new approach to designing efficacious multitargeted kinase inhibitors. Oncogene 25, 1340–1348 [DOI] [PubMed] [Google Scholar]