Abstract
Determining the localization, binding partners, and secondary modifications of individual proteins is crucial for understanding protein function. Several tags have been constructed for protein localization or purification under either native or denaturing conditions, but few tags permit all three simultaneously. Here, we describe a multifunctional tandem affinity purification (MAP) method that is both highly efficient and enables protein visualization. The MAP tag utilizes affinity tags inserted into an exposed surface loop of mVenus offering two advantages: (1) mVenus fluorescence can be used for protein localization or FACS-based selection of cell lines; and (2) spatial separation of the affinity tags from the protein results in high recovery and reduced variability between proteins. MAP purification was highly efficient in multiple organisms for all proteins tested. As a test case, MAP combined with liquid chromatography-tandem MS identified known and new candidate binding partners and modifications of the kinase Plk1. Thus the MAP tag is a new powerful tool for determining protein modification, localization, and interactions.
Many biological processes are orchestrated by imposing spatial and temporal regulation on interactions between proteins. Therefore, identification of post-translational modifications and interaction networks among proteins is an essential aspect of modern biology. The tandem affinity purification (TAP)1 method, originally developed for isolation of native protein complexes in yeast (1), allowed for diverse proteins to be easily purified using a single purification scheme. The TAP method works through expression of a fusion protein consisting of the TAP tag and the bait protein. The TAP tag contains two domains and motifs for tandem affinity purification steps; the rationale being that the use of the second step removes common contaminants that might be specific for the first step. Isolated protein complexes can then be used for biochemical experiments or analyzed by liquid-chromatography-tandem mass-spectrometry (LC-MS/MS).
The TAP method has been transferred to mammalian cells; however, the overall recovery of native complexes is low (2). Several modified TAP methods have been developed for mammalian cells, using various combinations of affinity tags. Some tags work well for native protein complexes (2–4), and others for purification under denaturing conditions (5). Other TAP systems incorporate fluorescent proteins to allow for protein localization and to facilitate FACS of individual cells expressing various levels of the tagged protein (6, 7). Each of these purification tags has strengths and weaknesses. Here we describe a fluorescent protein-based multifunctional affinity purification (MAP) tag that incorporates the advantages of multiple purification systems into a single tag. We show that the MAP tag allows protein visualization, rapid selection of cell lines, and efficient affinity purification for all proteins tested under either native or denaturing conditions in mammalian cells, Caenorhabditis elegans, and the fission yeast, Schizosaccharomyces pombe.
EXPERIMENTAL PROCEDURES
Plasmids
Fluorescent proteins EGFP, sfGFP, mVenus, mCherry, and mKate2 were modified by insertion of the His8-SBP-FLAG tandem tag and then inserted as an NotI/XbaI fragment into pcDNA4-TO-Hygromycin vector, which had the Zeocin selection marker in pcDNA4/TO (Invitrogen) replaced by hygromycin, (Kindly provided by Eric Campeau in University of Massachusetts Medical School) to generate the pMAP constructs. DNA fragments encoding human Cdc14B (CC14B), Cdc14A (CC14A), Plk1, Survivin (BIRC5), and Aurora B (AURKB) were inserted into the pMAP-mVenus vector to generate constructs for expression of MAP-tagged fusion proteins.
Establishment of Mammalian Stable Cell Lines and Cell Synchronization
U2OS (human osteosarcoma cells) were cultured at 37 °C in d-MEM supplemented with 10% fetal bovine serum. Stable cell lines were generated by transfection of U2OS cells at 40–60% confluence with pMAP plasmids using Lipofectamine 2000, followed by selection with 300 μg/ml hygromycin B. Inducible cell lines were transfected with pMAP and pcDNA6/TR plasmids then grown in 300 μg/ml hygromycin B and 5 μg/ml Blastcidin S. After 2 weeks in the selection medium, clones of cells expressing appropriate levels of fluorescent proteins were isolated by FACS sorting. After another 2 weeks, each clone was subcultured and examined for fluorescence. U2OS cells were synchronized by treatment with 2 mm Thymidine for 20 h (S-phase arrest) or Nocodazole (50 ng/ml) for 14–16 h (M-phase arrest). Cell synchrony was assayed using propidium iodide staining for DNA content analysis. Asynchronous, S-phase, and mitotic cells were collected and washed in phosphate-buffered saline (PBS). Then the cell concentration was adjusted to 2 × 106 cells per 100 μl in PBS. While vortexing gently, 900 μl of 95% ethanol was added dropwise to the cells. Cells were fixed at 4 °C for 24 h, then washed once with PBS and resuspended in a small amount of PBS. 1 ml of staining solution (900 μl 1x PBS, 2 mm MgCl2, 50 μl 1 mg/ml PI solution (Sigma) and 50 μl 1 mg/ml RNase solution (Sigma)) was added to each sample. After incubation in the dark at 37 °C for 20 min, the cells were stored on ice and analyzed within a few hours by FACS.
Generating Fusion Constructs and Single-copy Transgenes in C. elegans
MAP-tagged condensin subunits were constructed using the Multisite Gateway Three-fragment Construction Kit (Invitrogen, Carlsbad, CA). For each gene, its promoter (1 kb upstream of start) and 3′UTR (0.5 kb downstream of stop) were PCR amplified with primers containing appropriate Gateway att sites and recombined into pDONRP4-P1R and pDONRP2R-P3, respectively. To create N-terminal fusion protein ORFs, the MAP tag (minus stop codon) was PCR amplified with a primer containing a 24 nt overlap with the 5′ end of the gene to be tagged, and the genomic locus was amplified with a 24 nt overlap to the end of the MAP tag. These products were annealed and PCR extended with the overlap serving as primers, the full length product purified (Qiagen, Valencia, CA), then used in a final PCR amplification with outside primers containing Gateway sites and recombined into pDONR221. The three resulting entry vectors were recombined into the MosSCI destination vector, pCFJ150 (8) (Addgene, Cambridge, MA).
The direct insertion MosSCI technique was used to integrate a single copy of the fusion transgene construct into the ttTi6505 site on C. elegans chromosome II (8). Transgenes were outcrossed to wild-type, then crossed into null mutants to generate the following worm strains: KIR3 (stnSi1 [kle-2 promoter: mVenus-MAP::kle-2(+): kle-2 3′UTR cb-unc-119(+)]II; kle-2(ok1151) III) and KIR4 (stnSi2 [dpy-27 promoter: mVenus-MAP::dpy-27(+): dpy-27 3′UTR cb-unc-119(+)] II; dpy-27(tm3326) III].
Fluorescence Microscopy
The above stable cell lines were cultured in coverslips, fixed with 4% paraformaldehyde for 10 min, and permeabilized and blocked with 0.5% Triton X-100 and 5% normal goat serum for 20 min. Cells were subsequently incubated with antibodies against either tubulin (clone DM1A, Sigma) or gamma-tubulin (clone GTU-88, Sigma) for one hour at room temperature. They were washed three times in PBS and incubated with Alexa Fluor-conjugated secondary antibodies (Molecular Probes, Eugene, OR) for 45 min at room temperature. After three washes with PBS, nuclei were stained with DAPI. The coverslips were mounted on slides using ProLong Gold anti-fade reagent (Invitrogen), and viewed using a Nikon Eclipse E600 fluorescence microscope coupled to a cooled charge-coupled device camera (ORCA-ER; Hamamatsu, Bridgewater, NJ) and IPLab Spectrum software (Signal Analytics, Vienna, VA).
Protein Purification and Immunoblotting
U2OS cells stably expressing MAP tagged proteins were lysed on ice in lysis buffer (50 mm Tris-HCl, pH 7.5, 125 mm NaCl, 1 mm EDTA, 0.2% Nonidet P-40, 5% Glycerol) containing a protease inhibitor mixture (Roche) as specified by the manufacturer. Protein concentration was determined using the BCA assay (Pierce, Rockford, IL). Samples were incubated with Ni-NTA beads (Qiagen), streptavidin beads (Pierce), or anti-FLAG M2-beads (Sigma) for 2 h at 4 °C, and then the beads were washed with the lysis buffer and the bound protein was eluted by lysis buffer containing 300 mm imidazole, 2.5 mm d-Biotin, or 200 μg/ml FLAG peptide (Sigma) respectively. Protein samples were separated by SDS-PAGE followed by transfer to Immobilon-P membranes (Millipore, Billerica, MA) which were then incubated with specific primary antibodies followed by their detection with appropriate horseradish peroxidase-conjugated secondary antibodies and enhanced chemiluminescence substrate (Pierce). The primary antibodies and dilutions used for immunoblotting were mouse anti-GFP B-2 monoclonal antibody (Santa Cruz, Santa Cruz, CA; 1:1000) and rabbit anti-Cdc14B (Invitrogen, 1:500).
Tandem Affinity Purification Under Native Conditions
For MAP purifications, U2OS cells stably expressing MAP Tagged fusion proteins were seeded at 2.5 × 106 cells per 150-mm dish, cultured for 48 h and then synchronized by treatment with 2 mm Thymidine for 20 h (S-phase arrest) or Nocodazole (50 ng/ml) for 14–16 h (M-phase arrest). S-phase or mitotic cells were collected and packed cell volume was measured. All subsequent steps were performed at 4 °C with precooled buffer and equipment. Five packed cell volumes of lysis buffer (50 mm Tris-HCl, pH 7.5, 125 mm NaCl, 1 mm EDTA, 0.2% Nonidet P-40, 5% glycerol) containing the protease inhibitor mixture and phosphatase inhibitor mixture (Roche) was added to the pellet and agitated for 20 min. The extract was centrifuged for 10 min at 16,000 × g and each 1 ml of supernatant was incubated with 50 μl anti-FLAG M2-beads (Sigma) for 3 h. The beads were washed with 40 volumes of wash buffer (50 mm Tris-HCl, pH 7.5, 250 mm NaCl, 0.05% Nonidet P-40) in eppendorf tubes or on micro bio-spin column (Bio-Rad, Hercules, CA) and spun for 15 s at 100g. The bound proteins were eluted by 10 volumes of lysis buffer containing 200 μg/ml FLAG peptide with agitation for 15min. Each 1 ml eluted fraction was then incubated with 50 μl high capacity streptavidin beads (Pierce) for 3 h. After washing with 40 volumes of wash buffer, the bound proteins were incubated with agitation for 15 min in 10 volumes of elution buffer (25 mm Tris-HCl, pH 7.5, 125 mm NaCl) containing 2.5 mm d-Biotin. The final elution was either concentrated by Amicon Ultracel-10 Centrifugal Filters (Millipore) for Coomassie blue staining or precipitated with 20% trichloroacetic acid (TCA) for identification of interactive partners by LC-MS/MS.
For MAP purification in C. elegans, proteins were extracted from whole adult hermaphrodites by dounce homogenization followed by sonication in lysis buffer (50 mm HEPES-KOH, pH7.6, 150 mm KCl, 1 mm EDTA, 0.5% Nonidet P-40, 10% glycerol) containing protease inhibitor mixture (Roche). All steps were carried out at 4 °C. The extract was centrifuged for 20 min. at 16,000 × g, the supernatant was diluted to a total protein concentration of 12 mg/ml, and each 1 ml supernatant incubated with 100 μl anti-FLAG M2-beads (Sigma) for 2 h. Beads were spun for 1 min. at 500 × g and washed with 40 volumes of lysis buffer. Bound proteins were eluted by incubating beads for 15 min. in 8 volumes of streptavidin binding buffer (50 mm HEPES-KOH, pH7.6, 150 mm KCl, 1 mm EDTA, 0.2% Nonidet P-40, 4% glycerol) containing 200 μg/ml FLAG peptide. Each 0.8 ml eluted fraction was then incubated with 100 μl high capacity streptavidin beads (Pierce) for 2 h. Beads were washed twice with 10 volumes of streptavidin binding buffer then eluted by incubating in eight volumes of elution buffer (50 mm HEPES-KOH, pH 7.6, 150 mm KCl, 1 mm EDTA) containing 2.5 mm d-Biotin for 20 min. The final eluate was precipitated with 15% TCA for silver staining. Protein samples from each step were separated by SDS-PAGE and subjected to Western blot. Primary antibodies for immunoblotting were described previously (9) and used at 1:1000 dilution for anti-KLE-2, anti-HCP-6, anti-MIX-1, anti-SMC-4, and 1:500 dilution for anti-CAPG-2, anti-DPY-27.
For MAP purification in S. pombe, a strain expressing Rad25-mVenus-MAP was generated by knock-in at the C terminus of the chromosomal locus as previously described (10) using a PCR-based approach and a pFA6a based vector. Cells were grown in YE media and 20 OD of cells were collected by centrifugation. After washing the cell pellet with lysis buffer (150 mm NaCl, 2 mm EDTA, 6 mm Na2HPO4, 4 mm NaH2PO4, 1% TRITON, 1 mm phenylmethylsulfonyl fluoride, 0.5% yeast protease inhibitor mixture (Sigma), 100 μm NaVO4, and 50 mm NaF), cells were disrupted with glass beads in a multivortexer for 1min. Cell extracts were diluted to 1 ml in lysis buffer and centrifuged at 8000 RPM for 10 min at 4 °C to discard cell debris and intact cells. All purification steps were performed in batch. Bead incubation and washes were done in 1 ml total volume, and elutions in 10-bead volumes. Extracts from cells expressing Rad25-mVenus-MAP were first incubated with 25 μl anti-FLAG M2 agarose beads (SIGMA) for 3 h at 4 °C. Beads were centrifuged at 1500 RPM and washed twice with the same buffer. Elution from the FLAG beads was performed by competition with FLAG peptide at 200 μg/ml (SIGMA) for 30 min at 4 °C. The eluate was then incubated with 25 μl high capacity streptavidin agarose beads (Thermo Scientific) for 2 h at 4 °C. After washing the beads twice in lysis buffer, Rad25-mVenus-MAP was eluted by incubation with 2.5 mm biotin (Sigma) for 30 min at 4 °C.
Tandem Affinity Purification Under Denaturing Conditions
U2OS cells stably expressing MAP tagged fusion proteins were seeded at 2.5 × 106 cells per 150-mm dish and cultured for 48 h and then synchronized by treatment of Nocodazole (50 ng/ml) for 14–16 h. Mitotic cells were directly lysed in each dish in 0.5 ml full denaturing lysis buffer (20 mm Tris-HCl, pH 7.5, 125 mm NaCl, 0.2% Nonidet P-40, 8 m Urea) for 10 min. The extract was centrifuged for 10 min at 16,000 × g. Protein concentration was determined using the BCA assay. Samples were incubated overnight at 4 °C with 50 μl Ni-NTA resin per 1 ml lysate, and then the beads were washed with 40 volumes of wash buffer (50 mm Tris-HCl, pH 7.5, 250 mm NaCl, 0.05% Nonidet P-40) on micro bio-spin column and the bound proteins were eluted with 10 volumes of wash buffer containing 300 mm imidazole plus protease inhibitor mixture and phosphatase inhibitor mixture. Each 500 μl eluted fraction was diluted to 1 ml and then incubated with 50 μl streptavidin beads for 3 h at 4 °C. After washing with 40 volumes of wash buffer, the bound proteins were eluted with 10 Volumes of elution buffer (25 mm Tris-HCl, pH 7.5, 125 mm NaCl) containing 2.5 mm d-Biotin after agitation for 15 min. Protein samples were separated by SDS-PAGE and subjected to Western blot or 20% TCA precipitated for LC-MS/MS.
LC-MS/MS and MS Data Analysis
Purified proteins were denatured, reduced with Tris 2-carboxyethyl phosphine, alkylated with iodoacetamide, and digested overnight at 37 °C with Trypsin Gold (Promega) after diluting to 2 m urea with 50 mm Tris pH 8.5. The resulting peptides were subjected to two-dimensional LC-MS/MS (MudPIT) on a Thermo LTQ as previously detailed (11). All RAW files can be freely accessed on the Tranche project (proteomecommons.org) under the project name “MAP-Maetal2012,” which includes duplicate BIRC5-MAP, PLK1-MAP, and MAP (empty) 2D LC-MS/MS Thermo RAW files.
Thermo RAW files were converted to MZML files using Scansifter 2.0.13 (47) and spectra with fewer than 20 peaks were excluded from analysis. Protein identification was performed with the Myrimatch algorithm (v1.6.33, (12)) on a high performance computing cluster (Advanced Computing Center for Research and Education at Vanderbilt University) using the human IPI protein database, v3.65. Contaminant proteins (specifically, multiple keratin and IgG isoforms and bovine trypsin) were added and all sequences were reversed and concatenated to allow estimation of false discovery rates (total of 172904 entries). Myrimatch parameters were as follows: strict tryptic cleavage; modification of methionine (oxidation, dynamic modification, +16 Da), S/T/Y (phosphorylation, dynamic modification, +80 Da) and cysteine (carboxamidomethylation, static modification, +57 Da) were allowed; precursor ions were required to be within 0.6 m/z of the peptide monoisotopic mass; fragment ions were required to fall within 0.5 m/z of the expected monoisotopic mass; any number of missed cleavages were allowed. IDPicker (13) v2.6.165 was used to filter peptide matches with the following parameters: max. FDR per result 0.05, max. ambiguous IDs per result 2, min. peptide length per result 5, min. distinct peptides per protein 2, min. additional peptides per protein group 2, minimum number of spectra per protein 5, indistinct modifications M 15.994 Da, C 57.05 Da and distinct modifications S/T/Y 80 Da.
As reported in IDPicker, the calculated global protein-level FDRs were 2.0% and 4.3% for the Plk1 (S-phase and M-phase combined) and Survivin purifications, respectively. IDPicker results were processed in Excel (Microsoft®) to generate interactor lists. All isoforms of keratin, IgG, and trypsin (common contaminants) were excluded from protein identification lists. Some peptides matched to multiple protein isoforms; in these cases, all isoforms were reported and no detailed analysis of specific protein isoforms was completed or reported. Only proteins identified in both biological replicates are reported in the protein ID lists (Fig. 4, Table II, Supplemental Tables S2 and S3). The MAP purifications were normalized to the total spectral counts of bait recovered prior to comparison (Fig. 4, Table II and Supplemental Table S3). Spectra indicative of phosphopeptides were manually inspected in SeeMS and a related program called PTMDigger, software developed in-house (Surendra Dasari, Matthew Chambers, and David Tabb, Vanderbilt University Medical Center) and filtered according to the following criteria: (1) exhibit a prominent (often base) 98 Da (H3PO4) neutral loss peak at the MS2 level and (2) b and y ion intensities > 10% of the neutral loss peak (3) contained two or more sequential fragments (b and/or y) bracketing the phosphorylation site(s). Phosphorylation sites were assigned based on the presence of sequential fragment ions surrounding the modification; if these ions were missing, the phosphorylation site(s) were assigned to multiple sites ambiguously (Supplemental Table S4). Ion match tables and spectra for PLK1 phosphopeptides (Supplemental Table S5) were generated using software developed in-house (Zeqiang Ma, Surendra Dasari, Matthew Chambers, and David Tabb, Vanderbilt University Medical Center).
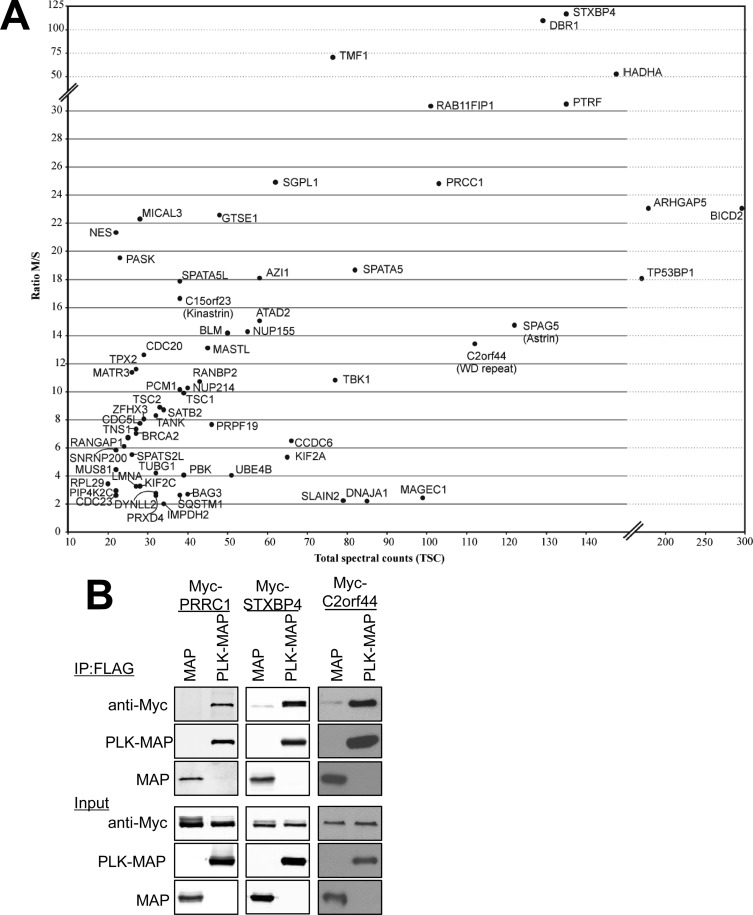
Fig. 4.
Identification and characterization of Plk1 binding partners. A, Scatterplot of the ratios of normalized spectral counts (see Material and Methods for details) in mitotic (M) verses S-phase (M/S) purifications for individual proteins is plotted versus total spectral counts (TSC, combined mitotic and S-phase purifications) for all unique proteins identified in M or S phase Plk1 purifications with more than 10 total spectral counts (TSC). B, The mVenus-MAP tag plasmid or the mVenus-MAP-tagged Plk1 plasmid were transfected along with Myc-tagged PRRC1, STXBP4, or the C2orf44 constructs into HEK 293 cells. Plk1-mVenus-MAP or mVenus-MAP were immunoprecipitated (IP) from cell lysates with anti-FLAG antibodies. The immunoprecipitates and cell lysates were subjected to immunoblot with anti-Myc and anti-FLAG antibody. The levels of PRRC1, STXBP4, and C2orf44 (Myc) as well as Plk1-mVenus-MAP and mVenus-MAP in the starting lysates are shown in the lower panels (Input). Note that the darker background in the C2orf44 panels is because these blots were developed with film for enhanced sensitivity instead of fluorescent imaging using an Odyssey system as was done for PRRC1 and STXBP4.
Table II. Mitotically-enriched proteins identified by LC-MS/MS in Plk1-MAP isolates. A list of all proteins (>10 TSC) enriched in mitotic Plk1 purifications by at least sevenfold is shown. Proteins in bold were not detected in S-phase and were artificially assigned 0.5 spectral counts (in S-phase) so they could be included in this list. IPI AC is the IPI accession number; UP, unique peptides; Norm, spectral counts divided by total PLK spectral counts times 1000; Ratio M/S, mitotic “Norm” divided by S-phase “norm”; TSC, total spectral counts for all PLK1-MAP purifications; Gene Symbol, Uniprot gene symbol; Proteins in bold were not detected in the S-phase purification and these proteins were assigned an arbitrary value of 0.5 spectral counts.
| IPI AC | Coverage | Ratio | TSC | UP | Gene symbol | References |
|---|---|---|---|---|---|---|
| IPI00217200 | 62 | 119.7 | 135 | 33 | STXBP4 | Kettenbach et al. 2011 |
| IPI00305545 | 42 | 117.4 | 124 | 19 | DBR1 | Kettenbach et al. 2011 |
| IPI00010586 | 42 | 72.1 | 79 | 44 | TMF1 | Svendsen et al. 2009; Kettenbach et al. 2011 |
| IPI00031522 | 62 | 52.7 | 174 | 34 | HADHA | |
| IPI00176903 | 54 | 30.5 | 135 | 20 | PTRF | |
| IPI00419433 | 43 | 30.3 | 101 | 38 | RAB11FIP1 | Kettenbach et al. 2011 |
| IPI00099463 | 45 | 24.9 | 62 | 21 | SGPL1 | |
| IPI00217053 | 28 | 24.8 | 103 | 10 | PRRC1 | Kettenbach et al. 2011 |
| IPI00178185 | 60 | 23.1 | 295 | 65 | BICD2 | Svendsen et al. 2009; Kettenbach et al. 2011 |
| IPI00013988 | 48 | 23.1 | 195 | 59 | ARHGAP5 | Kettenbach et al. 2011 |
| IPI00871779 | 36 | 22.6 | 48 | 19 | GTSE1 | Kettenbach et al. 2011 |
| IPI00852645 | 10 | 22.3 | 28 | 16 | MICAL3 | Kettenbach et al. 2011 |
| IPI00010800 | 14 | 21.3 | 22 | 16 | NES | |
| IPI00160901 | 37 | 21.3 | 45 | 19 | GTSE1 | Kettenbach et al. 2011 |
| IPI00398824 | 11 | 19.5 | 23 | 11 | PASK | Kettenbach et al. 2011 |
| IPI00651721 | 35 | 18.8 | 60 | 31 | AZI1 | Kettenbach et al. 2011 |
| IPI00329583 | 50 | 18.7 | 82 | 29 | SPATA5 | Kettenbach et al. 2011 |
| IPI00298883 | 36 | 18.1 | 58 | 31 | AZI1 | Kettenbach et al. 2011 |
| IPI00029778 | 41 | 18.1 | 191 | 60 | TP53BP1 | Lowery et al. 2007; van Vugt et al. 2010 |
| IPI00031608 | 34 | 17.9 | 38 | 17 | SPATA5L1 | |
| IPI00294680 | 44 | 16.6 | 38 | 11 | C15orf23 | |
| IPI00170548 | 27 | 15.1 | 58 | 31 | ATAD2 | |
| IPI00328118 | 57 | 14.7 | 122 | 47 | SPAG5 | Kettenbach et al. 2011 |
| IPI00026625 | 34 | 14.3 | 55 | 31 | NUP155 | Lowery et al. 2007 |
| IPI00004859 | 33 | 14.2 | 50 | 30 | BLM | Leng et al. 2006; Santamaria et al. 2010; Kettenbach et al. 2011 |
| IPI00018786 | 62 | 13.4 | 112 | 37 | C2orf44 | Svendsen et al. 2009; Kettenbach et al. 2011 |
| IPI00396230 | 30 | 13.1 | 45 | 20 | MASTL | Kettenbach et al. 2011 |
| IPI00329526 | 26 | 12.6 | 29 | 9 | CDC20 | Golan at al. 2002; Kettenbach et al. 2011 |
| IPI00008477 | 29 | 11.6 | 27 | 15 | TPX2 | De Luca et al. 2006; Lowery et al. 2007; Santamaria et al. 2010; Kettenbach et al. 2011 |
| IPI00017297 | 17 | 11.4 | 26 | 10 | MATR3 | |
| IPI00293613 | 46 | 10.8 | 77 | 25 | TBK1 | Kettenbach et al. 2011 |
| IPI00221325 | 13 | 10.7 | 43 | 29 | RANBP2 | Kettenbach et al. 2011 |
| IPI00183294 | 16 | 10.3 | 40 | 22 | NUP214 | |
| IPI00006213 | 20 | 10.2 | 38 | 26 | PCM1 | Kettenbach et al. 2011 |
| IPI00022043 | 21 | 9.9 | 39 | 17 | TSC1 | Astrinidis et al. 2006; Kettenbach et al. 2011 |
| IPI00879496 | 21 | 8.9 | 33 | 21 | TSC2 | Astrinidis et al. 2006; Kettenbach et al. 2011 |
| IPI00010433 | 43 | 8.7 | 34 | 21 | SATB2 | Kettenbach et al. 2011 |
| IPI00299166 | 48 | 8.3 | 32 | 13 | TANK | Zhang et al. 2010; Kettenbach et al. 2011 |
| IPI00014186 | 6 | 8.1 | 29 | 17 | ZFHX3 | Kettenbach et al. 2011 |
| IPI00465294 | 28 | 7.7 | 28 | 13 | CDC5L | |
| IPI00004968 | 40 | 7.7 | 46 | 11 | PRPF19 | |
| IPI00307545 | 16 | 7.3 | 27 | 16 | TNS1 | Santamaria et al. 2010 |
RESULTS
Development of the MAP Tag
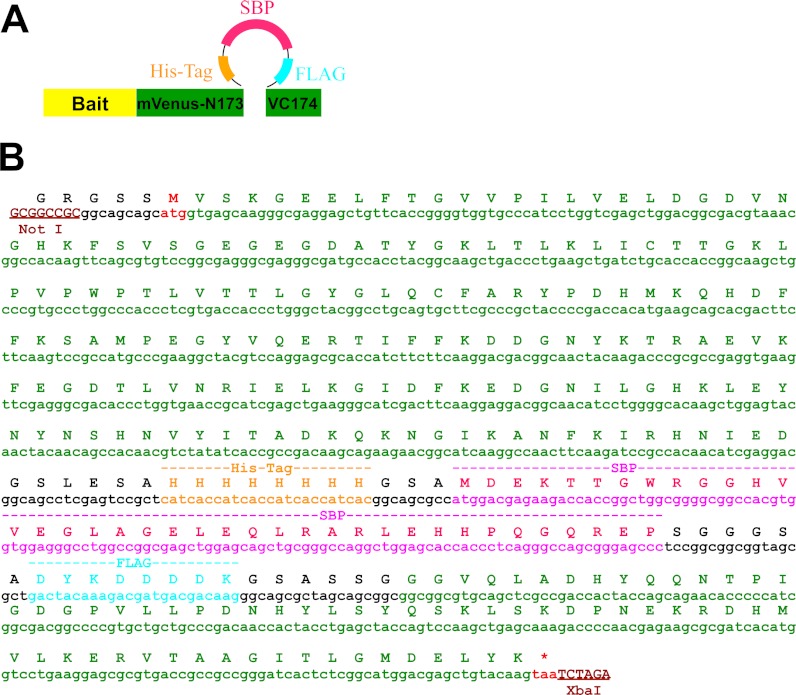
To optimize the accessibility of affinity tags for protein purification, we inserted a set of tandem affinity tags into the exposed surface loop between β8 and β9 of GFP-derived fluorescent proteins, a region previously targeted for this purpose (7, 14, 15). The inserted tag array consists of a His8-Tag (16), followed by a 38-amino-acid streptavidin binding peptide (SBP) (17), and an eight-amino-acid FLAG tag (18) (Fig. 1A, 1B). This combination of tags was chosen because it yielded fast and efficient purifications under both physiological and denaturing conditions (see below), and has been used successfully in previous studies (19). In particular, the His tag can be used for immobilized metal ion affinity chromatography under native or denaturing conditions and eluted by buffers containing imidazole (20). The SBP binds streptavidin with dissociation constants as low as 5 nm and can be eluted by buffers containing d-biotin (17), and finally the FLAG epitope can be isolated using anti-FLAG antibody and eluted efficiently by the FLAG octapeptide (21).
Fig. 1.
Outline of the fluorescent protein-based multifunctional tandem affinity purification method. A, Schematic illustration of multifunctional tandem affinity purification tag (MAP), in which His8, SBP, and FLAG-Tag was inserted in tandem into the surface loop between β8-β9 of mVenus. B, DNA and amino acid sequence of the mVenus-MAP tag.
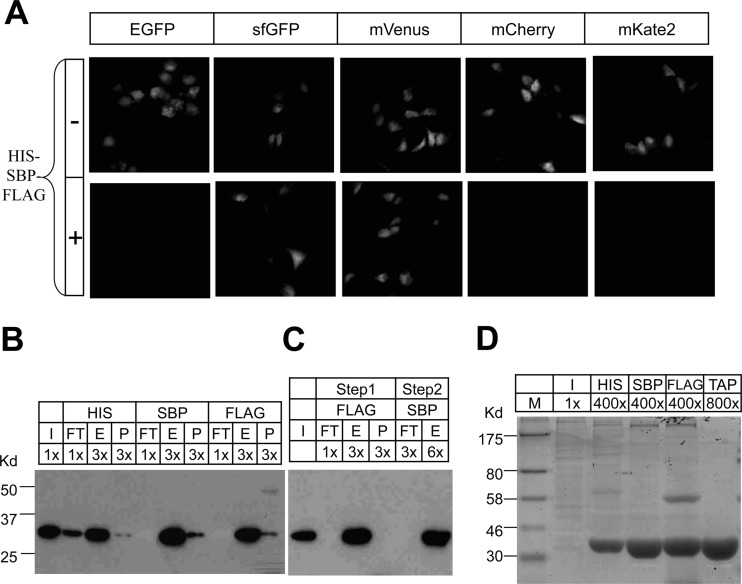
Initially, we inserted the His8-SBP-FLAG tandem tag in the loop between β8 and β9 of EGFP, however the fluorescence of EGFP after tag insertion was extremely low (Fig. 2A), consistent with earlier studies showing that insertion of random peptides up to 20 residues in length within loops of GFP have deleterious effects on GFP folding and fluorescence (14, 15, 22). We next tested two GFP variants: superfolder GFP (sfGFP) and mVenus because of their fast folding and chromophore formation kinetics (23, 24), and also attempted to generate red fluorescent versions using mCherry (25) and mKate2 (26). Both sfGFP and mVenus maintained fluorescence after insertion of the His8-SBP-FLAG tag, but insertion of the tag into mCherry and mKate2 disrupted their fluorescence (Fig. 2A).
Fig. 2.
Development of the MAP tag purification system. A, Plasmids encoding EGFP, superfolder GFP (sfGFP), mVenus, mCherry, and mKate2, with or without insertion of the His8-SBP-FLAG tag were transfected into U2OS cells and images were acquired 24 h after transfection. The same exposure was used for each fluorescent protein with and without the inserted tag, and all images are scaled the same. B–D, The levels of mVenus-MAP in the starting input lysate (I), the flow through (FT) after binding to beads, after elution from affinity beads (E), and the remaining protein in the bead pellet following elution (P) is shown. Equal volumes were loaded in each lane and the relative volume of each fraction compared with the starting lysate are shown (i.e. 1×, 3×….). B, The mVenus-MAP tag was purified using either the His, SBP, or FLAG tags followed by Western blot with anti GFP antibody. C, Tandem purification of the mVenus-MAP tag was carried out by sequential purification using the FLAG and SBP tags followed by Western blot with anti-GFP antibody. D, The mVenus-MAP tag that was purified in one step using the individual tags, or in two steps (TAP) using the FLAG and SBP tags and was subjected to SDS-PAGE separation and Coomassie Blue staining.
To test and develop purification schemes, we focused on the mVenus version of the MAP tag. A U2OS cell line stably expressing the mVenus-MAP protein was established, and the mVenus-MAP protein was purified using the affinity tags singly or in combination. The mVenus-MAP protein was successfully pulled down using all three tags (His8, SBP, FLAG) individually, but the pull down using the His8 tag was slightly less efficient than with SBP or FLAG (Fig. 2B). Therefore we used the FLAG/SBP combination for tandem affinity purification under native conditions (Fig. 2C–2D). For two-step purification, we first immunoprecipitated the mVenus-MAP protein using FLAG antibody and eluted with FLAG peptide. The mVenus-MAP protein could easily be purified from cells with high efficiency (Table I). Theoretically the FLAG and SBP steps can be done in either order, but FLAG was used first so that FLAG peptide would not be present in the final purification where it could interfere with MS. Analysis of mVenus-MAP purifications after tandem affinity purification using LC-MS/MS revealed relatively few copurifying proteins (Supplemental Table 1). Together these results show that the mVenus-MAP tag can be purified with high efficiency and low background.
Table I. mVenus-MAP tagging in diverse organisms.
| Functional (rescue) | Proper localization | Efficiency |
||
|---|---|---|---|---|
| FLAG | TAP | |||
| Mammal | ||||
| mVenus-MAP | / | + | 61% | 30% |
| Cdc14A | / | + | 95% | 52% |
| Cdc14B | / | + | 96% | 51% |
| Aurora B | / | + | 58% | 42% |
| Plk1 | / | + | 54% | 32% |
| Worm | ||||
| Kle-2 | + | N.T. | 77% | 43% |
| Dpy-27 | + | N.T. | 65% | 45% |
| Fission Yeast | ||||
| Rad25 | + | + | 80% | 22% |
The mVenus-MAP Tag Allows Efficient Purification of Various Tagged Proteins
We next tested how well the mVenus-MAP tag worked when fused to other proteins, in particular the human polo kinase Plk1. For creation of stable mammalian cell lines expressing Plk1-mVenus-MAP and other mVenus-MAP fusions, plasmids for expression of mVenus-MAP fusions were transfected into cells, stable cell lines were selected, and then clonal cell lines were isolated by FACS sorting using mVenus fluorescence. In general we selected lines with lower expression of bait proteins. In the case of Plk1, the Plk1-mVenus-MAP fusion was overexpressed 13-, 10-, and 3.5-fold compared with endogenous Plk1 in asynchronous, S-phase arrested, and M-phase arrested cells respectively. We expect that expression closer to physiological levels could be achieved using FACS if desired. In cases where cells cannot tolerate moderate overexpression of the fusion protein, the MAP tagged protein can be expressed inducibly by cotransfection of TetR expression plasmids, so that expression of the fusion protein is repressed until doxycycline is added (27).
Plk1-mVenus-MAP stably expressed in U2OS cells displayed a cell cycle localization pattern similar to that previously reported (28) (Fig. 3A). During interphase and early mitosis, Plk1 was observed at the centrosome whereas, during cytokinesis, Plk1 was observed at the midbody. Purification of Plk1 under native conditions was highly efficient using the mVenus-MAP tag, with 32% recovery of starting material (Fig. 3B, Table I). Similar results were observed with other mVenus-MAP fusions. For example, cell lines expressing several other mVenus-MAP tagged cell cycle regulators, including Cdc14A, Cdc14B, Aurora B, and Survivin, were created. In all cases the mVenus-MAP fusion proteins yielded between 32–52% recovery of starting bait (Table I).
Fig. 3.
Localization and purification of Plk1-mVenus-MAP. A, Plk1-mVenus-MAP expressing U2OS cells were stained with antibodies against either gamma-tubulin or alpha-tubulin and analyzed using immunofluorescence. For the Plk1-mVenus-MAP purifications shown in parts (B, C), Plk1-mVenus-MAP was purified from U2OS cells stably expressing the fusion protein and then subjected to SDS-PAGE separation and Western blot analysis by anti GFP antibody. The levels of Plk1-mVenus-MAP in the starting input lysate (I), the flow through (FT) after binding to beads, after elution from affinity beads (E), and the remaining protein in the bead pellet following elution (P) is shown. Equal volumes were loaded in each lane and the relative volume of each fraction compared with the starting lysate are shown (i.e. 1×, 3×….). B, Tandem affinity purification of Plk1-mVenus-MAP was performed by sequential purification using the FLAG and SBP tags. C, Plk1-mVenus-MAP was purified from lysates of cells stably expressing Plk1-mVenus-MAP using the His-Tag under denaturing conditions (8 m urea), followed by FLAG or SBP under renaturing conditions.
We next tested whether the mVenus-MAP tag could be used for purification under denaturing conditions. Native purifications are useful for identification of binding partners, but are often not ideal for characterization of post-translational modifications of the tagged protein. For instance, in mild buffer conditions, phosphatases and deubiquitinating enzymes have an opportunity to remove their respective modifications. Methods that involve preparation of cell lysates under protein denaturing conditions quickly inactivate enzymes and optimize identification of modifications (5). Therefore we developed a method for using the MAP tag under denaturing conditions. Plk1-mVenus-MAP was first purified on Ni-NTA beads through its His8 tag under denaturing conditions (8 m Urea), followed by FLAG or SBP purification under renaturing conditions (Fig. 3C). Using this scheme Plk1-mVenus-MAP was efficiently purified (28% recovery), showing that the mVenus-MAP tag worked well for purifications under both native and denaturing conditions.
Isolation of Survivin and Plk1 Binding Partners Using the MAP Tag
To test how well our method worked for co-purification of binding partners, we analyzed duplicate purifications of Survivin (BIRC5-MAP) from human cells using LC-MS/MS. Survivin is part of the chromosome passenger protein complex involved in chromosome segregation and cytokinesis (29). As expected, the chromosome passenger complex proteins, Borealin, and to a lesser extent INCENP and Aurora B (supplemental Table S2), were identified in the Survivin purifications. As another test of our method, we analyzed the composition of MAP-isolated Plk1 complexes; Plk1 is an essential regulator of cell cycle progression (30). Substrates and binding partners of Plk1 have been identified by various methods, including yeast two hybrid, phage display, immunoprecipitation, and in vitro pull downs (31–34). Although Plk1 is well known as a mitotic regulator, more recent analysis has revealed roles in maintenance of DNA integrity during S-phase (35). Thus we set out to identify Plk1 binding partners during both S-phase and M-phase using the U2OS cell line expressing Plk1-mVenus-MAP that had been synchronized in S-phase by thymidine block, or in prometaphase using nocodazole (supplemental Fig. S1).
LC-MS/MS of Plk1-mVenus-MAP duplicate purifications identified a large number of potential Plk1 binding proteins from both S-phase and mitotically arrested cells (supplemental Table S3). Consistent with the well-documented role of Plk1 in mitosis, many proteins were highly enriched in mitotic purifications compared with those from S-phase (Fig. 4A; Table II, supplemental Table S3). About 40 proteins, identified with >10 total spectral counts (TSC), were enriched at least sevenfold in both biological replicates (Table II). Thirty of these proteins were reported in previous studies as binding proteins and/or targets of Plk1 (34–43). The protein that was most highly enriched in our mitotic Plk1 purifications is STXBP4, which was recently identified by a high throughput study as a candidate Plk1 binding partner and substrate (37). STXBP4 as well as two other major Plk1 binding proteins identified by our analysis (PRRC1 (44) and C2orf44) were epitope tagged and co-expressed with Plk1-mVenus-MAP in HEK-293 cells. All three proteins were specifically recovered from Plk1-mVenus-MAP immunoprecipitations (Fig. 4B). Other proteins with mitotic functions such as the kinesins KIF2C/MCAK and KIF2A, and SPAG5/Astrin were also enriched in mitotic purifications. The identification of MASTL (greatwall kinase) is consistent with the known role of Plk1 in promoting mitotic entry. Although numerous proteins were enriched in S-phase preparations of Plk1 (supplemental Table S3), functions of Plk1 during S-phase are less well described making it difficult to evaluate their significance. Clearly this study along with earlier screens for Plk1 targets and binding partners reveals that Plk1 carries out its many functions in the cell through interactions with a plethora of binding partners. Proteins identified using the mVenus-MAP method and LC-MS/MS include multiple previously identified Plk1 interacting proteins as well as many uncharacterized interactors, further validating this method for identification of binding partners. Future analysis of Plk1 binding proteins identified here will likely yield additional insight into the many important functions of Plk1.
To test the utility of the mVenus-MAP tag for identifying post-translational modifications, we analyzed Plk1-mVenus-MAP purifications using LC-MS/MS to identify phosphorylation sites on Plk1 (93% sequence coverage) (supplemental Tables S4 and S5). This analysis clearly identified phosphorylation on serine 210, the major site of activating phosphorylation on Plk1 (45), two other previously identified phosphorylation sites (T214, T498) (46), as well as several other potentially novel sites of Plk1 phosphorylation (supplemental Tables S4 and S5). Overall our analysis of Plk1 showed that the mVenus-MAP tag works well for identifying both binding partners and post-translational modifications.
The mVenus-MAP Tag Works Efficiently in Diverse Organisms
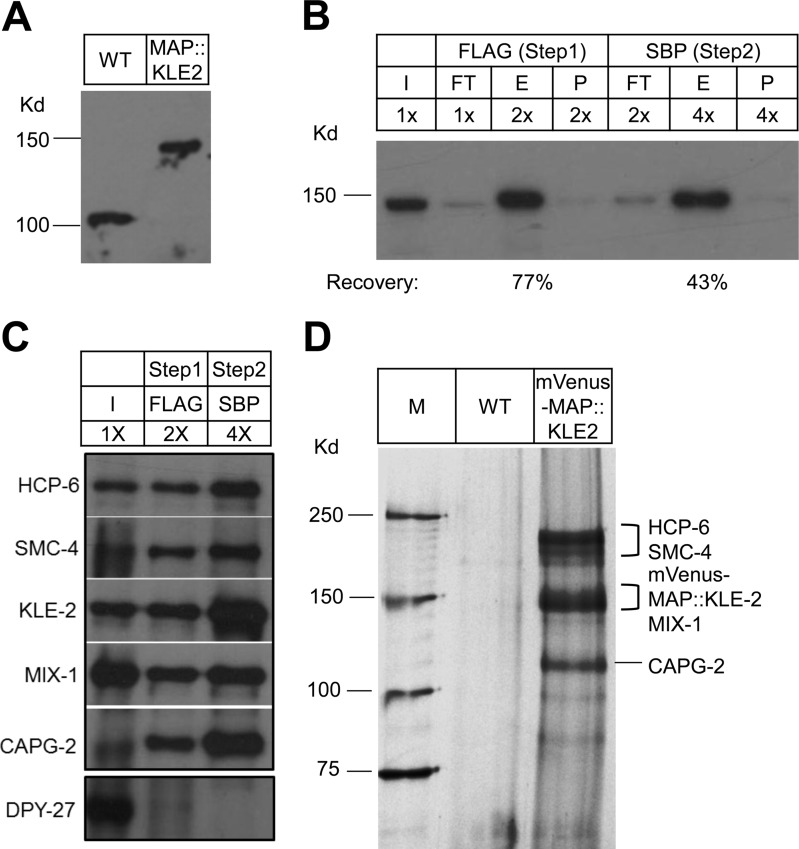
To assess the general utility of the mVenus-MAP tag system, we used it for purification of fusion proteins in C. elegans and S. pombe. The mVenus-MAP module was fused to the N terminus of the C. elegans condensin II subunit KLE-2 (9) and expressed under endogenous 5′ and 3′ genomic elements in a kle-2 null mutant. The single-copy transgene was expressed at an endogenous level and was fully functional as judged by its ability to rescue the kle-2(null) lethal phenotype (Fig. 5A, data not shown). Rescued worms were used for purification. The mVenus-MAP::KLE-2 fusion protein was efficiently purified, yielding 77% recovery after FLAG purification and 43% recovery after FLAG + SBP purification (Figs. 5B–5D, Table I). Along with mVenus-MAP::KLE-2, the purification equally recovered the other condensin II subunits, HCP-6, SMC-4, MIX-1, and CAPG-2, but not DPY-27, which is a member of a different condensin complex (Fig. 5C). In addition, purification from untagged worms yielded little to no protein detectable by silver staining, whereas purification from mVenus-MAP::KLE-2 worms yielded proteins that match the sizes of known condensin II subunits (Fig. 5D). Similarly efficient and specific recovery was observed when DPY-27, a subunit of the condensin IDC dosage compensation complex, was tagged (Table I, data not shown), further illustrating the utility of the mVenus-MAP tag for purification of intact complexes with low background.
Fig. 5.
Purification of C. elegans KLE-2 using the MAP tag. A, Western blot probed with anti-KLE-2 antibody shows similar levels of KLE-2 protein in 50 wild-type worms (WT, left) or 50 kle-2(null) worms rescued with MAP::KLE-2. B, MAP purification from MAP::KLE-2. Sequential purification of MAP::KLE-2 was performed using the FLAG and SBP tags, and subjected to SDS-PAGE separation and Western blot analysis with anti-KLE-2 antibody. The panel is labeled as in Fig. 2C. The percent recovery relative to input was estimated by measuring band intensity using ImageJ. C, Identification of condensin subunits co-purified with MAP::KLE-2. The purified MAP::KLE-2 complex was separated by SDS-PAGE and probed with antibodies against all five condensin II subunits (HCP-6, SMC-4, KLE-2, MIX-1, CAPG-2), or DPY-27, which is member of a different condensin complex. D, Tandem affinity purifications of protein complexes from wild-type (WT) or MAP::KLE-2 worms were TCA-precipitated, separated by SDS-PAGE, and subjected to silver-staining. The estimated positions of the five condensin II subunits are indicated. Kd = kilodalton size of marker proteins.
We next tested the mVenus-MAP tag system in S. pombe by tagging the chromosomal locus of the 14–3-3 protein Rad25 with the mVenus-MAP tag. The Rad25 fusion protein was purified using the FLAG and streptavidin binding sites. The Rad25-mVenus-MAP was efficiently recovered (22%) (supplemental Fig. S2 and Table I) and many specifically interacting proteins, including a known hetero-dimerization partner Rad24, were identified by LC-MS/MS (data not shown). The purification efficiency of Rad25-mVenus-MAP was also similar in phosphate and Tris-based buffers (results not shown), suggesting that MAP is a robust method that can be adapted to diverse experimental conditions. Furthermore because the mVenus-MAP tag was placed into the previously described kanMX6 tagging cassette (10), it can be used for PCR-based C-terminal tagging of the chromosomal locus of any gene in budding or fission yeast using standard primer sets. Together these results show that the mVenus-MAP method is a very useful and widely applicable and adaptable technique for identifying protein interactors and mapping post translational modifications in an array of organisms.
DISCUSSION
Central to the characterization of a protein's function is determination of its localization and binding partners. Although this can be accomplished through the use of antibodies against individual proteins, antibodies are expensive, time consuming to generate, and difficult to use for high-throughput approaches. The use of protein tags for protein purification and localization is a popular alternative strategy; however poor accessibility of affinity tags and protease cleavage sites might explain the low purification efficiency of some tagged proteins as well as the high variability of purification success between proteins. Protein to protein variability of purification efficiency is a major issue for high-throughput studies that rely on a single purification approach for a large number of proteins. The MAP tag system described here offers an ideal solution to these problems. The insertion of the affinity tags into the exposed surface loops of mVenus makes them readily accessible and appears to eliminate most of the variability associated with other purification systems. Of the eight different proteins that we have tagged with the mVenus-MAP tag in mammalian cells, C. elegans, and S. pombe, all have yielded between 22 and 52% recovery of the bait protein following purification, which compares quite favorably to other purification strategies (2). Furthermore the MAP tag scheme works well for identification of binding proteins as seen by our efficient purification of the condensin complex in C. elegans and the large number of known and novel binding proteins identified in the Plk1 and Survivin purifications from mammalian cells. Because the MAP tag allows both protein localization and efficient reproducible protein purification, it is an ideal tag for analysis of individual proteins as well as high throughput studies.
Acknowledgments
We thank Drs. Eric Campeau, Jiri Lukas, Eric Nigg, and Atsushi Miyawaki for plasmids, and Dr. Thoru Pederson for use of his microscope. We thank Ping Liang and Malwina Huzarska for performing LC-MS/MS experiments and David L. Tabb and members of his lab (especially Zeqiang Ma, Surendra Dasari, and Matt Chambers) for use and adaptation of software tools developed in their lab. J.R.M. was supported by NCI T32CA119925.
Footnotes
* This work was supported by the Howard Hughes Medical Institute of which K.L.G. is an investigator, and National Institutes of Health grants GM068786 to D. McCollum and GM076378 to K. Hagstrom.
 This article contains supplemental Figs. S1 and S2 and Tables S1 to S5.
This article contains supplemental Figs. S1 and S2 and Tables S1 to S5.
1 The abbreviations used are:
- TAP
- tandem affinity purification
- MAP
- multifunctional tandem affinity purification
- mVenus
- monomeric Venus
- FACS
- fluorescence-activated cell sorting
- LC-MS/MS
- liquid chromatography - tandem mass spectrometry
- Plk1
- polo-like kinase 1
- Cdc14B
- CDC14 cell division cycle 14 homolog B
- LAP
- localization and tandem affinity purification
- SBP
- streptavidin binding peptide
- sfGFP
- superfolder GFP
- EGFP
- enhanced green fluorescent protein
- INCENP
- inner centromere protein
- Aurora B
- aurora kinase B
- C. elegans
- Caenorhabditis elegans
- S. pombe
- Schizosaccharomyces pombe
- TSC
- total spectral counts
- BCA
- bicinchoninic acid
- TCA
- trichloroacetic acid.
REFERENCES
- 1. Rigaut G., Shevchenko A., Rutz B., Wilm M., Mann M., Séraphin B. (1999) A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17, 1030–1032 [DOI] [PubMed] [Google Scholar]
- 2. Bürckstummer T., Bennett K. L., Preradovic A., Schütze G., Hantschel O., Superti-Furga G., Bauch A. (2006) An efficient tandem affinity purification procedure for interaction proteomics in mammalian cells. Nat. Methods 3, 1013–1019 [DOI] [PubMed] [Google Scholar]
- 3. Gloeckner C. J., Boldt K., Schumacher A., Roepman R., Ueffing M. (2007) A novel tandem affinity purification strategy for the efficient isolation and characterisation of native protein complexes. Proteomics 7, 4228–4234 [DOI] [PubMed] [Google Scholar]
- 4. Glatter T., Wepf A., Aebersold R., Gstaiger M. (2009) An integrated workflow for charting the human interaction proteome: insights into the PP2A system. Mol. Syst. Biol. 5, 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ota K., Kito K., Iemura S., Natsume T., Ito T. (2008) A parallel affinity purification method for selective isolation of polyubiquitinated proteins. Proteomics 8, 3004–3007 [DOI] [PubMed] [Google Scholar]
- 6. Cheeseman I. M., Desai A. (2005) A combined approach for the localization and tandem affinity purification of protein complexes from metazoans. Sci. STKE 2005, pl1. [DOI] [PubMed] [Google Scholar]
- 7. Kobayashi T., Morone N., Kashiyama T., Oyamada H., Kurebayashi N., Murayama T. (2008) Engineering a novel multifunctional green fluorescent protein tag for a wide variety of protein research. PLoS One 3, e3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Frøkjaer-Jensen C., Davis M. W., Hopkins C. E., Newman B. J., Thummel J. M., Olesen S. P., Grunnet M., Jorgensen E. M. (2008) Single-copy insertion of transgenes in Caenorhabditis elegans. Nat. Genet. 40, 1375–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Csankovszki G., Collette K., Spahl K., Carey J., Snyder M., Petty E., Patel U., Tabuchi T., Liu H., McLeod I., Thompson J., Sarkeshik A., Yates J., Meyer B. J., Hagstrom K. (2009) Three distinct condensin complexes control C. elegans chromosome dynamics. Curr. Biol. 19, 9–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bähler J., Wu J. Q., Longtine M. S., Shah N. G., McKenzie A., 3rd, Steever A. B., Wach A., Philippsen P., Pringle J. R. (1998) Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14, 943–951 [DOI] [PubMed] [Google Scholar]
- 11. McDonald W., Ohi R., Miyamoto D., Mitchison T., Yates J. R., 3rd. (2002) Comparison of three directly coupled HPLC MS/MS strategies for identification of proteins from complex mixtures: single-dimension LC-MS/MS, 2-phase MudPIT, and 3-phase MudPIT. Int. J. Mass Spectrom. 219, 245–251 [Google Scholar]
- 12. Tabb D. L., Fernando C. G., Chambers M. C. (2007) MyriMatch: highly accurate tandem mass spectral peptide identification by multivariate hypergeometric analysis. J. Proteome Res. 6, 654–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ma Z. Q., Dasari S., Chambers M. C., Litton M. D., Sobecki S. M., Zimmerman L. J., Halvey P. J., Schilling B., Drake P. M., Gibson B. W., Tabb D. L. (2009) IDPicker 2.0: Improved protein assembly with high discrimination peptide identification filtering. J. Proteome Res. 8, 3872–3881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Peelle B., Lorens J., Li W., Bogenberger J., Payan D. G., Anderson D. C. (2001) Intracellular protein scaffold-mediated display of random peptide libraries for phenotypic screens in mammalian cells. Chem. Biol. 8, 521–534 [DOI] [PubMed] [Google Scholar]
- 15. Paramban R. I., Bugos R. C., Su W. W. (2004) Engineering green fluorescent protein as a dual functional tag. Biotechnol. Bioeng. 86, 687–697 [DOI] [PubMed] [Google Scholar]
- 16. Hochuli E., Bannwarth W., Döbeli H., Gentz R., Stüber D. (1988) Genetic approach to facilitate purification of recombinant proteins with a novel metal chelate adsorbent. Nat. Biotechnol. 6, 1321–1325 [Google Scholar]
- 17. Wilson D. S., Keefe A. D., Szostak J. W. (2001) The use of mRNA display to select high-affinity protein-binding peptides. Proc. Natl. Acad. Sci. U.S.A. 98, 3750–3755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hopp T., Prickett K., Price V., Libby R., March C., Cerretti D., Urdal D., Conlon P. (1988) A Short Polypeptide Marker Sequence Useful for Recombinant Protein Identification and Purification. Nat. Biotechnol. 6, 1204–1210 [Google Scholar]
- 19. Mak A. B., Ni Z., Hewel J. A., Chen G. I., Zhong G., Karamboulas K., Blakely K., Smiley S., Marcon E., Roudeva D., Li J., Olsen J. B., Wan C., Punna T., Isserlin R., Chetyrkin S., Gingras A. C., Emili A., Greenblatt J., Moffat J. (2010) A lentiviral functional proteomics approach identifies chromatin remodeling complexes important for the induction of pluripotency. Mol. Cell. Proteomics 9, 811–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hochuli E. (1990) Purification of recombinant proteins with metal chelate adsorbent. Genet. Eng. 12, 87–98 [DOI] [PubMed] [Google Scholar]
- 21. Einhauer A., Jungbauer A. (2001) The FLAG peptide, a versatile fusion tag for the purification of recombinant proteins. J. Biochem. Biophys. Methods 49, 455–465 [DOI] [PubMed] [Google Scholar]
- 22. Baird G. S., Zacharias D. A., Tsien R. Y. (1999) Circular permutation and receptor insertion within green fluorescent proteins. Proc. Natl. Acad. Sci. U.S.A. 96, 11241–11246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nagai T., Ibata K., Park E. S., Kubota M., Mikoshiba K., Miyawaki A. (2002) A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 20, 87–90 [DOI] [PubMed] [Google Scholar]
- 24. Pédelacq J. D., Cabantous S., Tran T., Terwilliger T. C., Waldo G. S. (2006) Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 24, 79–88 [DOI] [PubMed] [Google Scholar]
- 25. Shaner N. C., Campbell R. E., Steinbach P. A., Giepmans B. N., Palmer A. E., Tsien R. Y. (2004) Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22, 1567–1572 [DOI] [PubMed] [Google Scholar]
- 26. Shcherbo D., Murphy C. S., Ermakova G. V., Solovieva E. A., Chepurnykh T. V., Shcheglov A. S., Verkhusha V. V., Pletnev V. Z., Hazelwood K. L., Roche P. M., Lukyanov S., Zaraisky A. G., Davidson M. W., Chudakov D. M. (2009) Far-red fluorescent tags for protein imaging in living tissues. Biochem. J. 418, 567–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yao F., Svensjö T., Winkler T., Lu M., Eriksson C., Eriksson E. (1998) Tetracycline repressor, tetR, rather than the tetR-mammalian cell transcription factor fusion derivatives, regulates inducible gene expression in mammalian cells. Hum. Gene Ther. 9, 1939–1950 [DOI] [PubMed] [Google Scholar]
- 28. Berdougo E., Nachury M. V., Jackson P. K., Jallepalli P. V. (2008) The nucleolar phosphatase Cdc14B is dispensable for chromosome segregation and mitotic exit in human cells. Cell Cycle 7, 1184–1190 [DOI] [PubMed] [Google Scholar]
- 29. Altieri D. C. (2008) Survivin, cancer networks and pathway-directed drug discovery. Nat. Rev. Cancer 8, 61–70 [DOI] [PubMed] [Google Scholar]
- 30. Archambault V., Glover D. M. (2009) Polo-like kinases: conservation and divergence in their functions and regulation. Nat. Rev. Mol. Cell Biol. 10, 265–275 [DOI] [PubMed] [Google Scholar]
- 31. Casenghi M., Meraldi P., Weinhart U., Duncan P. I., Körner R., Nigg E. A. (2003) Polo-like kinase 1 regulates Nlp, a centrosome protein involved in microtubule nucleation. Dev. Cell 5, 113–125 [DOI] [PubMed] [Google Scholar]
- 32. Zhou T., Aumais J. P., Liu X., Yu-Lee L. Y., Erikson R. L. (2003) A role for Plk1 phosphorylation of NudC in cytokinesis. Dev. Cell 5, 127–138 [DOI] [PubMed] [Google Scholar]
- 33. Neef R., Gruneberg U., Kopajtich R., Li X., Nigg E. A., Sillje H., Barr F. A. (2007) Choice of Plk1 docking partners during mitosis and cytokinesis is controlled by the activation state of Cdk1. Nat. Cell Biol. 9, 436–444 [DOI] [PubMed] [Google Scholar]
- 34. Lowery D. M., Clauser K. R., Hjerrild M., Lim D., Alexander J., Kishi K., Ong S. E., Gammeltoft S., Carr S. A., Yaffe M. B. (2007) Proteomic screen defines the Polo-box domain interactome and identifies Rock2 as a Plk1 substrate. EMBO J. 26, 2262–2273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Svendsen J. M., Smogorzewska A., Sowa M. E., O'Connell B. C., Gygi S. P., Elledge S. J., Harper J. W. (2009) Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell 138, 63–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Santamaria A., Wang B., Elowe S., Malik R., Zhang F., Bauer M., Schmidt A., Silljé H. H., Körner R., Nigg E. A. (2011) The Plk1-dependent phosphoproteome of the early mitotic spindle. Mol. Cell. Proteomics 10, M110 004457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kettenbach A. N., Schweppe D. K., Faherty B. K., Pechenick D., Pletnev A. A., Gerber S. A. (2011) Quantitative phosphoproteomics identifies substrates and functional modules of Aurora and Polo-like kinase activities in mitotic cells. Sci. Signal 4, rs5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang W., Wang J., Zhang Y., Yuan Y., Guan W., Jin C., Chen H., Wang X., Yang X., He F. (2010) The scaffold protein TANK/I-TRAF inhibits NF-kappaB activation by recruiting polo-like kinase 1. Mol. Biol. Cell 21, 2500–2513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Astrinidis A., Senapedis W., Henske E. P. (2006) Hamartin, the tuberous sclerosis complex 1 gene product, interacts with polo-like kinase 1 in a phosphorylation-dependent manner. Hum. Mol. Genet. 15, 287–297 [DOI] [PubMed] [Google Scholar]
- 40. De Luca M., Lavia P., Guarguaglini G. (2006) A functional interplay between Aurora-A, Plk1 and TPX2 at spindle poles: Plk1 controls centrosomal localization of Aurora-A and TPX2 spindle association. Cell Cycle 5, 296–303 [DOI] [PubMed] [Google Scholar]
- 41. Golan A., Yudkovsky Y., Hershko A. (2002) The cyclin-ubiquitin ligase activity of cyclosome/APC is jointly activated by protein kinases Cdk1-cyclin B and Plk. J. Biol. Chem. 277, 15552–15557 [DOI] [PubMed] [Google Scholar]
- 42. Leng M., Chan D. W., Luo H., Zhu C., Qin J., Wang Y. (2006) MPS1-dependent mitotic BLM phosphorylation is important for chromosome stability. Proc. Natl. Acad. Sci. U.S.A. 103, 11485–11490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. van Vugt M. A., Gardino A. K., Linding R., Ostheimer G. J., Reinhardt H. C., Ong S. E., Tan C. S., Miao H., Keezer S. M., Li J., Pawson T., Lewis T. A., Carr S. A., Smerdon S. J., Brummelkamp T. R., Yaffe M. B. (2010) A mitotic phosphorylation feedback network connects Cdk1, Plk1, 53BP1, and Chk2 to inactivate the G(2)/M DNA damage checkpoint. PLoS Biol. 8, e1000287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kamakari S., Roussou A., Jefferson A., Ragoussis I., Anagnou N. P. (2005) Structural analysis and expression profile of a novel gene on chromosome 5q23 encoding a Golgi-associated protein with six splice variants, and involved within the 5q deletion of a Ph(-) CML patient. Leuk. Res. 29, 17–31 [DOI] [PubMed] [Google Scholar]
- 45. Seki A., Coppinger J. A., Jang C. Y., Yates J. R., Fang G. (2008) Bora and the kinase Aurora a cooperatively activate the kinase Plk1 and control mitotic entry. Science 320, 1655–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Daub H., Olsen J. V., Bairlein M., Gnad F., Oppermann F. S., Körner R., Greff Z., Kéri G., Stemmann O., Mann M. (2008) Kinase-selective enrichment enables quantitative phosphoproteomics of the kinome across the cell cycle. Mol. Cell 31, 438–448 [DOI] [PubMed] [Google Scholar]
- 47. Ma Z. Q., Tabb D. L., Burden J., Chambers M. C., Cox M. B., Cantrell M. J., Ham A. J., Litton M. D., Oreto M. R., Schultz W. C., Sobecki S. M., Tsui T. Y., Wernke G. R., Liebler D. C. (2011) Supporting tool suite for production proteomics. Bioinformatics 27, 3214–3215 [DOI] [PMC free article] [PubMed] [Google Scholar]