Abstract
Liquid chromatography coupled to tandem mass spectrometry in combination with stable-isotope labeling is an established and widely spread method to measure gene expression on the protein level. However, it is often not considered that two opposing processes are responsible for the amount of a protein in a cell—the synthesis as well as the degradation. With this work, we provide an integrative, high-throughput method—from the experimental setup to the bioinformatics analysis—to measure synthesis and degradation rates of an organism's proteome. Applicability of the approach is demonstrated with an investigation of heat shock response, a well-understood regulatory mechanism in bacteria, on the biotechnologically relevant Corynebacterium glutamicum. Utilizing a multilabeling approach using both heavy stable nitrogen as well as carbon isotopes cells are metabolically labeled in a pulse-chase experiment to trace the labels' incorporation in newly synthesized proteins and its loss during protein degradation. Our work aims not only at the calculation of protein turnover rates but also at their statistical evaluation, including variance and hierarchical cluster analysis using the rich internet application QuPE.
In contrast to the genome, which constitutes a rather static entity, the transcriptome as well as the proteome of a cell are highly dynamic, and depend on an organism's developmental stage and its environment. Because of the short lifetime of messenger RNA molecules, microarray studies provide a detailed picture of the current rate of gene expression, and thereby, one might assume, function as indicator for cellular protein amounts. However, when the abundances of proteins in a sample are directly measured and compared with transcriptome data, indeed the observed correlations are far from perfect with reported Pearson's correlation coefficients (R2) ranging between 0.09 and 0.87 with an average of approximately R2 = 0.4 (1–2). The reason for this discrepancy is simple: two important and opposing cellular processes—synthesis and degradation—dictate each protein's level in the cell. On the one hand, sequences of amino acids are assembled at the ribosome, whereby the rate of protein synthesis depends, inter alia, on mRNA expression, the availability of tRNA molecules, and the modulation of gene expression at the post-transcriptional level by small noncoding RNAs (3, 4). On the other hand, proteins are intracellularly degraded, controlled by multiple factors that determine this rate of degradation, e.g. the N-terminal's residue of a protein (5) or the C-terminal peptide tail (6–8).
One way to monitor protein synthesis is to impulsively introduce a label in the form of an essential nutrient in an organism or a cell, and then to chase the label's incorporation in newly translated proteins. Already in the late 1940s, Sprinson and Rittenberg employed 15N-labeled glycine as a diet to measure the utilization of nitrogen for protein synthesis (9). Using 35S-labeled methionine, Hecker and colleagues implemented such a pulse-labeling in combination with two-dimensional gel electrophoresis to compare the amount of total to newly synthesized protein (10, 11). Similar to protein synthesis, degradation can be investigated by setting the protein amounts before and after an induced pulse into relation. In this manner, Pratt et al. (12) used stable isotope labeling by amino acids in cell culture (SILAC)1 (13) and matrix assisted laser desorption/time-of-flight (MALDI-TOF) mass spectrometry to determine degradation rates for approx. 50 proteins in glucose-limited yeast cells grown in an aerobic chemostat at steady state. In a similar experiment Doherty et al. (14) used liquid chromatography/tandem mass spectrometry (LC-MS/MS) and 13C6-Arginine “to profile the intracellular stability of almost 600 proteins from human A549 adenocarcinoma cells.”
Jayapal et al. (15) combined both SILAC as well as the chemical labeling method iTRAQ (16) to estimate both protein synthesis and degradation rates in Streptomyces coelicolor. The transfer of 13C615N4-arginine-labeled cells into unlabeled medium allowed the tracing of newly synthesized proteins. Moreover, iTRAQ was utilized to tag all SILAC-labeled proteins at four time points after the chase, which in turn made it possible to monitor protein degradation.
Certainly, the highest possible labeling efficiency is provided by metabolic labeling with stable isotopes such as 13C or 15N. This enables the analysis of the entire proteome of an organism (17–20) as the success of the labeling does not depend e.g. on a specific amino acid that has to be present in an investigated peptide. In contrast to SILAC, it is moreover not necessary to make sure that the targeted organism is auxotrophic for a specific amino acid (21). The strategy, however, has one significant drawback: a peptide that contains an unknown number of heavy isotopes obviously is also subject to an unknown mass shift. Haegler et al. (22) proposed one of the first software tools to estimate this mass shift for partially labeled peptides. They introduced QuantiSpec, which is designed for the relative quantification of 14N to 15N peptide pairs measured by MALDI-TOF mass spectrometry. Recently, members of the same institute published ProTurnyzer (23), which facilitates the analysis of LC-MS/MS data in a high throughput manner. The Java-based software has particularly been designed for the quantification of samples that reveal such a low incorporation of heavy stable isotopes that, in principle, only the monoisotopic peak can certainly be assigned to an unlabeled peptide. All other peaks are, on the opposite, expected to be influenced by both the labeled and the unlabeled variant. Guan et al. (24) devised a further approach that constitutes an extensive pipeline for the calculation of protein turnover rates from 15N-labeled samples. Their algorithm was successfully used by Price et al. (25) to obtain turnover rates for the impressive number of 2,500 proteins from mice, which were fed with a diet of 15N-labeled algae. The comprehensive experiment included three different tissues from liver, blood, and brain. The method, however, has one drawback that complicates its unrestricted transfer to other experiments. It is required that the samples are highly comparable with respect to their retention time—a precondition that is difficult to be fulfilled in every experimental setup. For organisms that have a comparably fast protein turnover, which is especially the case for bacteria, it is safe to assume that in all cases either a fully labeled or a fully unlabeled peptide is available. This can then be used for protein identification. It is, therefore, possible to analyze each sample on its own, and hence not necessary to ensure highly stable retention times.
The work of Guan et al. (24) shows that there is a strong need for data and analysis pipelines to determine the components of protein turnover. Aiming at the calculation not only of synthesis but also degradation ratios, we extended the idea of metabolic labeling with stable isotopes, and utilized not only 15N but also 13C as traceable markers. We, therefore, developed a new approach to gain these protein turnover ratios from isotopically labeled LC-MS/MS data in a high-throughput manner which is, first, well suited for fast-growing organisms such as bacteria and, second, does not impose any restrictions on sample handling and chromatographic setup. Moreover, it was our aim to provide an integrated, user-friendly, and instantly accessible software solution, which allows not only the calculation of synthesis and degradation ratios but also their in-depth statistical evaluation.
EXPERIMENTAL PROCEDURES
Growth and Cell Lysis of C. glutamicum
Strain ATCC 13032 served as Corynebacterium glutamicum wild type. C. glutamicum cells were grown either in Brain Heart Infusion (BHI) medium (Roth, Karlsruhe, Germany) or in the minimal media MMI (26) with 4% glucose, as well as MME-SN, a slightly modified medium of MMES from Koch et al. (27) that is used for cultivation with 15NH4Cl or 13C glucose. Cultivation was done in shaking flasks at 30 °C or 40 °C and 145 r.p.m. (Certomat MO, B. Braun Biotech International, Melsungen, Germany). The cell lysis was carried out as follows: The complete 100 ml culture of a shaking flask was harvested by centrifugation at 3700 × g and 4 °C for 10 min, washed once with precooled 1 × PBS buffer (137 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, 1.8 mm KH2PO4, pH 7.4 adjusted with HCl) and resuspended in 20 ml PBS lysis buffer. The lysis buffer consisted of PBS, 1 ml protease inhibitors (Sigma Aldrich, Germany, product number P8465) per 6 g of cells, 200U DNaseI per ml buffer, 10 mm MgCl2 and 5 mm MnCl2. The disruption of C. glutamicum cells and the membrane preparation were done according to Haussmann et al. (28). The supernatant that contains the cytosolic proteins was decanted and stored at −70 °C.
For validation of the quantification procedure C. glutamicum cells were precultured in 10 ml BHI medium at 30 °C for approx. 8 h. Afterward, three different overnight cultures in 50 ml MME-SN containing 5 g/L NH4Cl were prepared with: (a) ∼100% 14NH4Cl, (b) 55% 14NH4Cl/45% 15NH4Cl, and (c) 30% 14NH4Cl/70% 15NH4Cl. Next day, these precultures were used for inoculation of three different shaking flasks containing 100 ml MME-SN with 1 g/L NH4Cl in the above mentioned 14N/15N ratio to an OD600 of one. After reaching the mid-exponential growth phase (OD600 ∼11) the complete samples were taken for cell lysis.
To analyze protein turnover both for 30 °C and 40 °C four biological replicates were investigated. For all experiments C. glutamicum cells were precultered in 10 ml BHI medium at 30 °C for approx. 8 h and subsequently used for inoculation of 50 ml MME-SN medium, containing only 15NH4Cl as nitrogen source. After approx. 16 h, the culture was used to inoculate four shaking flasks each containing 100 ml fresh MME-SN medium with 15NH4Cl to an OD600 of one. The cells were cultivated to log phase (OD600 6 - 8). After 285 min the cultures were harvested by centrifugation at RT for 10 min and 5800 × g, and the cells were washed once with 50 ml MMI containing 14N nitrogen sources. Subsequently, for setting the pulse the cells were resuspended in 100 ml pre-warmed 14N-MMI (30 °C or 40 °C) and cultivated either at 30 °C or at 40 °C to perform the heat-shock. Samples were taken 30, 60, 120, and 210 min after inoculation of the fresh media.
Additionally, a 13C internal standard was generated to calculate the proteins' degradation. According to the above mentioned protocol, C. glutamicum was precultured in BHI medium and this culture was used to inoculate MME-SN medium containing 2.5% 13C glucose as carbon source and additionally 100 μm protocatechuate instead of citric acid as well as Na2CO3. Next day, the cells were transferred to fresh MME-SN medium containing 13C glucose and incubated at 30 °C. After reaching the mid-exponential growth phase a sample was taken and the temperature was shifted to 40 °C. After 60 min a second sample was taken and after washing the cells with PBS buffer the protein preparation was carried out as described.
Digestion Conditions
The cytosolic fractions were processed using the FASP II protocol (29) or directly. Directly means that after inactivation of the protease inhibitor (60 °C, 1 h) 100 μg of soluble proteins were incubated over night at 60 °C with 4 μg sequencing grade trypsin (Promega, Madison, WI). For the direct analysis of the membrane fraction samples comprising 200 μg protein were washed twice with 100 mm ammonium carbonate to remove soluble proteins. The resulting pellet was dissolved in 60 μl methanol by ultrasonication and subsequently 40 μl 25 mm ammonium bicarbonate buffer (pH = 8.6) was added. Afterward, 2 μg sequencing grade trypsin was added and the preparation was incubated at 37 °C over night. After removal of membranes by centrifugation (100,000 × g; 4 °C; 35 min) the supernatants were used for analysis. All samples were desalted by using Spec PT C18AR solid phase extraction pipette tips (Varian, Lake Forest, CA).
For the validation experiment only the cytoplasmic fractions were used and digested according to the FASP II protocol. Therefore, sample “a” (with ∼100% 14N) was pooled with sample “b” (55% 14N/45% 15N) as well as sample “c” (30% 14N/70% 15N) in the ratios 6:1, 1:1, and 1:6 resulting in 100 μg total protein amount.
To analyze the protein turnover four biological replicates were investigated at the four distinct time points either grown at 30 °C or at 40 °C. Both the cytoplasmic and the membrane proteome were investigated. Cytoplasmic fractions were digested directly.
For the calculation of protein degradation using the 13C internal standard only the cytoplasmic fractions were analyzed. Fifty micrograms of the 13C standard were spiked to 50 μg of the soluble fraction of the pulsed samples (including 14N as well as 15N). Subsequently, these samples were digested directly.
Protein Identification Via 1D-nLC-ESI/MS
All desalted samples were resuspended in buffer A (0.1% formic acid in water) and subjected to 1D-nLC-ESI-MS/MS using an autosampler. An UPLC BEH C18 column (1.7 μm, 75 μm x 150 mm, Waters, Milford, MA) and an UPLC Symmetry C18 trapping column (5 μm, 180 μm x 20 mm, Waters) for LC as well as a PicoTip Emitter (SilicaTip, 30 μm, New Objective, Woburn, MA) were used in combination with the nanoACQUITY gradient UPLC pump system (Waters) coupled to an LTQ Orbitrap XL (analyzing both the validation and synthesis samples) or LTQ Orbitrap Velos (analyzing the degradation samples) mass spectrometer (Thermo Fisher Scientific Inc., Waltham, MA). The analytical column oven was set to 45 °C. For elution of the peptides a multiple step gradient of buffer A to buffer B (0.1% formic acid in acetonitrile) was applied (0–5min: 1% buffer B; 5–10min: 5% buffer B; 10–175min: 40% buffer B; 175–200min: 99% buffer B; 200–210min: 1% buffer B) at a flow rate of 0.4 μl/min and a spray voltage of 1.5–1.8 kV. The Orbitrap XL was operated by instrument method files of Xcalibur Rev. 2.0.7, the Orbitrap Velos by instrument method files of Xcalibur Rev. 2.1.0. The linear ion trap and Orbitrap were operated in parallel, i.e. during a full MS scan on the Orbitrap in the range of 300–2000 m/z (XL) or 150–2000 m/z (Velos) at a resolution of 60,000, MS/MS spectra of the four (XL) or 10 most (Velos) intense precursors were detected in the ion trap. The heated desolvation capillary was set to 200 °C (XL) or 275 °C (Velos). The relative collision energy for collision-induced dissociation was set to 35%. Dynamic exclusion was enabled with a repeat count of one and a 1 min exclusion duration window. More than triply charged ions and ions of unknown charge state were rejected from MS/MS.
Database Searches
All searches were performed in a C. glutamicum database (30) using the SEQUEST algorithm, either embedded in BioworksTM (Rev. 3.3.1 SP1, Thermo Electron © 1998–2007), according to Haussmann et al. (28), or embedded in ProteomeDiscoverer 1.2.0.208 (Thermo Electron © 2008–2010). Additionally, for the determination of the false discovery rate a reversed database generated in BioworksTM Browser was used. Separate searches were performed for each raw file according to the utilized labels 14N, 15N, and, if appropriate, 13C. Because of the selected enzyme, only tryptic peptides with up to two missed cleavages were accepted. No fixed modifications were considered. Oxidation of methionine was permitted as variable modification. The mass tolerance for precursor ions was set to 10 ppm (XL) or 6 ppm (Velos); the mass tolerance for fragment ions was set to 1 amu (XL) or 0.8 amu (Velos).
For protein identification using BioworksTM software (Thermo Electron) DTA and OUT files were generated from the 14N, the corresponding 15N, and, if appropriate, the 13C search results-(srf)-files. All including hits were further filtered using the software DTASelect 1.9 (31) with the following parameters: minimum ΔCn was set to 0.08, minimum XCorr was set to 2.5 (+2), 3.5 (+3), duplicate spectra for each sequence were retained (-t 0) and at least two different peptides per protein were required. These filter criteria were chosen in such a way that using a reversed database a false discovery rate of 0% was achieved (32). For protein identification using ProteomeDiscoverer (Thermo Electron) the mass spec format-(msf)-files were filtered with peptide confidence “high” and two peptides per protein. ExcelTM files (Microsoft Corporation, Redmond, WA, USA) were generated from the 14N and the corresponding 15N msf-files to be used subsequently in QuPE.
Quantification Procedure
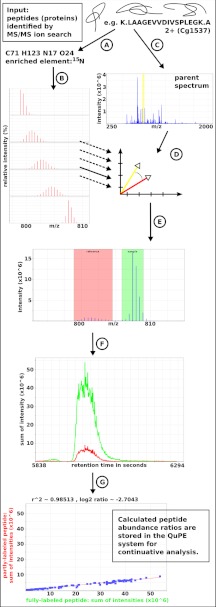
To determine the components of protein turnover, an algorithm had to be developed that aims at the calculation of the ratio between the abundances of two differentially labeled peptides, first, a fully 15N-labeled and a partially labeled peptide, i.e. synthesized either before or after the pulse-chase, second, two peptides that are either labeled with 13C or 15N. The overall idea is to compute extracted ion chromatograms (XICs) for each of the two isotopologous peptides, which are then compared with each other. Therefore, several computational steps are necessary, which are described in the following section (see Fig. 1 for a workflow of the algorithm).
Fig. 1.
Workflow of the algorithm: A, Required input are peptides identified by an MS/MS ion search along with information about their amino acid sequence, charge state, modifications and the associated protein accession number. B, Based on each peptide's molecular composition theoretical isotopic distributions are computed for varying rates of 15N incorporation. C, The parent spectrum is retrieved and D, both, the theoretical as well as the “real” isotopic distributions are compared using the dot product. E, The best match of a theoretical distribution (e.g. at 30% enrichment) determines the m/z ranges for the extraction of ion chromatograms (XIC) for each the partly- and the fully-labeled peptide (F). Both XICs are compared by perpendicular linear regression (G), where the slope of the regression line leads to the peptide abundance ratio (e.g. –2.704).
Required input of the algorithm is a list of identified peptides including their sequence, charge state, and any modification observed. Apart from the associated protein accession number further information comprises the sample or run the peptides were found in as well as their spectrum and retention time. The algorithm was implemented using the QuPE API (33), which provides comprehensive methods to, on the one hand, import and access all required data in a variety of formats, and on the other hand, to store the computational results for continuative analysis. The API facilitates a modular design of the algorithm, and enables the processing of this computationally intensive task on a compute cluster.
Inherent to our applied pulse-chase approach with 15N is the idea that in the same spectrum one peptide will always be found fully labeled, at least at a fixed enrichment rate of about 98%, whereas its partner is only partly labeled at an unknown rate of enrichment. The crucial task is to determine this incorporation level.
In most cases only one of the two peptides has been identified by an MS/MS scan, which is also the reason why, in contrast to techniques such as iTRAQ (16), the full MS scan has to be used for quantification (Fig. 1C). For the construction of XICs, it is however crucial to know the m/z positions of both peptides. Computation therefore starts with the calculation of a range of theoretical isotope distributions for each observed peptide. Based on the sequence, charge state, and any present modification the molecular composition of each peptide is determined (Fig. 1A). First, isotopic distributions are calculated (34) using the naturally occurring atomic weights and isotope probabilities (35), second distributions are calculated for variable rates of enrichment of the isotope that has been used as label, e.g. starting at a high rate of 15N and ending with a distribution in which almost all 15N isotopes are replaced by their light counterpart (Fig. 1B).
Following the recommendations of Stein and Scott (36), who investigated mass spectral library search algorithms, the scalar product has been chosen to determine the similarity between any theoretical isotope distribution and the peptide's associated mass spectrum (Fig. 1D). Assuming a spectrum S consists of p discrete peaks, each described by its m/z value m and intensity i, formally m = {m1 … mp} and i = {i1 … ip}, and analogously, q peaks belong to a calculated theoretical distribution T, ranging from the lowest m/z value μ1 to the highest m/z value μq with intensities ι, a similarity is computed as dS × T = arccos = arccos , wherein ı̃ is derived from i to first hold the necessary condition ‖ı̃‖ = ‖ι‖, and second to reduce noise, e.g. from overlapping peptides. Using a small-sized value ε, for all peaks of T with m/z values μ = {μ1 … μq}, a new vector of intensity values ĩ = {ı̃1 … ı̃q} is defined with each ı̃j = ∑ik, ∀k : μj − ε ≤ mk ≤ μj + ε, j ∈ {1 … q}. Calculated for any theoretical isotopic distribution the highest value of dS × T determines the rate of enrichment. This is also used to verify that a peptide has correctly been identified: If the theoretical isotope distributions do not match the given mass spectrum, i.e. all calculated similarities fall below a threshold, the peptide is omitted from further calculation.
The gained knowledge about the m/z values of each of the two peptides directly leads to their intensities, and the intensities' ratio to a measurement of relative abundance (Fig. 1E). For quantification the monoisotopic or the most abundant peak of the isotopic distribution may be used. However, as isotopic patterns of differentially labeled peptides vary heavily in their form (see Fig. 2 and supplemental Fig. S4 for an example), in this approach the complete isotopic distribution (with all intensities above a distinct threshold) is considered instead.
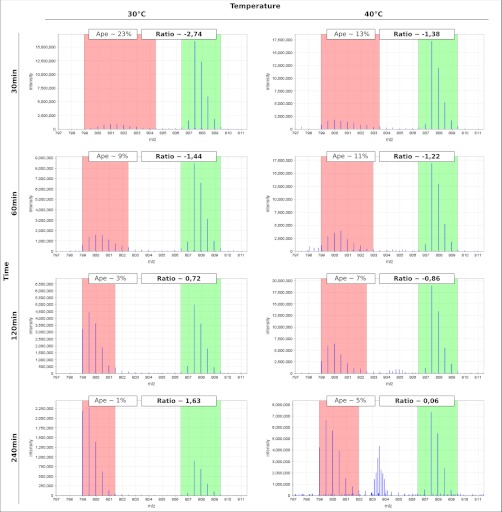
Fig. 2.
The Figure displays selected MS level one spectra of the peptide K. LAAGEVVDIVSPLEGK.A (charge 2+) from the protein PtsG (Cg1537), a component of the phosphotransferase system. Calculated incorporation rates (ape) of the partly labeled peptide and the ratio of the abundances of both peptides are reported in the boxes above each spectrum plot. It can be seen that in the time span after the 14N pulse more and more protein is synthesized that does not fully incorporate the heavy stable isotope of nitrogen. As a clear difference between the samples taken at 30 °C and those taken at 40 °C the turnover rate is significantly slower when the bacteria are exposed to heat stress. The colors mark the two m/z ranges that are used for the extraction of ion chromatograms for the partly (red) and fully (green) labeled peptide.
Certainly, it is expected that a peptide does not only elute at the time point at which it was detected, but possibly within a distinct time interval. Based on the now given exact theoretic ion masses of each of the two peptides and using the window size ε, XICs are computed for both the partially and the fully labeled peptide (Fig. 1F). To calculate a ratio between the abundances either the area under the two elution peaks, one for each peptide, may be integrated and compared, or alternatively, MacCoss et al. (37) and others (38, 39) proposed a linear regression approach for this purpose. Beforehand, however, it is necessary to find the first and last time point a peptide eluted at the borders of the peptide's elution peaks in the two XICs. Yang, He, and Yu (40) found an algorithm based on continuous wavelet transform having the best performance for the purpose of peak detection in chromatographic data. In our application, this is furthermore limited to the most abundant peak at a time point close to the retention time of the given peptide. The algorithm of Du et al. (41) utilizes a Mexican Hat wavelet, as it approximately describes the form of a peak. Given a time point τ the wavelet function ψ (τ) is defined as ψ (τ) = . If furthermore an XIC is defined as a function of retention time, c (τ), a continuous wavelet transformation can be defined as , where a is a scaling factor to shrink or stretch the width of the wavelet, in analogy to the width of a peak. Convolution operations are conducted for a range of scales, each resulting in a list of wavelet coefficients. The maximal observed coefficient indicates the apex of an eluting peptide's peak, and the borders of the peak can be deduced from the corresponding scaling factor and the roots of the folded function.
Finally, the ion current ratio is estimated using the resulting elution peak borders. The sections of the two chromatograms c1 (τ) and c2 (τ), attributable to the fully and the partially labeled peptide, are plotted against each other (Fig. 1G). A function c1 (τ) = a + bc2(τ) is fit to the data (‖c1‖ = ‖c2‖). The slope of the regression line b gives an estimate of the ratio of the abundances. Instead of vertical offsets, which are commonly used in least squares fitting, perpendicular offsets were utilized to allow for uncertainties of the data points along both axis. Hence, the term needs to be minimized. Calculated abundance ratios that exceed a given threshold of r are then stored in the QuPE database. In addition, for each ratio a signal-to-noise (S/N) value is computed that sets the overall peak intensity in relation to the mean signal intensity in a range before and after the detected peak borders.
RESULTS
Aiming at a proteome-wide applicable method to analyze the components of protein turnover—protein synthesis and degradation—we designed and implemented a pulse-chase experiment based on a multilabeling strategy that incorporates both 15N as well as 13C. To this end, it was necessary to develop an algorithm that allows comparing the abundances of two differentially labeled peptides, i.e. a partially labeled peptide and its fully labeled or unlabeled counterpart. Furthermore, it was our aim not only to determine the changes in total protein turnover but also to provide in-depth statistical analyses and a thorough evaluation strategy for this new type of data. Often neglected by other software solutions, we placed particular emphasis on the accessibility and usability of our solution. Our implemented quantification procedure is, therefore, integrated into a web browser-based application, the rich internet application QuPE (33). It is, hence, directly accessible to the public without the need of installation, and offers fellow researchers, from various disciplines, to gain detailed insight into a rather underdeveloped dimension in proteomics.
To demonstrate our approach we analyzed the effect of heat-stress in the Gram-positive, nonpathogenic soil bacterium Corynebacterium glutamicum, employing an extensively investigated cellular adaptation process that is easy to implement as environmental stressor (42). C. glutamicum was, in particular, selected because of its high labeling efficiency and the presence of an established labeling protocol employing 15NH4Cl (28).
Validation of the Quantification Procedure
We performed a benchmark experiment to test the accuracy and validity of our implemented quantification procedure. Therefore, samples of C. glutamicum were prepared with different incorporation rates of stable nitrogen isotopes (see materials and methods for details), and mixed in distinct ratios. Overall six data sets were analyzed combining in ratios of 1:1, 1:6, and 6:1 each one sample with natural abundances of nitrogen isotopes and one sample having been labeled with either 45% or 70% 15N. Detailed quantification results can be found in Table I. For all six data sets the calculated incorporation rates match the employed enrichments of 15N. Despite a small but systematic bias of about M = 0.5, which has presumably been introduced during sample preparation, the calculated abundance values adequately reflect the true ratios of the data. In a real world experiment, this error would need to be compensated for, e.g. by normalization of the data. In summary, the results verify that our implemented procedure allows gaining abundance ratios of a partially labeled peptide in relation to a fully unlabeled peptide.
Table I. Summarizes the quantification results achieved with our implemented algorithm on a benchmark experiment consisting of six different data sets. Each two samples were mixed in distinct ratios of 1:1, 1:6, and 6:1. While one sample was unlabeled, i.e. with natural occurring nitrogen isotopes, one sample was partially-labeled with a fixed incorporation rate of 15N—either 45% or 70%. The first three columns of the table demonstrate the employed ratio and ape, as well, the expected ratio for each sample mixture. In the following column, the overall number of quantified proteins is printed. Determined incorporation rates (mean, standard deviation, median) can be found in the next three columns, followed by the calculated ratios' mean value, standard deviation, and median.
| Employed ratio | Employed ape | Expected ratio | Number of quantified proteins | Calculated ape (mean) | Calculated ape (sd) | Calculated ape (median) | Calculated ratio (mean) | Calculated ratio (sd) | Calculated ratio (median) |
|---|---|---|---|---|---|---|---|---|---|
| Ratio 1:1 | 0.45 | 0 | 160 | 0.45 | 0.068 | 0.44 | 0.46 | 0.51 | 0.41 |
| Ratio 1:6 | 0.45 | −2.585 | 54 | 0.44 | 0.024 | 0.44 | −2.26 | 0.39 | −2.28 |
| Ratio 6:1 | 0.45 | 2.585 | 117 | 0.49 | 0.133 | 0.44 | 3.12 | 0.80 | 3.22 |
| Ratio 1:1 | 0.70 | 0 | 191 | 0.69 | 0.053 | 0.68 | 0.66 | 0.60 | 0.65 |
| Ratio 1:6 | 0.70 | −2.585 | 73 | 0.68 | 0.019 | 0.68 | −2.21 | 0.54 | −2.21 |
| Ratio 6:1 | 0.70 | 2.585 | 122 | 0.70 | 0.096 | 0.68 | 3.22 | 0.96 | 3.40 |
Application Study: Investigation of Heat Shock Response in C. glutamicum
To demonstrate the applicability of our newly developed method we designed and performed two experiments investigating the response of C. glutamicum to heat-shock (Fig. 3). First, targeting protein synthesis, samples were grown in 15N-enriched medium and transferred to unlabeled medium. In summary, four biological replicates were investigated at 30 °C and 40 °C. Including a membrane as well as a cytoplasmic fraction this comprises in total 60 runs. A second experiment with 30 runs was conducted to analyze protein degradation. Here, we extended the idea of the first experiment and grew—in addition to the pulse-chase approach with nitrogen isotopes—C. glutamicum in 13C-enriched medium. The extracted proteins of this 13C-labeled internal standard were then spiked to each of the four biological replicates of our turnover experiment. In this second experiment, only the cytoplasmic fraction was analyzed.
Fig. 3.

The growth curves of C. glutamicum incubated at 30 °C (open squares, □) or 40 °C (open triangles, ▿) as well as the time point of the 14N pulse and the heat shock, respectively, are shown. The time-points of the samples for the proteome analysis are also indicated (arrows with time specification). The samples were taken 30, 60, 120, and 210 min after the pulse. Error bars indicate the standard deviation.
To have a detailed look at our experimental results, all data is available online in the rich internet application QuPE (see supplemental material S1 for a brief guide on using the software).
Analysis of Protein Synthesis
Protein Identification
From 3058 protein coding regions that have been predicted for the genome of C. glutamicum ATCC 13032 (30), we could identify in total 805 proteins in all 60 MS runs of the first experiment. This covers about 25% of the whole proteome including a high fraction of membrane proteins. For each run, first, a database search was configured to identify all fully 15N-labeled peptides. As the precultivation was carried out in minimal medium containing only this heavy isotope of nitrogen, each protein already existing before the pulse was expected to be identified in this search. Second, newly synthesized 14N-labeled peptides were searched for (see Material and Methods for further details). Starting with the time point of the 14N pulse initiation, the newly synthesized proteins should incorporate an increasing amount of this light isotope. In this connection, it has to be denoted that the utilized database search engine does not support the identification of partially labeled peptides. This is reflected by the number of proteins found in the database searches as shown in supplemental Fig. S1. Although approximately equal numbers of identifications were reached in both the 30 °C and the 40 °C approach, only at the later time points and, furthermore, mainly for the lower temperature 14N-labeled peptides could have been identified. Whereas, for example, in the membrane fractions of the 30 °C grown cells, 67 and 150 proteins were identified after 120 and 210 min, respectively, only four and 22 proteins were detectable in the corresponding samples of the heat-shocked cells. Although the cells incubated at 40 °C and 30 °C reach the same OD 120 min after the pulse/heat-shock (Fig. 3), it can thus be assumed that synthesis of proteins is slowed down at the elevated temperature.
In Fig. 2 selected mass spectra of a peptide of the protein PtsG are presented. In all cases, the heavy isotopolog of this peptide is particularly well evident, with a high intensity observed, in particular, for the first two samples taken after 30 and 60 min, respectively. In contrast, the intensity of the light isotopolog is constantly increasing over time, whereas the detected incorporation of 15N (ape) is decreasing.
Quantification Results
Protein synthesis rates were calculated based on each protein's abundance ratio of newly (partly 14N-labeled) to formerly (fully 15N-labeled) synthesized protein amounts. Aiming to ensure a high quality of the results, the algorithm was configured with rather strict parameters (r > 0.6, similarity > 0.9, S/N > 3.0), which led to the successful quantification of 698 proteins representing approx. 85% of the protein identifications.
In order to get an overview of the data, we first investigated the distribution of all calculated abundance ratios at each of the two temperatures over time. Using box- and whisker plots, median, lower, and upper quartiles, and extreme values of the data are visualized in Fig. 4, and allow to get an impression of the overall synthesis rates at each of the two temperatures. A two-factorial (time, temperature) analysis of variance (ANOVA) confirmed that protein abundances differ significantly (p < 0.001) between 30 °C and 40 °C. To further quantitatively measure the synthesis rates separately for both temperatures, linear regression analyses using time after pulse as the independent variable were performed on all values. At the lower temperature, it was found a significant raise in protein synthesis (p < 0.001) with a regression coefficient of about β = 1.4 compared with a significant lower increase (p < 0.001) of only β = 0.82 at the higher temperature. This observation is also supported by the determined incorporation rates of 15N as summarized in supplemental Fig. S2. After the pulse, the ratio of 14N to 15N is considerably faster increasing in newly synthesized peptides at 30 °C than in the cells affected by the heat shock, and in the latter case, even after 210 min, this ratio has still not reached 100%.
Fig. 4.
An overview of all protein synthesis ratios. The values are measured as the relative abundances between the partly 14N-labeled peptide of a protein and its fully 15N-labeled counterpart. It is observable that for both temperatures proteins are steadily synthesized whereas the rate is higher for 30 °C. Using linear regression these were determined as β = 0.82 for 40 °C compared with β = 1.4 for 30 °C.
Aiming to detect those proteins that are differentially synthesized because of heat shock, next a two-factorial (time, temperature) ANOVA was conducted per protein as described in Albaum et al. (43). As this includes several hundreds of individual significance tests, resulting p values were adjusted using the method of Holm (44) to account for this multiple testing situation and to give control of the family wise error rate. To further ensure the validity of our results, it was additionally tested whether the prerequisites to unreservedly interpret the results of the ANOVA were fulfilled. Therefore, a Fligner-Killeen test (45) was performed on all protein measurements to reveal inhomogeneous variances as well as a Shapiro-Wilks test (46) to verify the normal distribution assumption. In summary, 95 of initially 152 proteins complied with these strict requirements and can therefore be regarded as significantly differentially synthesized regarding the factor temperature (p ≤ 0.05). Among these proteins are several heat-shock-related as well as ribosomal proteins (see supplemental Table S1).
As our approach measures synthesis rates of proteins, one may assume a moderate to strong correlation to corresponding transcriptome data. Barreiro et al. (47) conducted a microarray study to analyze the response of C. glutamicum to moderate (and severe) heat shock. In their experiment, bacteria were grown at 30 °C and shifted to 40 °C after 60 min. To investigate the agreement between our measured synthesis ratios and the mRNA abundances determined by this microarray experiment, we utilized Pearson's correlation coefficient. However, as the microarray signal intensities directly set the transcript abundances at moderate heat into relation to normal temperature conditions, it was necessary to transform our data: Let p30 be our ratio calculated at 30 °C and p40 the corresponding value at 40 °C, then for each of the four points in time an overall ratio p was calculated as . The highest correlation of r = .58 between all proteins and genes found in both the transcriptome and the proteome data set was observed with the protein synthesis ratios recorded 60 min after the pulse. Interestingly, a correlation coefficient of r = .80 was determined, if only significantly differentially synthesized proteins are taken into account.
Cluster Analysis
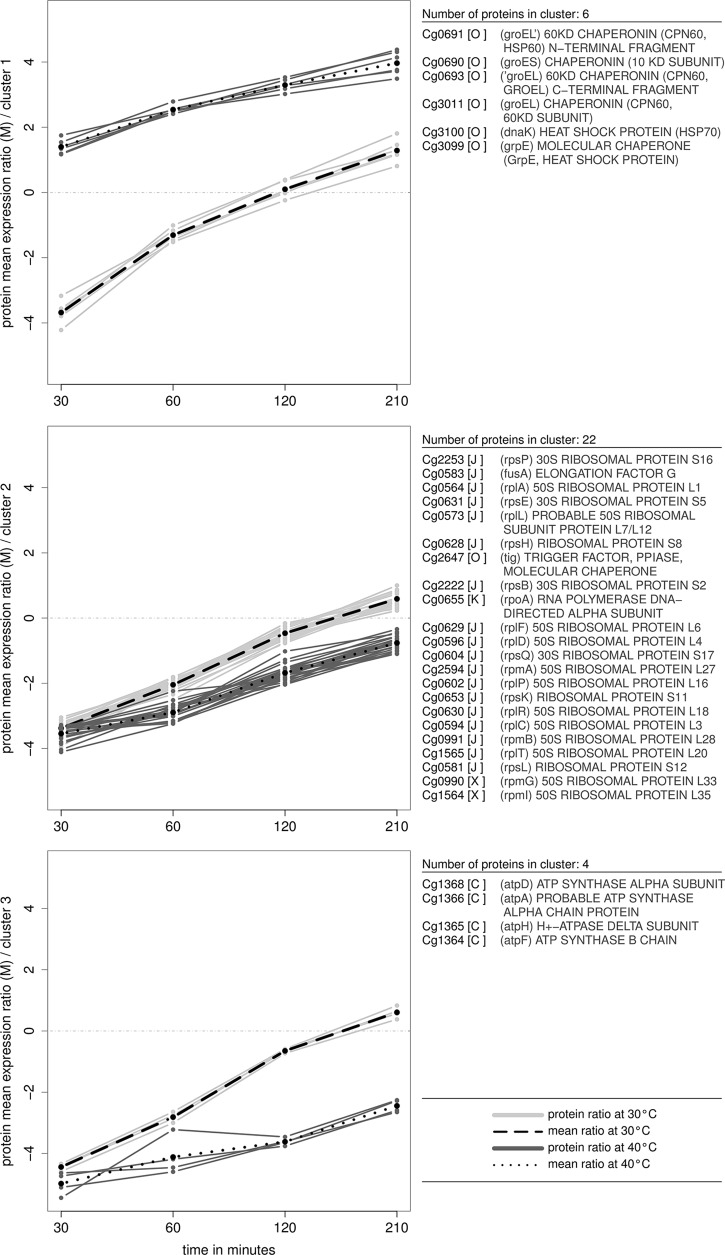
The next interesting question was whether there are groups of proteins that show similar synthesis ratios over time. Therefore, a cluster analysis was performed on the data. For this purpose, we determined the average synthesis ratios for each protein at each of the eight different temperature/time combinations using the arithmetic mean. In this connection, it has to be denoted that because of missing values a synthesis ratio could not be acquired for each single time step and temperature. It was decided, to include all proteins in the analysis having for at least six of the eight combinations two or more peptide synthesis ratios available. Missing values were then replaced by the protein's overall mean synthesis ratio. The Figure of Merit (48) provides a mean to assess the predictive power of different cluster algorithms, and revealed Ward's hierarchical cluster algorithm using Euclidean distances as the method (49) that best predicts the correct number of clusters in the data set. The results—238 proteins satisfied the above mentioned criteria—are displayed in the form of a heat map (see supplemental Fig. S3). Additionally, we employed the cluster index of Krzanowski and Lai (50) to determine the clustering that, at best, groups strongly similar proteins in the same cluster whereas the clusters themselves are different (43). This was found in 36 clusters. In Fig. 5, three of these clusters are shown. Interestingly, the heat shock proteins (HSPs) GroEL, GroES, and DnaK cluster together, showing a considerably higher synthesis rate at 40 °C over time (Fig. 5, cluster 1). Compared with 30 °C, synthesis ratios are already notably increased directly at the beginning of the sampling, which means 30 min after the pulse. Furthermore, ATPase subunits and actors of the translationary machinery such as several ribosomal proteins reveal similar synthesis rates—in both cases rates are reduced at 40 °C (Fig. 5, clusters 2 and 3).
Fig. 5.
Shows parts of the result of a hierarchical cluster analysis using Ward's method and Euclidean distances. Calculated peptide abundance ratios were combined per protein by their mean value. Missing values were replaced if for at least six of the overall eight time points enough (at least two) values were present. Using the cluster index of Krzanowski and Lai (1988) a clustering of the data in 23 groups was chosen. Here, three selected clusters are presented. Turnover values regarding the temperature 30 °C are drawn with light grey, values at 40 °C are dark grey. The black, dashed and dotted lines show the mean turnover rates for each temperature. On the right side the proteins belonging to each of the three clusters are listed, COG functional categories (53) are given in square brackets. Interestingly, one cluster (cluster 2) contains only ribosomal proteins (COG functional category J), which show a similar behavior disregarding the temperature. In another cluster (cluster 1) six heat shock proteins clearly indicate an up-regulation at higher degrees, while the third cluster mainly shows down-regulated proteins of the category energy production and conversion.
Analysis of Protein Degradation
Protein Identification
In our second experiment, which in addition to the pulse-chase approach with nitrogen isotopes utilizes a 13C-enriched medium to monitor protein degradation, in total 542 proteins—covering about 17% of the proteome—could be identified using the previously described filter criteria for all four time points and the two temperatures 30 °C and 40 °C.
Quantification Results
Based on the peptide identifications we calculated the differences in abundance between each fully 15N- and fully 13C-labeled peptide to determine protein degradation. Using the same parameters as described above 464 proteins could be quantified in at least one time point.
Similar to the first experiment targeting protein synthesis, we utilized a two-factorial (time, temperature) ANOVA to reveal differentially degraded proteins regarding the temperature. After correcting for the multiple testing situation using the method of Holm and checking the preconditions of this statistical test, only nine proteins (initially 14 proteins) were reported as significantly differentially regulated (p ≤ 0.05) including Cg1368 (ATP synthase), Cg1790 (3-phosphoglycerate kinase), and the dehydrogenase Cg3219 (see supplemental Table S2 for further results).
Being interested in those proteins that show an extraordinary rate of degradation, and putting forward the hypothesis that—at least for C. glutamicum as a fast growing organism—most proteins are stable under normal growth conditions (51, 52), we devised the following procedure: each calculated degradation ratio, the ratio between the abundances of the fully-15N- and the fully 13C-labeled peptide partners, was normalized using the mean degradation ratio of all values observed for the same temperature and time point. The two box- and whisker plots in Fig. 6 show the distribution of all calculated degradation ratios for the two temperatures over time, before (upper plot) and after normalization (lower plot).
Fig. 6.
Allows getting an impression of the overall protein degradation ratios. The values are measured as the relative abundances between the two fully labeled 15N and 13C variants of a peptide. Calculated relative protein degradation ratios were normalized on each time point's median value. It can be noted that the overall rate of protein degradation is relatively low for both temperatures.
Having a look at individual proteins, we first filtered out all proteins with two or more missing measurements at any time point, which resulted in a list of 158 proteins at 30 °C, and 156 proteins at 40 °C. We next utilized linear regression analysis to determine proteins showing surpassing degradation, i.e. having a negative slope of the regression line of β ≤ −0.01. Using this criterion for the temperature 30 °C 50% of all analyzed proteins (= 79) could be identified as potentially degraded, compared with 59.6% (= 93 proteins) at 40 °C (see supplemental Document S2 for detailed results). Proteins that show significantly (p < = 0.05) increased degradation rates at both temperatures include, inter alia, Cg3299 (thiol-disulfide isomerase), and Cg2464 (conserved hypothetical). This was similarity determined for the proteins Cg0924 (ABC-type transport systems), and Cg1290 (methionine synthase II), yet in these two cases protein degradation was much higher at 40 °C. On the whole, it is noticeable that for several proteins a significantly increased degradation could be found in response to the heat shock (e.g. Cg0048). This was, however, rarely observed for the lower temperature.
DISCUSSION
We devised and applied an integrative solution to analyze the components of protein turnover. A pulse-chase approach using heavy stable isotopes of nitrogen as well as of carbon was utilized to trace nitrogen incorporation in newly synthesized proteins and to estimate protein degradation. Instead of merely determining the changes in protein amounts themselves, we could thereby gain detailed knowledge about the dynamic process of protein turnover—once regarded “a missing dimension in proteomics” (12). It was our aim to develop a method that, first, allows proteome-wide synthesis to degradation ratios to be calculated in a high-throughput manner, particularly, for fast-growing organisms such as bacteria. Second, we wanted our algorithm to be fully automatic and require only a minimal level of manual intervention, especially concerning parameters such as the type of labeling. We considered it important that, third, no preconditions were imposed on the labeled samples, e.g. in terms of retention time and MS signal variance. Special emphasis was, fourth, placed on a user-friendly and directly accessible solution that does not require a bioinformatician for its execution. Fifth, we did not want to end with the calculated lists of ratios but, furthermore, test and approve appropriate multivariate statistical analysis methods. In this connection, the ANOVA proves successful for the identification of up- or down-regulated proteins. We propose hierarchical cluster analysis using Ward-linkage and Euclidean distances to find groups of co-regulated proteins. The inclusion of external information turned out to be a key element, as for example proteins of the COG functional category “Post-translational modification, protein turnover, chaperones” (53) showed similar synthesis rates regarding the two temperatures 30° and 40 °C.
Employing a pulse-chase approach using metabolic labeling with 15N or 13C required the consideration that each tryptic fragment of a protein may have an unknown incorporation rate of heavy isotopes. This inherently results in a variable mass shift in the mass spectrometry data, which is not known a priori. Moreover, partly labeled peptides reveal a rather complex isotopic distribution pattern with the monoisotopic peak rarely corresponding to the first peak (see Fig. 2 and supplemental Fig. S4). Although several bioinformatics software applications have been introduced to calculate relative abundance ratios from metabolically labeled samples using stable isotopes, these tools can only process a mixture of both a fully labeled and an entirely unlabeled sample. Whereas not complete, the list includes RelEx (37), ASAPRatio (38), ProRata (39), Census (54), and QN (55). Doping a cell culture with stable isotopes to analyze protein turnover, however, demanded an algorithm that is able to quantify sample mixtures with one peptide counterpart being only partly labeled at an unknown rate of incorporation. In their approach Rao et al. (20) compared the isotope distributions of all identified peptides, and developed a model to estimate the average m/z ranges based on the number of N-atoms in a peptide. Cargile et al. (19) utilized a Poisson distribution model to predict the isotope distribution patterns of labeled isotopes. However, manual estimation of parameters and preprocessing of data is time-consuming, tedious and error-prone— high-throughput experiments obviously demand a fully automated out-of-the-box solution.
Quantispec (22) and ProTurnyzer (23) constitute two solutions to this problem. Whereas the first mentioned has exclusively been created for MALDI-TOF data, the second software tool was, in particular, designed for the quantification of LC-MS/MS samples, which show a low incorporation of heavy stable isotopes. An approach that is similar to our algorithm was recently introduced by Guan et al. (24), which, however, demands samples to be highly comparably with respect to their retention time. As we did not want to guarantee the fulfillment of this precondition in our experiment, the approach was not applicable on our data.
Quantification Procedure
In this work, an integrative algorithm has been developed that targets LC-MS/MS data and includes the temporal dimension of eluting peptides. The algorithm is highly modular in a way that individual components can easily be extended, optimized, or even replaced. This applies, for example, to the calculation of theoretic isotopic distributions. Currently, a polynomial approach is utilized for this purpose. Although the extraction of ion chromatograms is, by far, the most expensive part of the algorithm (depending on the configuration, this takes up to 90% of the overall running time), this implementation could be replaced in a future version of the algorithm by a more efficient method based, for instance, on Fourier transform (56–58) or dynamic programming (59). In contrast to other approaches, which consider for instance only the most abundant peak of a peptide, our algorithm features the use of (sufficiently) exact isotopic distributions to extract all isotope peaks of the real data that potentially belong to an investigated peptide. This reflects the highly different patterns of isotopic distributions that occur particularly for partially labeled peptides, and in addition, ensures that only relevant peaks are considered in the calculation of the ratio. The influence of peaks from overlapping peptides and background noise is substantially reduced. The most crucial part of the algorithm is the determination of the similarity between a theoretical isotope distribution and the real peptide's mass spectrum to finally identify the correct incorporation rate of stable isotopes for a peptide. Guan et al. (24) utilized a nonnegative least-square algorithm for this purpose. In our hands the—on first sight comparably simple—scalar product provided the best results (36). To accurately predict the elution peak of a peptide within its XIC, a continuous wavelet transform approach revealed the best performance for our application, not least because its practicability was already confirmed in several studies (40). In contrast to other methods, such as a simple top-down approach that searches for a peak's apex and its ascending and descending flanks, the impact of instrumental error is minimized and, moreover, the application of a smoothing filter, e.g. Savitzky-Golay (60) can be omitted. The final step of the quantification algorithm concerns the calculation of the difference in abundance between, for instance, each partially and fully 15N-labeled peptide pair. Instead of the area-under-the-curve, we employed a linear regression approach similar to those used in the RelEx tool (37). However, we replaced vertical with perpendicular offsets, thereby allowing for uncertainties in the intensity measurements of both the fully- as well as the partially-labeled peptide. Following the standard practice, the ratio is, afterward, logarithmically transformed contributing the input for all further statistical analyses.
Application Study: Investigation of Heat Shock Response in C. glutamicum
To demonstrate the applicability of our newly developed tool in real-life, we investigated the response of C. glutamicum to heat stress, a well-understood regulatory mechanism in bacteria. In contrast to the cell culture's growth, which was scarcely affected by a moderate increase in temperature, the results of our newly developed algorithm show a drastic impact of heat stress on the turnover. When cells are exposed to heat the synthesis of most proteins is expeditiously reduced. The exception are the heat shock proteins (HSPs), which show a considerably increased synthesis at 40 °C to compensate the heat impact (42, 61). Synthesis of HSPs was found well synchronized over time, as shown in Fig. 5. At the onset of heat shock induction the HSPs undergo a quasi jump-wise increase during the first 30 min of the applied heat shock stressor. Obviously, as protein turnover can be assumed to be identical prior to the applied heat shock for all samples, and quite similar during the final phase from 30 to 210 min, the main increase in synthesis has to take place during the initial 30 min. During the initial phase of our applied 14N pulse, i.e. the first 30 min, the amount of newly synthesized proteins containing 14N does, however, not allow for reliable peak detection and quantification. To overcome this limitation, amino acid analogues could be used for peptide labeling (62, 63). But because of the fact that these molecules negatively influence the cell metabolism we refrained from their usage. We speculate that, in future, more sensitive MS technology will surmount existing limitations in the quantification of low abundant peptides using our algorithm. Actually, for the majority of targets quantified in this study (Fig. 5 and supplemental Fig. S2) (e.g. ATPase subunits, ribosomal proteins etc.) decreased synthesis rates were calculated at 40 °C. The rate of translation of many proteins is lower under heat stress because the essential sigma factor SigA, which is responsible for transcription of the housekeeping genes under normal growth conditions (64), is replaced by the alternative sigma factors SigH and SigM (65–66). The expression of translationary machinery genes are repressed under moderate heat stress (47) and, accordingly, we found lower synthesis for the corresponding proteins as shown in Fig. 5 as well as supplemental Fig. S2. In addition, regulation of several other proteins was in agreement with previous transcriptome and proteome studies (47, 67–69). The alteration in abundance of the 95 significantly regulated proteins concerning the temperature in this study matches, to a large extent, with the 787 regulated genes (358 up and 420 down) of published transcriptome analyses (47), with a correlation of r = .80. Interestingly, the over-all correlation between the whole proteome and transcriptome data was merely at r = .58, which is nevertheless in line with a correlation of .65 between mRNA and protein dynamics in the actinomycete Streptomyces coelicolor (15).
In the present work we also analyzed the decrease in intensity of the fully labeled 15N protein species relative to the 13C standard to get an impression, which targets may be degraded under the chosen treatment. In contrast to a variety of studies on protein synthesis, there exist only few publications dealing with the degradation of proteins. In our analysis, we found 59.6% of all quantified proteins revealing a slightly increased proteolysis in the heat shocked cells in comparison to 50% under optimal growth conditions. It is assumed that heat stress leads to an elevated degradation of proteins (70), which our results can confirm.
CONCLUSION
With our work we can now provide a comprehensive method to fully understand the dynamics of an organism's proteome. Aiming to understand the biological processes that cause the discrepancies between proteome and transcriptome, it is indispensable to investigate protein turnover, being set by the two opposing processes of protein synthesis and degradation. Utilizing a multilabeling strategy that employs both 15N and 13C we rely on the gold standard for quantitative mass spectrometry analysis. As the metabolic label is introduced at an early stage of an experiment (22), errors because of sample handling or processing have an effect on both—the labeled as well as the unlabeled variant—of any protein (21).
With our method, we can, moreover, address a severe problem of metabolic labeling approaches: often it is impossible to reach a high rate, let alone a complete, incorporation of stable isotopes, which is in particular true for higher organisms such as Eukaryotes (71). Although initially developed for pulse-chase experiments, our algorithm is also capable of successfully quantifying proteins having an uncertain and diversified incorporation rate of isotopic labels such as heavy stable nitrogen or carbon.
All presented methods are integrated into the rich internet application QuPE (33), which is accessible from any place an Internet connection is available. The provided workflow comprises not only the calculation of protein synthesis and degradation ratios but also a comprehensive analysis strategy for this type of data including analysis of variance to identify significantly differently synthesized proteins and cluster analysis to find groups of proteins having similar patterns of abundance over time.
Supplementary Material
Acknowledgments
Furthermore, the authors wish to thank Frank ter Veld for critical reading of the manuscript, Tanja Bojarzyn for preparing and measuring the 13C samples, Andrea Lünenschloβ for initial heat shock experiments, and the BRF system administrators for expert technical support.
Footnotes
* This work was supported by the Federal Ministry of Education and Research (BMBF, 0313812A, QuantPro) and the Deutsche Forschungsgemeinschaft (DFG, PO1033/3-1).
 This article contains supplemental Files, Figs. S1 to S4, and Tables S1 and S2.
This article contains supplemental Files, Figs. S1 to S4, and Tables S1 and S2.
christian.troetschel@rub.de, alu@cebitec.uni-bielefeld.de, daniel.wolff@rub.de, schroeder.simon@googlemail.com, agoesman@cebitec.uni-bielefeld.de, tnattkem@techfak.uni-bielefeld.de, ansgar.poetsch@rub.de
Conflict of interest: The authors declare that they have no conflict of interest.
1 The abbreviations used are:
- SILAC
- stable isotope labeling by amino acids in cell culture
- LC-MS/MS
- liquid chromatography-tandem MS
- XIC
- extraction of ion chromatograms.
REFERENCES
- 1. Maier T., Guell M., Serrano L. (2009) Correlation of mRNA and protein in complex biological samples. FEBS Lett. 583, 3966–3973 [DOI] [PubMed] [Google Scholar]
- 2. de Sousa Abreu R., Penalva L. O., Marcotte E. M., Vogel C. (2009) Global signatures of protein and mRNA expression levels. Mol. Biosyst. 5, 1512–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Eddy S. R. (2001) Non-coding RNA genes and the modern RNA world. Nat. Rev. Genet. 2, 919–929 [DOI] [PubMed] [Google Scholar]
- 4. Storz G. (2002) An expanding universe of noncoding RNAs. Science 296, 1260–1263 [DOI] [PubMed] [Google Scholar]
- 5. Tobias J. W., Shrader T. E., Rocap G., Varshavsky A. (1991) The N-end rule in bacteria. Science 254, 1374–1377 [DOI] [PubMed] [Google Scholar]
- 6. Gottesman S., Roche E., Zhou Y., Sauer R. T. (1998) The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev. 12, 1338–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Herman C., Thevenet D., Bouloc P., Walker G. C., D'Ari R. (1998) Degradation of carboxy-terminal-tagged cytoplasmic proteins by the Escherichia coli protease HflB (FtsH). Genes Dev. 12, 1348–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lies M., Maurizi M. R. (2008) Turnover of endogenous SsrA-tagged proteins mediated by ATP-dependent proteases in Escherichia coli. J. Biol. Chem. 283, 22918–22929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sprinson D. B., Rittenberg D. (1949) The rate of interaction of the amino acids of the diet with the tissue proteins. J. Biol. Chem. 180, 715–726 [PubMed] [Google Scholar]
- 10. Bernhardt J., Buttner K., Scharf C., Hecker M. (1999) Dual channel imaging of two-dimensional electropherograms in Bacillus subtilis. Electrophoresis 20, 2225–2240 [DOI] [PubMed] [Google Scholar]
- 11. Gerth U., Kock H., Kusters I., Michalik S., Switzer R. L., Hecker M. (2008) Clp-dependent proteolysis down-regulates central metabolic pathways in glucose-starved Bacillus subtilis. J. Bacteriol. 190, 321–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pratt J. M., Petty J., Riba-Garcia I., Robertson D. H., Gaskell S. J., Oliver S. G., Beynon R. J. (2002) Dynamics of protein turnover, a missing dimension in proteomics. Mol. Cell. Proteomics 1, 579–591 [DOI] [PubMed] [Google Scholar]
- 13. Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 14. Doherty M. K., Hammond D. E., Clague M. J., Gaskell S. J., Beynon R. J. (2009) Turnover of the human proteome: determination of protein intracellular stability by dynamic SILAC. J. Proteome Res. 8, 104–112 [DOI] [PubMed] [Google Scholar]
- 15. Jayapal K. P., Sui S., Philp R. J., Kok Y. J., Yap M. G., Griffin T. J., Hu W. S. (2010) Multitagging proteomic strategy to estimate protein turnover rates in dynamic systems. J. Proteome Res. 9, 2087–2097 [DOI] [PubMed] [Google Scholar]
- 16. Ross P. L., Huang Y. N., Marchese J. N., Williamson B., Parker K., Hattan S., Khainovski N., Pillai S., Dey S., Daniels S., Purkayastha S., Juhasz P., Martin S., Bartlet-Jones M., He F., Jacobson A., Pappin D. J. (2004) Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics 3, 1154–1169 [DOI] [PubMed] [Google Scholar]
- 17. Fischer F., Poetsch A. (2006) Protein cleavage strategies for an improved analysis of the membrane proteome. Proteome Sci. 4, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Oda Y., Huang K., Cross F. R., Cowburn D., Chait B. T. (1999) Accurate quantitation of protein expression and site-specific phosphorylation. Proc. Natl. Acad. Sci. U. S. A. 96, 6591–6596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cargile B. J., Bundy J. L., Grunden A. M., Stephenson J. L., Jr. (2004) Synthesis/degradation ratio mass spectrometry for measuring relative dynamic protein turnover. Anal. Chem. 76, 86–97 [DOI] [PubMed] [Google Scholar]
- 20. Rao P. K., Rodriguez G. M., Smith I., Li Q. (2008) Protein dynamics in iron-starved Mycobacterium tuberculosis revealed by turnover and abundance measurement using hybrid-linear ion trap-Fourier transform mass spectrometry. Anal. Chem. 80, 6860–6869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gouw J. W., Krijgsveld J., Heck A. J. (2010) Quantitative proteomics by metabolic labeling of model organisms. Mol. Cell. Proteomics 9, 11–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Haegler K., Mueller N. S., Maccarrone G., Hunyadi-Gulyas E., Webhofer C., Filiou M. D., Zhang Y., Turck C. W. (2009) QuantiSpec–Quantitative mass spectrometry data analysis of (15)N-metabolically labeled proteins. J. Proteomics 71, 601–608 [DOI] [PubMed] [Google Scholar]
- 23. Zhang Y., Reckow S., Webhofer C., Boehme M., Gormanns P., Egge-Jacobsen W. M., Turck C. W. (2011) Proteome Scale Turnover Analysis in Live Animals Using Stable Isotope Metabolic Labeling. Anal. Chem. 83, 1665–1672 [DOI] [PubMed] [Google Scholar]
- 24. Guan S., Price J. C., Prusiner S. B., Ghaemmaghami S., Burlingame A. L. (2011) A data processing pipeline for Mammalian proteome dynamics studies using stable isotope metabolic labeling. Mol. Cell. Proteomics 10, M111 010728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Price J. C., Guan S., Burlingame A., Prusiner S. B., Ghaemmaghami S. (2010) Analysis of proteome dynamics in the mouse brain. Proc. Natl. Acad. Sci. U. S. A. 107, 14508–14513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kase H., Nakayama K. (1972) Production of L-Threonine by Analog-Resistant Mutants. Ag. Biol. Chem. 36, 1611–1621 [Google Scholar]
- 27. Koch D. J., Ruckert C., Rey D. A., Mix A., Puhler A., Kalinowski J. (2005) Role of the ssu and seu genes of Corynebacterium glutamicum ATCC 13032 in utilization of sulfonates and sulfonate esters as sulfur sources. Appl. Environ. Microbiol. 71, 6104–6114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Haussmann U., Qi S. W., Wolters D., Rogner M., Liu S. J., Poetsch A. (2009) Physiological adaptation of Corynebacterium glutamicum to benzoate as alternative carbon source - a membrane proteome-centric view. Proteomics 9, 3635–3651 [DOI] [PubMed] [Google Scholar]
- 29. Wisniewski J. R., Zougman A., Nagaraj N., Mann M. (2009) Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362 [DOI] [PubMed] [Google Scholar]
- 30. Kalinowski J., Bathe B., Bartels D., Bischoff N., Bott M., Burkovski A., Dusch N., Eggeling L., Eikmanns B. J., Gaigalat L., Goesmann A., Hartmann M., Huthmacher K., Kramer R., Linke B., McHardy A. C., Meyer F., Mockel B., Pfefferle W., Puhler A., Rey D. A., Ruckert C., Rupp O., Sahm H., Wendisch V. F., Wiegrabe I., Tauch A. (2003) The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of L-aspartate-derived amino acids and vitamins. J. Biotechnol. 104, 5–25 [DOI] [PubMed] [Google Scholar]
- 31. Tabb D. L., McDonald W. H., Yates J. R., 3rd (2002) DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res. 1, 21–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Higdon R., Hogan J. M., Van Belle G., Kolker E. (2005) Randomized sequence databases for tandem mass spectrometry peptide and protein identification. OMICS 9, 364–379 [DOI] [PubMed] [Google Scholar]
- 33. Albaum S. P., Neuweger H., Franzel B., Lange S., Mertens D., Trotschel C., Wolters D., Kalinowski J., Nattkemper T. W., Goesmann A. (2009) Qupe–a Rich Internet Application to take a step forward in the analysis of mass spectrometry-based quantitative proteomics experiments. Bioinformatics 25, 3128–3134 [DOI] [PubMed] [Google Scholar]
- 34. Nolting D. (2008) Isotopic Pattern Calculator [http://isotopatcalc.sourceforge.net]
- 35. Coursey J. S., Schwab D. J., Dragoset R. A. (2005) Atomic weights and isotopic compositions [http://physics.nist.gov/Comp]
- 36. Stein S. E., Scott D. R. (1994) Optimization and testing of mass spectral library search algorithms for compound identification. J. Am. Soc. Mass Spectrom. 5, 859–866 [DOI] [PubMed] [Google Scholar]
- 37. MacCoss M. J., Wu C. C., Liu H., Sadygov R., Yates J. R., 3rd (2003) A correlation algorithm for the automated quantitative analysis of shotgun proteomics data. Anal. Chem. 75, 6912–6921 [DOI] [PubMed] [Google Scholar]
- 38. Li X. J., Zhang H., Ranish J. A., Aebersold R. (2003) Automated statistical analysis of protein abundance ratios from data generated by stable-isotope dilution and tandem mass spectrometry. Anal. Chem. 75, 6648–6657 [DOI] [PubMed] [Google Scholar]
- 39. Pan C., Kora G., Tabb D. L., Pelletier D. A., McDonald W. H., Hurst G. B., Hettich R. L., Samatova N. F. (2006) Robust estimation of peptide abundance ratios and rigorous scoring of their variability and bias in quantitative shotgun proteomics. Anal. Chem. 78, 7110–7120 [DOI] [PubMed] [Google Scholar]
- 40. Yang C., He Z., Yu W. (2009) Comparison of public peak detection algorithms for MALDI mass spectrometry data analysis. BMC Bioinformatics 10, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Du P., Kibbe W. A., Lin S. M. (2006) Improved peak detection in mass spectrum by incorporating continuous wavelet transform-based pattern matching. Bioinformatics 22, 2059–2065 [DOI] [PubMed] [Google Scholar]
- 42. Ventura M., Canchaya C., Zhang Z., Bernini V., Fitzgerald G. F., van Sinderen D. (2006) How high G+C Gram-positive bacteria and in particular bifidobacteria cope with heat stress: protein players and regulators. FEMS Microbiol. Rev. 30, 734–759 [DOI] [PubMed] [Google Scholar]
- 43. Albaum S. P., Hahne H., Otto A., Haussmann U., Becher D., Poetsch A., Goesmann A., Nattkemper T. W. (2011) A guide through the computational analysis of isotope-labeled mass spectrometry-based quantitative proteomics data: an application study. Proteome Sci. 9, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Holm S. (1979) A simple sequentially rejective multiple test procedure. Scand. J. Statistics 6, 65–70 [Google Scholar]
- 45. Fligner M. A., Killeen T. J. (1976) Distribution-free two-sample tests for scale. J. Am. Statistical Assoc. 71, 210–213 [Google Scholar]
- 46. Shapiro S. S., Wilk M. B. (1965) An analysis of variance test for normality (Complete Samples). Biometrika 52, 591–611 [Google Scholar]
- 47. Barreiro C., Nakunst D., Huser A. T., de Paz H. D., Kalinowski J., Martin J. F. (2009) Microarray studies reveal a ‘differential response’ to moderate or severe heat shock of the HrcA- and HspR-dependent systems in Corynebacterium glutamicum. Microbiology 155, 359–372 [DOI] [PubMed] [Google Scholar]
- 48. Yeung K. Y., Haynor D. R., Ruzzo W. L. (2001) Validating clustering for gene expression data. Bioinformatics 17, 309–318 [DOI] [PubMed] [Google Scholar]
- 49. Ward J. H., Jr. (1963) Hierarchical grouping to optimize an objective function. J. Am. Statistical Assoc. 58, 236–244 [Google Scholar]
- 50. Krzanowski W. J., Lai Y. T. (1988) A criterion for determining the number of groups in a data set using sum-of-squares clustering. Biometrics 44, 23–34 [Google Scholar]
- 51. Plotkin J. B. (2011) Cell biology. The lives of proteins. Science 331, 683–684 [DOI] [PubMed] [Google Scholar]
- 52. Gottesman S., Maurizi M. R. (1992) Regulation by proteolysis: energy-dependent proteases and their targets. Microbiol. Rev. 56, 592–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tatusov R. L., Galperin M. Y., Natale D. A., Koonin E. V. (2000) The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 28, 33–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Park S. K., Venable J. D., Xu T., Yates J. R., 3rd (2008) A quantitative analysis software tool for mass spectrometry-based proteomics. Nat. Methods 5, 319–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Andreev V. P., Li L., Rejtar T., Li Q., Ferry J. G., Karger B. L. (2006) New algorithm for 15N/14N quantitation with LC-ESI-MS using an LTQ-FT mass spectrometer. J. Proteome Res. 5, 2039–2045 [DOI] [PubMed] [Google Scholar]
- 56. Rockwood A. L., Van Orden S. L. (1996) Ultrahigh-speed calculation of isotope distributions. Anal. Chem. 68, 2027–2030 [DOI] [PubMed] [Google Scholar]
- 57. Sperling E., Bunner A. E., Sykes M. T., Williamson J. R. (2008) Quantitative analysis of isotope distributions in proteomic mass spectrometry using least-squares Fourier transform convolution. Anal. Chem. 80, 4906–4917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Fernandez-de-Cossio J. (2010) Computation of the isotopic distribution in two dimensions. Anal. Chem. 82, 6726–6729 [DOI] [PubMed] [Google Scholar]
- 59. Snider R. K. (2007) Efficient calculation of exact mass isotopic distributions. J. Am. Soc. Mass Spectrom. 18, 1511–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Savitzky A., Golay M. J. E. (1964) Smoothing and Differentiation of Data by Simplified Least Squares Procedures. Anal. Chem. 36, 1627–1639 [Google Scholar]
- 61. Yura T., Nakahigashi K. (1999) Regulation of the heat-shock response. Curr. Opin. Microbiol. 2, 153–158 [DOI] [PubMed] [Google Scholar]
- 62. Dieterich D. C., Link A. J., Graumann J., Tirrell D. A., Schuman E. M. (2006) Selective identification of newly synthesized proteins in mammalian cells using bioorthogonal noncanonical amino acid tagging (BONCAT). Proc. Natl. Acad. Sci. U.S.A. 103, 9482–9487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kramer G., Sprenger R. R., Back J., Dekker H. L., Nessen M. A., van Maarseveen J. H., de Koning L. J., Hellingwerf K. J., de Jong L., de Koster C. G. (2009) Identification and quantitation of newly synthesized proteins in Escherichia coli by enrichment of azidohomoalanine-labeled peptides with diagonal chromatography. Mol. Cell. Proteomics 8, 1599–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Helmann J. D., Chamberlin M. J. (1988) Structure and function of bacterial sigma factors. Annu. Rev. Biochem. 57, 839–872 [DOI] [PubMed] [Google Scholar]
- 65. Ehira S., Teramoto H., Inui M., Yukawa H. (2009) Regulation of Corynebacterium glutamicum heat shock response by the extracytoplasmic-function sigma factor SigH and transcriptional regulators HspR and HrcA. J. Bacteriol. 191, 2964–2972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Nakunst D., Larisch C., Huser A. T., Tauch A., Puhler A., Kalinowski J. (2007) The extracytoplasmic function-type sigma factor SigM of Corynebacterium glutamicum ATCC 13032 is involved in transcription of disulfide stress-related genes. J. Bacteriol. 189, 4696–4707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Muffler A., Bettermann S., Haushalter M., Horlein A., Neveling U., Schramm M., Sorgenfrei O. (2002) Genome-wide transcription profiling of Corynebacterium glutamicum after heat shock and during growth on acetate and glucose. J. Biotechnol. 98, 255–268 [DOI] [PubMed] [Google Scholar]
- 68. Barreiro C., Gonzalez-Lavado E., Brand S., Tauch A., Martin J. F. (2005) Heat shock proteome analysis of wild-type Corynebacterium glutamicum ATCC 13032 and a spontaneous mutant lacking GroEL1, a dispensable chaperone. J. Bacteriol. 187, 884–889 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 69. Barreiro C., Gonzalez-Lavado E., Patek M., Martin J. F. (2004) Transcriptional analysis of the groES-groEL1, groEL2, and dnaK genes in Corynebacterium glutamicum: characterization of heat shock-induced promoters. J. Bacteriol. 186, 4813–4817 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 70. Araki T. (1992) An analysis of the effect of changes in growth temperature on proteolysis in vivo in the psychrophilic bacterium Vibrio sp. strain ANT-300. J. Gen. Microbiol. 138, 2075–2082 [DOI] [PubMed] [Google Scholar]
- 71. Frank E., Kessler M. S., Filiou M. D., Zhang Y., Maccarrone G., Reckow S., Bunck M., Heumann H., Turck C. W., Landgraf R., Hambsch B. (2009) Stable isotope metabolic labeling with a novel N-enriched bacteria diet for improved proteomic analyses of mouse models for psychopathologies. PLoS One 4, e7821. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.