Abstract
To perform differential studies of complex protein mixtures, strategies for reproducible and accurate quantification are needed. Here, we evaluated a quantitative proteomic workflow based on nanoLC-MS/MS analysis on an LTQ-Orbitrap-VELOS mass spectrometer and label-free quantification using the MFPaQ software. In such label-free quantitative studies, a compromise has to be found between two requirements: repeatability of sample processing and MS measurements, allowing an accurate quantification, and high proteomic coverage of the sample, allowing quantification of minor species. The latter is generally achieved through sample fractionation, which may induce experimental bias during the label-free comparison of samples processed, and analyzed independently. In this work, we wanted to evaluate the performances of MS intensity-based label-free quantification when a complex protein sample is fractionated by one-dimensional SDS-PAGE. We first tested the efficiency of the analysis without protein fractionation and could achieve quite good quantitative repeatability in single-run analysis (median coefficient of variation of 5%, 99% proteins with coefficient of variation <48%). We show that sample fractionation by one-dimensional SDS-PAGE is associated with a moderate decrease of quantitative measurement repeatability while largely improving the depth of proteomic coverage. We then applied the method for a large scale proteomic study of the human endothelial cell response to inflammatory cytokines, such as TNFα, interferon γ, and IL1β, which allowed us to finely decipher at the proteomic level the biological pathways involved in endothelial cell response to proinflammatory cytokines.
With recent advances in mass spectrometry, label-free quantitative proteomic approaches have progressed and are now considered as reliable and efficient methods to study protein expression level changes in complex mixtures. These approaches, which have been reviewed recently (1, 2), are based on the measurement either of the MS/MS sampling rate of a particular peptide or of its MS chromatographic peak area, these values being directly related to peptide abundance. The increase of instrument sequencing speed has benefited MS/MS spectral counting approaches by improving MS/MS sampling of peptide mixtures, whereas the introduction of high resolution analyzers such as FT-Orbitrap has boosted the use of methods based on peptide intensity measurements by greatly facilitating the matching of peptide peaks in different complex maps acquired independently. The most obvious advantage of these methods over isotopic labeling techniques is their ease of use at the sample preparation step, because they do not require any preliminary treatment to introduce a label into peptides or proteins. Being more straightforward, they also do not present the classical drawbacks of labeling methods, i.e., cost, applicability to limited types of samples (mostly cultured cells in the case of metabolic labeling) and the limited number of conditions that can be compared. On the other hand, the use of label-free strategies is hampered by two main difficulties: 1) the variability of all sample processing steps before MS analysis and of the analytical measurement itself, because the samples to be compared are processed and analyzed individually, and 2) the complexity of the MS data analysis step, which requires proper realignment, normalization, and peptide peaks matching across different nanoLC-MS runs.
Many bioinformatic tools have been developed in recent years for the quantification of MS data generated in label-free experiments, either by spectral counting (3–5) or by peptide MS signal intensity measurement (6–9). In the later field, a lot of emphasis has been put on peptide pattern-based methods, in which the software performs feature detection in LC-MS maps through analysis of the characteristic isotopic pattern of a peptide ion in the m/z dimension, and on its chromatographic elution peak in the retention time (RT)1 dimension. The total ion current integrated under this MS feature can then be used as a quantitative measurement of the peptide concentration. The primary advantage of this approach is that any signal detected by the mass spectrometer in the MS survey scan can be in principle analyzed and quantified, whether or not the peak has been selected for MS/MS sequencing. Bioinformatic programs based on peptide feature detection as the starting step for label-free analysis include among others SuperHirn (8), MSInspect (6), OpenMS (9), Decon2LS (7), or the commercial software Progenesis LC-MS. Although they offer an attractive and powerful analysis of the data, algorithms based on recognition of peptide features and LC-MS maps alignment require intensive computer calculation, making the quantification time-consuming and difficult to perform on a large number of LC-MS files. In addition, integration of MS features quantitative data with MS/MS identification results from search engines occurs as a second step, and depending on the bioinformatic tool used, retrieving quantitative values for the list of identified and validated peptides, and then for the associated list of proteins, can be difficult to implement. Finally, because the LC-MS maps are usually analyzed individually, low intensity features near the cut-off value set for the recognition process are detected in an irreproducible way, and most of the available software generates quantitative data sets containing many missing values, which complicates further statistical analysis of the results.
On the other hand, another approach to extract quantitative data from MS survey scans is based on the reverse process, i.e., making use initially of peptide identification results to go back in the MS scans to obtain peptide intensity values. For each peptide ion identified from MS/MS sequencing, experimentally measured RT and monoisotopic m/z values can be used as a starting point to retrieve the associated extracted ion chromatograms (XIC) of this ion. In that case, confident extraction of a peptide signal (versus chemical noise) is supported by the identification result, and because the charge state of the ion is known, definition of isotopic patterns and extraction of intensity values for the different isotopes of a same peptide ion is facilitated. Such a method, which is in principle more simple and rapid, has been used in a few software packages such as Serac (10), Quoil (11), Ideal-Q (12), and MFPaQ (13, 14). A drawback of this method, however, is that only identified peptides can be quantified. For analysis of highly complex peptide mixtures, MS/MS undersampling thus limits the number of identified and quantified proteins. Depending on the software, this problem can be alleviated by a cross-assignment of peptide signals across different replicate LC-MS/MS runs: if a peptide ion is identified in only one or a few runs, its signal can be extracted in the other analytical runs by using a predicted RT value, even if identification results are missing for this particular peptide in these runs because of MS/MS undersampling. Thus, acquisition of multiple replicate runs allows to increase the number of identified and thus quantified peptides and proteins. Nevertheless, the performances of identity-based methods are still strictly linked to the number of identifications and to the depth of the proteomic analysis on highly complex samples.
In a study focusing on label-free quantitative analysis of clinical samples (14), we previously described an approach based on the use of the MFPaQ software to circumvent the undersampling problem. Following extensive proteomic analysis of cerebrospinal fluid after treatment with combinatorial libraries of peptide ligands and one-dimensional SDS-PAGE fractionation, we generated an identification database containing sequences of identified peptides, along with their m/z and retention time-associated values that were then used to extract the XIC of these peptides in the one-shot analytical runs of unfractionated samples. This method was well suited for the analysis of clinical series in which very limited or no fractionation at all is performed on the samples, because of the large number of analyses (number of patients and technical replicates), and we showed that it indeed allowed significant increase of the number of proteins correctly quantified in replicate runs of individual samples. However, not all of the peptides from the database could be retrieved in the individual runs, because of the limited dynamic range of the instrument during the one-shot analysis of complex peptide mixtures. To overcome also the dynamic range limitation, a commonly used and efficient approach is to prefractionate individually the samples to be compared and perform nanoLC-MS/MS analysis of each fraction separately. Although it requires longer analytical time, this shotgun type of analysis clearly offers an improved coverage of the sample and allows the detection of low abundance proteins that remain undetected when the whole sample is analyzed in one run. To that aim, one-dimensional SDS-PAGE is often selected as a robust and simple method to fractionate most kinds of protein samples, even membrane ones, and is particularly used on SILAC or ICAT labeled proteomes, because the two samples to be compared are gathered and can be processed simultaneously. However, when label-free quantification is to be performed, parallel processing steps such as electrophoretic migration, gel cutting, and in-gel digestion represent different sources of variability that may alter the final quantitative comparison of the samples.
In the present study, our objective was to perform an in-depth quantitative analysis of the endothelial cell (EC) proteome using a label-free approach. First, we thus checked whether SDS-PAGE fractionation of the individual samples, which gives the best dynamic range on a global analysis, is compatible with accurate label-free quantitation based on peptide signal intensity measurement. We evaluated the performances of a label-free quantitative workflow in terms of repeatability and number of quantified proteins, with or without protein fractionation by one-dimensional SDS-PAGE, for the analysis of a complex cellular proteome. We applied the MFPaQ software, which uses an identity-based extraction approach, to quantify the data obtained from the nanoLC-MS/MS analysis of a total lysate of primary cultured human vascular ECs. New data normalization and integration procedure dedicated to shotgun experiments were introduced in the software, allowing integration at the protein level the quantitative data from different fractions and correction of errors related to nonreproducible electrophoretic migration of proteins. We showed that the approach based on peptide XIC extraction provides good quality quantitative data on the identified proteome and that high repeatability is obtained on proteins quantified in single run analysis (median CV of 5%, 99% proteins with CV values of <48%). When the protein sample is fractionated by one-dimensional SDS-PAGE, the repeatability of the quantitative measurement decreases, although in a moderate way (median CV of 7%, 99% proteins with CV values of <62%), and concomitantly the depth of proteomic coverage is largely increased. We then applied the method for a large scale proteomic study of the response of ECs to proinflammatory treatments with TNFα/IFNγ or IL1β. It allowed us to identify and quantify more than 5400 unique proteins, providing an in-depth analysis of the endothelial cell proteome and a detailed characterization of the proteomic variations associated with the inflammatory response.
MATERIALS AND METHODS
EC Culture and Cytokine Stimulation
Primary human umbilical vein ECs (HUVECs) were purchased from Clonetics, grown in ECGM medium (Promocell, Heidelberg, Germany), and used after four passages for proteomic analyses. Cytokine treatment was performed by incubating the ECs for 12 h in OptiMEM medium (Invitrogen) with a combination of TNFα (25 ng/ml; R & D Systems) and IFNγ (50 ng/ml; R & D Systems) or with IL1β (5 ng/ml; R & D Systems).
Protein Sample Processing
The cells were lysed in a buffer containing 2% of SDS and sonicated, and protein concentration was determined by detergent-compatible assay (DC assay; Bio-Rad). Protein samples were reduced in Laemmli buffer (final composition: 25 mm DTT, 2% SDS, 10% glycerol, 40 mm Tris, pH 6.8) for 5 min at 95 °C. Cysteine residues were alkylated by addition of iodoacetamide at a final concentration of 90 mm and incubation for 30 min at room temperature in the dark. During the alkylation reaction, the pH of the samples was adjusted using small amounts of 1 m Tris, pH 8. Protein samples were loaded on a homemade one-dimensional SDS-PAGE gel (separating gel 1.5 mm × 5 cm, 12% acrylamide polymerized in SDS 0.1%, 375 mm Tris, pH 8.8, and stacking gel 1.5 mm × 1.5 cm, 4% acrylamide polymerized in 0.1% SDS, 125 mm Tris. For one-shot analysis of the entire mixture, no fractionation was performed, and the electrophoretic migration was stopped as soon as the protein sample (15 μg) entered the separating gel. The gel was briefly stained with Coomassie Blue, and a single band, containing the whole sample, was cut. For shotgun analysis, electrophoretic migration was performed to fractionate the protein sample (100 μg) into 12 gel bands. For replicate and comparative analyses, the samples were processed on adjacent migration lanes that were cut simultaneously with a long razor blade. To evaluate gel to gel repeatability, different gels were prepared and migrated in parallel, and the same number of homogeneous gel slices were cut successively on the separate gels, following the same cutting pattern. Gel slices were washed by two cycles of incubation in 100 mm ammonium bicarbonate for 15 min at 37 °C, followed by 100 mm ammonium bicarbonate/acetonitrile (1:1) for 15 min at 37 °C. The proteins were digested by 0.6 μg of modified sequencing grade trypsin (Promega) in 50 mm ammonium bicarbonate, overnight at 37 °C. The resulting peptides were extracted from the gel by incubation in 50 mm ammonium bicarbonate for 15 min at 37 °C and twice in 10% formic acid/acetonitrile (1:1) for 15 min at 37 °C. The three collected extractions were pooled with the initial digestion supernatant, dried in a SpeedVac, and resuspended with 17 μl of 5% acetonitrile, 0.05% TFA.
NanoLC-MS/MS Analysis
The Resulting peptides were analyzed by nanoLC-MS/MS using an Ultimate3000 system (Dionex, Amsterdam, The Netherlands) coupled to an LTQ-Orbitrap Velos mass spectrometer (Thermo Fisher Scientific, Bremen, Germany). Five μl of each sample were loaded on a C-18 precolumn (300-μm inner diameter × 5 mm; Dionex) at 20 μl/min in 5% acetonitrile, 0.05% TFA. After 5 min of desalting, the precolumn was switched online with the analytical C-18 column (75 μm inner diameter × 15 cm; PepMap C18, Dionex) equilibrated in 95% solvent A (5% acetonitrile, 0.2% formic acid) and 5% solvent B (80% acetonitrile, 0.2% formic acid). The peptides were eluted using a 5 to 50% gradient of solvent B during 80 min at 300 nl/min flow rate. The LTQ-Orbitrap Velos was operated in data-dependent acquisition mode with the XCalibur software. Survey scan MS were acquired in the Orbitrap on the 300–2000 m/z range with the resolution set to a value of 60,000. The 10 most intense ions per survey scan were selected for CID fragmentation, and the resulting fragments were analyzed in the linear trap (LTQ). Dynamic exclusion was employed within 60 s to prevent repetitive selection of the same peptide.
Database Search and Data Validation
The Mascot Daemon software (version 2.3.2; Matrix Science, London, UK) was used to perform database searches, using the Extract_msn.exe macro provided with Xcalibur (version 2.0 SR2; Thermo Fisher Scientific) to generate peaklists. The following parameters were set for creation of the peaklists: parent ions in the mass range 400–4500, no grouping of MS/MS scans, and threshold at 1000. A peaklist was created for each analyzed fraction (i.e., gel slice), and individual Mascot (version 2.3.01) searches were performed for each fraction. The data were searched against Homo sapiens entries in Uniprot protein database (release 2010_09, September 21, 2010; 1,215,533 sequences). Carbamidomethylation of cysteines was set as a fixed modification, and oxidation of methionine and protein N-terminal acetylation were set as a variable modifications. Specificity of trypsin digestion was set for cleavage after Lys or Arg, and two missed trypsin cleavage sites were allowed. The mass tolerances in MS and MS/MS were set to 5 ppm and 0.6 Da, respectively, and the instrument setting was specified as “ESI-Trap.” To calculate the false discovery rate (FDR), the search was performed using the “decoy” option in Mascot. Peptide identifications extracted from Mascot result files were validated at a final peptide FDR of 5%. Peptide matches were validated if their score was greater than the Mascot homology threshold (when available, otherwise the Mascot identity threshold was used) for a given Mascot p value. The FDR at the peptide level was calculated as described in Navarro and Vázquez (15). Using this method, the p value was automatically adjusted to obtain a FDR of 5% at the peptide level. Validated peptides were assembled into proteins groups following the principle of parsimony (Ocam's razor), which involves the creation of the minimal list of protein groups explaining the list of peptide spectrum matches. Protein groups were then rescored for the protein validation process. For each peptide match belonging to a protein group, the difference between its Mascot score and its homology threshold (or identity threshold) was computed for a given p value (automatically adjusted to increase the discrimination between target and decoy matches), and these “score offsets” were then summed to obtain the protein group score. Protein groups were validated based on this score to obtain a FDR of 1% at the protein level (FDR = number of validated decoy hits/(number of validated target hits + number of validated decoy hits) × 100). In the case of sample fractionation on one-dimensional SDS-PAGE, the MFPaQ software was used to create a unique nonredundant protein list from the identification results of each fraction by clustering protein groups containing sequences matching the same set of peptides. If a final group was composed of several TrEMBL and SwissProt entry names, a SwissProt entry was singled out, and the associated protein description was reported in the final lists (supplemental Tables I and II).
Data Quantification
Quantification of proteins was performed using the label-free module implemented in the MFPaQ v4.0.0 software (http://mfpaq.sourceforge.net/). For each sample, the software uses the validated identification results and XICs of the identified peptide ions in the corresponding raw nanoLC-MS files, based on their experimentally measured RT and monoisotopic m/z values The time value used for this process is retrieved from Mascot result files, based on an MS2 event matching to the peptide ion. If several MS2 events were matched to a given peptide ion, the software checks the intensity of each corresponding precursor peak in the previous MS survey scan. The time of the MS scan that exhibits the highest precursor ion intensity is attributed to the peptide ion and then used for XIC extraction as well as for the alignment process. Peptide ions identified in all the samples to be compared were used to build a retention time matrix to align LC-MS runs. If some peptide ions were sequenced by MS/MS and validated only in some of the samples to be compared, their XIC signal was extracted in the nanoLC-MS raw file of the other samples using a predicted RT value calculated from this alignment matrix by a linear interpolation method. Quantification of peptide ions was performed based on calculated XIC areas values. To perform normalization of a group of comparable runs, the software computed XIC area ratios for all the extracted signals between a reference run and all the other runs of the group and used the median of the ratios as a normalization factor. To perform protein relative quantification in different samples, a protein abundance index was calculated, defined as the average of XIC area values for at most three intense reference tryptic peptides identified for this protein (the three peptides exhibiting the highest intensities across the different samples were selected as reference peptides, and these same three peptides were used to compute the PAI of the protein in each sample; if only one or two peptides were identified and quantified in the case of low abundant proteins, the PAI was calculated based on their XIC area values). In the case of SDS-PAGE fractionation, integration of quantitative data across the fractions was performed as indicated in the text, by summing the PAI values for fractions adjacent to the fraction with the best PAI (the same three consecutive fractions for all the samples to be compared). For differential studies, a Student's t test on the PAI values was used for statistical evaluation of the significance of expression level variations. For proteins specifically detected in one condition and not in the other, the t test p value was calculated by assigning a noise background value to the missing PAI values. A 2-fold change and p value of 0.05 were used as combined thresholds to define biologically regulated proteins.
Quantitative PCR Experiments
Total RNA from HUVEC cells (mock treated, TNFα + IFNγ-treated, or IL1β-treated) was isolated using the Absolute RNA kit from Stratagene (Agilent Technologies, Santa Clara, CA), and cDNAs were synthesized using SuperSript III First strand cDNA synthesis system for RT-PCR (Invitrogen) according to the manufacturer's instructions. Quantitative PCR was performed using the ABI7300 Prism SDS real time PCR detection system (Applied Biosystems, Foster City, CA) with a SYBR Green PCR Master Mix kit (Applied Biosystems) and a standard temperature protocol. The results are expressed as relative quantities and calculated by the 2-ΔΔCT method. Actin was used as a control gene for normalization. Three separate experiments were performed. Primers used were purchased from Qiagen (QuantiTect primer assay), except Actin, GAPDH, NFKB2, ICAM1, and VCAM1 (from Sigma Genosys).
RESULTS
Analytical Workflow
A total lysate of cultured primary human vascular ECs was used for all experiments and processed in all cases through one-dimensional SDS-PAGE, as shown in Fig. 1. When the samples were to be analyzed in one analytical nanoLC-MS/MS run (no fractionation), the electrophoretic migration was stopped immediately after the protein samples entered the separating part of the gel, so that the whole sample was isolated into a unique gel band and subsequently in-gel digested. In our hands, processing in this way, the total cell lysate for tryptic digestion gave slightly better proteomic coverage than digestion in solution. For sample fractionation and shotgun analysis, migration was performed so that 12 gel bands could be cut afterward along the migration lanes. Gel cutting was performed systematically with a long razor blade to simultaneously cut all the corresponding gel bands for the different samples to be compared, perpendicularly to the migration direction. All in gel digestion steps were manually performed in parallel. The resulting tryptic digests were analyzed by nanoLC-MS/MS on an Orbitrap-Velos instrument with high sequencing speed to improve the MS/MS sampling and analytical coverage of the samples. MS scans were recorded in the Orbitrap, and MS/MS CID spectra were recorded in the ion trap using a classical parallel acquisition mode to obtain high resolution MS1 data for peptide quantification while optimizing the number of MS2 sequencing events to increase peptide identifications. Database searches using MS/MS sequencing data were performed through Mascot, and the results files were parsed and validated based on target decoy calculated FDRs, set at 5% for peptides and 1% for proteins. After realignment in time of nanoLC-MS runs, the software uses the m/z and time values associated to validated peptides ions of validated proteins, to extract the XIC of each of them. If some peptide ions were sequenced by MS/MS and validated only in some of the samples to be compared, their XIC signal was extracted in the nanoLC-MS raw file of the other samples using a predicted RT value and a time tolerance window. For protein quantification, a PAI was calculated, defined as the average of XIC area values of at most three intense reference tryptic peptides identified for this protein.
Fig. 1.
Experimental design to estimate the accuracy of label-free quantification with or without sample fractionation. The same endothelial cell lysate was loaded on a one-dimensional SDS gel and either collected in a single band or fractionated into 12 gel bands cut along the migration lane, and to assess how repeatability is affected by each step of the analytical process, we compared either nanoLC-MS/MS injection replicates or gel replicates. Four experiments were performed. A, the protein sample (15 μg) was collected in a single band and digested, and the corresponding peptide digest was analyzed three times by nanoLC-MS/MS. B, three identical protein samples (15 μg each) were loaded on the gel and collected in three bands, and after digestion, one-third of each corresponding peptide digest was analyzed once by nanoLC-MS/MS. C, the protein sample (100 μg) was fractionated by electrophoresis into 12 gel fractions, the 12 bands were digested, and each of the corresponding peptide digests was consecutively analyzed three times by nanoLC-MS/MS. D, three identical protein samples (150 μg each) were loaded on the gel and fractionated into 12 gel bands, and after digestion, the corresponding molecular weight bands of each gel lane were consecutively analyzed once by nanoLC-MS/MS (one-third of resulting peptide digests for each band).
Repeatability of the Label-free Quantification without Sample Fractionation
We first evaluated the repeatability of the label-free analytical workflow by comparing replicate LC-MS analyses of the same sample, without any fractionation. The first experiment consisted in triplicate nanoLC-MS/MS injections of the tryptic digest prepared from one gel band containing the whole protein mixture (Fig. 1A). In that case, sources of errors in the final quantitative results include only the variability of the nanoLC separation, of the mass spectrometry measurement, as well as of potential inconsistencies related to bioinformatic extraction of peptide XICs by the software. To evaluate the additional variability related to upstream sample processing steps (gel loading, gel migration, manual band cutting, in-gel trypsin digestion and peptide extraction), three nanoLC-MS/MS analyses were then performed on the tryptic digests obtained from triplicate gel bands containing each the same sample loaded on the gel (Fig. 1B). In both cases, the number of proteins identified from the three analytical runs was very similar (respectively 718 and 715 proteins for injection replicates or gel replicates; supplemental data 1). Although some of these proteins were identified by MS/MS in only one or two of the triplicates, the cross-assignment procedure used in MFPaQ allowed extraction of their MS signal in the runs, which did not contain any identification data for these particular proteins. As shown in supplemental data 2, this method generated a very modest number of missing values for quantification, at both the peptide and protein levels, leading to quantification of 715 and 686 proteins in these two experiments. To evaluate repeatability, the CVs of the PAI values obtained for these proteins were calculated. As shown in Fig. 2, the distribution of CVs for proteins quantified in the three gel replicates is very similar to that of CVs obtained with three injection replicates. The median CV is 5 and 6%, respectively, for the two experiments, and the interquartile range of the CV distribution is slightly increased in the case of gel replicates compared with injection replicates. Experimental steps such as gel migration or gel band processing may account for this little decrease of quantification accuracy observed for gel replicates. However, when the sample is isolated in only one band, such processes are supposed to be quite reproducible. Indeed, they seem to bring only a little additional variability, because the results show that a high percentage of the protein population is still correctly quantified (99% of proteins have CVs under 50%), with a relatively small absolute number of outlier proteins with extreme CV values. These results also confirm that label-free quantification using the identity-based signal extraction procedure in MFPaQ allows an accurate quantification of more than 600 proteins on a complex sample analyzed in a single run. This can also be seen from the correlation plots and the distribution of protein PAI ratios calculated between replicate nanoLC-MS/MS analyses (supplemental data 3).
Fig. 2.
Coefficients of variation for protein PAI values between triplicate LC-MS measurements. The results are shown for experiments without fractionation (one gel band analyzed three times or three replicate gel bands analyzed once by LC-MS) or with one-dimensional SDS-PAGE fractionation (each of the 12 molecular weight fractions from one gel lane analyzed three times consecutively or analysis of three gel lanes). A, histogram of CVs distribution in the different experiments. B, box and whisker plots showing the dispersion of protein CVs near the median value. The bottoms and tops of the boxes correspond to the 25th and 75th percentiles of the CV distribution, and whiskers correspond to the lowest and highest values within 1.5× interquartile range of these limits. Extreme values falling out of the box plots correspond to outliers. C, number of proteins and quantitative repeatability in each experiment.
Label-free Quantification after One-dimensional SDS-PAGE Shotgun Analysis
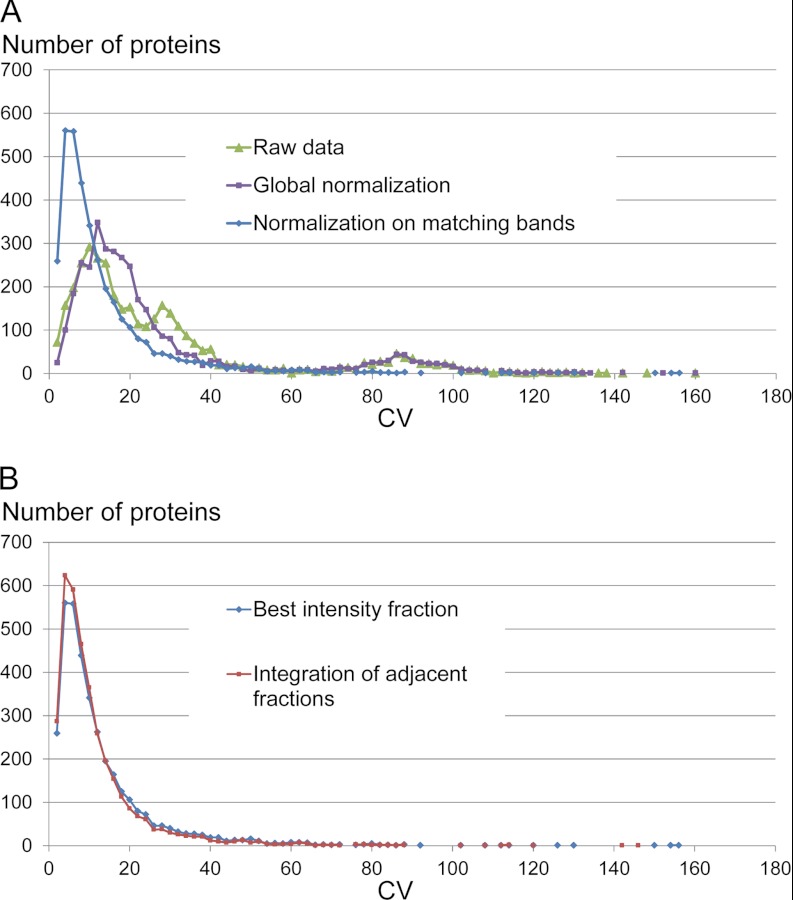
The sample was then submitted to one-dimensional SDS-PAGE and fractionated into 12 gel bands. Again, to assess how repeatability is affected by each step of the analytical process, we performed either three LC-MS/MS analyses of the 12 gel bands from the same migration lane or LC-MS/MS analyses of the gel bands from three replicate migration lanes of the same sample loaded on the gel. In this latter case, the bands within a particular molecular weight from the three lanes were analyzed successively, and peptide identifications from each of them were used to extract XICs in the corresponding bands, by cross-assignment of peptide signals in replicate LC-MS/MS runs. As expected, after fractionation the analytical coverage of the protein mixture was greatly improved, because more than 3500 unique proteins were identified in both experiments (supplemental data 1). Even on this larger population, the signal extraction performed by the software allowed retrieval of quantitative data for almost 99% of the proteins after triplicate sample fractionation through one-dimensional gel (supplemental data 2). Preprocessing of the raw quantitative data was performed to remove the systematic effects and variations because of the measurement process. As for one-shot analysis, a normalization step was used to take into account variability of the nanoLC ESI-MS signal, and in the case of gel replicates, unequal amounts of protein were loaded on the gel and gel processing variability. When this normalization procedure was performed at the scale of the whole experiment (i.e., by comparing signal intensity of all the peptides detected all along the migration lane in replicate experiments), a global correction factor was calculated and used to correct the protein PAI values of replicates experiments against a reference. As shown in Fig. 3A, this process improves to some extent the CVs of PAI values for proteins detected in replicate gel lanes but only in a limited way. A significantly better correction was achieved by comparing intensities of peptides detected in matching gel bands, allowing derivation of 12 different normalization factors applied separately to correct quantitative values in each group of molecular weight gel fractions replicates. Obviously, this approach is best suited to correct LC-MS variability, because it compares samples that were measured within a shorter lapse of time and that contain similar protein subpopulations. An automatic normalization procedure to correct peptide intensities of matching fractions in the case of fractionation experiments was thus included in MFPaQ and used in this study.
Fig. 3.
Effect of normalization and integration procedures on the quantitative results after SDS-PAGE fractionation. A, histograms of CVs for 1) nontransformed protein PAI values calculated on raw data, 2) normalized protein PAI values transformed with a global correction factor (ratio of summed PAI values for all proteins along each migration lane in replicate experiments), or 3) protein PAI values calculated from normalized XIC signals for each group of matching gel bands. B, histograms of CVs for protein PAI values calculated after normalization of XIC signals in matching gel bands, taking into account only the fraction with the best PAI for proteins identified in several gel bands, or after integration by summing PAI values on three consecutive gel bands near the best intensity fraction.
In addition, to correct for gel migration variability from lane to lane, an integration procedure was also included to sum up the signal of proteins detected in several fractions over the SDS-PAGE lane. However, bands along the migration lane will be corrected with different normalization factors, and integration of signal from very distant gel bands can generate quantitative errors. To evaluate gel migration variability, MFPaQ was used to retrieve the apex of the electrophoretic gel migration pattern for each peptide identified in each replicate experiment. Table I shows the apex count distribution, reflecting the number of peptides that were detected at their maximal intensity in nonmatching fractions in the three different replicates. As expected, the apex of the vast majority of peptide ions was detected in the same gel band, but in the case of replicate gel lanes, for 1402 of the peptide ions (∼4% of the total number of precursors) the “best” gel band is identical in only two replicates of three, and for a small minority of them (91 peptide ions, 0.25%), it is different in all three replicates. To some extent, these figures include cases that may be explained by LC-MS variability: indeed, in the case of LC-MS replicates, there is also a small degree of disparity between apex fractions (364 peptide ions, 1% of total, for which the maximal intensity is measured in a nonreproducible way in one injection replicate). However, variability of the electrophoretic migration of proteins along the gel lanes and manual cutting of the fractions account for the majority of the discrepancies in the case of replicate gel lanes. Of the 1493 peptide ions for which conflicts were detected, as shown in Table I, the maximal distance between apex fractions is 1 for 1269 precursors, i.e., the maximal intensity is measured in matching gel bands or in an adjacent band for all three replicates. In many cases, this is probably due to gel cutting inside protein migration patterns and unequal partitioning of these proteins into adjacent gel bands depending on the migration lane. In a small number of other cases, the apex fractions are more distant, probably because of migration problems, irreproducible degradation or precipitation of some proteins, or wrong signal extraction by the software. To correct the most frequent artifacts associated with the SDS-PAGE fractionation process, without introducing additional errors, we thus decided to integrate quantitative data by summing the PAI values for fractions adjacent to the fraction with the best PAI (the same three consecutive fractions for all the replicates to be compared). Fig. 3B shows the result of this integration procedure on the CVs of PAI values for proteins detected in replicate gel lanes, compared with CVs calculated by retrieving only the best PAI in one fraction (identified across all the replicates, and the same matching fraction for all of them). Although the distributions are globally very similar, integration brings a small improvement on the CVs, in particular by reducing the number of extreme values (89 proteins of 3585 are measured with a CV higher than 50% when the PAI is retrieved from the best intensity fraction, versus 66 proteins when integration is performed). Thus, PAI values were summed up from three consecutive fractions in the case of fractionation experiments.
Table I. Statistics of peptide migration across the SDS-PAGE fractions.
Peptide apex count distribution indicates the number of peptide precursor ions that were detected at their maximal intensity in the same matching fractions across all three replicates (one fraction), in the same fraction in only two replicates (two fractions), or in different fractions in the three replicates (three fractions). Peptide apex distance distribution illustrates the maximal gap between apex fractions when peptides were detected in nonmatching fractions across the replicates.
| Peptide apex | No. of precursor ions |
|
|---|---|---|
| Three migration lanes | Three injections of one lane | |
| Count distribution | ||
| One fraction | 33,932 | 34,769 |
| Two fractions | 1,402 | 364 |
| Three fractions | 91 | 0 |
| Distance distribution | ||
| 0 | 33,932 | 34,769 |
| 1 | 1,269 | 312 |
| 2 | 53 | 22 |
| 3 | 21 | 10 |
| 4 | 12 | 4 |
| 5 | 11 | 1 |
| 6 | 10 | 3 |
| 7 | 9 | 6 |
| 8 | 54 | 1 |
| 9 | 38 | 4 |
| 10 | 15 | 1 |
| 11 | 1 | 0 |
Finally, as shown in Fig. 2, in the case of sample fractionation, the number of quantified proteins clearly increases, but this is associated with a higher number of extreme values falling of the normal distribution of CVs for both experiments, a significant number of proteins quantified with CVs above 50%, and a higher interquartile range for gel replicates. In the case of replicate injections, quantitative errors occur again from the same causes than in the first one-shot experiment (variations in the nanoLC peptide separation, MS analysis, and bioinformatic processing), and globally, the accuracy of the quantification is thus similar (median CV of ∼5% and comparable interquartile ranges). However, the presence of extreme values can be explained by the higher number of low abundance species that are quantified compared with one-shot measurements. Indeed, by increasing the depth of proteome analysis, the fractionation strategy generates quantitative data on low intensity signals that may be subject to larger fluctuations from one run to another or that may be incorrectly extracted by the software. This is illustrated by CV to PAI plots, which reflect a significant decrease of quantitative repeatability for lower PAI values (supplemental data 4). On the other hand, when the 12 gel bands from the three different migration lanes are analyzed independently, additional errors related to the one-dimensional SDS-PAGE fractionation process (migration and gel band cutting), which was expected to be the most important source of variability, are introduced. The distribution of CVs is shifted compared with what was obtained for injection replicates and now has a median of 7%. Thus, the gel fractionation process contributes to the variability of the measurement. However, even in that case, still 99% of the protein population has CVs for PAI values under 70%. These values illustrate the variability of the gel fractionation process when samples are loaded on adjacent lanes, on the same gel. To further evaluate gel to gel repeatability, which may be an important parameter when numerous samples have to be processed, we also performed triplicate fractionation experiments on different gels. As shown in supplemental data 5, the median CV of proteins PAI shifts from 7% when they are fractionated and quantified on one gel, to 9% when they are quantified from samples fractionated on different gels, and the distribution of CVs is slightly broader. In conclusion, sample fractionation largely improves the depth of proteome coverage, although this is obtained at the expense of quantification accuracy. However, the repeatability of the method is still acceptable for a differential quantitative study, performed with statistical analysis of replicate gel migration lanes.
Large Scale Label-free Quantitative Proteomic Analysis of Human Primary ECs under Inflammatory Conditions
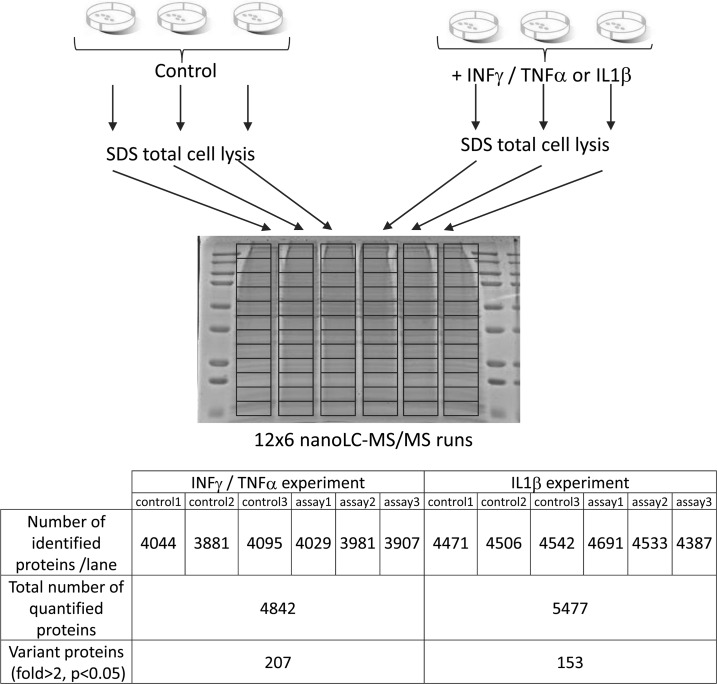
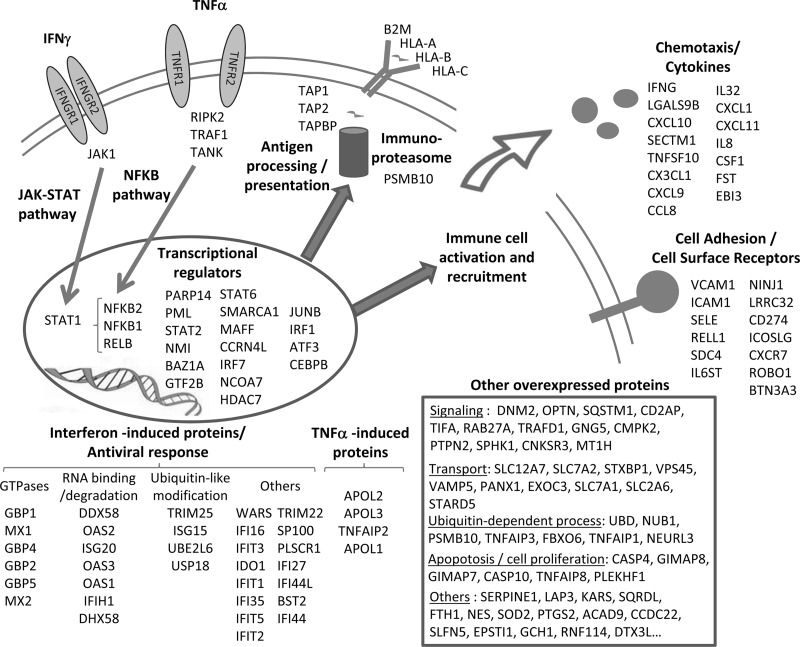
The workflow was then used in the context of a real differential biological analysis, in which we stimulated primary HUVECs with TNFα/IFNγ or IL1β, which represent potent proinflammatory cytokines that trigger inflammatory and immunological responses. The cells were lysed directly in SDS buffer and sonicated, and the resulting protein extract was loaded on a one-dimensional gel. Three biological experiments were performed, with three control samples and three stimulated samples fractionated independently on six migration lanes (Fig. 4). Using the fractionation workflow, we could identify and quantify 4842 and 5477 proteins, respectively, from ECs in the TNFα/IFNγ and IL1β experiments (supplemental data 6 and 7). Statistical analysis was performed on protein PAI values calculated after normalization and integration, as described above. For defining expression changes, two criteria were applied to derive confident data sets of modulated proteins: Student's t test p value <0.05 and expression fold change >2, as described in previous studies (16). Based on these cut-off values, 207 proteins were found to exhibit a significant variation following TNFα/IFNγ stimulation (175 up-regulated and 32 down-regulated) (supplemental data 6). Endothelial cell response to IL1β stimulation was slightly more restricted, because we measured 153 modulated proteins (119 over-regulated and 34 down-regulated) (supplemental data 7). Functional analysis of modulated proteins using the ProteinCenter software shows an important enrichment of functional categories related to inflammation and immune response (supplemental data 8). Fig. 5 shows the volcano plot representing statistical significance in function of protein variation between treated and control ECs in the case of TNFα/IFNγ stimulation. Among the most induced proteins, we found many well known cell surface membrane proteins involved in leukocyte recognition and recruitment (E-selectin, ICAM-1, V-CAM1, and ICOSLG), proteins involved in antigen processing and presentation through the class I major histocompatibility complex, but also inflammatory mediators, such as signaling molecules and transcription factors downstream the TNFα pathway (TRAF1, NF-κB, and RELB) or IFNγ pathway (JAK1 and STAT transcription factors), as well as many characteristic interferon-induced proteins involved in antiviral response, as illustrated in Fig. 6. Interestingly, 42 proteins were found to be up-regulated both by IL1β and TNFα/IFNγ (supplemental data 9). Most of them have been described as NF-κB target genes, confirming the role of NF-κB as a key mediator of IL1 and TNFα pathways. To corroborate the results obtained using our quantitative workflow, we tested by quantitative PCR the up-regulation of a series of genes corresponding to modulated proteins identified by the proteomic approach. All of the genes tested confirmed the results of the proteomic study, including strongly induced genes coding for proteins well known to be involved in the inflammatory process and also other genes moderately up-regulated, corresponding to proteins less described in the literature to be part of endothelial cell response to cytokines, such as ROBO1 (supplemental data 10). Altogether, this study shows that the quantitative label-free workflow used here can successfully identify the pathways activated under inflammatory conditions, and it provided a detailed proteomic characterization of the response triggered by inflammatory cytokines in ECs.
Fig. 4.
Experimental design and identification results of the large scale quantitative proteomic study of endothelial cells. Three independent biological experiments were performed by stimulating HUVECs with inflammatory cytokines in culture (either a combination of TNFα and IFNγ or IL1β). Total cell lysates from control and stimulated samples were loaded and fractionated on six parallel gel lanes and cut into 12 gel [GRAPHIC]bands. The table indicates the number of proteins identified for each gel lane, and the total number of proteins identified and quantified in each experiment, as well as the number of proteins detected as differentially expressed.
Fig. 5.
Quantitative analysis of endothelial cells proteome following TNFα-IFNγ stimulation. The volcano plot of the statistical significance of expression level changes (t test p value) as a function of protein expression ratio between control and inflamed endothelial cells. The red and blue dots indicate up-regulated and down-regulated genes, respectively.
Fig. 6.
Biological pathways activated upon inflammatory response of endothelial cells after TNFα/IFNγ stimulation. Proteins found to be up-regulated from the proteomic analysis are illustrated and classified in function of the biological processes in which they are involved or their subcellular location, according to literature data.
DISCUSSION
Global analysis and quantitative comparison of large proteomes is a fruitful approach to get insights into molecular mechanisms of complex biological systems. To obtain a comprehensive picture of such systems, proteomic analysis must be as deep as possible, to map and quantify a large range of protein species, even low abundant ones. Although they have been greatly improved in recent years, the dynamic range and the sequencing speed of mass spectrometers still represent limiting factors for discovery-based proteomics, and in classical experimental LC-MS designs, they restrict the list of proteins that can be detected and quantified in a single-run analysis. To extend the list of identified proteins and obtain quantitative data on minor species, sample prefractionation is thus generally combined to nanoLC-MS analysis, either at the protein level (mainly by SDS-PAGE) or at the peptide level (often by SCX or isoelectric focusing). In recent studies, several thousand proteins could be identified from eukaryotic cells following sample fractionation (1, 17–19). This upstream separation step is often performed on isotopically labeled and mixed samples, ensuring accurate quantification. Here, we evaluated the repeatability of an analytical workflow combining SDS-PAGE fractionation and label-free quantification based on MS signal analysis. Some features of the label-free quantification performed through the MFPaQ software in this study were 1) extraction in raw MS files of XICs from identified and carefully validated peptides, 2) use of a global index for relative quantification at the protein level, derived from the intensity values of at most three intense peptides, and 3) integration of the quantitative data from different fractions and overview of the shotgun experiment through the MFPaQ interface. The identity-based approach allowed extraction of signal for confidently identified peptides in an automated batch mode, directly on the 72 raw files of the comparative experiment (two conditions, three replicates, and twelve fractions). Quantitative data can be viewed at the peptide level through the MFPaQ interface, which displays the XICs of the peptide ions in all of the raw files corresponding to matched fractions but was also directly integrated by the software at the protein level, by computing the mean value of at most three intense peptides per protein. In the case of relatively abundant proteins identified with more than three peptides, this allowed calculation of PAI values on the highest quality signals, for a more accurate quantification. However, minor species identified with less than three peptides were also quantified based on the available peptide signals. Finally, data normalization and integration procedures were used in MFPaQ to correct LC-MS variability and errors related to nonreproducible electrophoretic migration of proteins in the case of sample fractionation.
Overall, our approach proved to behave in a robust way for the quantification of a complex proteome. Our results show that for one-shot analysis, label-free quantification can be achieved with good accuracy (median CV of 5%, 99% proteins with CV values < 48%). Sample fractionation largely improved the depth of proteomic coverage, and this was associated with a moderate decrease of quantitative measurement repeatability (median CV of 7%, 99% proteins with CV values of <62%). Thus, prefractionation by SDS-PAGE appears to be compatible with label-free quantitation for the extensive analysis of complex proteomes. In the present study, it provided a detailed characterization of the proteomic variations associated with the inflammatory response in human primary ECs. Although they can be maintained for some time in culture, these primary cells are not easily amenable to SILAC labeling, and label-free methods were particularly convenient for their quantitative analysis. For each condition (control or stimulated cells), triplicate samples were fractionated by SDS-PAGE, and analysis of each gel lane led to the identification of up to 4600 unique proteins, based on a protein FDR of 1%. Globally, analysis of the six different gel lanes by nanoLC-MS/MS and cross-assignment of peptide signals between samples led to the identification and quantitation of more than 5400 unique proteins in the IL1β experiment. In a recent study, the use of very long LC-MS gradients on 50-cm-long columns was described for in-depth analysis of complex proteomes without prefractionation (20). Such experimental strategies are probably not yet routinely applicable because they generally require high pressure chromatography devices and generate very large raw files that may be difficult to process with most current bioinformatic tools. However, they appear to be a promising approach that in principle could combine the advantages of extensive proteomic coverage and quantitative accuracy that can be obtained in single-run analysis. However, the quantitative repeatability of these several-hours-long LC-MS gradients is still to be assessed on replicate analytical runs. Regarding analytical time, analysis of 12 fractions of a one-dimensional gel lane in 2-h-long LC-MS gradients on conventional LC systems is three times longer, but technically easier to implement, than analysis of the whole sample on a long column with an 8-h gradient. On the other hand, sample prefractionation still probably represents up to now the most efficient way to get the deepest analytical coverage of a complex proteome, and the present study shows that the additional variability associated with this upstream process does not preclude quantitative analysis. Thus, although there is a trade-off between analytical time, quantitative accuracy, and proteomic coverage, putting the emphasis on this last parameter would probably require both sample fractionation and extensive peptide separation with long gradients for very extensive characterization of complex proteomes like the human one. Here, by using only a shotgun approach based on one-dimensional protein fractionation, sufficient depth was obtained here to detect changes on very low abundance proteins such as some transcription factors and signaling molecules. Although this label-free approach requires more analytical time than a SILAC-based experiment, because the samples are injected separately, it also avoids possible quantitation errors caused by superposition of one peptide of the SILAC pair with other different isotopic peptide patterns. In our hands, it also yielded a higher number of identified proteins, because the same MS/MS sequencing time is spent on less complex mixtures, containing half the number of peaks compared with isotopically labeled and mixed samples. Thus, MS intensity-based label-free quantification associated with SDS-PAGE fractionation appears as a valuable strategy for the differential analysis of complex proteomes.
The shotgun approach used in our study provided an in-depth characterization of the EC proteome and the label-free quantitative proteomic workflow allowed deciphering the inflammatory response of these cells. TNFα and IFNγ are potent pleiotropic cytokines that exert a number of biological effects and trigger a set of complex molecular programs in response to microbial or viral infection. IFNγ is produced mainly by NK cells and T helper type I cells and, through binding to its specific type II IFN receptor, activates the JAK-STAT signaling pathway, to induce the expression of a large number of genes (21, 22). In this large scale proteomic experiment, we measured an up-regulation of the JAK1 kinase and STAT1 transcription factor, which are known to mediate IFNγ response and regulate genes downstream of γ-activated sequence elements. We could also detect an increase of proteins involved in the TNFα signal transduction pathway, such as TRAF1, TANK, and RIPK2, converging to the activation of the NF-κB transcription factor (23). Accordingly, we measured overexpression of NF-κB subunits (NFKB2, NFKB1, and RELB) and a decrease of the inhibitor of NF-κB (IKBB), which controls nuclear translocation of NF-κB and undergoes proteasomal degradation upon TNFα signaling (24). In addition, several other proteins involved in transcriptional regulation were shown to be up-regulated after stimulation by the two cytokines (Fig. 6), such as members of the STAT family (STAT2 and STAT6); the PARP-14 protein, which enhances STAT6-dependent transcription (25); or the IRF1 secondary transcription factor, which is induced by STAT1 and plays a key role in orchestrating the IFN-induced inflammatory response (26).
One major biological process that makes part of this response in ECs is recruitment and activation of leukocytes to the inflammatory site. ECs line the blood vessel walls, and upon stimulation by cytokines, they secrete chemokines, which are chemoattractants for lymphocytes and monocytes, and express at their surface adhesion molecules that capture circulating leukocytes. We measured in our analysis the strong up-regulation of a panel of chemokines, such as Fractalkine, IL8, CXCL10, CXCL11, CXCL9, CXCL1, or CCL8, and of other secreted signaling molecules such as IL27b, or IL32. Indeed, IL32 was recently shown to be a critical regulator of EC function, which is strongly increased upon IL1β or TNFα stimulation and mediates in particular the expression of cell surface adhesion molecules involved in lymphocytes binding such as VCAM1 (27). Although our analysis was performed on a whole cell lysate and not focused on membrane proteins, we could clearly measure the overexpression of several cell surface proteins involved in cell-cell interactions. Leukocyte adhesion molecules such as E-selectin, ICAM1, and VCAM1 were the among the most strongly induced gene products and represent major players in the initial rolling and arrest step of leukocytes-EC interaction along the blood vessel walls (28). Simultaneously, molecules known to promote procoagulant activity at the EC surface such as plasminogen activator inhibitor 1 (Serpine 1) were also induced (29). Other cell surface proteins were shown to be overexpressed in response to TNFα/IFNγ treatment, such as the ICOS-ligand protein, which is an important costimulator in EC-mediated T cell activation (30); the ROBO1 receptor that may play a role in leukocyte migration (31); or the programmed cell death 1 receptor ligand PDL1, involved in immune regulation (32). Additionally, the expression of cell surface class I MHC molecules was also increased upon stimulation by inflammatory cytokines. ECs constitutively express class I MHC molecules in vivo, which are significantly decreased during cell culture but can be restored upon IFNγ or TNFα treatment (33). Following stimulation, we observed concomitantly the induction of all the machinery for antigen processing and presentation, including the immunoproteasome responsible for degradation of cytoplasmic endogenous or viral proteins; TAP proteins involved in antigenic peptide transport to the endoplasmic reticulum; and Tapasin, which binds to the TAP complex and allows antigen loading to assembled MHC molecules (34). Finally, a wide range of interferon-induced proteins were detected as strongly up-regulated, such as small GTPases (guanylate-binding proteins, Mx1, and Mx2) and the 2′-5′-oligoadenylate synthase family, which play an essential role on viral RNA degradation and the innate immune response to viral infection (21).
The EC response to IL1β stimulation, as characterized from the second large scale proteomic experiment, shared many features with the response induced by the TNFα/IFNγ treatment. Major biological processes of EC inflammatory activation were again highlighted, i.e., secretion of chemoattractant molecules and other cytokines (CXCL6, interleukin 8, CXCL1, CXCL2, CCL2, granulocyte colony-stimulating factor, macrophage colony-stimulating factor, interleukin 27, and interleukin 32), expression of cell surface leukocyte ligands (ICAM1, VCAM1, selectin, ICOS ligand, Syndecan-4), as well as antigen processing and presentation through MHC class I molecules (immunoproteasome subunits, TAP1, Tapasin, and HLA molecules). As IL1β signals through the NF-κB pathway, many proteins induced by IL1β were also induced by TNFα (see supplemental data 9). Both cytokine are, for example, endogenous pyrogens that cause fever, and in both experiments, we found an up-regulation of the prostaglandin G/H synthase 2 of the prostaglandin G/H synthase 2 (cyclooxygenase-2, COX2), which is responsible for synthesis of prostaglandin E2 prostaglandin, the key molecule for activation of thermosensitive neurons in the hypothalamus (35, 36). Additionally, in the IL1β experiment, we detected the induction of phospholipase A2, which hydrolyzes glycerophospholipids to produce arachidonic acid, the rate-limiting step in the synthesis of prostaglandin E2 by COX2. In this experiment, we could also specifically detect the induction of the cysteine protease caspase 1, which is directly involved in cleavage of proactive IL1β into its mature form, as well new regulatory molecules such as TC1, which has been described as a novel endothelial inflammatory regulator that is up-regulated by IL1β and amplifies NF-κB signaling via a positive feedback (37).
Many proteins could be identified that were not previously described as activated in ECs, deserving further studies to determine their exact function in the inflammatory process. For example, the ROBO1 receptor protein has been described to be involved in axon guidance and neuronal precursor cell migration (38), but its potential role in mediating cell-cell interactions at the endothelial surface under inflammatory conditions has been poorly described (39). Here, we show that this protein is overexpressed in HUVECs after TNFα/IFNγ stimulation, and the induction of the corresponding gene was confirmed by quantitative PCR for both TNFα/IFNγ and IL1β treatment. Another example is the circadian deadenylase Nocturnin, which was found to be significantly induced by both TNFα/IFNγ and IL1β stimulations. This protein, that is under circadian regulation, can also mediate immediate early gene responses, and it has been hypothesized that it could be involved in the post-transcriptional regulation of both rhythmic and acutely inducible mRNAs, by controlling mRNA decay through poly(A) tail removal (40). Indeed, very recently it was shown that Nocturnin can be induced by endotoxin lipopolysaccharide and that it stabilizes the proinflammatory transcript inducible nitric-oxide synthase (40), suggesting that Nocturnin could play a role in the circadian response to inflammatory signals. The proteomic data obtained here indicate that it is also induced in endothelial cells upon stimulation with TNFα/IFNγ and IL1β and thus support the idea that this protein could play a general role in the regulation of cytokine-induced inflammatory response.
In conclusion, this is the most extensive proteomic study of EC to date, performed on the widely used in vitro primary endothelial cell model HUVEC. It allowed identification in these endothelial cells of more than 5400 proteins, adding some more depth to a large scale data set previously published (41), in which ∼3800 proteins were identified and 1300 proteins could be quantified by 18O labeling, following treatment with the proangiogenic factor vascular endothelial growth factor. The present study provides the first complete characterization at the proteomic level of the EC response to inflammatory cytokines such as TNFα, IFNγ, and particularly IL1β. The list of proteins modulated by these factors, as characterized here in a global way, can thus represent a reference to study the function of other newly discovered interleukins of the IL1 family that may trigger similar responses but also some specific pathways.
Supplementary Material
Footnotes
* This work was supported by grants from the Agence Nationale de la Recherche (Programme Plates-formes Technologiques du Vivant); Fondation pour la Recherche Médicale (Programme Grands Equipements); Ibisa (Infrastructures en Biologie, Santé, et Agronomie); and FEDER (Fonds Européen de Développement Régional); and a fellowship from Région Midi-Pyrénées (to N. D.).
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- RT
- retention time
- XIC
- extracted ion chromatogram
- EC
- endothelial cells
- IFNγ
- interferon γ
- HUVEC
- human umbilical vein endothelial cells
- FDR
- false discovery rate
- PAI
- protein abundance index
- CV
- coefficient of variation
- MHC
- major histocompatibility complex.
REFERENCES
- 1. Wiśniewski J. R., Zougman A., Nagaraj N., Mann M. (2009) Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362 [DOI] [PubMed] [Google Scholar]
- 2. Vaudel M., Sickmann A., Martens L. (2010) Peptide and protein quantification: A map of the minefield. Proteomics 10, 650–670 [DOI] [PubMed] [Google Scholar]
- 3. Park S. K., Venable J. D., Xu T., Yates J. R., 3rd (2008) A quantitative analysis software tool for mass spectrometry-based proteomics. Nat. Methods 5, 319–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Searle B. C. (2010) Scaffold: A bioinformatic tool for validating MS/MS-based proteomic studies. Proteomics 10, 1265–1269 [DOI] [PubMed] [Google Scholar]
- 5. Heinecke N. L., Pratt B. S., Vaisar T., Becker L. (2010) PepC: Proteomics software for identifying differentially expressed proteins based on spectral counting. Bioinformatics 26, 1574–1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bellew M., Coram M., Fitzgibbon M., Igra M., Randolph T., Wang P., May D., Eng J., Fang R., Lin C., Chen J., Goodlett D., Whiteaker J., Paulovich A., McIntosh M. (2006) A suite of algorithms for the comprehensive analysis of complex protein mixtures using high-resolution LC-MS. Bioinformatics 22, 1902–1909 [DOI] [PubMed] [Google Scholar]
- 7. Jaitly N., Mayampurath A., Littlefield K., Adkins J. N., Anderson G. A., Smith R. D. (2009) Decon2LS: An open-source software package for automated processing and visualization of high resolution mass spectrometry data. BMC Bioinformatics 10, 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mueller L. N., Rinner O., Schmidt A., Letarte S., Bodenmiller B., Brusniak M. Y., Vitek O., Aebersold R., Müller M. (2007) SuperHirn: A novel tool for high resolution LC-MS-based peptide/protein profiling. Proteomics 7, 3470–3480 [DOI] [PubMed] [Google Scholar]
- 9. Sturm M., Bertsch A., Gröpl C., Hildebrandt A., Hussong R., Lange E., Pfeifer N., Schulz-Trieglaff O., Zerck A., Reinert K., Kohlbacher O. (2008) OpenMS: An open-source software framework for mass spectrometry. BMC Bioinformatics 9, 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Old W. M., Meyer-Arendt K., Aveline-Wolf L., Pierce K. G., Mendoza A., Sevinsky J. R., Resing K. A., Ahn N. G. (2005) Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol. Cell. Proteomics 4, 1487–1502 [DOI] [PubMed] [Google Scholar]
- 11. Hoffert J. D., Wang G., Pisitkun T., Shen R. F., Knepper M. A. (2007) An automated platform for analysis of phosphoproteomic datasets: Application to kidney collecting duct phosphoproteins. J. Proteome Res. 6, 3501–3508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tsou C. C., Tsai C. F., Tsui Y. H., Sudhir P. R., Wang Y. T., Chen Y. J., Chen J. Y., Sung T. Y., Hsu W. L. (2010) IDEAL-Q, an automated tool for label-free quantitation analysis using an efficient peptide alignment approach and spectral data validation. Mol. Cell. Proteomics 9, 131–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bouyssié D., Gonzalez de Peredo A., Mouton E., Albigot R., Roussel L., Ortega N., Cayrol C., Burlet-Schiltz O., Girard J. P., Monsarrat B. (2007) Mascot file parsing and quantification (MFPaQ), a new software to parse, validate, and quantify proteomics data generated by ICAT and SILAC mass spectrometric analyses: Application to the proteomics study of membrane proteins from primary human endothelial cells. Mol. Cell. Proteomics 6, 1621–1637 [DOI] [PubMed] [Google Scholar]
- 14. Mouton-Barbosa E., Roux-Dalvai F., Bouyssié D., Berger F., Schmidt E., Righetti P. G., Guerrier L., Boschetti E., Burlet-Schiltz O., Monsarrat B., Gonzalezde Peredo A. (2010) In-depth exploration of cerebrospinal fluid by combining peptide ligand library treatment and label-free protein quantification. Mol. Cell. Proteomics 9, 1006–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Navarro P., Vázquez J. (2009) A refined method to calculate false discovery rates for peptide identification using decoy databases. J. Proteome Res. 8, 1792–1796 [DOI] [PubMed] [Google Scholar]
- 16. Sun N., Pan C., Nickell S., Mann M., Baumeister W., Nagy I. (2010) Quantitative proteome and transcriptome analysis of the archaeon Thermoplasma acidophilum cultured under aerobic and anaerobic conditions. J. Proteome Res. 9, 4839–4850 [DOI] [PubMed] [Google Scholar]
- 17. Graumann J., Hubner N. C., Kim J. B., Ko K., Moser M., Kumar C., Cox J., Schöler H., Mann M. (2008) Stable isotope labeling by amino acids in cell culture (SILAC) and proteome quantitation of mouse embryonic stem cells to a depth of 5,111 proteins. Mol. Cell. Proteomics 7, 672–683 [DOI] [PubMed] [Google Scholar]
- 18. Geiger T., Cox J., Mann M. (2010) Proteomic changes resulting from gene copy number variations in cancer cells. PLoS Genet. 6, pii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Luber C. A., Cox J., Lauterbach H., Fancke B., Selbach M., Tschopp J., Akira S., Wiegand M., Hochrein H., O'Keeffe M., Mann M. (2010) Quantitative proteomics reveals subset-specific viral recognition in dendritic cells. Immunity 32, 279–289 [DOI] [PubMed] [Google Scholar]
- 20. Thakur S. S., Geiger T., Chatterjee B., Bandilla P., Froehlich F., Cox J., Mann M. (2011) Deep and highly sensitive proteome coverage by LC-MS/MS without pre-fractionation. Mol. Cell. Proteomics 10.1074/mcp.M110.003699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Boehm U., Klamp T., Groot M., Howard J. C. (1997) Cellular responses to interferon-γ. Annu. Rev. Immunol. 15, 749–795 [DOI] [PubMed] [Google Scholar]
- 22. Stark G. R., Kerr I. M., Williams B. R., Silverman R. H., Schreiber R. D. (1998) How cells respond to interferons. Annu. Rev. Biochem. 67, 227–264 [DOI] [PubMed] [Google Scholar]
- 23. Baud V., Karin M. (2001) Signal transduction by tumor necrosis factor and its relatives. Trends Cell Biol. 11, 372–377 [DOI] [PubMed] [Google Scholar]
- 24. Traenckner E. B., Pahl H. L., Henkel T., Schmidt K. N., Wilk S., Baeuerle P. A. (1995) Phosphorylation of human IκB-α on serines 32 and 36 controls IκB-α proteolysis and NF-κB activation in response to diverse stimuli. EMBO J. 14, 2876–2883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mehrotra P., Riley J. P., Patel R., Li F., Voss L., Goenka S. (2011) PARP-14 functions as a transcriptional switch for Stat6-dependent gene activation. J. Biol. Chem. 286, 1767–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Taniguchi T., Ogasawara K., Takaoka A., Tanaka N. (2001) IRF family of transcription factors as regulators of host defense. Annu. Rev. Immunol. 19, 623–655 [DOI] [PubMed] [Google Scholar]
- 27. Nold-Petry C. A., Nold M. F., Zepp J. A., Kim S. H., Voelkel N. F., Dinarello C. A. (2009) IL-32-dependent effects of IL-1β on endothelial cell functions. Proc. Natl. Acad. Sci. U.S.A. 106, 3883–3888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Springer T. A. (1994) Traffic signals for lymphocyte recirculation and leukocyte emigration: The multistep paradigm. Cell 76, 301–314 [DOI] [PubMed] [Google Scholar]
- 29. van Hinsbergh V. W., Kooistra T., van den Berg E. A., Princen H. M., Fiers W., Emeis J. J. (1988) Tumor necrosis factor increases the production of plasminogen activator inhibitor in human endothelial cells in vitro and in rats in vivo. Blood 72, 1467–1473 [PubMed] [Google Scholar]
- 30. Khayyamian S., Hutloff A., Büchner K., Gräfe M., Henn V., Kroczek R. A., Mages H. W. (2002) ICOS-ligand, expressed on human endothelial cells, costimulates Th1 and Th2 cytokine secretion by memory CD4+ T cells. Proc. Natl. Acad. Sci. U.S.A. 99, 6198–6203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Prasad A., Qamri Z., Wu J., Ganju R. K. (2007) Slit-2/Robo-1 modulates the CXCL12/CXCR4-induced chemotaxis of T cells. J. Leukocyte Biol. 82, 465–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Singh A. K., Stock P., Akbari O. (2011) Role of PD-L1 and PD-L2 in allergic diseases and asthma. Allergy 66, 155–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lapierre L. A., Fiers W., Pober J. S. (1988) Three distinct classes of regulatory cytokines control endothelial cell MHC antigen expression. Interactions with immune γ interferon differentiate the effects of tumor necrosis factor and lymphotoxin from those of leukocyte α and fibroblast β interferons. J. Exp. Med. 167, 794–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li S., Paulsson K. M., Chen S., Sjögren H. O., Wang P. (2000) Tapasin is required for efficient peptide binding to transporter associated with antigen processing. J. Biol. Chem. 275, 1581–1586 [DOI] [PubMed] [Google Scholar]
- 35. Dinarello C. A. (1999) Cytokines as endogenous pyrogens. J. Infect. Dis. 179, (Suppl. 2) S294–S304 [DOI] [PubMed] [Google Scholar]
- 36. Dinarello C. A., Gatti S., Bartfai T. (1999) Fever: Links with an ancient receptor. Current biology 9, R147–R150 [DOI] [PubMed] [Google Scholar]
- 37. Kim J., Kim Y., Kim H. T., Kim D. W., Ha Y., Kim J., Kim C. H., Lee I., Song K. (2009) TC1(C8orf4) is a novel endothelial inflammatory regulator enhancing NF-κB activity. J. Immunol. 183, 3996–4002 [DOI] [PubMed] [Google Scholar]
- 38. Wong K., Park H. T., Wu J. Y., Rao Y. (2002) Slit proteins: Molecular guidance cues for cells ranging from neurons to leukocytes. Curr. Opin. Genet. Dev. 12, 583–591 [DOI] [PubMed] [Google Scholar]
- 39. Legg J. A., Herbert J. M., Clissold P., Bicknell R. (2008) Slits and roundabouts in cancer, tumour angiogenesis and endothelial cell migration. Angiogenesis 11, 13–21 [DOI] [PubMed] [Google Scholar]
- 40. Niu S., Shingle D. L., Garbarino-Pico E., Kojima S., Gilbert M., Green C. B. (2011) The circadian deadenylase Nocturnin is necessary for stabilization of the iNOS mRNA in mice. PLoS One 6, e26954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jorge I., Navarro P., Martínez-Acedo P., Núñez E., Serrano H., Alfranca A., Redondo J. M., Vázquez J. (2009) Statistical model to analyze quantitative proteomics data obtained by 18O/16O labeling and linear ion trap mass spectrometry: Application to the study of vascular endothelial growth factor-induced angiogenesis in endothelial cells. Mol. Cell. Proteomics 8, 1130–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.