Abstract
Selected reaction monitoring (SRM), also called multiple reaction monitoring, has become an invaluable tool for targeted quantitative proteomic analyses, but its application can be compromised by nonoptimal selection of transitions. In particular, complex backgrounds may cause ambiguities in SRM measurement results because peptides with interfering transitions similar to those of the target peptide may be present in the sample. Here, we developed a computer program, the SRMCollider, that calculates nonredundant theoretical SRM assays, also known as unique ion signatures (UIS), for a given proteomic background. We show theoretically that UIS of three transitions suffice to conclusively identify 90% of all yeast peptides and 85% of all human peptides. Using predicted retention times, the SRMCollider also simulates time-scheduled SRM acquisition, which reduces the number of interferences to consider and leads to fewer transitions necessary to construct an assay. By integrating experimental fragment ion intensities from large scale proteome synthesis efforts (SRMAtlas) with the information content-based UIS, we combine two orthogonal approaches to create high quality SRM assays ready to be deployed. We provide a user friendly, open source implementation of an algorithm to calculate UIS of any order that can be accessed online at http://www.srmcollider.org to find interfering transitions. Finally, our tool can also simulate the specificity of novel data-independent MS acquisition methods in Q1–Q3 space. This allows us to predict parameters for these methods that deliver a specificity comparable with that of SRM. Using SRM interference information in addition to other sources of information can increase the confidence in an SRM measurement. We expect that the consideration of information content will become a standard step in SRM assay design and analysis, facilitated by the SRMCollider.
A major goal of MS-based proteomics is to accurately and reliably identify and quantify peptides derived from a biological sample. This is most frequently accomplished by LC-MS/MS. Chromatography is used to fractionate tryptic peptides derived from a protein sample before ionization and injection into the mass spectrometer, and collision-induced dissociation fragments selected peptide ions in the collision cell of the mass spectrometer. In shotgun proteomics, a specific precursor ion is chosen according to a data-dependent acquisition algorithm. At each time point a full fragment ion spectrum is acquired and then used to infer the nature of the original peptide ion (1, 2). In contrast, targeted proteomics methods such as those using selected reaction monitoring (SRM; also referred to as multiple reaction monitoring)1 measure combinations of precursor and fragment ions continuously over time to produce extracted ion chromatograms (3). Shotgun proteomics has proven highly valuable for discovery-driven experiments because new peptides can be identified and a sample can be deeply analyzed by extensive fractionation (4). However, the method can also suffer from limited sensitivity, low reproducibility across samples, sampling bias, and ambiguity in spectra assignments to peptides (5).
Targeted proteomics methods, such as SRM, provide an increase in sensitivity, signal to noise ratio, dynamic range, and reproducibility compared with shotgun proteomics. These properties have been beneficial in a range of quantitative proteomic studies (3, 6–15). However, current SRM instruments are limited to monitoring hundreds, if retention time scheduling is used, up to thousands of transitions in one run (3, 16). Therefore the selection of peptides to monitor and the corresponding transitions needs to be done a priori, and choosing the right transitions is crucial to the outcome of the experiment. Several tools and resources are available that help the researcher to select optimal transitions and target peptides for SRM, including spectral libraries and peptide observation counts from previous experiments (3, 17–26). Additionally, large scale peptide synthesis efforts have been conducted to provide reference spectra for several peptides per protein in yeast, ultimately for the whole proteome (19).2 Using reference spectra can be very helpful when designing SRM assays; however, they do not consider the selectivity of the assay in a particular sample matrix.
Sherman et al. (28, 29) recently brought forward the argument that information content could also be used as a criterion to select suitable SRM transitions. This argument is based on the fact that only a small subset of all possible transitions of a peptide is measured, and thus the problem of ambiguity and redundancy may arise: because SRM instruments work with limited resolution (± 0.1 – 1.0 Th), other peptides with transitions that are close in Q1 and Q3 to the values used to identify the targeted peptide can interfere with the detection of the target peptide and lead to ambiguity in the measurement. Sherman et al. introduce the concept of unique ion signatures (UIS; see Fig. 1) to denote combinations of ions that map exclusively to one peptide in the proteome to be analyzed. They show that it is not only possible but highly likely that more than one peptide map to a given SRM assay if few transitions are used. In these cases, the selection of assays with minimal redundancy and minimal interferences with other peptides could help to avert this problem.
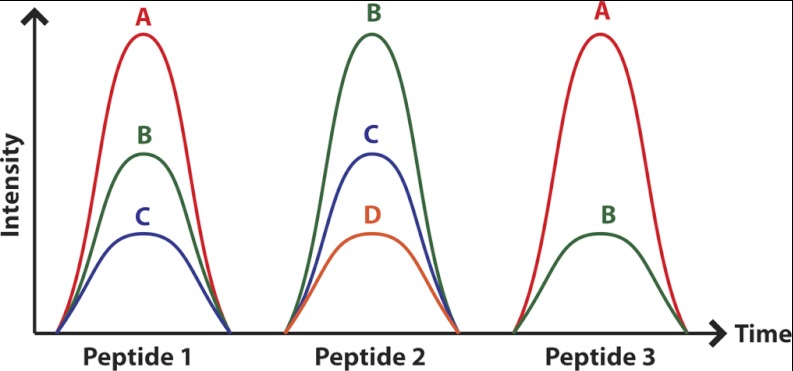
Fig. 1.
The UIS concept explained using three peptides that have some transitions in common. UISn is defined as a set of n transitions that maps exclusively to one peptide in the proteome to be analyzed. Assuming a proteome consisting of three peptides that resolve on the chromatography, there is no UIS1, one UIS2 (A and C), and one UIS3 (A–C) for peptide 1. Peptide 2, on the other hand, has one UIS1 (D), two UIS2 (D and C and D and B), and one UIS3 (B–D). Note that the transition pair (B and C) is not a unique ion signature because this signal can be explained by either peptide 1 or peptide 2.
Here we further investigated the problem of SRM assay redundancy and specificity and present a tool that predicts and simulates proteome-wide UIS, given user-defined proteolysis criteria. In contrast to previous work, we also used predicted retention times and empirical fragment ion intensities for the simulations and applied our algorithm to whole proteomes. Furthermore, we provide free access to our source code and a website where simulations can be carried out, thus helping scientists to easily apply our approach to their projects. In silico simulations of SRM experiments allowed us to reproduce and extend the results of Sherman et al. (29) by calculating unique ion signatures up to order 5 for all peptides in the proteome of human and yeast. We then show the added benefit of incorporating retention time constraints into our simulations. It reduces the number of false positive reports and relates closely to time-scheduled SRM experiments performed by experimentalists. Our platform can also be used to simulate the specificity aspect of data-independent acquisition methods, and we compare their specificity to traditional SRM methods. The results of our tool can be directly incorporated with the information derived from the SRMAtlas (19),2 which allows the researcher to combine two orthogonal methods of SRM assay development: information content-based and fragment intensity-based selection. Finally, we extend the concept of UIS using “coelution groups” (a set of transitions that coelute and are not necessarily produced by the same peptide).
We also validate our conclusions experimentally on 30 proteins of the yeast TCA cycle using 173 peptides in a mix of 14N and 15N Saccharomyces cerevisiae samples. We observed agreement of our UIS predictions with the experimental data in the majority of the MS-observable peptides.
EXPERIMENTAL PROCEDURES
Computational Methods
Unique ion signatures were calculated as described in Sherman et al. (29) with some minor differences: S. cerevisiae and Homo sapiens protein sequences were downloaded from http://www.ensembl.org, release 57_1j and 56_37a. We then generated theoretical precursor ions using trypsin for proteolysis (number of missed cleavages set to 0), carbamidomethyl on cysteine as fixed modification, and charge states 2+ and 3+ for parent ions and considered up to three heavy isotopes (+0, …, +3 atomic mass units). For each peptide we used SSRCalc version 3.0 to predict retention times (30, 31). For each precursor ion we generated the set of fragment ions (all b and y ions of charge 1+ and 2+). Of those, we generated assays for all monoisotopic doubly charged precursors using singly charged fragment ions between 400 and 1,400 Th. Note that all of these parameters are adjustable, e.g., it is possible to use a different algorithm for RT predictions, the number of missed cleavages and heavy isotopes can be chosen freely, and oxidation of methionines and deamidation of asparagine can be added as variable modifications. Furthermore, it is also possible to use other fragment ion series in the background, namely b - H2O, b - NH3, y - H2O, y - NH3, b + H2O, a, a - NH3, c, x, z, M - H2O, and M - NH3.
We use MySQL to store the data and the scripting language Python to analyze the data. Our algorithm is described in detail in supplemental Section S1, but briefly, it does the following: it finds for each query peptide all other background peptides within the specified retention time and Q1 windows and computes their transitions. The interfering peptides can either be obtained via MySQL or via a range tree that is implemented using CGAL, a C++ library for graphical algorithms (32). All transitions of the query peptide are then compared against those of the background peptides, and if the two transitions are within a predefined Q3 window, this is recorded on a per peptide basis. UIS are calculated by finding all n-tuples of transitions that do not occur in any other peptide (or transition group) all at once, where n is the order of the UIS. Unless otherwise indicated, simulations used no retention time window, a Q1 window of 1.2 Th, and a Q3 window of 2.0 Th. The complexity to compare one peptide against a background of size n is O(log2n + k) with k as the number of precursors that fall into the same Q1 window, and because k grows linearly with n, the algorithm has complexity O(n).
For the integration with spectral libraries, the collisions are computed on a per peptide basis against all potentially interfering precursors in the background (the background was calculated as described above). Then the n most intense transitions from the query peptide are evaluated whether they form a UISn, and the minimal n for which this is true is recorded.
For the extended UIS (eUIS), first all UIS combinations of the given order are calculated for each peptide and then checked whether they also form an eUIS. For each transition, the retention times of all the peptides that interfere with this transition are recorded, thus producing c arrays with retention times for a peptide with c target transitions. For an eUIS of order n, the algorithm then checks whether there exists an n-tuple, with each value drawn from a different array, such that all the values are within a certain bound Δx. If such a combination is found, it is deleted from the list of UIS. All of the eUIS simulations were done with Δx = 0.25 arbitrary SSRCalc units, considering only peptides within a window of 10 SSRCalc units around the target and using Q1 and Q3 windows of 1.2 Th and 2.0 Th as above. The source code for the SRMCollider and the web interface can be accessed at http://www.srmcollider.org/srmcollider/download.html and is available under LGPL v2.1.
Experimental Methods
S. cerevisiae strain YSBN6 was grown on YNB without AA and ammonium sulfate, 2% glucose, 10 mm KH-phthalate buffer, pH 5, and 5 g/liter ammonium sulfate containing either 14N or 15N. Proteins were sampled at optical density values of 0.97, 5.0, 5.6, and 7.1 and lysed in buffer using physical disruption. The samples were then mixed such that the protein contribution from each sample point was equal, aliquoted, precipitated with acetone, and frozen at −80 °C. The pellets were resolubilized in 150 μl of denaturing buffer containing 8 m urea and 0.1 m ammonium bicarbonate. The 14N and 15N samples were pooled, giving 300 μl of sample containing 1 mg of total protein.
Triplicate samples were treated and analyzed as described in Selevsek et al. (33). Briefly, the samples were reduced in denaturing buffer with 5 mm tris(2-carboxyethyl)phosphine at 30 °C for 30 min and alkylated with 40 mm iodoacetamide at room temperature in the dark for 30 min. The samples were diluted with 0.05 m ammonium bicarbonate to a final concentration of 1.5 m urea, and the proteins were digested overnight with sequencing grade porcine trypsin (Promega, Madison, WI) using a enzyme:substrate ratio of 1:100. Digestion was stopped by adding trifluoroacetic acid to a final concentration of 1%. Peptide mixtures were desalted using reversed phase cartridges Sep-Pak tC18 (Waters, Milford, MA) according to the following procedure: wet cartridge with 1 × 1,500 μl of 100% methanol, wash with 1 × 1,500 μl of 80% ACN, equilibrate with 5 × 1,500 μl of 0.1% trifluoroacetic acid, 2% ACN, load acidified digest, add flow through again, wash with 5 × 1,500 μl of 0.1% trifluoroacetic acid, 2% ACN, and elute with 3 × 400 μl of 40% ACN in 0.1% trifluoroacetic acid. The peptides were dried using a vacuum centrifuge, resolubilized in 500 μl of 0.1% formic acid, and frozen at −80 °C to give a final concentration of 2 μg/μl. Concentrations were confirmed with a Nanodrop (ND-1000) to be within a range of 1.2 and 1.5 mg/ml.
Tryptic peptides were analyzed on a TSQ Quantum UltraTM (Thermo Fisher, San Jose, CA). The instrument was equipped with a nanoelectrospray ion source. A spray voltage of 1.3 keV was used with a heated ion transfer tube set at a temperature of 280 °C. Chromatographic separations of peptides were performed on a NanoLC-2Dplus HPLC system (Eksigent, Dublin, CA) coupled with a 10-cm fused silica emitter, 75-μm diameter, packed with a Magic C18 AQ 5 μm resin (Michrom BioResources, Auburn, CA).
The peptides were loaded on the column from a cooled (4 °C) Eksigent autosampler and separated with a linear gradient of ACN/water, containing 0.1% formic acid, at a flow rate of 300 nl/min. A gradient from 5 to 35% ACN in 40 min was used. One microliter of each sample was injected. The mass spectrometer was operated in SRM mode. For SRM acquisitions, the first quadrupole (Q1) and the third quadrupole (Q3) were operated at 0.7 unit mass resolution. For all precursors (heavy and light, both charge states when possible), the six most intense transitions according to the yeast spectral library by Picotti et al.2 were acquired. A dwell time of 100 ms was chosen, and acquisitions occurred over the whole gradient of 40 min. Argon was used as the collision gas at a nominal pressure of 1.5 mTorr. Collision energies for each transition were calculated according to the following equations: CE = 0.034 × (m/z) + 3.314 and CE = 0.044 × (m/z) + 3.314 (where CE indicates collision energy, and m/z indicates the mass to charge ratio) for doubly and triply charged precursor ions, respectively.
Skyline (34) was used to analyze the data. A peak was considered a true signal if the heavy and light peptide traces coeluted and the intensity ratio of the two were between 0.5 and 2. The analysis was then repeated, using only the set of light transitions predicted to be unique by the SRMCollider.
RESULTS
We developed a Python-based software, the SRMCollider, that supports the simulation of a typical SRM experiment performed on a triple quadrupole instrument. It extends previous work (29) by incorporating empirical fragment ion intensities and predicted retention time information. The tool can be accessed by command line or over a website at http://www.srmcollider.org and allows the user to search a number of peptide transitions against different background databases. The parameters of the search can be selected dynamically (e.g., which mass windows to use, background proteome, number of missed cleavages and modifications in the background proteome, ion series for the background, and how many isotopes of the precursor to consider). The search will produce interfering peptides for each transition of the query peptide and unique ion signatures (UIS) of the query peptide up to order 5 (see supplemental Fig. S1 for an example). UIS are sets of transitions from the query peptide that are not found in any other peptide in the background; see Fig. 1 and Sherman et al. (29) for a more detailed explanation. The source code of our tool is published under the GNU Lesser General Public License v2.1 (LGPL v2.1).
Assay Redundancy Simulations
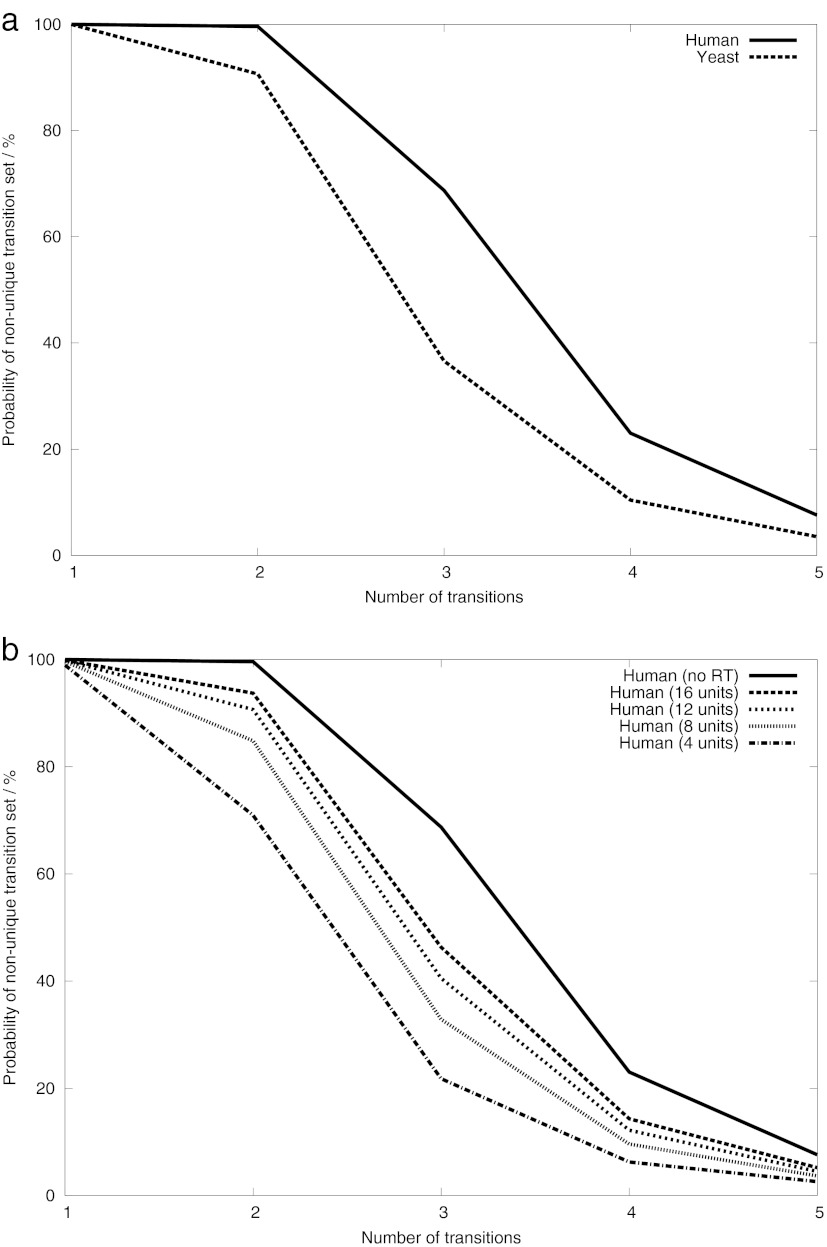
The simulation was performed to investigate the amount of redundancy in a typical SRM assay. We computed the fraction of unique assays (compared with all possible assays) for each peptide in the yeast and human proteomes, simulating the probability of selecting a unique ion signature when choosing transitions at random. Assays resulting from all monoisotopic doubly charged precursors using singly charged fragment ions between 400 and 1,400 Th were evaluated against the corresponding yeast or human background. The background was generated by in silico tryptic digestion of the respective proteome as described in the methods section. For both proteomes, we found a high rate of redundancy for single transitions, clearly illustrating the problem of transition specificity. In yeast, each transition was shared among 31.86 peptides on average; in some extreme cases over 350 peptides shared one transition. However, when using combinations of two or more transitions, the probability of selecting a redundant set of transitions decreased, confirming the results of Sherman et al. (29) (Fig. 2a). We argue that the confidence in an SRM assay can be increased by using only transitions that form a UIS.
Fig. 2.
Effects that influence the SRM assay redundancy. Displayed is the probability of picking a redundant set of SRM transitions when selecting a random set of transitions from a random peptide. a, simulating the effect of proteome size (sample complexity) using yeast and human proteomes. b, simulating the effect of using retention time separation on the probability of picking a redundant SRM assay. For each peptide, only precursor ions within a retention time window (no window, 16, 12, 8, or 4 arbitrary units) around the query peptide were considered for interference. SSRCalc (31) is used for RT predictions, and arbitrary units are displayed (on a 30-min gradient, one SSRCalc unit would correspond roughly to 30 s). All of the data were calculated for sets of transitions of size 1–5 for the complete respective proteomes (22,600 genes for human and 6,698 genes for yeast). A range of 400–1,400 Th was used for precursor and fragment ions; retention time information was not used unless indicated. Q1 and Q3 windows were 1.2 and 2.0 Th, respectively.
Influence of Retention Time
The extracted ion chromatogram covers only a fraction of the retention time space in a typical scheduled SRM experiment, and thus some of the peptides with interfering transitions are never even recorded in this mode. To avoid false positives and make our simulations more realistic, we simulated time-scheduled acquisition experiments by using SSRCalc to predict retention times for all peptides in the respective background (30, 31). Using SSRCalc allowed us to exclude ∼80% of the peptides eluting outside the acquisition window of a time-scheduled SRM experiment while retaining 80% of the peptides inside the window (see the supplemental Section S6). The number of unique combinations of transitions per peptide increased significantly when a smaller retention time window was used (Fig. 2b). Assuming that we only allow for a window of four SSRCalc units, corresponding (on a 30-min gradient) roughly to a 2-min retention time window in which the peptide of interest will elute, we find that using retention time information is similar in effect to adding one more transition in an assay. Implementing relatively narrow elution time windows is possible if standardized peptides with known retention times are measured in each run to recalibrate the retention times of the sample peptides.
Simulations for Data-independent Acquisition Methods
Data-independent acquisition methods can bridge the gap between discovery-driven proteomics and targeted proteomics (35–38). These methods fragment all or a part of the precursors and record a complete MS2 scan of the fragments over time, thus acquiring a complete scan of all species and their fragments in a sample. Some techniques, such as PAcIFIC (precursor acquisition independent from ion count), use window sizes similar to SRM (2.5 Th), whereas others such as MSE use ultra performance liquid chromatography and fragment the whole mass range of MS-accessible ions (36, 37). Here, we use our tool to compare different data-independent methods with respect to the number of interfering signals. Specifically, we wanted to design a Q1 acquisition window and a Q3 (full MS/MS) extraction window for data-independent SWATH (sequential windowed acquisition of all theoretical fragment ion mass spectra) acquisition that would perform similarly to SRM in terms of interfering signals (38). We thus ran simulations for different points in the Q1–Q3 space and recorded the number of UIS of different orders for each point for a given background scenario of all theoretical tryptic yeast peptides with no variable modifications or missed cleavages. We could thus theoretically provide optimal parameters that would exploit the speed gain of the SWATH method without sacrificing specificity in terms of increased number of interferences. Fig. 3 shows that parameters of 25 Th for the acquisition window and 50 m Th for the extraction window in the MS2 scan offer similar or even better specificity than traditional SRM with 1 Th on Q1 and 1 Th on Q3.
Fig. 3.
Simulations displaying the fraction of UIS2 for different points in Q1–Q3 space. The log-log plot of the Q1–Q3 space shows the fraction of UIS2 (unique transition pairs) per peptide compared with all possible sets of two transitions as a proxy for the number of unique assays. The simulation was performed in a tryptic yeast proteome using no separation in RT. Whenever one value is held constant, a clear trend toward more redundancy (red color) is observed when increasing the size of the other value. We can derive roughly the equivalence of certain configurations from this data, e.g., that 25 Th acquisition with an extraction window of 50 mTh is similar in specificity to 1 Th acquisition on the Q1 with a 0.5 Th acquisition on the Q3.
In Fig. 3 we provide a map for the Q1–Q3 space showing the fraction of free UIS2 (compared with all possible assays of size 2) for different values of Q1 and Q3. It is clearly apparent that, as expected, increasing either Q1 or Q3 leads to fewer unique assays. It is also apparent that it is possible even for very wide Q1 windows to retain a reasonable number of collision-free assays if the Q3 window size is adjusted accordingly. We found that introducing variable modifications or missed cleavages increases the absolute number of interferences but does not alter the relative values of interferences, and thus our overall conclusions also hold when more complex backgrounds are considered (data not shown).
Integrating Information from Spectral Libraries
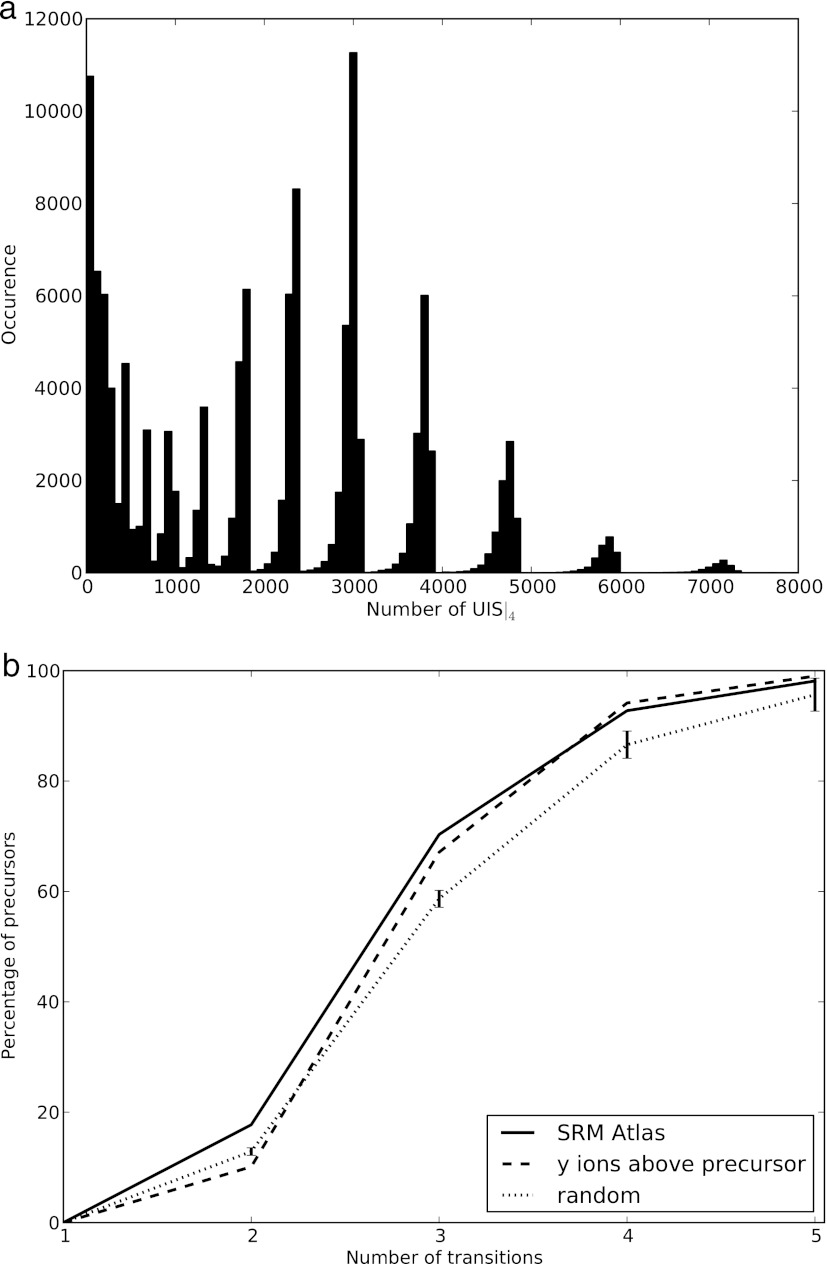
The SRMCollider considers assay redundancy and the relative intensities of the transitions to generate meaningful assays because they are experimentally extremely important and can be reproducibly obtained. The recent availability of high quantities of high quality reference spectra allowed us to integrate our tool with spectral libraries published in the SRMAtlas (19).2 Often there are many unique sets of transitions for a target peptide—for assays using four transitions in yeast, for example, the median number of possibilities is 2,163 per peptide—too many to make an informed decision on which set of transitions to use (Fig. 4a). This means that sensitivity is lost unnecessarily if a unique set of transitions is used but does not contain the most intense transitions. We thus formulated an algorithm that takes the fragment ion intensities into account when generating query SRM assays by selecting those assays that contain the most intense transitions and are thus experimentally relevant.
Fig. 4.
Combining information content-based and relative intensity-based approaches for SRM assay selection. a, distribution of the number of UIS4 for peptides with more than 7 amino acids in a yeast background. Many peptides have thousands of possible UIS4, thus demanding an informed choice on which set of transitions to use. b, by integrating relative intensity information from spectral libraries, the set of transitions to use is determined by the most intense transitions as inferred from the library. We show the minimal number of transitions needed to create a unique assay for all precursors in the SRMAtlas and compare them with a selection according to the “y ions above precursor” rule and to random transitions (average of 100 runs; error bars represent two times the standard deviation). We used no SSRCalc window, 1.2 Th on the Q1 and 2.0 Th on the Q3.
We calculated the minimal number of transitions n necessary to form a UIS where the transitions were selected with decreasing intensity from a reference spectrum. For example, in yeast the four most intense transitions (y5, y6, y3, and y4) of the precursor AIAGHLVEFFR/3 (A) form a UIS, whereas the three most intense transitions (y5, y6, and y3) do not (Table I). These are shared with the peptide LLQLNNDDTSK/3 (B), where the y5 of A interferes with the b6 of B, the y6 of A interferes with the b7 of B, and the y3 of A interferes with the b4 of B. In this case, some UIS of lower order also exist, and it would be possible to measure the UIS (y6 and y4) (second most abundant ion with the fourth most abundant ion). However, the most abundant and the third most abundant transitions could still be used for quantification and should also be measured. In this example, by measuring the four most abundant ions, one thus gets a theoretical guarantee of uniqueness for the measured signal (under the given assumptions) without sacrificing any sensitivity. We calculated the minimal number of transitions necessary for each precursor ion in the yeast SRMAtlas, and we find that for most precursors, it is sufficient to measure the three or four most intense transitions. In Fig. 4b, we compare the number of necessary transitions when using the most intense transitions to a random selection of transition and the “all y ions above the precursor” rule.
Table I. All peptides interfering with more than two transitions of AIAGHLVEFFR.
Listed are the 13 peptides that share three or more transitions with the target peptide (320 peptides sharing less than three transitions are not shown here). Target transitions are ordered by signal intensity, and signal interference is indicated by ×. For each peptide, the distance to the target peptide in retention time (predicted by SSRCalc) and Q1 is given as well as the number of observations in PeptideAtlas (PA). Although many possible unique ion signatures exist, the experimentally relevant ones are those that contain the most abundant transition signals (indicated by transition rank). The results are valid for SRM settings of 1.2 Th on Q1 and 2.0 Th on Q3; the last four peptides interfere with a higher isotope with the target peptide. Charge states of all precursors are 3+. Transitions are in their order of signal strength: y5+, y6+, y3+, y4+, y7++, y9++, b5+, y7+, y8+, and y8++.
| Transition rank | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | ΔSSRCalc | ΔQ1 | PA |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SSSVISTPVASPK | × | × | × | 13.07 | 0.00 | 0 | |||||||
| RPSENPFFHK | × | × | × | 12.43 | 0.35 | 0 | |||||||
| ALHFEYPPGTK | × | × | × | 11.78 | 0.02 | 0 | |||||||
| GEAELTFIPQR | × | × | × | 5.32 | 0.32 | 0 | |||||||
| KPSSYVMVPRP | × | × | × | 13.69 | 0.33 | 0 | |||||||
| GQQSITNEDLR | × | × | × | 16.73 | 0.31 | 0 | |||||||
| SILNMLSVIDR | × | × | × | 9.08 | 0.34 | 8 | |||||||
| SPQQQQGHPPR | × | × | × | × | 29.12 | 0.02 | 0 | ||||||
| LLQLNNDDTSK | × | × | × | × | 15.95 | 0.32 | 1 | ||||||
| APSDTTFDLYK | × | × | × | 9.59 | 0.31 | 7 | |||||||
| YSTAHLNKPPK | × | × | × | 22.26 | 0.33 | 2 | |||||||
| LPWYVLSSYK | × | × | × | 1.18 | 0.34 | 0 | |||||||
| AIYTSLLHLAR | × | × | × | 0.6 | 0.32 | 4 |
Extended UIS
There are certain cases in which a unique ion signature is not sufficient to produce a single, unique peak in SRM because of a violation of some assumptions used to construct the UIS. One reason for this can be a case of two coeluting peptides, where both peptides contribute one or more transitions and thus produce a second peak of coeluting transitions (in addition to the target peptide). This would not be captured by using UIS because they are only unique in respect to other individual peptides but not necessarily in respect to multiple coeluting peptides. We wanted to investigate whether we could incorporate this phenomenon into our simulations and find combinations of transitions that did not have that problem.
For the eUIS analysis, we simulated a proteome containing all synthetic yeast peptides. For each combination of transitions from the target peptide, our algorithm searched for a set of coeluting peptides (eluting within a retention time window of 2Δx) that together contain the transitions of the target peptide. If no such set of peptides existed, we considered the eUIS criteria to be fulfilled. The parameter Δx needs to be chosen such that it is lower than the time resolution of the SRM experiment to simulate coelution of peptides. The set of transitions from the coeluting peptides was termed a “coelution group”; note that if it only consists of transitions from one peptide (i.e., if Δx = 0), the analysis reduces to regular UIS analysis (see supplemental Section S3 for an example and a more detailed explanation of the eUIS concept).
We simulated acquisition with eUIS using a very strict definition of a coelution group (Δx = 0.25 arbitrary SSRCalc units window, considering only coeluting peptides within a window of 10 SSRCalc units) corresponding to two peaks whose apices have shifted at most 15 s on a 30-min gradient. For a whole yeast tryptic digest, we find that the probability to choose an eUIS set of transitions drops (compared with UIS) from 41.72 to 13.35% for order 2 and from 83.52 to 27.78% for order 3 and from 95.2% to 41.36% for order 4 and from 98.2% to 52.67% for order 5.
Unfortunately, the eUIS simulations are not of predictive value on an individual peptide level because the resolution of SSRCalc is below the used value. However, the global analysis is still valid, assuming that the SSRCalc values accurately represent the distribution of retention times.
Experimental Validation
We validated our predictions experimentally by targeting the tricarboxylic acid cycle in S. cerevisiae. Applying the spectral library approach described earlier, we selected 30 target enzymes associated with the TCA cycle using the Kyoto Encyclopedia of Genes and Genomes (39) and retrieved reference spectra from the spectral library (see supplemental Section S7). The six most intense fragments were chosen for SRM monitoring, and the corresponding transitions were measured on a yeast tryptic digest sample over the whole retention time range using a TSQ Quantum UltraTM instrument. An internal heavy 15N reference confirmed the identity of the peptides in question. We then calculated the minimal number of transitions n necessary to form a UIS where the transitions were selected with decreasing intensity from the reference spectrum and used only those transitions to identify a peptide. We used our experimental parameters of 0.7 and 0.7 Th for the Q1 and Q3 window as well as a background of mixed 14N and 15N peptides. Using triplicates, we were able to exclude noise from the data, e.g., cases where the noise present in one replicate created an apparent coelution. The relative intensity information in the data was not used here because we wanted to study the coelution of transitions. We found that 87% of the peptides and 100% of the studied proteins could be observed with the minimal number of transitions predicted by the SRMCollider.
From 30 proteins, we could detect 23 with certainty, whereas none of the 7 other peptides could be detected. 117 peptides from 23 proteins were analyzed whose presence was asserted using the heavy 15N reference. 102 peptides (87.2%, belonging to 23 proteins) had predicted UIS that were unique over the whole retention time range using a background that allowed for one missed cleavage and variable deamidation of asparagine and oxidation of methionine (only 93 peptides were unique considering a fully tryptic background without variable modifications). 3 peptides (2.5%) had interferences that were not predicted by the SRMCollider and required one or two additional transitions to be uniquely identified. Finally, 8 peptides (7%) could only be identified using the coelution information of the 15N reference, and the last 4 (3.4%) peptides could not even be identified using the heavy reference. The proteins that could not be detected were Sdh4, YJL045W, Sdh3, YMR118C, Idp3, Irc15, and Cit3. Most of these 7 proteins had zero observed peptides in PeptideAtlas or very few observations under special fractionation conditions and were thus not expected to be observable using standard extraction and MS protocols.
DISCUSSION
Although it is generally accepted in the proteomics community that one transition per peptide is not sufficient to create a conclusive SRM assay, the underlying reason for this fact might be underappreciated, namely that assay redundancy can become a hurdle when designing and measuring SRM assays for a given peptide in a highly complex background. Here we present a computational tool that shows that traditional methods to select SRM assays can lead to ambiguous assays because of interferences with peptides sharing transitions with the target peptide. However, when interferences are considered in the design phase of a study, unique assays may be obtained, often requiring fewer transitions per peptide than would be chosen by traditional criteria. We provide an open source program to assist in the design of unique assays that successfully integrates spectral library-based SRM assay design and information content-based SRM assay design. We thus extend previous work and furthermore show how our tool can be applied to make meaningful comparisons between different data-independent acquisition methods. Given certain assumptions, our tool is able to provide predicted SRM interferences and unique assays for target peptides conveniently over a web interface. Large scale analysis, where manual inspection of each SRM trace is infeasible, may especially benefit from using SRM interference analysis in tandem with other large scale SRM analysis platforms, such as mProphet (40).
Unique ion signatures occur frequently in human and yeast proteomes. 90% of all peptides in yeast (85% in human) possess at least one UIS of order 3 (Fig. 2a), as earlier studies on this topic have shown (29). Using our standalone tool, which is freely available for download online, UIS up to order 5 can be computed for all human peptides in less than 24 h on a regular laptop computer. For casual users who quickly want to determine the unique sets of transitions for a peptide before measurement, a webtool is available that allows fast and easy evaluation of single peptides and sets of peptides against several, freely selectable background scenarios. UIS-based analysis may also be advisable after measuring the data because sometimes transitions have to be removed from the data set, or they are missing in the best peak group. UIS-based analysis can help to decide whether the remaining transitions are sufficient to assign a peak group to a peptide, i.e., whether the remaining transitions still form a UIS. If ambiguity exists, it can be resolved by measuring additional transitions of the query peptide and of the supposed interfering peptide to establish presence or absence of either one.
By integrating the widely used SSRCalc (30, 31) retention time predictions into our analysis, we were able to simulate time-scheduled SRM experiment acquisition (Fig. 2b). The retention time constraint significantly decreases the number of interferences and mirrors experimental conditions of scheduled SRM measurements more closely. SSRCalc is used by several other SRM assay design and simulation programs (17, 41), and we validate our use of SSRCalc in the supplementary discussion with a large data set and also discuss other predictors based on support vector machines (27). We showed that the effect of using retention time scheduling with a window size of 4 SSRCalc units (corresponding to roughly 2 min on a 30-min gradient) can be comparable with adding one more transition to an assay. Acquisition windows of 2 min are within the range of achievable window sizes reported in other studies (15), but also larger windows offer considerable reduction in interferences (Fig. 2b). This underscores the importance of accurate retention time scheduling for SRM: not only can a higher number of assays be measured in one run, but the assays are also more specific.
Recent advancements in building spectral libraries make large resources of high quality reference spectra available to the scientific community. We used this data to circumvent the combinatorial explosion of possible UISn for any reasonable n by integrating spectral library-based and information content-based SRM assay design, thus creating assays that are biologically meaningful and still optimal in terms of information content. A simple algorithm creates a set of transitions that both is unique for the query peptide and also contains the most intense transitions. We are thus able to report the minimal number of transitions needed when using the SRMAtlas (19).2 Here we investigated whether any bias exists in terms of unique ion signatures when using the transitions reported in the SRMAtlas, and we found that there is a slight benefit when using the transitions from the SRMAtlas compared with random transitions in terms of specificity (Fig. 4b). Furthermore, SRMAtlas derived transitions are preferable in terms of signal intensity. Further improvements could include a reduction of the peptides in the background; for example, one could use only peptides that were observed in the PeptideAtlas repository (see Table I on how this could influence the results).
The SRMCollider can also be used in simulations of new acquisition methods such as data-independent schemata. A map of a selected metric (fraction of available UIS2 as a proxy for collision occurrence) of interferences is provided in the Q1–Q3 space (Fig. 3). This allows theoretical evaluation of traditional SRM settings (in the space up to a few Th) and data-independent acquisition methods (in the space of very wide Q1 windows and very narrow Q3 windows), as well as comparisons among these. The simulations can only give a first idea on the feasibility of such a method in terms of assay redundancy whereas other problems also have to be considered (speed, limit of detection, dynamic range, etc.).
We also showed experimentally that the predictions of SRMCollider translate well to real world SRM applications. We were able to measure 100% of the MS-observable proteins of the TCA cycle in S. cerevisiae (23 of 30) using only the minimal number of transitions predicted by the SRMCollider. Over 87% of the peptides used could be observed using only the minimal number of transitions predicted by the SRMCollider (without any use of retention time or relative fragment intensity information). Using the minimal number of transitions may also translate to reduced cost in terms of measurement time, depending on which strategy would have been used otherwise. Still, the fact that more than 10% of the predictions were incorrect shows that there are some phenomena that are not accounted for in our simulation.
Finally, we propose an extension to UIS termed eUIS by introducing the concept of a “coelution group” (all transition that coelute without necessarily belonging to the same peptide). In supplemental Section S3, we show that there are experimental conditions under which UIS are not sufficient to predict uniqueness because of the coelution of two peptides (which is caused by a violation of the assumptions underlying UIS). We also globally simulated the occurrence of eUIS in the yeast proteome and showed that for 73.25% of all peptides in yeast, at least one eUIS of order 3 exists. Unfortunately, we can only make global statements about the occurrence of eUIS in a proteome. To predict eUIS for single peptides, the exact retention times of all peptides in the background would have to be known, and current retention time predictions are not accurate enough to substitute for experimental measurements.
It is important to understand that all of the results presented here are only valid in the context of the assumptions made in our simulations. This means that there will be false positive hits, i.e., interferences that are never seen because the protein, peptide, or transition is not present, and false negative hits, i.e., interferences that occur but were not predicted because of phenomena that are not captured by our simulations. These reflect our limited understanding of the physical processes behind sample preparation and LC-MS/MS acquisition. They may result from contaminations, unknown post-translational modifications, unknown fragment ions, or miscleavages. Thus the accuracy of the prediction will depend strongly on the correlation between the simulated background matrix and the actual, experimental background. The number of false negative hits can be decreased by including a higher number of species in the background, e.g., allowing for modifications, missed cleavages, or ion series other than the b and y ion series in the background. There is a tradeoff, however, because many of these species will not be present and thus create false positive interferences predictions. We have designed our tool such that the user can add or remove certain constraints flexibly and easily add more species to the background proteome, thus adjusting the false positive rate. Furthermore, automated assay design can be hindered when no spectral library is available for the particular instrument because most SRM design pipelines (including this one) require a priori knowledge of the expected transition intensities. Without abundance models for the peptides and accurate predictions of fragment intensity, those problems will not be resolved very soon. Also, the retention time prediction suffers from certain limitations, and a more accurate predictor would be most beneficial to our approach; still, a true positive rate of 80% is similar to what other studies have reported (26) and allowed us to exclude a considerable amount of the potentially interfering peptides.
For SRM to become a high throughput targeted proteomics technology, the assay design and the data analysis need to be completely automated and free of manual steps. We present a tool that can be used in the assay design phase to create unique SRM assays or in the analysis phase to validate the uniqueness of the measurement. In combination with other design and analysis tools, we expect that the SRMCollider can contribute to a more streamlined and automated SRM design and analysis process. We also expect that the SRMCollider will help in incorporating information content in standard SRM analysis and thus increase confidence in the assignment of peak groups to peptides.
Supplementary Material
Footnotes
 This article contains supplemental material.
This article contains supplemental material.
2 Picotti, P., et al., A complete mass spectrometric reference map for the analysis of the yeast proteome, submitted for publication.
1 The abbreviations used are:
- SRM
- selected reaction monitoring
- UISn
- unique ion signature composed of n transitions that maps exclusively to one peptide in the proteome to be analyzed
- eUIS
- extended UIS
- Th
- Thomson (1 Th = 1(u/e) = 1(Da/e)).
REFERENCES
- 1. Aebersold R., Mann M. (2003) Mass spectrometry-based proteomics. Nature 422, 198–207 [DOI] [PubMed] [Google Scholar]
- 2. Domon B., Aebersold R. (2006) Mass spectrometry and protein analysis. Science 312, 212–217 [DOI] [PubMed] [Google Scholar]
- 3. Lange V., Picotti P., Domon B., Aebersold R. (2008) Selected reaction monitoring for quantitative proteomics: A tutorial. Mol. Syst. Biol. 4, 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. de Godoy L. M., Olsen J. V., Cox J., Nielsen M. L., Hubner N. C., Fröhlich F., Walther T. C., Mann M. (2008) Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature 455, 1251–1254 [DOI] [PubMed] [Google Scholar]
- 5. Domon B., Aebersold R. (2010) Options and considerations when selecting a quantitative proteomics strategy. Nat. Biotechnol. 28, 710–721 [DOI] [PubMed] [Google Scholar]
- 6. Gerber S. A., Rush J., Stemman O., Kirschner M. W., Gygi S. P. (2003) Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc. Natl. Acad. Sci. U.S.A. 100, 6940–6945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kuhn E., Wu J., Karl J., Liao H., Zolg W., Guild B. (2004) Quantification of C-reactive protein in the serum of patients with rheumatoid arthritis using multiple reaction monitoring mass spectrometry and 13C-labeled peptide standards. Proteomics 4, 1175–1186 [DOI] [PubMed] [Google Scholar]
- 8. Lin S., Shaler T. A., Becker C. H. (2006) Quantification of intermediate-abundance proteins in serum by multiple reaction monitoring mass spectrometry in a single-quadrupole ion trap. Anal. Chem. 78, 5762–5767 [DOI] [PubMed] [Google Scholar]
- 9. Wolf-Yadlin A., Hautaniemi S., Lauffenburger D. A., White F. M. (2007) Multiple reaction monitoring for robust quantitative proteomic analysis of cellular signaling networks. Proc. Natl. Acad. Sci. U.S.A. 104, 5860–5865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lange V., Malmström J. A., Didion J., King N. L., Johansson B. P., Schäfer J., Rameseder J., Wong C. H., Deutsch E. W., Brusniak M. Y., Bühlmann P., Björck L., Domon B., Aebersold R. (2008) Targeted quantitative analysis of Streptococcus pyogenes virulence factors by multiple reaction monitoring. Mol. Cell. Proteomics 7, 1489–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Costenoble R., Picotti P., Reiter L., Stallmach R., Heinemann M., Sauer U., Aebersold R. (2011) Comprehensive quantitative analysis of central carbon and amino-acid metabolism in Saccharomyces cerevisiae under multiple conditions by targeted proteomics. Mol. Syst. Biol. 7, 464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Redding-Johanson A. M., Batth T. S., Chan R., Krupa R., Szmidt H. L., Adams P. D., Keasling J. D., Lee T. S., Mukhopadhyay A., Petzold C. J. (2011) Targeted proteomics for metabolic pathway optimization: Application to terpene production. Metabolic Engineering 13, 194–203 [DOI] [PubMed] [Google Scholar]
- 13. Anderson L., Hunter C. L. (2006) Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol. Cell. Proteomics 5, 573–588 [DOI] [PubMed] [Google Scholar]
- 14. Keshishian H., Addona T., Burgess M., Kuhn E., Carr S. A. (2007) Quantitative, multiplexed assays for low abundance proteins in plasma by targeted mass spectrometry and stable isotope dilution. Mol. Cell. Proteomics 6, 2212–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Picotti P., Bodenmiller B., Mueller L. N., Domon B., Aebersold R. (2009) Full dynamic range proteome analysis of S. cerevisiae by targeted proteomics. Cell 138, 795–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Stahl-Zeng J., Lange V., Ossola R., Eckhardt K., Krek W., Aebersold R., Domon B. (2007) High sensitivity detection of plasma proteins by multiple reaction monitoring of N-glycosites. Mol. Cell. Proteomics 6, 1809–1817 [DOI] [PubMed] [Google Scholar]
- 17. Cham Mead J. A., Bianco L., Bessant C. (2010) Free computational resources for designing selected reaction monitoring transitions. Proteomics 10, 1106–1126 [DOI] [PubMed] [Google Scholar]
- 18. Mallick P., Schirle M., Chen S. S., Flory M. R., Lee H., Martin D., Ranish J., Raught B., Schmitt R., Werner T., Kuster B., Aebersold R. (2007) Computational prediction of proteotypic peptides for quantitative proteomics. Nat. Biotechnol. 25, 125–131 [DOI] [PubMed] [Google Scholar]
- 19. Picotti P., Lam H., Campbell D., Deutsch E. W., Mirzaei H., Ranish J., Domon B., Aebersold R. (2008) A database of mass spectrometric assays for the yeast proteome. Nat. Methods 5, 913–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Deutsch E. W., Lam H., Aebersold R. (2008) PeptideAtlas: A resource for target selection for emerging targeted proteomics workflows. EMBO Rep. 9, 429–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Martin D. B., Holzman T., May D., Peterson A., Eastham A., Eng J., McIntosh M. (2008) MRMer, an interactive open source and cross-platform system for data extraction and visualization of multiple reaction monitoring experiments. Mol. Cell. Proteomics 7, 2270–2278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mead J. A., Bianco L., Ottone V., Barton C., Kay R. G., Lilley K. S., Bond N. J., Bessant C. (2009) MRMaid, the web-based tool for designing multiple reaction monitoring (MRM) transitions. Mol. Cell. Proteomics 8, 696–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Prakash A., Tomazela D. M., Frewen B., Maclean B., Merrihew G., Peterman S., Maccoss M. J. (2009) Expediting the development of targeted SRM assays: Using data from shotgun proteomics to automate method development. J. Proteome Res. 8, 2733–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Walsh G. M., Lin S., Evans D. M., Khosrovi-Eghbal A., Beavis R. C., Kast J. (2009) Implementation of a data repository-driven approach for targeted proteomics experiments by multiple reaction monitoring. J. Proteomics 72, 838–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sherwood C. A., Eastham A., Lee L. W., Peterson A., Eng J. K., Shteynberg D., Mendoza L., Deutsch E. W., Risler J., Tasman N., Aebersold R., Lam H., Martin D. B. (2009) MaRiMba: A software application for spectral library-based MRM transition list assembly. J. Proteome Res. 8, 4396–4405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bertsch A., Jung S., Zerck A., Pfeifer N., Nahnsen S., Henneges C., Nordheim A., Kohlbacher O. (2010) Optimal de novo design of MRM experiments for rapid assay development in targeted proteomics. J. Proteome Res. 9, 2696–2704 [DOI] [PubMed] [Google Scholar]
- 27. Pfeifer N., Leinenbach A., Huber C. G., Kohlbacher O. (2007) Statistical learning of peptide retention behavior in chromatographic separations: A new kernel-based approach for computational proteomics. BMC Bioinformatics 8, 468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sherman J., McKay M. J., Ashman K., Molloy M. P. (2009) How specific is my SRM?: The issue of precursor and product ion redundancy. Proteomics 9, 1120–1123 [DOI] [PubMed] [Google Scholar]
- 29. Sherman J., McKay M. J., Ashman K., Molloy M. P. (2009) Unique ion signature mass spectrometry, a deterministic method to assign peptide identity. Mol. Cell. Proteomics 8, 2051–2062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Krokhin O. V. (2006) Sequence-specific retention calculator. Algorithm for peptide retention prediction in ion-pair RP-HPLC: Application to 300- and 100-A pore size C18 sorbents. Anal. Chem. 78, 7785–7795 [DOI] [PubMed] [Google Scholar]
- 31. Krokhin O. V., Ying S., Cortens J. P., Ghosh D., Spicer V., Ens W., Standing K. G., Beavis R. C., Wilkins J. A. (2006) Use of peptide retention time prediction for protein identification by off-line reversed-phase HPLC-MALDI MS/MS. Anal. Chem. 78, 6265–6269 [DOI] [PubMed] [Google Scholar]
- 32.Cgal, Computational Geometry Algorithms Library. http://www.cgal.org
- 33. Selevsek N., Matondo M., Sanchez Carbayo M., Aebersold R., Domon B. (2011) Systematic quantification of peptides/proteins in urine using selected reaction monitoring. Proteomics. 11, 1135–1147 [DOI] [PubMed] [Google Scholar]
- 34. MacLean B., Tomazela D. M., Shulman N., Chambers M., Finney G. L., Frewen B., Kern R., Tabb D. L., Liebler D. C., MacCoss M. J. (2010) Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Venable J. D., Dong M. Q., Wohlschlegel J., Dillin A., Yates J. R. (2004) Automated approach for quantitative analysis of complex peptide mixtures from tandem mass spectra. Nat. Methods 1, 39–45 [DOI] [PubMed] [Google Scholar]
- 36. Plumb R. S., Johnson K. A., Rainville P., Smith B. W., Wilson I. D., Castro-Perez J. M., Nicholson J. K. (2006) UPLC/MSE: A new approach for generating molecular fragment information for biomarker structure elucidation. Rapid Commun. Mass Spectrom. 20, 1989–1994 [DOI] [PubMed] [Google Scholar]
- 37. Panchaud A., Scherl A., Shaffer S. A., von Haller P. D., Kulasekara H. D., Miller S. I., Goodlett D. R. (2009) Precursor acquisition independent from ion count: How to dive deeper into the proteomics ocean. Anal. Chem. 81, 6481–6488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gillet J., Navarro P., Tate S., Röst H., Selevsek N., Reiter L., Bonner R., Aebersold R. (2012) Targeted data extraction of the MS/MS spectra generated by data independent acquisition: A new concept for consistent and accurate proteome analysis. Mol. Cell. Proteomics 10.1074/mcp.O111.016717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kanehisa M., Goto S., Furumichi M., Tanabe M., Hirakawa M. (2010) KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Research. 38, D355–D360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Reiter L., Rinner O., Picotti P., Hüttenhain R., Beck M., Brusniak M. Y., Hengartner M. O., Aebersold R. (2011) mProphet: Automated data processing and statistical validation for large-scale SRM experiments. Nat. Methods 8, 430–435 [DOI] [PubMed] [Google Scholar]
- 41. Geromanos S. J., Hughes C., Golick D., Ciavarini S., Gorenstein M. V., Richardson K., Hoyes J. B., Vissers J. P., Langridge J. I. (2011) Simulating and validating proteomics data and search results. Proteomics 11, 1189–1211 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.