Abstract
Hematopoietic stem cells (HSCs) and leukemic stem cells (LSCs) are both capable of self-renewal, with HSCs sustaining multiple blood lineage differentiation and LSCs indefinitely propagating leukemia. The GABP complex, consisting of DNA binding GABPβ subunit and transactivation GABPβ subunit, critically regulates HSC multipotency and self-renewal via controlling an essential gene regulatory module. Two GABPβ isoforms, GABPβ1L and GABPβ2, contribute to assembly of GABPα2β2 tetramer. We demonstrate that GABPβ1L/β2 deficiency specifically impairs HSC quiescence and survival, with little impact on cell cycle or apoptosis in differentiated blood cells. The HSC-specific effect is mechanistically ascribed to perturbed integrity of the GABP-controlled gene regulatory module in HSCs. Targeting GABPβ1L/β2 also impairs LSC self-renewal in p210BCR-ABL-induced chronic myelogenous leukemia (CML) and exhibits synergistic effects with tyrosine kinase inhibitor imatinib therapy in inhibiting CML propagation. These findings identify the tetramer-forming GABPβ isoforms as specific HSC regulators and potential therapeutic targets in treating LSC-based hematological malignancy.
INTRODUCTION
HSCs have two cardinal features, self-renewal and multipotency, and are responsible for sustained production of multiple blood lineages throughout an individual’s lifetime (Orkin and Zon, 2008). The HSC counterparts in leukemias, LSCs, are also endowed with unlimited self-renewal, generating the bulk leukemic blasts (Huntly and Gilliland, 2005). The transcriptional programs in HSCs have been greatly elucidated through transcriptomic analysis and genome-wide mapping of binding locations of key transcription factors (Novershtern et al., 2011; Wilson et al., 2010). Although information on regulation of LSCs is still limited, existing data indicate that both HSCs and LSCs share some transcription factors such as Foxo3a and similar pathways such as Pten for intrinsic control of their self-renewal capacity (Miyamoto et al., 2007; Naka et al., 2010; Tothova et al., 2007; Yilmaz et al., 2006; Zhang et al., 2006). Among the key transcription factors and pathways identified to regulate HSC biological activities, most have recurring roles in at least a subset of differentiated blood cells (Novershtern et al., 2011; Orkin and Zon, 2008; Wilson et al., 2010). Such pleiotropic effects limit their potential use as therapeutic targets.
The GA binding protein (GABP) complex, consisting of DNA-binding GABPα subunit and transactivation GABPβ subunit, has been known to critically regulate cell cycle control, protein synthesis, and cellular metabolism (Rosmarin et al., 2004), as evidenced by early lethality upon germline deletion of GABPα (Ristevski et al., 2004; Xue et al., 2004). Conditional targeting studies also revealed that GABPα has cell type-specific roles in myeloid cells, as well as T and B lymphocytes (Xue et al., 2007; Yang et al., 2011; Yu et al., 2010). Whereas GABPα is encoded by a single Gabpa gene, GABPβ exists in three different isoforms: GABPβ1L and GABPβ1S are splice variants from the Gabpb1 gene, and GABPβ2 is produced from the Gabpb2 gene (de la Brousse et al., 1994; LaMarco et al., 1991). All GABPβ isoforms contain an N-terminal ankyrin repeat domain that mediates interactions with GABPα (Figure 1A). However, only GABPβ1L and GABPβ2 have highly homologous C-terminal leucine-zipper domains that mediate their homo- or heterodimerization (de la Brousse et al., 1994). These dimerizing GABPβ isoforms thus contribute to assembly of GABPα2β2 tetramer, when two or more consensus GABPα binding motifs are adjacent or brought into proximity via chromatin looping (Batchelor et al., 1998; Graves, 1998). On the other hand, GABPβ1L and GABPβ1S share identical 332 amino acids in the N-termini, but GABPβ1S does not contain the C-terminal leucine-zipper structure. Thus, GABPβ1S cannot form dimers with other GABPβ isoforms and do not contribute to tetramer assembly.
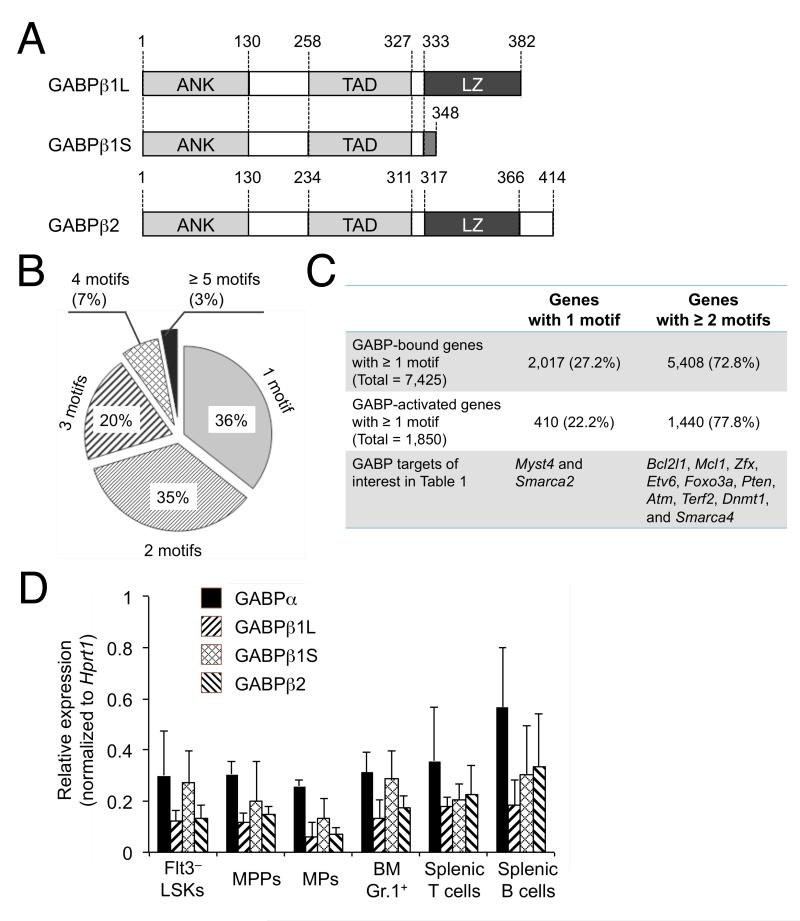
Figure 1. Analysis of GABP motif distribution in HSC genome indicates involvement of GABPα2β2 tetramer.
(A) Diagram showing the structure of GABPβ isoforms. The known functional domains of GABPβ proteins include ankyrin repeat domain (ANK), transactivation domain (TAD), and leucine-zipper structure (LZ). The amino acid boundaries of each domain are shown. Note that GABPβ1L and GABPβ1S are identical from amino acids 1-332, and GABPβ2 is highly homologous but not identical to GABPβ1 isoforms.
(B) Number of GABP motif occurrences within genome-wide GABPα binding locations in HSCs. Genome-wide GABP occupancy was mapped by ChIP-Seq, and 13,614 out of the total 15,767 GABPα binding locations in HSCs contained at least one consensus GABP motif (Yu et al., 2011). The pie chart shows the distribution of the number of GABP motifs contained within the 13,614 GABPα binding locations.
(C) Distribution of GABP motifs in GABP-bound and GABP-activated genes. Also listed are GABP-activated direct targets that have known critical roles in HSCs and are measured for expression as shown in Table 1.
(D) The expression of GABP subunits/isoforms in hematopoietic stem/progenitor cells and differentiated blood cells. Indicated cell populations were isolated from BM cells or splenocytes of wild-type C57BL/6 mice by cell sorting. The expression of GABPα, GABPβ1L, GABPβ1S, and GABPβ2 was measured by quantitative RT-PCR. The relative expression of each transcript was normalized to Hprt1, whose expression was arbitrarily set to 1 in each cell population. Data are means ± standard deviation (for Flt3− LSKs, MPPs, and MPs, n = 9 from 4 experiments, and for the rest, n = 4 from 2 independent experiments).
It has been demonstrated that the GABP complex can interface with other transcription factors or cofactors via either the GABPα or GABPβ subunit. GABPα can physically interact with Sp1, HNF4a, and FOXA2 transcription factors or recruit the CBP/p300 coactivator (Bush et al., 2003; Galvagni et al., 2001; Kang et al., 2008; Ravel-Chapuis et al., 2007; Wallerman et al., 2009). GABPβ1 can interact with non-DNA binding cofactors including HCF, YEAF1, and YAF2 (Sawa et al., 2002; Vogel and Kristie, 2000). All these GABPα- or GABPβ-interacting factors appear to act through the GABP complex, rather than functioning in lieu of the β or α subunit, respectively. Because GABPα is the sole DNA binding subunit, inactivation of GABPα disrupts the activity of entire GABP complex and abrogates its interaction with other cooperating factors, accounting for early embryonic lethality in Gabpa-targeted animals (Ristevski et al., 2004; Xue et al., 2004). On the other hand, the GABPβ subunit has three co-expressed isoforms. Germline targeting of GABPβ1L or GABPβ2 individually did not cause apparent abnormalities in embryogenesis or lymphoid lineage development (Jing et al., 2008; Xue et al., 2008), suggesting that GABPβ isoforms have partly redundant functions and may confer fine-tuned regulatory activities of the GABP complex.
Coupled with functional and transcriptomic analyses of GABPα-deficient HSCs, we previously mapped genome-wide GABPα occupancy and constructed a GABP-controlled gene regulatory module, which includes key molecules regulating HSC survival, quiescence, and self-renewal (Yu et al., 2011). Due to a dominant role of GABPα in HSC survival, it was not feasible to use GABPα-targeted animals to assess other functional requirements of the GABP complex in HSCs inferred from GABP target genes. By crossing GABPβ1L- or GABPβ2-targeted strains, we have obtained double deficient animals in which the capacity of GABPα2β2 tetramer formation is completely abrogated. Loss of the tetramer-forming GABPβ isoforms impaired HSC self-renewal and repopulation capacity, specifically perturbing survival and quiescence of HSCs without affecting more differentiated blood cells. We further demonstrated that the tetramer-forming GABPβ isoforms critically regulated self-renewal of LSCs in a CML model and that targeting these proteins synergized with the tyrosine kinase inhibitor imatinib in treating CML. These data suggest that GABPα2β2 tetramer is an HSC-specific regulator and may be explored as a therapeutic target for eradication of LSCs.
RESULTS
Targeting GABPβ1L and GABPβ2 diminished the HSC pool but did not affect HSC differentiation to myeloid or lymphoid progenitors
We have previously mapped genome-wide GABP occupancy in HSCs by ChIP-Seq and found that more than 85% GABP binding locations contained the core consensus motif “(a/c)GGAA(g/a)” (Yu et al., 2011) (Figure S1). Further motif analysis revealed that 64% of these GABP binding locations contained 2 or more GABP motifs (Figure 1B). In our previous studies, genes harboring GABP binding within 2 kb of their transcription initiation sites (TISs) were defined as “GABP-bound genes”, and among these, genes that are positively regulated by GABP were defined as “GABP-activated genes” (Yu et al., 2011). We found that approximately 3/4 of both sets of genes harbored 2 or more GABP motifs (Figure 1C). It has been shown that GABPα and GABPβ subunits spontaneously form αβ heterodimers, but require the presence of at least two GABP motifs in DNA to assemble into GABPα2β2 tetramers (Chinenov et al., 2000). Therefore, ChIP-seq with GABPα identifies all target genes for the GABP complex, and number of GABP motifs in each target determines whether an αβ dimer of an α2β2 tetramer is assembled. Our motif analysis thus suggests that regulation of most if not all of the GABP target genes may involve assembly of GABPα2β2 tetramers. For the remaining 1/4 of GABP target genes that contain only one GABP motif within 2 kb of their TISs, tetramer formation may still occur when distal GABP motifs are brought into close proximity through chromatin looping.
To specifically address a requirement of GABPα2β2 tetramer-forming capacity for regulating HSCs, we crossed germline GABPβ1L- and GABPβ2-targeted mice to obtain double deficient animals (dKO) in which tetramer assembly is abrogated. Inactivation of GABPβ1L or GABPβ2 alone did not compromise normal embryogenesis or lymphopoiesis (Jing et al., 2008; Xue et al., 2008), and the dKO mice remained viable, suggesting that targeting the tetramer-forming GABPβ isoforms did not drastically disrupt the activity of GABP complex. In contrast to massive cell death in HSCs upon induced inactivation of GABPα, the GABPβ1L/β2-targeted animals allowed detailed functional characterization of the GABP complex in regulating HSCs. By quantitative RT-PCR, we confirmed that all GABP subunits/isoforms were abundantly expressed in bone marrow (BM) Lin−Sca1+c-Kit+Flt3− cells (Flt3–LSKs) that contained both long-term and short-term HSCs (LT- and ST-HSCs, Figure 1C). Similarly, all these transcripts were detected in more differentiated progenitors including Lin−Sca1+c-Kit+Flt3+ multi-potential progenitors (MPPs), Lin−Sca1−c-Kit+ myeloid progenitors (MPs), and mature blood lineage cells, including BM Gr.1+ granulocytes, splenic CD3+ T cells and B220+ B cells (Figure 1D).
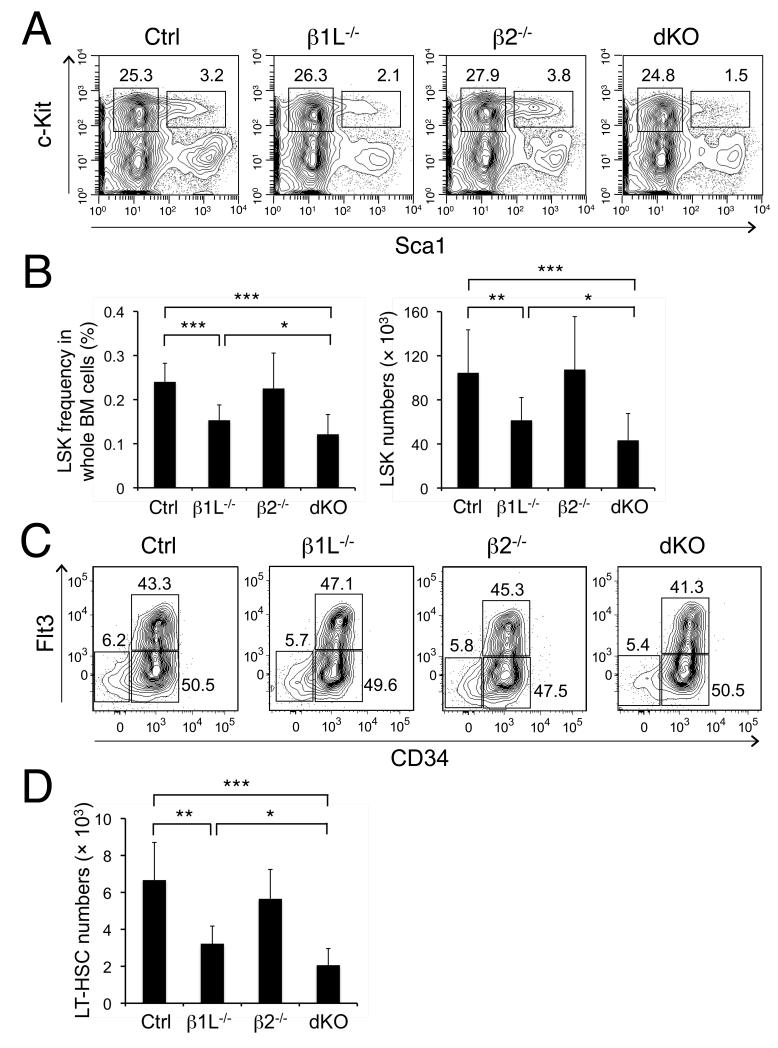
The dKO mice did not exhibit apparent defects in HSCs by the age of 6 weeks. When observed at 8 weeks or older, GABPβ1L−/− and dKO mice showed approximately 15% and 25% reductions in total BM cells, respectively (Table S1), and exhibited more pronounced decreases in LSK frequency and absolute counts (Figure 2A and 2B). Flt3 and CD34-based immunophenotypic analysis of LSKs indicates that deficiency in GABPβ1L and/or GABPβ2 did not alter differentiation of LT-HSCs (CD34−Flt3− LSKs) to ST-HSCs (CD34+Flt3− LSKs) and MPPs (Figure 2C), whereas LT-HSCs were decreased in absolute counts in GABPβ1L−/− and dKO mice (Figure 2D). Similarly, LTHSCs defined by SLAM family receptors (CD150+CD48− LSKs) were diminished in frequency in dKO Lin− BM cells (Figure S2A). On the other hand, the frequencies of SLAM LT-HSCs within the LSK subset were similar between dKO and control animals, further corroborating that HSC differentiation was not detectably affected based on SLAM markers (Figure S2B). Taken together, these observations indicate that tetramer-forming GABPβ isoforms have critical roles in maintaining a pool of HSCs and MPPs under a steady state without affecting HSC differentiation.
Figure 2. The tetramer-forming GABPβ isoforms are required for HSC maintenance but not for HSC differentiation.
(A) Detection of MPs and LSKs. BM cells from indicated mice were surface-stained, and percentages of MPs and LSKs are shown in representative contour plots (n = 10-12 from 8 experiments). (B) LSK frequency and numbers in whole BM cells. The absolute counts are from 2 tibias and 2 femurs from each mouse. Data are means ± s.d. (n ≥ 10). (C) Detection of LT-, ST-HSCs, and MPPs. The BM LSK cells were further fractionated based on CD34 and Flt3 expression. The percentage of each subset is shown. (D) LT-HSC numbers in whole BM cells. The absolute counts are from 2 tibias and 2 femurs. Data are means ± s.d. (n = 5-6 from 4 independent experiments). *, p < 0.05; **, p < 0.01; ***, p < 0.001.
In contrast to decreased LSK frequency in whole BM cells, the frequency of MPs (Figure 2A) and Lin−IL-7Rα+c-KitmedSca1med common lymphoid progenitors (CLPs, Figure S3A) was similar among animals of all genotypes (Table S1). Further differentiation of MPs in the BM, T cell development in the thymus, and B cell development in the BM was not detectably perturbed in either single knockout or dKO strains (Figure S3B-S3D). Moderate decreases in cell counts of MPs and some differentiated cells were observed in dKO mice (Table S1), which were likely secondary to diminished HSC and MPP pools. These data further support that tetramer-forming GABPβ isoforms are not required for HSC differentiation, but rather have a more specific role in HSC maintenance.
Targeting GABPβ1L and GABPβ2 impaired HSC self-renewal and repopulation capacity
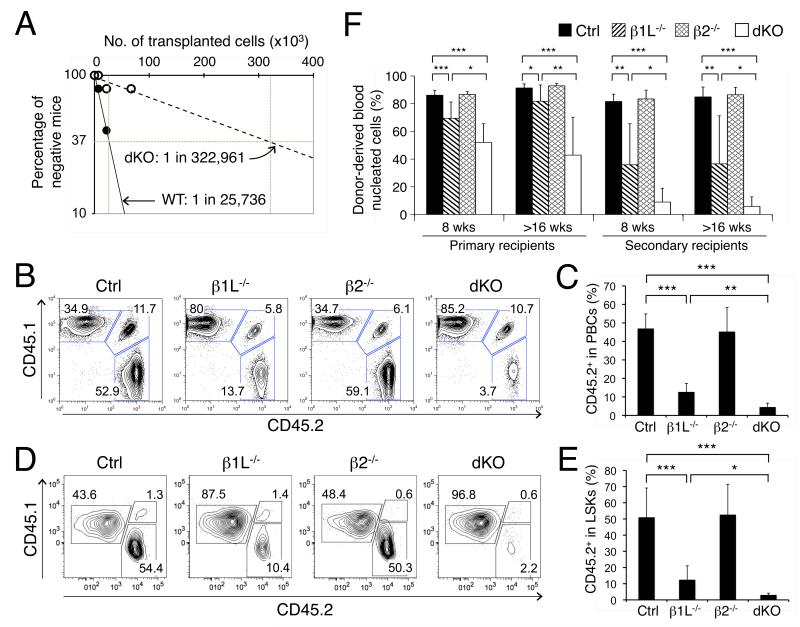
In addition to immunophenotypic analysis, we measured functional HSCs by limiting dilution assays, which detected HSCs at >10 fold lower frequency in dKO mice than controls (Figure 3A), indicating more severely impaired HSC functionality caused by GABPβ1L/β2 deficiency. We next performed a competitive repopulation assay by transplanting the test (CD45.2+) and competitor (CD45.1+) BM cells at an LSK ratio of 1:1 (Figure S4A). After ≥ 10 weeks, we found that control and GABPβ2−/− BM cells contributed similarly as the competitors to peripheral blood nucleated cells (PBCs). However, GABPβ1L−/− and dKO BM cells were much less competitive, with dKO exhibiting the most diminished contribution (Figure 3B and 3C). The same trends were evident in thymocytes, splenic T and B cells, BM B cells and granulocytes (Figure S4BS4F). Importantly, GABPβ1L−/−-derived LSKs in recipient BM were greatly diminished, and dKO-derived LSKs were further reduced (Figure 3D and 3E), thus accounting for their decreased contributions to blood lineage reconstitution. We further measured HSC self-renewal by performing serial transplantations using sort-purified LSKs lacking GABPβ1L and/or β2. Whereas BM cells from each genotype can reconstitute all blood lineages both short- and long-term (8 wks and ≥ 16 wks, respectively), the contribution from GABPβ1L−/−, especially dKO LSKs was moderately decreased in the primary recipients (Figure 3F). In secondary recipients, however, the multi-lineage contribution from GABPβ1L−/− and dKO LSKs was more markedly reduced, with dKO cells constituting <10% of blood nucleated cells (Figure 3F). Together, these results together revealed a critical requirement for tetramer-forming GABPβ isoforms in HSC self-renewal and blood lineage reconstitution capacity.
Figure 3. The tetramer-forming GABPβ isoforms are essential for repopulation capacity and self-renewal of HSCs.
(A) Detection of functional HSCs by limiting dilution assay. Graded numbers of test BM cells (CD45.2+) were mixed with 2 × 105 protector BM cells (CD45.1+) and transplanted into irradiated congenic recipients (CD45.1+). Plotted are the percentages of recipient mice containing less than 1% CD45.2+ blood nucleated cells at 16 weeks after transplantation. Frequency of functional HSCs was calculated according to Poisson statistics. (B) to (E) Competitive repopulation assay. Test (CD45.2+) and competitor (CD45.1+) whole BM cells were mixed at 1:1 LSK ratio and transplanted into irradiated CD45.1+CD45.2+ recipients. After ≥ 10 weeks, their relative contributions to blood nucleated cells (B and C) and BM LSKs (D and E) were determined. The percentages of test-, competitor-, and host-derived cells are shown in representative contour plots from 3 independent experiments (B and D). The contribution of CD45.2+ test cells to PBCs and LSKs in the hosts was collectively summarized in (C) and (E), respectively. (F) Serial transplantation assay. LSK cells were sorted from the original gene-targeted mice or primary recipients and injected into irradiated CD45.1+CD45.2+ recipients. Blood nucleated cells were analyzed at 8 and >16 weeks post-transplantation. Data are means ± s.d. (n = 5-10 from 2 independent experiments). *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Targeting GABPβ1L and GABPβ2 specifically affected HSC survival and quiescence through perturbation of the GABP gene regulatory module
We next investigated the underlying mechanisms accounting for the specific regulation of HSC activity by the tetramer-forming GABPβ isoforms. Our data indicate that, compared with GABPβ2, GABPβ1L exhibited a predominant role in HSC self-renewal and repopulation capacity. HSCs lacking both factors exhibited more severe defects than those deficient in GABPβ1L alone. This is analogous to the roles of Myc and Foxo transcription factors in HSCs. Whereas N-Myc is dispensable for normal hematopoiesis, c-Myc deletion specifically impaired HSC differentiation without affecting HSC self-renewal (Wilson et al., 2004). Targeting both N-Myc and c-Myc revealed an unexpected requirement of Myc proteins in maintaining HSC survival and proliferation (Laurenti et al., 2008). Among the Foxo transcription factors, Foxo3a appear to have a more predominant effect, but triple deletion of Foxo1/3a/4 was necessary to uncover their roles in coping with oxidative stress in HSCs (Miyamoto et al., 2007; Tothova et al., 2007). For the GABP complex, it is thus necessary to target both GABPβ1L and GABPβ2 to reveal the complete spectrum of regulatory activity of the GABPα2β2 tetramer in HSCs. We therefore focused on dKO mice in the following analyses.
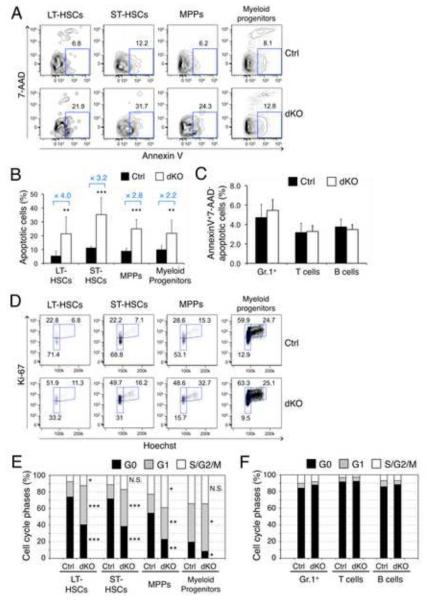
Consistent with an absolute requirement of GABPα in HSC survival (Yu et al., 2011), the frequencies of AnnexinV+7-AAD− apoptotic cells in the LT-HSC pool and in other LSK subsets were increased approximately 3-4 fold in dKO mice (Figure 4A and 4B), demonstrating a phenocopy between GABPα and GABPβ1L/β2 deficiencies. In contrast, the frequencies of apoptotic cells were not increased in dKO BM Gr.1+, splenic T and B cells (Figure 4C), supporting a specific role of the tetramer-forming GABPβ isoforms in survival of HSCs and progenitor cells. Our previously defined GABP-controlled gene regulatory module in HSCs contained 2,224 GABP-activated direct target genes (Figure S1) (Yu et al., 2011). These included pro-survival Bcl-2 family members (Bcl-2, Bcl-XL, and Mcl-1) and transcription factors Zfx and Etv6, all of which are known to critically regulate HSC survival (Galan-Caridad et al., 2007; Hock et al., 2004b; Opferman, 2007). In sorted LT-/ST-HSCs, we observed consistent decreases in these transcripts in dKO mice, ranging between 15-34% (Table 1). In contrast, consistent expression decreases were only observed for Bcl-2 in dKO BM Gr.1+ cells, among all the transcripts tested in BM Gr.1+, and splenic T and B cells (Table 1). These observations thus lend molecular support to a specific role for GABPβ1Lβ2 isoforms in HSC survival.
Figure 4. Deficiency in GABPβ1L/β2 specifically impairs HSC survival and quiescence.
(A) Detection of apoptosis in HSCs and progenitors. BM cells from dKO and control mice were surface-stained to identify LSKs and MPs, and CD34 and Flt3 expression was used to define LT-, ST-HSCs, and MPPs within the LSK subset. These cells were further stained for AnnexinV and 7-AAD. The percentage of AnnexinV+7-AAD− apoptotic cells in each subset was shown in representative contour plots. (B) Cumulative data on frequency of apoptotic HSCs or progenitors. Data are means ± s.d. (n = 5-6 from 4 experiments). (C) Apoptosis in differentiated blood cells. The frequencies of apoptotic BM granulocytes, splenic T and B cells are shown as means ± s.d. (n = 4 from 2 independent experiments). No significant differences were observed between control and dKO cells. (D) Cell cycle analysis of HSCs and progenitor cells. BM cells were surface-stained as in (A) and further stained intracellularly for Ki-67 and Hoechst33342. The percentages of Ki-67lowHoechst− G0, Ki-67highHoechst− G1, and Ki-67highHoechst+ S/G2/M phase cells are shown in representative dot plots. The gating of Ki-67 positivity was based on isotype control staining. (E) Cumulative data of cell cycle status in each LSK subset and myeloid progenitors. n = 5 from 3 independent experiments. *, p<0.05; **, p < 0.01; ***, p < 0.001; N.S., not significant. (F) Cell cycle status in differentiated blood cells. No significant differences were observed between control and dKO cells.
Table 1.
Expression of GABP target genes in HSCs, progenitors, and differentiated blood cells lacking GABPβ1L/GABPβ2
| Gene symbol | Flt3- LSKs (LT- & ST-HSCs) |
MPPs | Myeloid progenitors |
BM granulocytes |
Splenic T cells |
Splenic B cells |
|---|---|---|---|---|---|---|
| HSC Survival | ||||||
|
| ||||||
| Bcl2 | 0.66±0.20*** | 0.74±0.17** | 1.32±0.31* | 0.75±0.18* | 1.88±0.71* | 1.25±0.95N.S. |
| Bcl2l1 | 0.85±0.24* | 0.65±0.07*** | 0.91±0.37N.S. | 1.13±0.41N.S. | 0.92±0.28N.S. | 1.30±0.32N.S. |
| Mcl1 | 0.76±0.27* | 0.87±0.19N.S. | 0.95±0.14N.S. | 1.36±0.29* | 1.01±0.21N.S. | 0.88±0.26N.S. |
| Zfx | 0.72±0.17** | 0.92±0.09N.S. | 0.86±0.10* | 1.03±0.24N.S. | 1.01±0.28N.S. | 0.98±0.20N.S. |
| Etv6 | 0.79±0.24* | 0.67±0.38** | 0.91±0.15N.S. | 1.23±0.41N.S. | 0.98±0.30N.S. | 0.99±0.31N.S. |
|
| ||||||
| HSC quiescence | ||||||
|
| ||||||
| Foxo3a | 0.64±0.24*** | 0.61±0.11** | 1.06±0.33N.S. | 1.40±0.37* | 0.98±0.42N.S. | 1.08±0.48N.S. |
| Pten | 0.71±0.20** | 0.77±0.18* | 1.24±0.30N.S. | 1.10±0.25N.S. | 0.95±0.31N.S. | 0.89±0.43N.S. |
| Cdkn1a § | 0.91±0.38N.S. | 0.91±0.11N.S. | 0.77±0.22* | N.R.D. | 0.89±0.49N.S. | N.R.D. |
| Cdkn1b § | 0.78±0.39N.S. | 1.00±0.16N.S. | 0.83±0.26N.S. | 1.34±0.45N.S. | 1.33±0.66N.S. | N.R.D. |
|
| ||||||
| Other HSC regulators | ||||||
|
| ||||||
| Atm | 0.82±0.24* | 0.85±0.30N.S. | 1.27±0.26* | 1.29±0.38N.S. | 1.22±0.14* | 0.80±0.37N.S. |
| Terf2 | 0.79±0.17** | 0.74±0.13** | 0.89±0.07* | 1.47±0.73N.S. | 1.25±0.60N.S. | 1.34±0.92N.S. |
| Dnmt1 | 0.81±0.25* | 0.57±0.15*** | 0.96±0.09N.S. | 1.30±0.34N.S. | 1.30±0.29N.S. | 0.97±0.17N.S. |
| Dnmt3a ¶ | 0.68±0.24** | 0.63±0.16** | 0.91±0.29N.S. | 1.22±0.29N.S. | 0.92±0.05** | 0.93±0.09N.S. |
| Dnmt3b ¶ | 0.80±0.26*. | 0.71±0.13** | 0.83±0.16* | 1.95±0.60* | 0.77±0.25N.S. | 0.65±0.22* |
| Crebbp ¶ | 0.73±0.32* | 0.69±0.19** | 1.11±0.23N.S. | 1.22±0.34N.S. | 1.11±0.27N.S. | 0.86±0.20N.S. |
| Myst4 | 0.70±0.38* | 0.71±0.27* | 1.04±0.20N.S. | 1.18±0.17* | 1.23±0.33N.S. | 0.86±0.18N.S. |
| Smarca2 | 0.62±0.22*** | 0.63±0.29** | 0.94±0.10N.S. | 1.19±0.23N.S. | 1.04±0.12N.S. | 0.77±0.18* |
| Smarca4 | 0.79±0.26* | 0.65±0.10*** | 0.90±0.25N.S. | 0.99±0.22N.S. | 1.06±0.32N.S. | 0.98±0.20N.S. |
| Gabpa | 1.12±0.37N.S. | 1.21±0.40N.S. | 1.28±0.12** | 1.82±0.49** | 1.90±0.60N.S. | 1.04±0.20N.S. |
Quantitative RT-PCR was performed on each sorted population on indicated transcripts (all normalized to Hprt1). Data are ratios of dKO vs. control cells expressed as means ± standard deviation (n = 9 from 4 independent experiments for Flt3−LSKs, MPPs, and MPs, n = 4 from 2 independent experiments for BM granulocytes, splenic T and B cells).
, non-GABP target genes as unaffected controls;
, non-GABP direct target genes but affected in expression by GABPα deficiency in HSCs. N.R.D., not reliably detectable; N.S., not statistically significant;
, p < 0.001;
p<0.01;
p<0.05 by Student’s t-test. Significant and consistent changes in HSCs and MPPs are highlighted in red, and those in myeloid progenitors and differentiated blood cells are in blue for direct comparison.
The GABP direct targets in the gene regulatory module also predict a role of the GABP complex in maintaining HSC quiescence (Yu et al., 2011). Cell cycle analysis with Hoechst 33342 and Ki67 showed that whereas the majority of control LT-HSCs were maintained in the Ki67−Hoechstlow G0 phase, dKO LT-HSCs exhibited substantial reduction in the G0 dormant state and concomitant increase in G1 and S/G2/M phases with active cycling (Figure 4D and 4E). The dKO ST-HSCs and MPPs showed similarly increased cycling, but this was not observed in mature myeloid or lymphoid cells (Figure 4E and 4F). By 5-bromo-2′-deoxyuridine (BrdU) pulsing in vivo, we found that all the LSK subsets (in particular LT-HSCs), but not myeloid progenitors or mature blood cells, exhibited a pronounced increase in proliferation in dKO mice (Figure S5). Two GABP direct target genes, Foxo3a and Pten, are known to restrain HSCs from hyper-proliferation (Miyamoto et al., 2007; Tothova et al., 2007; Yilmaz et al., 2006; Zhang et al., 2006). Transcripts of both genes showed approximately 30% reduction in dKO LT-/ST-HSCs and MPPs, but were not decreased in MPs or differentiated blood cells (Table 1). The non-GABP direct targets, cyclin-dependent kinase inhibitors p21Cip/Waf1 and p27Kip1 (encoded by Cdkn1a and Cdkn1b, respectively), are known to have critical roles in maintaining HSC quiescence (Cheng et al., 2000; Walkley et al., 2005). Their expression was not consistently altered in dKO cells (Table 1). These findings indicate a specific role of the tetramer-forming GABPβ isoforms in HSC cell cycle regulation and preserving a dormant pool of HSCs.
We also examined other GABP direct target genes that have been reported to regulate various aspects of HSC biology, including Atm in DNA break repair, Terf2 in telomere maintenance, Dnmt1 in DNA methylation, Myst4 in histone acetylation, as well as Brg1 and Brm (encoded by Smarca4 and Smarca2 respectively) in chromatin remodeling. We also included Dnmt3a and 3b DNA methyltransferases, and CBP histone acetyltransferase (encoded by Crebbp) whose expression is dependent on GABPα but not directly regulated by GABP at the transcription initiation sites (Yu et al., 2011). All these genes were reduced in expression in the range of 18-38% in dKO LT-/ST-HSCs as well as MPPs (except for Atm in MPPs). The reductions were specific to dKO LT-/ST-HSCs and MPPs, as such consistent decreases in gene expression were only occasionally observed in MPs or other differentiated blood cells (Table 1). In contrast, GABPα expression itself was not decreased in dKO HSCs, demonstrating that deficiency in GABPβ1L/β2 did not diminish global gene expression in HSCs non-specifically. In sum, the molecular characterization revealed that loss of GABPβ1L/β2 resulted in moderate but consistent changes in the expression of multiple targets, leading to perturbation of the overall integrity of the GABP gene regulatory module. This evidence also corroborates a molecular phenocopy between GABPα and GABPβ1L/β2 deficiencies. The regulatory effect of GABPβ1L/β2 was only evident in HSCs but not in MPs or more differentiated blood lineage cells. One possible explanation is that the differentiated cells may express additional regulatory factors that compensate for the loss of GABPβ1L/β2. Global mapping of histone modification status has revealed stark differences between the multipotent HSCs and differentiated erythrocyte precursors (Cui et al., 2009). It is thus possible that the histone marks and/or chromatin remodeling are modified in a manner specific to differentiated cells, so that they become less sensitive to the absence of tetramer-forming GABPβ isoforms. These possibilities merit further investigation.
The tetramer-forming GABPβ isoforms critically regulated LSC self-renewal
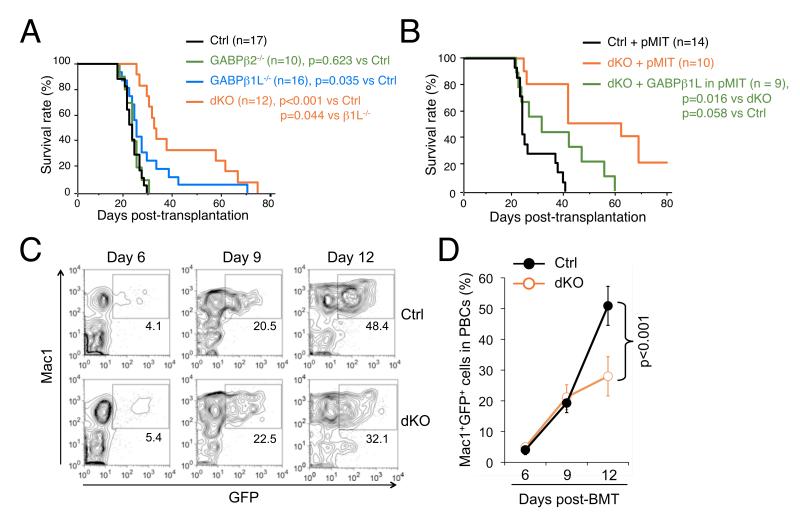
Given their specific regulatory role in HSC self-renewal, we next investigated if GABPβ1L/β2 isoforms have a role in maintaining LSCs. CML is a paradigmatic stem cell disorder that involves chromosomal translocation between BCR and ABL in humans, giving rise to the BCR-ABL fusion protein with constitutive tyrosine kinase activity (Wang and Dick, 2005). This disease can be recapitulated in mice by retrovirally introducing the p210 form of BCR-ABL (p210BCR-ABL) into cycling hematopoietic stem/progenitor cells followed by transplantation into irradiated recipients (Pear et al., 1998). We thus introduced p210BCR-ABL into GABPβ1L- and/or GABPβ2-deficient Lin− BM cells, and transplanted these cells into irradiated syngeneic recipients to induce CML. We confirmed that delivery of p210BCR-ABL via retroviral infection into BM cells was not negatively affected by deficiency in GABPβ1L and/or GABPβ2 (Figure S6A). In addition, GABP subunits/isoforms were neither enriched nor diminished in expression in p210BCR-ABL-transformed LSCs (Figure S6B). Within 20-30 days of bone marrow transplantation (BMT), the recipients of both control and GABPβ2−/− donor cells developed symptoms of CML-like myeloproliferative disease with increased myeloid cells in peripheral blood (Figure 5A). Deficiency in GABPβ1L slightly prolonged recipient survival, and double deficiency in GABPβ1L/β2 further delayed CML initiation and promoted recipient survival (Figure 5A). Additionally, introducing GABPβ1L into dKO Lin− BM cells re-sensitized CML induction by p210BCR-ABL (Figure 5B). These observations suggest that both GABPβ isoforms are important for efficiently establishing LSCs, mirroring their roles in regulating normal HSC self-renewal (Figure 3F).
Figure 5. The tetramer-forming GABPβ isoforms are critical for CML initiation and propagation.
(A) Kaplan-Meier survival curves for recipients of p210BCR-ABL-transduced Lin− BM cells. Lin− BM cells from mice of indicated genotypes were transduced with a bicistronic retrovirus that expresses p210BCR-ABL along with GFP. The infected cells (each containing 6,000 GFP+ LSK cells) along with 2 × 105 protector BM cells were transplanted into irradiated syngeneic recipients followed by observation of CML progression. Data in (A) and (B) are pooled results from 2 independent experiments, and the p values were obtained by log-rank (Mantel-Cox) statistical analysis. (B) Complementation of dKO BM cells with the GABPβ1L gene restores sensitivity to CML induction. Lin− BM cells from dKO mice were infected with p210BCR-ABL-GFP and a pMIT retrovirus expressing GABPβ1L. WT and dKO BM cells were also infected with p210BCR-ABL-GFP along with empty pMIT retrovirus as controls. Recipients of these transduced cells were monitored for CML progression. (C) and (D) Sustained generation of transformed myeloid cells depends on GABPβ1L/β2. Control or dKO Lin− BM cells were retrovirally transduced with p210BCR-ABL and transplanted into irradiated syngeneic recipients as in (A). GFP+Mac1+ cells were tracked in PBCs on indicated days post-BMT. The frequency of GFP+Mac1+ cells is marked in representative contour plots in (C) and collectively summarized in (D). Data are from two independent experiments (n = 22 for control, and n = 15 for dKO).
As demonstrated above, GABPβ1L/β2 are intrinsically required for maintaining HSC homeostasis and functionality, and thus delayed CML onset in dKO recipients might be alternatively interpreted as a result of reduced chimerism and ensuing generation of transformed myeloid cells. To test this, we tracked GFP+Mac1+ cells in PBCs during early stages of transplantation of p210BCR-ABL-infected BM cells. As shown in Figure 5C and 5D, both WT and dKO-derived GFP+Mac1+ cells contributed similarly to the PBCs on days 6 and 9 post-BMT. When the recipients were examined on day 12 post-BMT, GFP+Mac1+ cells derived from WT BM cells continued to expand, but those from dKO BM cells failed to maintain the same growth rate. In the CML model, 2×105 WT BM cells are transplanted together with p210BCR-ABL-transduced cells to confer radioprotection. These protector BM cells also compete for engraftment, especially with the transduced dKO BM cells (Figure 3B to 3E). To reduce such competition and thus maximize the dKO BM engraftment, we performed a parallel experiment in which the same amounts of p210BCR-ABL-transduced WT and dKO BM cells were transplanted, but the protectors for recipients of dKO cells were reduced to 1×105. As shown in Figure S6C, this approach did improve engraftment of the transduced dKO cells, and dKO-derived GFP+Mac1+ cells persisted at similar frequencies in PBCs as those derived from WT cells till up to day 14 post-BMT. In spite of the similar chimerism to a much later time point, recipients of the transduced dKO BM cells exhibited delayed onset of CML and prolonged survival (Figure S6D). These observations indicate that the diminished sensitivity to CML induction in the absence of GABPβ1L/β2 cannot be solely explained by reduced chimerism upon bone marrow transplantation. Whereas we cannot entirely exclude the possibility that GABPβ1L/β2-deficient LSCs did not engraft as efficiently as control LSCs in this system, the experimental evidence presented above suggests that the tetramer-forming GABPβ isoforms contribute to maintaining p210BCR-ABL-transformed LSCs and hence sustained production of leukemic cells. In line with this notion, we used an in vitro serial plating assay where p210BCR-ABL-conferred self-renewing capacity was measured by colony formation in methylcellulose media, without implications of the engraftment issue in the transplantation setting. We have observed consistent reduction in dKO-derived colonies after both first and second plating (Figure S6E), which lends additional support to an intrinsic role of GABPβ1L/β2 in the maintenance of LSCs.
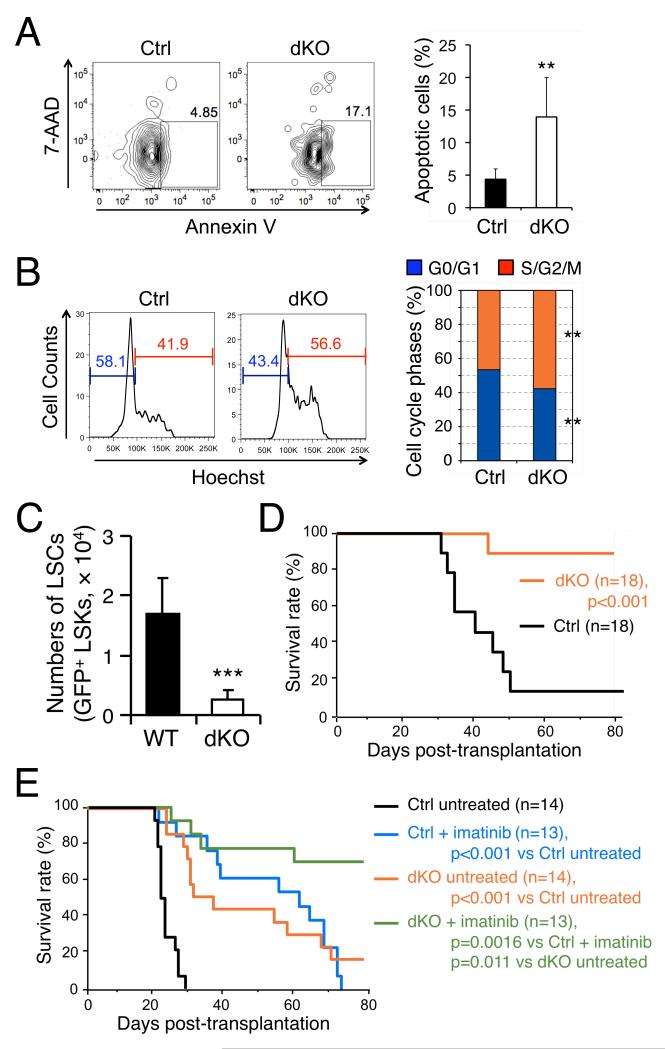
To further explore the mechanism by which deficiency in GABPβ1L/β2 affects LSCs, we examined survival and cell cycle progression of BM LSCs during days 15-20 post-BMT. We found that dKO LSCs are more susceptible to apoptosis (Figure 6A) and that an increased fraction of dKO LSCs are in actively cycling S/G2/M phase (Figure 6B). Thus, the reduced production of leukemic cells and slow onset of CML in dKO recipients are most likely ascribed to intrinsic defects in LSCs lacking the tetramer-forming GABPβ isoforms. Consistent with these observations, enumeration of LSCs (GFP+ LSK cells) in recipient mouse BM revealed a substantial decrease in accumulation of dKO LSCs compared with controls (Figure 6C). To directly assess LSC self-renewal in vivo, we first induced CML in primary recipients as described above, isolated LSCs from the BM by cells sorting, and transplanted 10,000 LSCs into another cohort of irradiated syngeneic mice. The control LSCs propagated CML in ≥ 90% of the secondary recipients. In striking contrast, only 10% of the secondary recipients of dKO LSCs were affected by CML (Figure 6D and Figure S6F). These findings collectively indicate that GABPβ1L/β2 proteins have essential roles in self-renewal of LSCs as well as HSCs.
Figure 6. Targeting GABPβ1L/β2 impairs LSC self-renewal and synergizes with imatinib therapy to eradicate CML.
(A) Intrinsic requirement of GABPβ1L/β2 for LSC survival. Fifteen to twenty days after transplantation of p210BCR-ABL-infected cells, BM cells from the primary recipients were surface-stained to identify LSCs (GFP+ LSKs) and further stained for AnnexinV and 7-AAD. The percentage of AnnexinV+7-AAD− apoptotic cells is shown in representative contour plots and summarized in bar graph on the right (n = 4 for control, and n = 8 for dKO). **, p<0.01. (B) LSCs lacking GABPβ1L/β2 exhibit more active cycling. BM cells isolated from the primary recipients as in (A). To avoid quenching of GFP in LSCs, the cells were first incubated with Hoechst33342 at 37oC for 45 min, followed by two washes with HBSS and surface staining on ice. Percentages of cells in G0/G1 or S/G2/M phases are shown in representative histograms and summarized in bar graph on the right (n = 4 for Ctrl, and n = 8 for dKO). **, p<0.01. (C) Enumeration of LSCs in primary recipients. Primary recipients of p210BCR-ABL-transduced BM cells were sacrificed on day 15 post-BMT, and GFP+ LSK cells in the BM were determined. Representative data from 3 independent experiments are shown. ***, p<0.001. (D) CML propagation in secondary recipients. GFP+ LSK cells were sorted from primary recipients of p210BCR-ABL-transduced BM cells on day 15 post-BMT. Ten thousand sorted cells were injected into another cohort of syngeneic mice along with 2 × 105 protectors, and CML progression was monitored. Data in (D) and (E) are pooled results from 2 independent experiments, and statistical significance was assessed using log-rank test. (E) Synergistic effect of GABPβ1L/β2 deficiency and imatinib therapy in controlling CML in vivo. Primary recipients of p210BCR-ABL-transduced BM cells were untreated or treated with imatinib (100 mg/kg body weight, b.i.d.) from day 8 to day 80 post-BMT, with CML progression monitored.
The tyrosine kinase inhibitor imatinib preferentially targets actively dividing leukemic cells, whereas the quiescent LSCs are resistant to imatinib and therefore account for CML relapse in human patients (Holtz et al., 2007). We induced CML in primary recipients as above, and initiated imatinib treatment on day 8 post-BMT. For the recipients of p210BCRABL-infected WT BM cells, the imatinib treatment prolonged their survival compared with the untreated group, although all the treated recipients eventually succumbed to the disease (Figure 6E). Significantly, the imatinib treatment of recipients of p210BCR-ABL-infected dKO BM cells further prolonged their survival, and protected ≥ 70% of them (Figure 6E). These observations suggest that targeting self-renewing LSCs by interfering with GABPβ activity can achieve better control of leukemia in synergy with drugs targeting bulk leukemic blasts.
DISCUSSION
Transcription factors are intrinsic determinants of HSC self-renewal and multi-lineage differentiation capacity (Orkin and Zon, 2008). Genome-wide mapping of transcription factor binding locations and various histone marks in HSCs has greatly advanced our understanding of the extensive crosstalk among transcription factors and their interplay with epigenetic states (Cui et al., 2009; Wilson et al., 2010). Many transcription factors are known to have recurring roles in both multipotent HSCs and differentiated blood cells in different lineages. For example, deletion of PU.1, an Ets family transcription factor, greatly diminished LSKs in fetal livers as well as adult bone marrow and completely abrogated generation of CMPs and CLPs from HSCs (Dakic et al., 2005; Iwasaki et al., 2005). In addition, graded levels of PU.1 regulate macrophage versus B cell generation, with higher concentrations promoting a macrophage fate, whereas lower concentrations favor production of B cells (DeKoter and Singh, 2000). Similarly, the transcriptional repressor Gfi1 is required for restraining excessive HSC proliferation and preserving its self-renewal capacity (Hock et al., 2004a), as well as normal T cell development (Yucel et al., 2003). Interestingly, Gfi1 was recently found to directly repress PU.1 expression to promote B cell differentiation at the expense of macrophages (Spooner et al., 2009). The pleiotropic effect and interconnected nature of transcription factors thus limit their potentials to be utilized as therapeutic targets in enhancing HSC engraftments or treating hematological malignancy. In the case of GABP complex, the GABPα subunit has similar limitations. We have recently demonstrated that induced deletion of GABPα subunit severely impaired HSC survival and differentiation (Yu et al., 2011). In addition, previous studies by our group and others revealed that GABPα is required for B cell development by directly regulating Pax5 (Xue et al., 2007), and for myeloid differentiation via direct regulation of Gfi1 (Yang et al., 2011). Thus, it was quite unexpected that targeting the tetramer-forming GABPβ isoforms, GABPβ1L and GABPβ2, would specifically affect HSC homeostasis and functionality. Deficiency in GABPβ1L/β2 compromised HSC survival and quiescence, but had no detectable effect on apoptosis or cell cycle regulation in differentiated blood cells. However, LT-HSCs lacking GABPβ1L/β2, albeit reduced in absolute counts, were capable of differentiation to ST-HSCs and MPPs within the LSK population and generation of CMPs and CLPs at the similar frequency as control LT-HSCs. These findings suggest that the activity of the GABP complex can be modulated via its GABPβ isoforms to dissociate HSC survival/self-renewal from HSC differentiation.
Such a specific role for the tetramer-forming GABPβ isoforms in HSCs is substantiated at the gene expression level. The GABPβ1L/β2 double deficient mouse model allowed us to determine how target genes for the GABP complex in HSCs are regulated through these tetramer-forming GABPβ isoforms. Out of the GABP-controlled gene regulatory module in HSCs, we sampled dozens of GABP direct or indirect target genes that have known critical roles in regulating different aspects of HSC biology. Whereas acute deletion of GABPα in HSCs led to >10-fold reduction in some target genes for the GABP complex, including Bcl-2, Zfx, Pten, Atm and Brm (Yu et al., 2011), targeting GABPβ1L/β2 isoforms resulted in more subtle decreases in all these genes in LT/STHSCs and MPPs. Such moderate gene expression reductions in dKO HSCs/MPPs were consistent and widespread in almost all GABP targets examined. This may also help explain our inability to identity HSC biology-relevant, validatable targets for GABPβ1L/β2 via microarray-based transcriptomic analysis of dKO and control HSCs (data not shown). Although the qPCR-mediated gene expression profiling of dKO HSCs is not necessarily exhaustive, our data suggest that the tetramer-forming GABPβ isoforms confer a fine-tuned digital regulation of target genes for the GABP complex. Rather than causing drastic changes in one or a few particular target genes, deficiency in GABPβ1L/β2 perturbs the integrity of the GABP-controlled gene regulatory module. The combinatorial effects of small changes on multiple gene targets thus lead to functional impairments of HSC survival, quiescence, self-renewal, and repopulation capacity. Significantly, the impact on gene expression is highly specific for HSCs and MPPs. Consistent gene expression reductions were rarely seen in myeloid progenitors or granulocytes in the BM, or splenic T or B cells, thus offering a molecular explanation for the specific regulation of HSC activity by the GABPβ1L/β2 isoforms.
The role of GABPβ1S in the GABP complex has been controversial. When fused with a GAL4 DNA-binding domain, both GABPβ1L and GABPβ1S were found to be equally proficient in activating transcription (Gugneja et al., 1995); however, others reported that GABPβ1S failed to activate transcription using in vitro reporter assays (Sawa et al., 1996). Additionally, a C-terminal truncated form of GABPβ1 encompassing amino acids 1-330, which is common to both GABPβ1L and GABPβ1S, has been shown to have dominant negative effect against the GABP complex (Briguet and Ruegg, 2000; Schaeffer et al., 1998). In GABPβ1L/β2 double deficient mice, GABPβ1S is the only remaining GABPβ isoform and forms GABPα/GABPβ1S heterodimer with GABP α2β2 tetramer assembly abrogated. If GABPβ1S were to have a dominant effect, the GABPβ1L/β2 dKO mice would be expected to have similar phenotypes as in GABPα-targeted animals, such as early embryonic lethality. The fact that dKO mice remain viable indicates that the GABP α2β2 tetramer assembly is dispensable for embryogenesis and that the GABPα/GABPβ1S heterodimer has essential regulatory roles in transcription activation/repression rather than functioning as a dominant negative. However, the GABPα/GABPβ1S heterodimer was not sufficient to optimally activate transcription of critical GABP target genes in the context of HSCs, because the loss of GABPβ1L/β2 impaired HSC survival and quiescence. Previous biochemical and structural studies have established that the GABP α2β2 tetramer binds to target DNA sequences with increased affinity and stability compared with αβ dimers (Batchelor et al., 1998; Chinenov et al., 2000; Graves, 1998). Our motif analysis of genome-wide GABP binding locations revealed that approximately 3/4 of GABP target genes harbor 2 or more GABP binding motifs in their proximal regulatory sequences. Whereas heterodimers between GABPα and individual GABPβ isoforms may maintain the ability to activate GABP targets, the presence of 2 or more motifs within one GABP binding location should facilitate the assembly of GABP α2β2 tetramer, which activates GABP target genes to an optimal level that meets functional requirements in HSCs. Although our data cannot actively exclude the possibility that GABPβ1L and β2 have tetramer-independent function, it is more likely that GABP α2β2 tetramer forms naturally in vivo wherever 2 or more GABP motifs are present in or recruited to proximal gene regulatory sequences. Overall, the optimal expression of GABP-controlled gene regulatory module in HSCs requires assembly of GABP α2β2 tetramer. GABPβ2−/− HSCs did not exhibit detectable defects, indicating that loss of α2(β2)2 tetramers is compensated for by the α2(β1L)2 tetramers. On the other hand, GABPβ1L−/− HSCs retained the ability of assembling α2(β2)2 tetramers yet exhibited impaired repopulation capacity and self-renewal. A possible explanation could be that α2(β1L)2 tetramers have a more potent regulatory role than α2(β2)2 tetramers, analogous to c-Myc among the Myc proteins and Foxo3a among the Foxo transcription factors (Laurenti et al., 2008; Tothova et al., 2007).
Of particular interest is that the essential roles of tetramer-forming GABPβ isoforms in HSC self-renewal are extended to LSCs. Understanding molecular wiring in LSCs is of importance given the promise of eradicating leukemia by targeting LSCs. Although molecular characterization of LSCs is currently less extensive compared with that of HSCs, existing evidence indicates that LSCs from different hematological malignancies share common transcriptional regulators for their self-renewal. For example, a requirement of β-catenin has been demonstrated in both CML and acute myelogenous leukemias (Wang et al., 2010; Zhao et al., 2007). Additionally, LSCs and HSCs share some common regulators for their self-renewal, including two GABP direct targets, Pten and Foxo3a (Miyamoto et al., 2007; Naka et al., 2010; Tothova et al., 2007; Yilmaz et al., 2006; Yu et al., 2010; Zhang et al., 2006). Our data demonstrate that targeting GABPβ1L/β2 impairs LSC initiation and propagation in a CML model, indicating that both HSCs and LSCs depend on the tetramer-forming GABPβ isoforms for self-renewal. It is of note that targeting GABPβ1L/β2 more strongly impaired LSC initiation in primary recipients compared with targeting other factors, such as promyelocytic leukemia protein or Foxo3a, where suppression of LSC self-renewal became evident only in secondary or tertiary recipients (Ito et al., 2008; Naka et al., 2010). We further showed that combination of GABPβ1L/β2 deficiency and imatinib therapy synergistically controlled CML initiation. These observations provide key rationale for exploring the tetramer-forming GABPβ isoforms as therapeutic targets to eliminate LSCs without severely compromising the function of differentiated blood cells, particularly the immune cells. Because these GABPβ isoforms are also required for normal HSCs and hence hematopoiesis, upon completion of the LSC-targeted chemotherapy, allogeneic HSC transplantation would be necessary to restore a sustained supply of blood cells.
EXPERIMENTAL PROCEDURES
Mice
GABPβ1L−/− and GABPβ2−/− mice were described previously (Jing et al., 2008; Xue et al., 2008). All mouse experiments were performed under protocols approved by the Institutional Animal Use and Care Committee of the University of Iowa.
BM Reconstitution Assays
For limiting dilution assay, graded numbers of test (dKO or control) BM cells (7 × 103, 2.2 × 104, 6.7 × 104, and 2 × 105) were mixed with 2 × 105 protector B6.SJL BM cells and transplanted into lethally irradiated B6.SJL recipients. Sixteen weeks later, contribution of test cells to multi-lineage blood reconstitution was determined by flow cytometry. For serial transplantation assay, 1,500-2,500 sorted LSKs were transplanted into lethally irradiated CD45.1+CD45.2+ mice, and 8 weeks later, a portion of the primary recipient mice was sacrificed, and CD45.2+ LSKs were sorted again and transplanted into secondary recipients at 1,500 cells/mouse.
CML Model and imatinib Therapy
p210BCR-ABL retrovirus was packaged and used to infect Lin− BM cells. The infected cells containing 6,000 GFP+ LSK cells along with 2 × 105 protector BM cells were transplanted into lethally irradiated C57BL/6 recipients to induce CML (Chen et al., 2009; Ito et al., 2010). The recipients were then evaluated daily for lethargy, splenomegaly, and signs of morbidity. For imatinib treatment, the drug was administered at 100 mg/kg body weight by oral gavage twice a day, during 8-80 days after transplantation of p210BCR-ABL-infected Lin− BM cells.
Supplementary Material
Highlights.
GABPβ1L and GABPβ2 specifically control HSC survival and quiescence
GABPβ1L and GABPβ2 critically regulate HSC and LSC self-renewal
Targeting GABPβ1L and GABPβ2 synergizes with imatinib in eradicating CML
GABP-controlled gene regulatory module is a potential therapeutic target for LSCs
ACKNOWLEDGEMENTS
We thank Dr. Jothi Raja (NIEHS) for analyzing distribution of GABP motifs in genome-wide GABP binding locations, Dr. Justin Fishbaugh at the Flow Cytometry Core for assistance with the use of LSR II flow cytometer, Heath Vignes and George Rasmussen for cell sorting, and Amanda Kalen at the University of Iowa Radiation Core Facility for assistance with mouse irradiation. We thank Dr. Warren S. Pear (University of Pennsylvania) for providing the BCR-ABL plasmid, Jennifer Barr (University of Iowa), Drs. Shaoguang Li (University of Massachusetts Medical School) and Reya Tannishtha (University of California, San Diego) for advice on establishing the CML model. This study is supported by Oberley Seed Grant from Holden Comprehensive Cancer Center, the University of Iowa, and NIH grants AI080966 and HL095540 (HHX).
Footnotes
SUPPLEMENTAL INFORMATION Supplemental information includes Supplemental Experimental Procedures, 6 supplemental figures, and 1 supplemental table.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no competing financial interests.
REFERENCES
- Batchelor AH, Piper DE, de la Brousse FC, McKnight SL, Wolberger C. The structure of GABPalpha/beta: an ETS domain-ankyrin repeat heterodimer bound to DNA. Science. 1998;279:1037–1041. doi: 10.1126/science.279.5353.1037. [DOI] [PubMed] [Google Scholar]
- Briguet A, Ruegg MA. The Ets transcription factor GABP is required for postsynaptic differentiation in vivo. J Neurosci. 2000;20:5989–5996. doi: 10.1523/JNEUROSCI.20-16-05989.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush TS, St Coeur M, Resendes KK, Rosmarin AG. GA-binding protein (GABP) and Sp1 are required, along with retinoid receptors, to mediate retinoic acid responsiveness of CD18 (beta 2 leukocyte integrin): a novel mechanism of transcriptional regulation in myeloid cells. Blood. 2003;101:311–317. doi: 10.1182/blood.V101.1.311. [DOI] [PubMed] [Google Scholar]
- Chen Y, Hu Y, Zhang H, Peng C, Li S. Loss of the Alox5 gene impairs leukemia stem cells and prevents chronic myeloid leukemia. Nat Genet. 2009;41:783–792. doi: 10.1038/ng.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng T, Rodrigues N, Shen H, Yang Y, Dombkowski D, Sykes M, Scadden DT. Hematopoietic stem cell quiescence maintained by p21cip1/waf1. Science. 2000;287:1804–1808. doi: 10.1126/science.287.5459.1804. [DOI] [PubMed] [Google Scholar]
- Chinenov Y, Henzl M, Martin ME. The alpha and beta subunits of the GA-binding protein form a stable heterodimer in solution. Revised model of heterotetrameric complex assembly. J Biol Chem. 2000;275:7749–7756. doi: 10.1074/jbc.275.11.7749. [DOI] [PubMed] [Google Scholar]
- Cui K, Zang C, Roh TY, Schones DE, Childs RW, Peng W, Zhao K. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell. 2009;4:80–93. doi: 10.1016/j.stem.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dakic A, Metcalf D, Di Rago L, Mifsud S, Wu L, Nutt SL. PU.1 regulates the commitment of adult hematopoietic progenitors and restricts granulopoiesis. J Exp Med. 2005;201:1487–1502. doi: 10.1084/jem.20050075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Brousse FC, Birkenmeier EH, King DS, Rowe LB, McKnight SL. Molecular and genetic characterization of GABP beta. Genes Dev. 1994;8:1853–1865. doi: 10.1101/gad.8.15.1853. [DOI] [PubMed] [Google Scholar]
- DeKoter RP, Singh H. Regulation of B lymphocyte and macrophage development by graded expression of PU.1. Science. 2000;288:1439–1441. doi: 10.1126/science.288.5470.1439. [DOI] [PubMed] [Google Scholar]
- Galan-Caridad JM, Harel S, Arenzana TL, Hou ZE, Doetsch FK, Mirny LA, Reizis B. Zfx controls the self-renewal of embryonic and hematopoietic stem cells. Cell. 2007;129:345–357. doi: 10.1016/j.cell.2007.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvagni F, Capo S, Oliviero S. Sp1 and Sp3 physically interact and cooperate with GABP for the activation of the utrophin promoter. J Mol Biol. 2001;306:985–996. doi: 10.1006/jmbi.2000.4335. [DOI] [PubMed] [Google Scholar]
- Graves BJ. Inner workings of a transcription factor partnership. Science. 1998;279:1000–1002. doi: 10.1126/science.279.5353.1000. [DOI] [PubMed] [Google Scholar]
- Gugneja S, Virbasius JV, Scarpulla RC. Four structurally distinct, non-DNA-binding subunits of human nuclear respiratory factor 2 share a conserved transcriptional activation domain. Mol Cell Biol. 1995;15:102–111. doi: 10.1128/mcb.15.1.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hock H, Hamblen MJ, Rooke HM, Schindler JW, Saleque S, Fujiwara Y, Orkin SH. Gfi-1 restricts proliferation and preserves functional integrity of haematopoietic stem cells. Nature. 2004a;431:1002–1007. doi: 10.1038/nature02994. [DOI] [PubMed] [Google Scholar]
- Hock H, Meade E, Medeiros S, Schindler JW, Valk PJ, Fujiwara Y, Orkin SH. Tel/Etv6 is an essential and selective regulator of adult hematopoietic stem cell survival. Genes Dev. 2004b;18:2336–2341. doi: 10.1101/gad.1239604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtz M, Forman SJ, Bhatia R. Growth factor stimulation reduces residual quiescent chronic myelogenous leukemia progenitors remaining after imatinib treatment. Cancer Res. 2007;67:1113–1120. doi: 10.1158/0008-5472.CAN-06-2014. [DOI] [PubMed] [Google Scholar]
- Huntly BJ, Gilliland DG. Leukaemia stem cells and the evolution of cancer-stem-cell research. Nat Rev Cancer. 2005;5:311–321. doi: 10.1038/nrc1592. [DOI] [PubMed] [Google Scholar]
- Ito K, Bernardi R, Morotti A, Matsuoka S, Saglio G, Ikeda Y, Rosenblatt J, Avigan DE, Teruya-Feldstein J, Pandolfi PP. PML targeting eradicates quiescent leukaemia-initiating cells. Nature. 2008;453:1072–1078. doi: 10.1038/nature07016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Kwon HY, Zimdahl B, Congdon KL, Blum J, Lento WE, Zhao C, Lagoo A, Gerrard G, Foroni L, et al. Regulation of myeloid leukaemia by the cell-fate determinant Musashi. Nature. 2010;466:765–768. doi: 10.1038/nature09171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki H, Somoza C, Shigematsu H, Duprez EA, Iwasaki-Arai J, Mizuno S, Arinobu Y, Geary K, Zhang P, Dayaram T, et al. Distinctive and indispensable roles of PU.1 in maintenance of hematopoietic stem cells and their differentiation. Blood. 2005;106:1590–1600. doi: 10.1182/blood-2005-03-0860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing X, Zhao DM, Waldschmidt TJ, Xue HH. GABPbeta2 is dispensible for normal lymphocyte development but moderately affects B cell responses. J Biol Chem. 2008;283:24326–24333. doi: 10.1074/jbc.M804487200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HS, Nelson ML, Mackereth CD, Scharpf M, Graves BJ, McIntosh LP. Identification and structural characterization of a CBP/p300-binding domain from the ETS family transcription factor GABP alpha. J Mol Biol. 2008;377:636–646. doi: 10.1016/j.jmb.2008.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaMarco K, Thompson CC, Byers BP, Walton EM, McKnight SL. Identification of Ets- and notch-related subunits in GA binding protein. Science. 1991;253:789–792. doi: 10.1126/science.1876836. [DOI] [PubMed] [Google Scholar]
- Laurenti E, Varnum-Finney B, Wilson A, Ferrero I, Blanco-Bose WE, Ehninger A, Knoepfler PS, Cheng PF, MacDonald HR, Eisenman RN, et al. Hematopoietic stem cell function and survival depend on c-Myc and N-Myc activity. Cell Stem Cell. 2008;3:611–624. doi: 10.1016/j.stem.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto K, Araki KY, Naka K, Arai F, Takubo K, Yamazaki S, Matsuoka S, Miyamoto T, Ito K, Ohmura M, et al. Foxo3a is essential for maintenance of the hematopoietic stem cell pool. Cell Stem Cell. 2007;1:101–112. doi: 10.1016/j.stem.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Naka K, Hoshii T, Muraguchi T, Tadokoro Y, Ooshio T, Kondo Y, Nakao S, Motoyama N, Hirao A. TGF-beta-FOXO signalling maintains leukaemia-initiating cells in chronic myeloid leukaemia. Nature. 2010;463:676–680. doi: 10.1038/nature08734. [DOI] [PubMed] [Google Scholar]
- Novershtern N, Subramanian A, Lawton LN, Mak RH, Haining WN, McConkey ME, Habib N, Yosef N, Chang CY, Shay T, et al. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 2011;144:296–309. doi: 10.1016/j.cell.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opferman JT. Life and death during hematopoietic differentiation. Curr Opin Immunol. 2007;19:497–502. doi: 10.1016/j.coi.2007.06.002. [DOI] [PubMed] [Google Scholar]
- Orkin SH, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132:631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pear WS, Miller JP, Xu L, Pui JC, Soffer B, Quackenbush RC, Pendergast AM, Bronson R, Aster JC, Scott ML, et al. Efficient and rapid induction of a chronic myelogenous leukemia-like myeloproliferative disease in mice receiving P210 bcr/abl-transduced bone marrow. Blood. 1998;92:3780–3792. [PubMed] [Google Scholar]
- Ravel-Chapuis A, Vandromme M, Thomas JL, Schaeffer L. Postsynaptic chromatin is under neural control at the neuromuscular junction. Embo J. 2007;26:1117–1128. doi: 10.1038/sj.emboj.7601572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristevski S, O’Leary DA, Thornell AP, Owen MJ, Kola I, Hertzog PJ. The ETS transcription factor GABPalpha is essential for early embryogenesis. Mol Cell Biol. 2004;24:5844–5849. doi: 10.1128/MCB.24.13.5844-5849.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosmarin AG, Resendes KK, Yang Z, McMillan JN, Fleming SL. GA-binding protein transcription factor: a review of GABP as an integrator of intracellular signaling and protein-protein interactions. Blood Cells Mol Dis. 2004;32:143–154. doi: 10.1016/j.bcmd.2003.09.005. [DOI] [PubMed] [Google Scholar]
- Sawa C, Goto M, Suzuki F, Watanabe H, Sawada J, Handa H. Functional domains of transcription factor hGABP beta1/E4TF1-53 required for nuclear localization and transcription activation. Nucleic Acids Res. 1996;24:4954–4961. doi: 10.1093/nar/24.24.4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa C, Yoshikawa T, Matsuda-Suzuki F, Delehouzee S, Goto M, Watanabe H, Sawada J, Kataoka K, Handa H. YEAF1/RYBP and YAF-2 are functionally distinct members of a cofactor family for the YY1 and E4TF1/hGABP transcription factors. J Biol Chem. 2002;277:22484–22490. doi: 10.1074/jbc.M203060200. [DOI] [PubMed] [Google Scholar]
- Schaeffer L, Duclert N, Huchet-Dymanus M, Changeux JP. Implication of a multisubunit Ets-related transcription factor in synaptic expression of the nicotinic acetylcholine receptor. EMBO J. 1998;17:3078–3090. doi: 10.1093/emboj/17.11.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spooner CJ, Cheng JX, Pujadas E, Laslo P, Singh H. A recurrent network involving the transcription factors PU.1 and Gfi1 orchestrates innate and adaptive immune cell fates. Immunity. 2009;31:576–586. doi: 10.1016/j.immuni.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tothova Z, Kollipara R, Huntly BJ, Lee BH, Castrillon DH, Cullen DE, McDowell EP, Lazo-Kallanian S, Williams IR, Sears C, et al. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell. 2007;128:325–339. doi: 10.1016/j.cell.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Vogel JL, Kristie TM. The novel coactivator C1 (HCF) coordinates multiprotein enhancer formation and mediates transcription activation by GABP. EMBO J. 2000;19:683–690. doi: 10.1093/emboj/19.4.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walkley CR, Fero ML, Chien WM, Purton LE, McArthur GA. Negative cell-cycle regulators cooperatively control self-renewal and differentiation of haematopoietic stem cells. Nat Cell Biol. 2005;7:172–178. doi: 10.1038/ncb1214. [DOI] [PubMed] [Google Scholar]
- Wallerman O, Motallebipour M, Enroth S, Patra K, Bysani MS, Komorowski J, Wadelius C. Molecular interactions between HNF4a, FOXA2 and GABP identified at regulatory DNA elements through ChIP-sequencing. Nucleic Acids Res. 2009;37:7498–7508. doi: 10.1093/nar/gkp823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JC, Dick JE. Cancer stem cells: lessons from leukemia. Trends Cell Biol. 2005;15:494–501. doi: 10.1016/j.tcb.2005.07.004. [DOI] [PubMed] [Google Scholar]
- Wang Y, Krivtsov AV, Sinha AU, North TE, Goessling W, Feng Z, Zon LI, Armstrong SA. The Wnt/beta-catenin pathway is required for the development of leukemia stem cells in AML. Science. 2010;327:1650–1653. doi: 10.1126/science.1186624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A, Murphy MJ, Oskarsson T, Kaloulis K, Bettess MD, Oser GM, Pasche AC, Knabenhans C, Macdonald HR, Trumpp A. c-Myc controls the balance between hematopoietic stem cell self-renewal and differentiation. Genes Dev. 2004;18:2747–2763. doi: 10.1101/gad.313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson NK, Foster SD, Wang X, Knezevic K, Schutte J, Kaimakis P, Chilarska PM, Kinston S, Ouwehand WH, Dzierzak E, et al. Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell. 2010;7:532–544. doi: 10.1016/j.stem.2010.07.016. [DOI] [PubMed] [Google Scholar]
- Xue HH, Bollenbacher J, Rovella V, Tripuraneni R, Du YB, Liu CY, Williams A, McCoy JP, Leonard WJ. GA binding protein regulates interleukin 7 receptor alpha-chain gene expression in T cells. Nat Immunol. 2004;5:1036–1044. doi: 10.1038/ni1117. [DOI] [PubMed] [Google Scholar]
- Xue HH, Bollenbacher-Reilley J, Wu Z, Spolski R, Jing X, Zhang YC, McCoy JP, Leonard WJ. The transcription factor GABP is a critical regulator of B lymphocyte development. Immunity. 2007;26:421–431. doi: 10.1016/j.immuni.2007.03.010. [DOI] [PubMed] [Google Scholar]
- Xue HH, Jing X, Bollenbacher-Reilley J, Zhao DM, Haring JS, Yang B, Liu C, Bishop GA, Harty JT, Leonard WJ. Targeting the GA binding protein beta1L isoform does not perturb lymphocyte development and function. Mol Cell Biol. 2008;28:4300–4309. doi: 10.1128/MCB.01855-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZF, Drumea K, Cormier J, Wang J, Zhu X, Rosmarin AG. GABP transcription factor is required for myeloid differentiation, in part, through its control of Gfi-1 expression. Blood. 2011;118:2243–2253. doi: 10.1182/blood-2010-07-298802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz OH, Valdez R, Theisen BK, Guo W, Ferguson DO, Wu H, Morrison SJ. Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature. 2006;441:475–482. doi: 10.1038/nature04703. [DOI] [PubMed] [Google Scholar]
- Yu S, Cui K, Jothi R, Zhao DM, Jing X, Zhao K, Xue HH. GABP controls a critical transcription regulatory module that is essential for maintenance and differentiation of hematopoietic stem/progenitor cells. Blood. 2011;117:2166–2178. doi: 10.1182/blood-2010-09-306563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Zhao DM, Jothi R, Xue HH. Critical requirement of GABPalpha for normal T cell development. J Biol Chem. 2010;285:10179–10188. doi: 10.1074/jbc.M109.088740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yucel R, Karsunky H, Klein-Hitpass L, Moroy T. The transcriptional repressor Gfi1 affects development of early, uncommitted c-Kit+ T cell progenitors and CD4/CD8 lineage decision in the thymus. J Exp Med. 2003;197:831–844. doi: 10.1084/jem.20021417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Grindley JC, Yin T, Jayasinghe S, He XC, Ross JT, Haug JS, Rupp D, Porter-Westpfahl KS, Wiedemann LM, et al. PTEN maintains haematopoietic stem cells and acts in lineage choice and leukaemia prevention. Nature. 2006;441:518–522. doi: 10.1038/nature04747. [DOI] [PubMed] [Google Scholar]
- Zhao C, Blum J, Chen A, Kwon HY, Jung SH, Cook JM, Lagoo A, Reya T. Loss of beta-catenin impairs the renewal of normal and CML stem cells in vivo. Cancer Cell. 2007;12:528–541. doi: 10.1016/j.ccr.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.