Full text
PDF


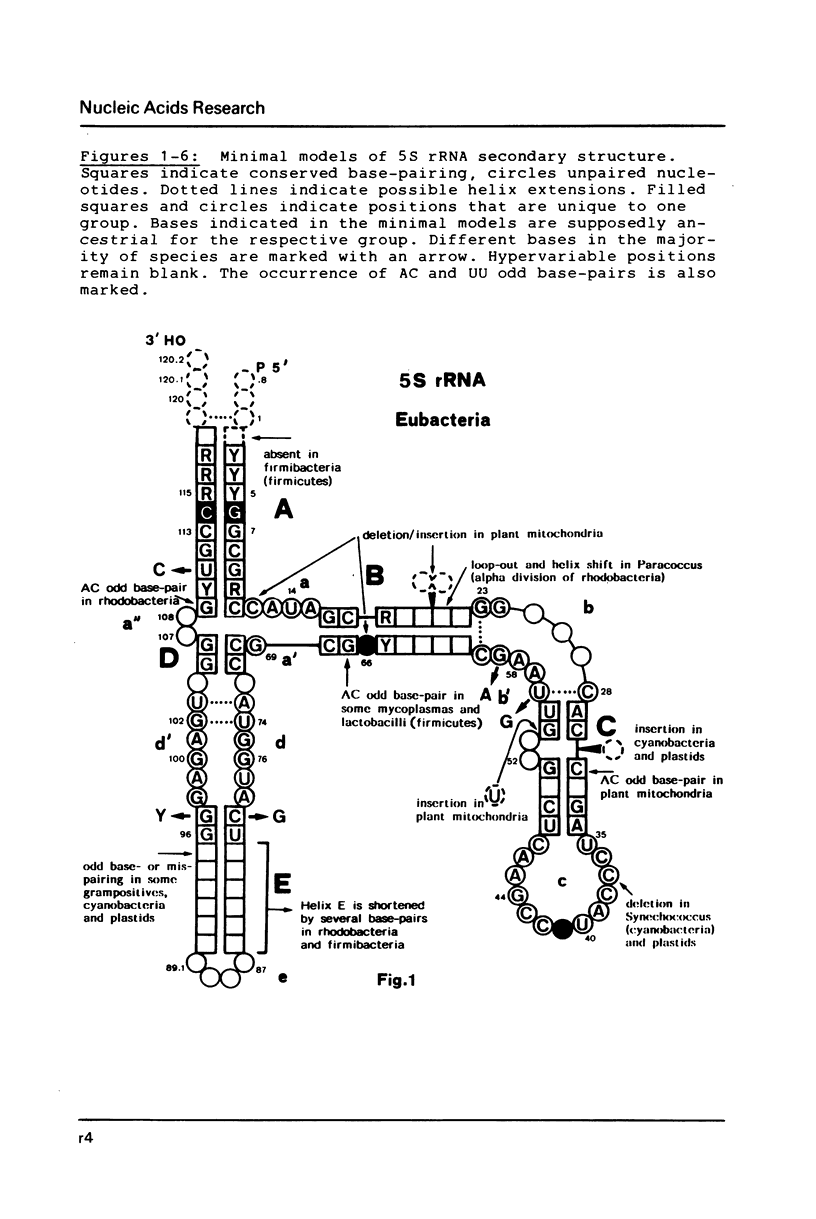
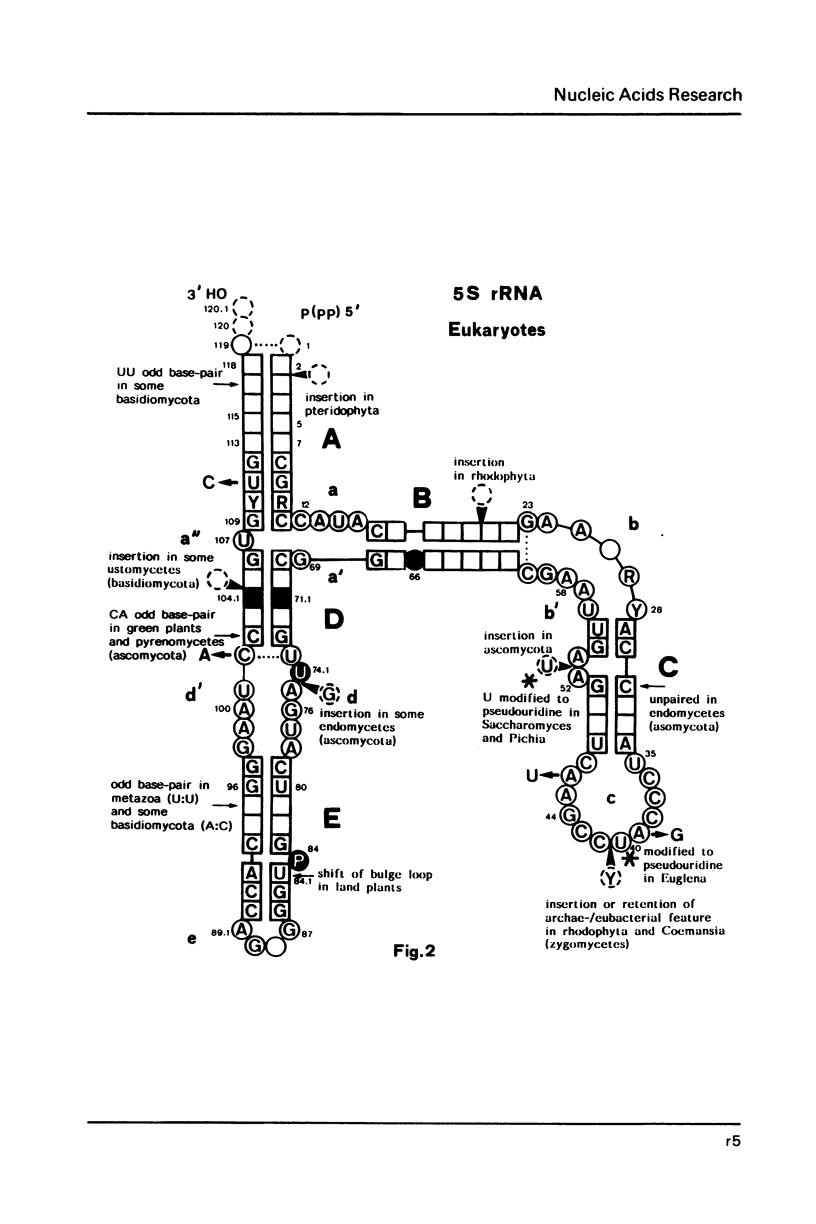
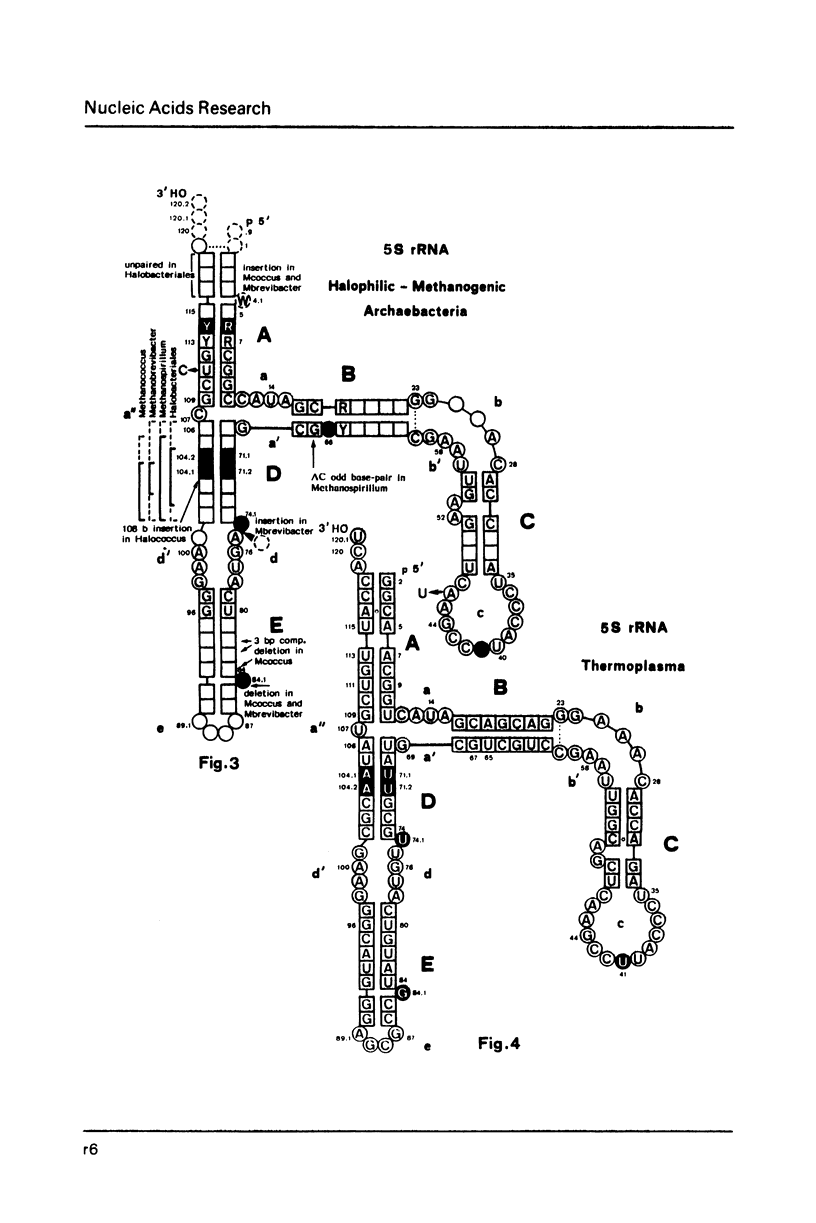
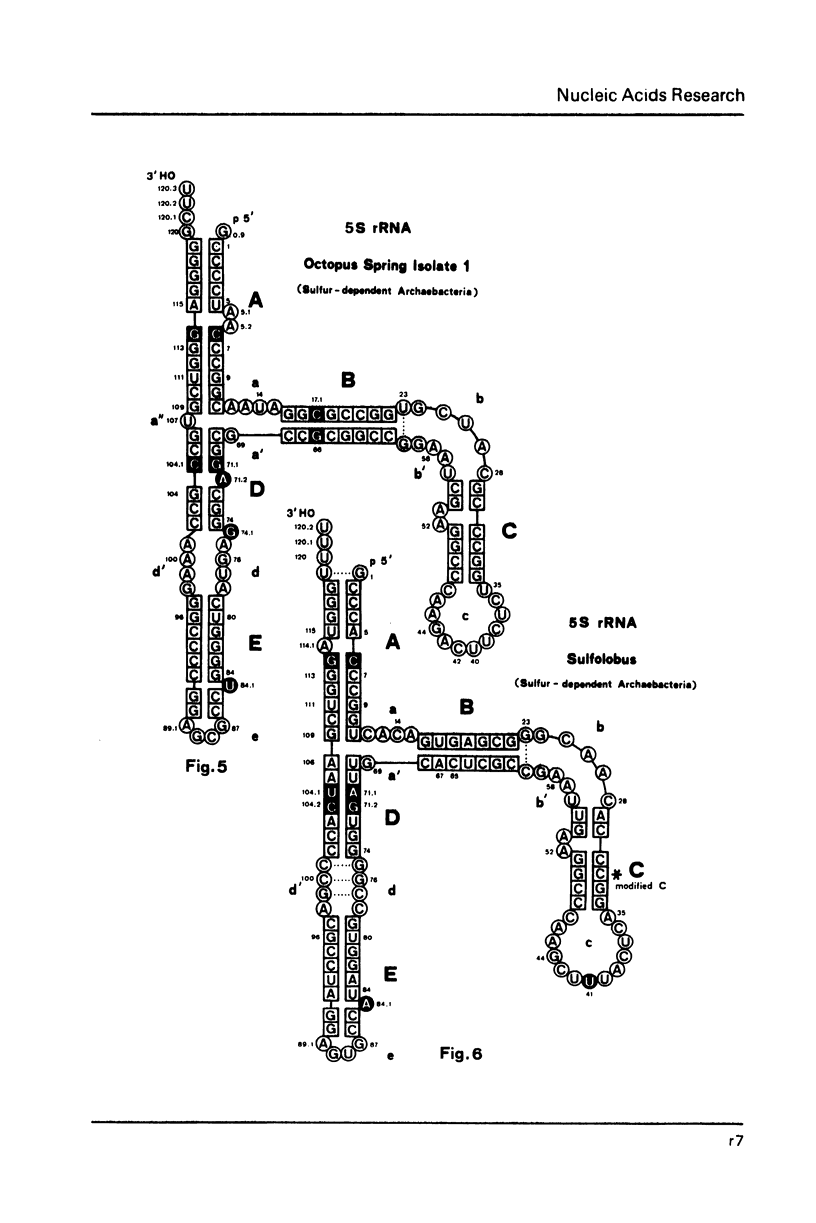




























Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cornish-Bowden A. Nomenclature for incompletely specified bases in nucleic acid sequences: recommendations 1984. Nucleic Acids Res. 1985 May 10;13(9):3021–3030. doi: 10.1093/nar/13.9.3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wachter R., Chen M. W., Vandenberghe A. Conservation of secondary structure in 5 S ribosomal RNA: a uniform model for eukaryotic, eubacterial, archaebacterial and organelle sequences is energetically favourable. Biochimie. 1982 May;64(5):311–329. doi: 10.1016/s0300-9084(82)80436-7. [DOI] [PubMed] [Google Scholar]
- Delihas N., Andersen J. Generalized structures of the 5S ribosomal RNAs. Nucleic Acids Res. 1982 Nov 25;10(22):7323–7344. doi: 10.1093/nar/10.22.7323. [DOI] [PMC free article] [PubMed] [Google Scholar]
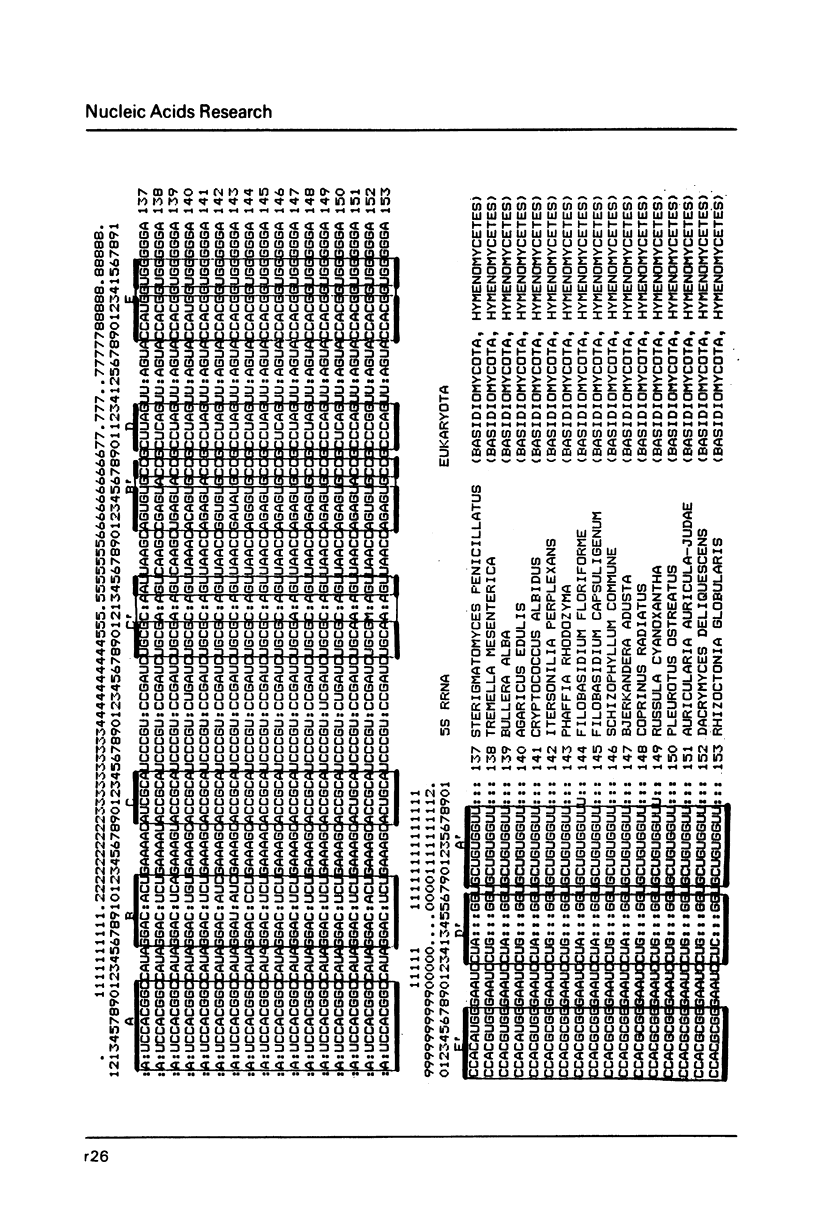
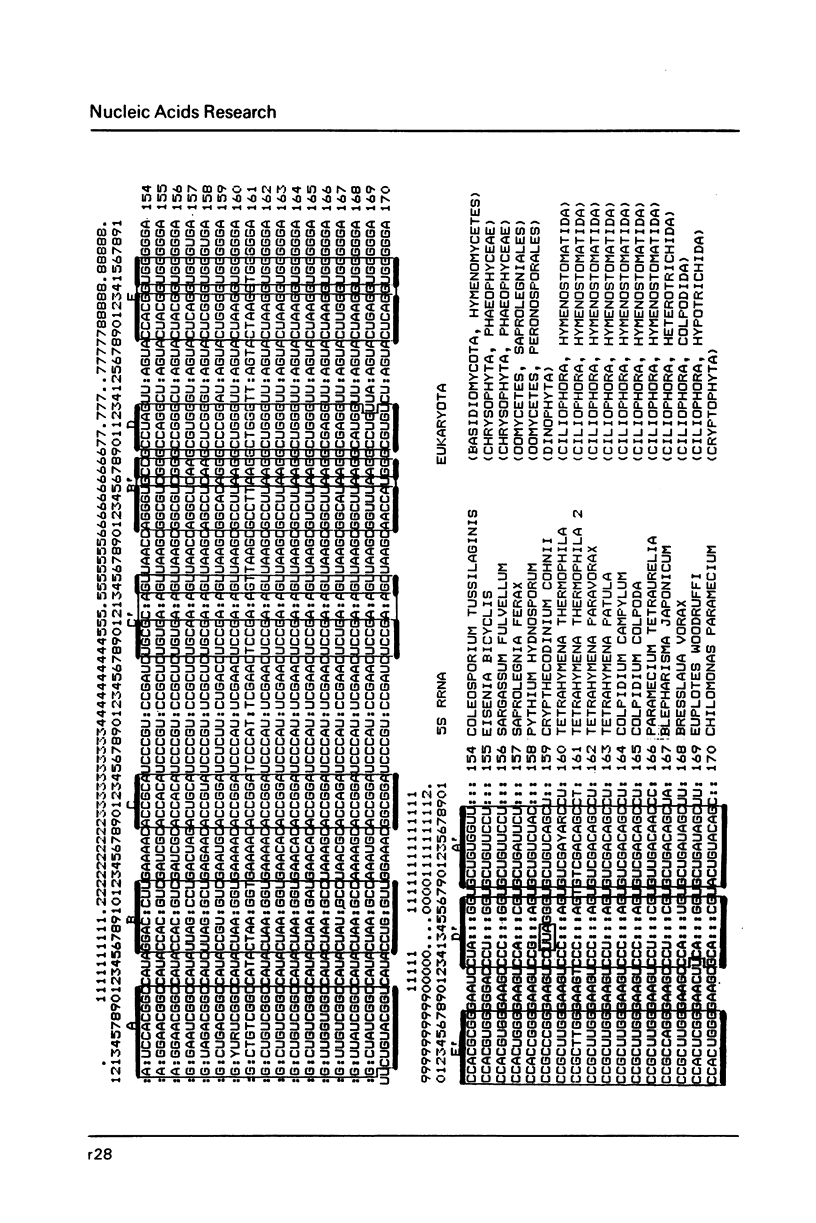
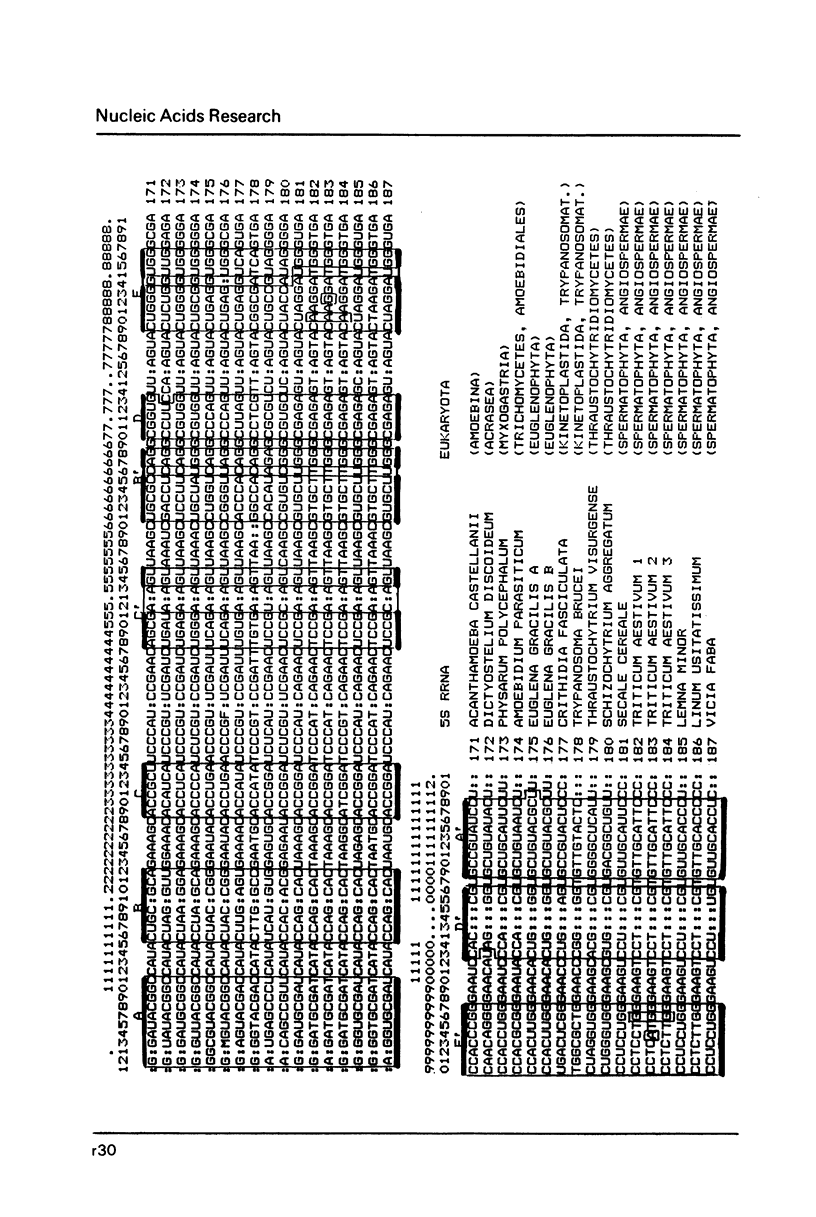
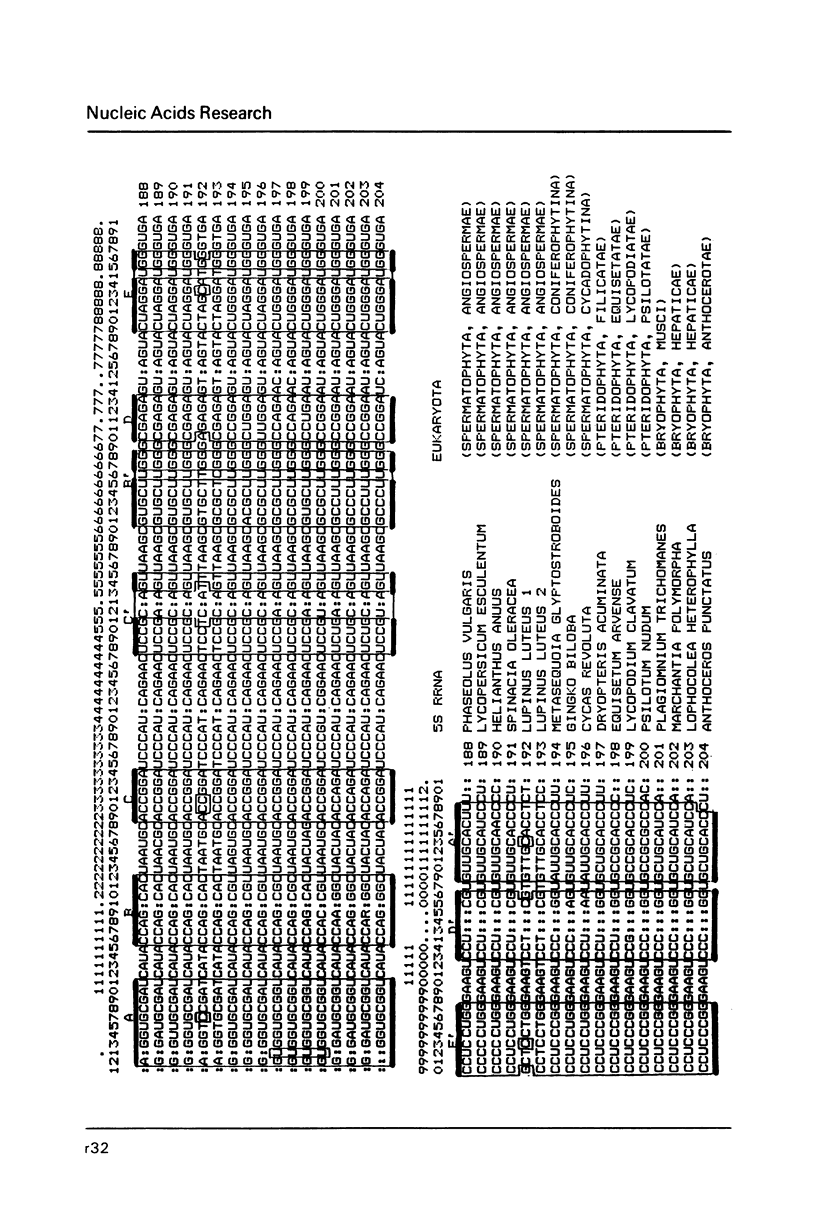
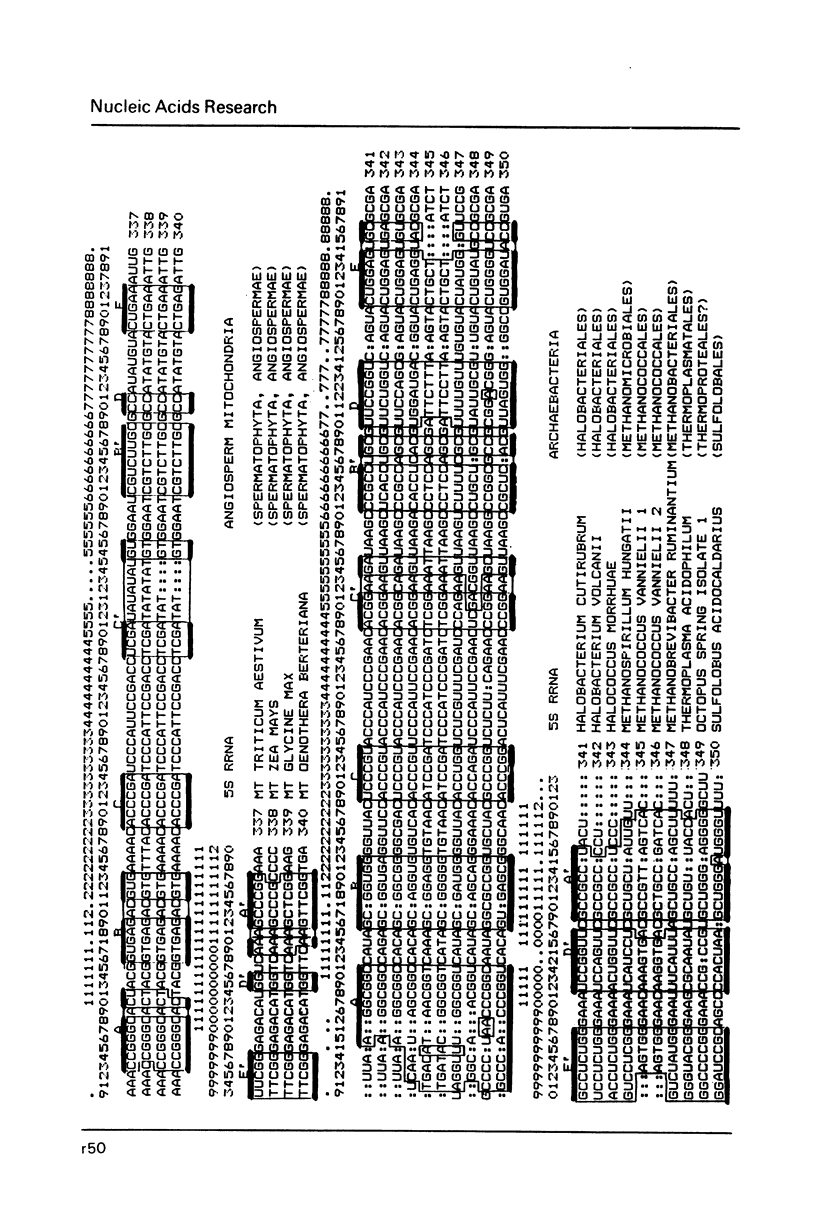
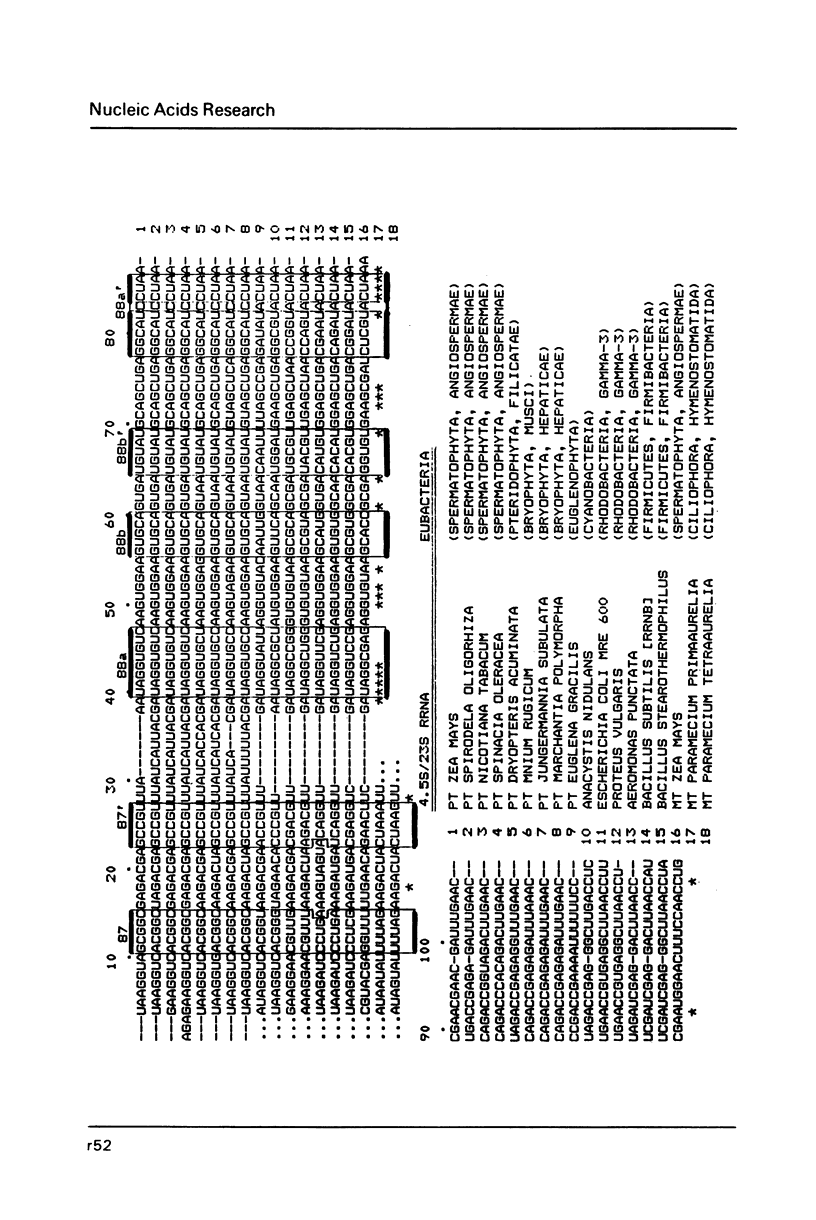
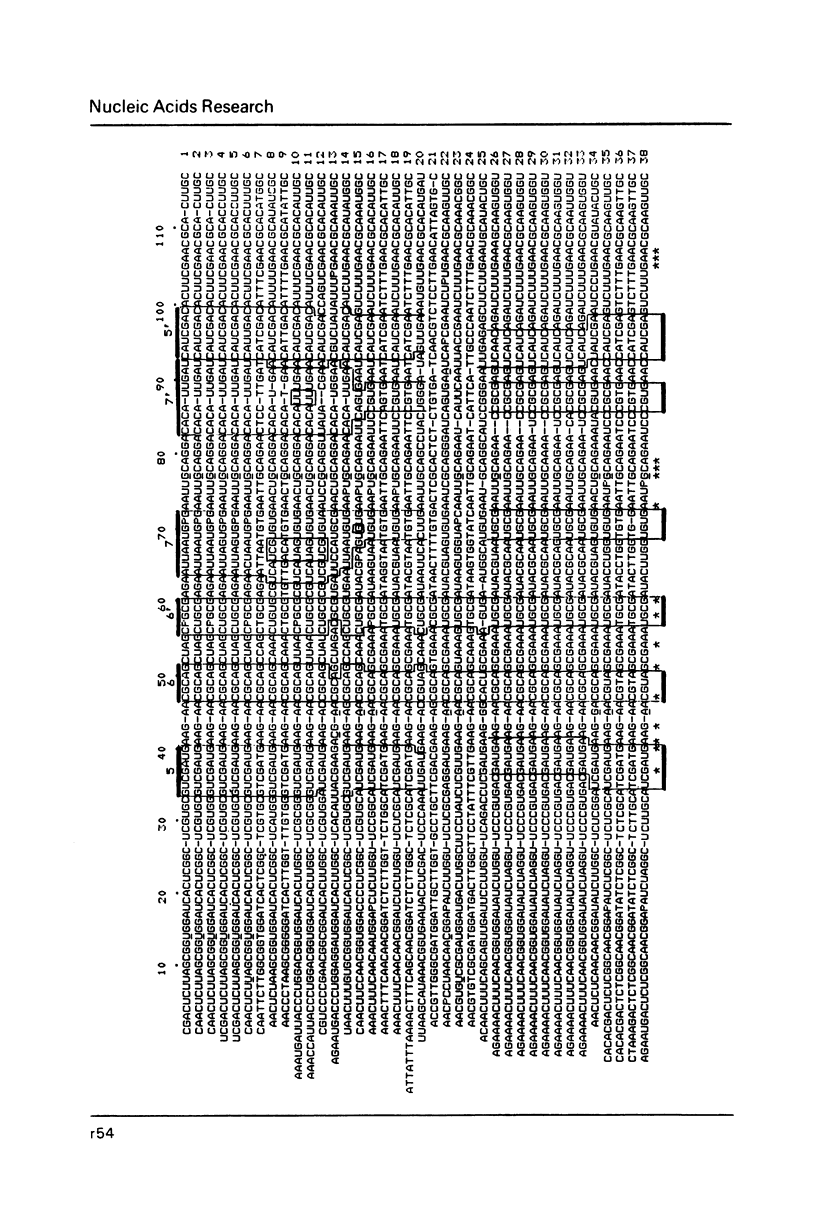
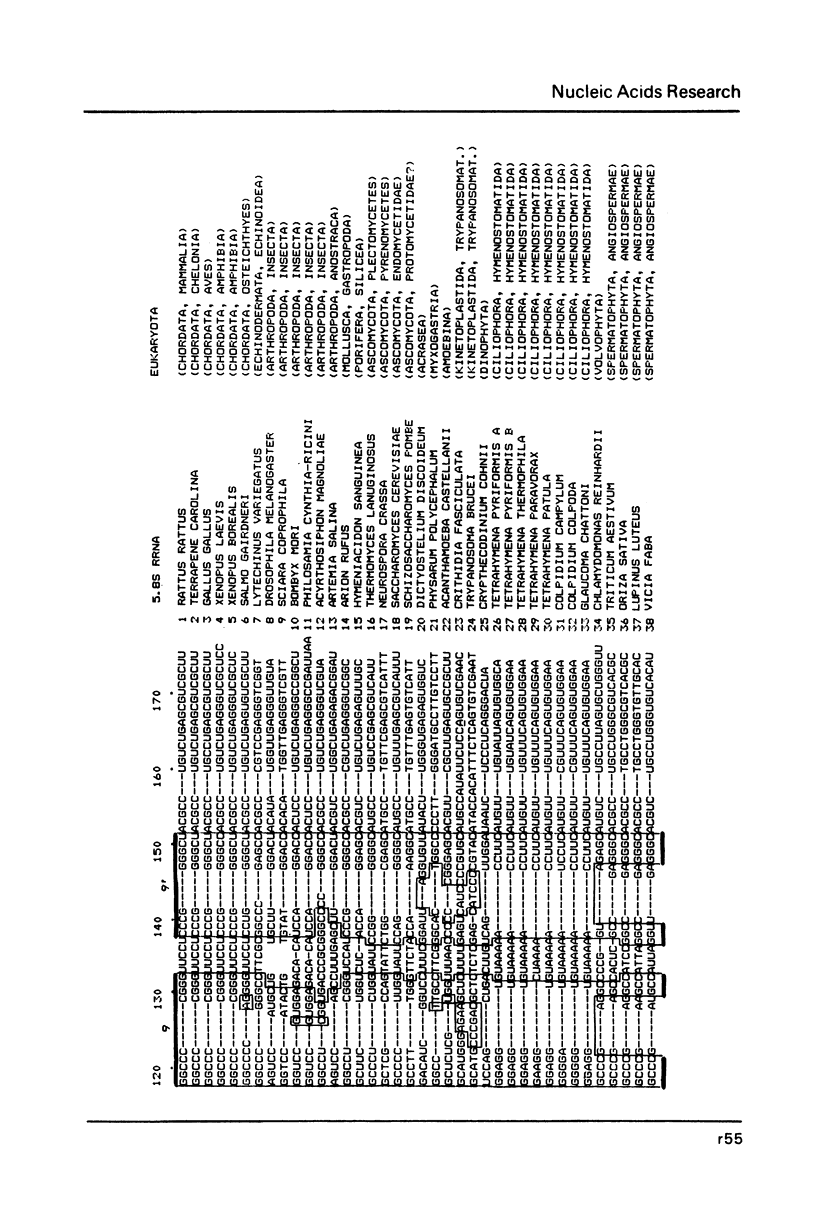
- Erdmann V. A., Wolters J., Huysmans E., De Wachter R. Collection of published 5S, 5.8S and 4.5S ribosomal RNA sequences. Nucleic Acids Res. 1985;13 (Suppl):r105–r153. doi: 10.1093/nar/13.suppl.r105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox G. E., Woese C. R. 5S RNA secondary structure. Nature. 1975 Aug 7;256(5517):505–507. doi: 10.1038/256505a0. [DOI] [PubMed] [Google Scholar]
- Hori H. Evolution of 5sRNA. J Mol Evol. 1975 Dec 31;7(1):75–86. doi: 10.1007/BF01732181. [DOI] [PubMed] [Google Scholar]
- Hori H. Molecular evolution of 5S RNA. Mol Gen Genet. 1976 May 7;145(2):119–123. doi: 10.1007/BF00269583. [DOI] [PubMed] [Google Scholar]
- Hori H., Osawa S. Evolutionary change in 5S RNA secondary structure and a phylogenic tree of 54 5S RNA species. Proc Natl Acad Sci U S A. 1979 Jan;76(1):381–385. doi: 10.1073/pnas.76.1.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Küntzel H., Piechulla B., Hahn U. Consensus structure and evolution of 5S rRNA. Nucleic Acids Res. 1983 Feb 11;11(3):893–900. doi: 10.1093/nar/11.3.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine N. D., Corliss J. O., Cox F. E., Deroux G., Grain J., Honigberg B. M., Leedale G. F., Loeblich A. R., 3rd, Lom J., Lynn D. A newly revised classification of the protozoa. J Protozool. 1980 Feb;27(1):37–58. doi: 10.1111/j.1550-7408.1980.tb04228.x. [DOI] [PubMed] [Google Scholar]
- MacKay R. M., Spencer D. F., Schnare M. N., Doolittle W. F., Gray M. W. Comparative sequence analysis as an approach to evaluating structure, function, and evolution of 5S and 5.8S ribosomal RNAs. Can J Biochem. 1982 Apr;60(4):480–489. doi: 10.1139/o82-057. [DOI] [PubMed] [Google Scholar]
- Nishikawa K., Takemura S. Nucleotide sequence of 5 S RNA from Torulopsis utilis. FEBS Lett. 1974 Mar 15;40(1):106–109. doi: 10.1016/0014-5793(74)80904-x. [DOI] [PubMed] [Google Scholar]
- Studnicka G. M., Eiserling F. A., Lake J. A. A unique secondary folding pattern for 5S RNA corresponds to the lowest energy homologous secondary structure in 17 different prokaryotes. Nucleic Acids Res. 1981 Apr 24;9(8):1885–1904. doi: 10.1093/nar/9.8.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Stackebrandt E., Macke T. J., Fox G. E. A phylogenetic definition of the major eubacterial taxa. Syst Appl Microbiol. 1985;6:143–151. doi: 10.1016/s0723-2020(85)80047-3. [DOI] [PubMed] [Google Scholar]


