Abstract
MicroRNAs (miRNAs) are 19- to 25-nt-long non-coding RNAs that regulate gene expression by base-pairing with target mRNAs and reducing their stability or translational efficiency. Mammalian miRNAs function in association with four closely related Argonaute proteins, AGO1–4. All four proteins contain the PAZ and the MID domains interacting with the miRNA 3′ and 5′ termini, respectively, as well as the PIWI domain comprising an mRNA ‘slicing’ activity in the case of AGO2 but not AGO1, AGO3 and AGO4. However, the slicing mode of the miRNA-programmed AGO2 is rarely realized in vivo and the four Argonautes are thought to play largely overlapping roles in the mammalian miRNA pathway. Here, we show that the average length of many miRNAs is diminished during nervous system development as a result of progressive shortening of the miRNA 3′ ends. We link this modification with an increase in the fractional abundance of Ago2 in the adult brain and identify a specific structural motif within the PAZ domain that enables efficient trimming of miRNAs associated with this but not the other three Argonautes. Taken together, our data suggest that mammalian Argonautes may define the length and possibly biological activity of mature mammalian miRNAs in a developmentally controlled manner.
INTRODUCTION
MicroRNAs (miRNAs) are ∼22-nt regulatory RNAs that base pair with mRNA targets and modulate their stability and translational efficiency (1,2). Human and mouse genomes encode hundreds distinct miRNAs, each potentially targeting an extensive set of mRNAs (3). A number of miRNAs have been shown to play critical roles in diverse aspects of development and physiology (4–7).
Metazoan miRNA genes are transcribed as long primary miRNAs (pri-mRNAs), which are normally processed into miRNA precursors (pre-miRNAs) and mature miRNAs by the endoribonucleases Drosha and Dicer, respectively (2,8). Mature miRNAs form a functional complex with the Argonaute subunit of the RNA-induced gene silencing complex (9). Notably, vertebrates encode four closely related Argonaute paralogs referred to as AGO1–4 in human and Ago1–4 in mouse (10–12). All of these proteins are characterized by the presence of the N, PAZ, MID and PIWI domains. The PAZ domain binds the miRNA 3′ ends and the MID domain participates in the 5′ end recognition (13,14).
Of the four paralogs, AGO2 is unique in that its PIWI domain can cleave mRNAs base-paired with perfectly complementary miRNAs or small interfering RNAs (siRNAs) (9,15). This ‘slicing’ activity of AGO2 is also required for Dicer-independent processing of the precursor of the evolutionarily conserved miRNA miR-451 (16–18). Aside from these two specialized functions, all mammalian Argonautes appear to function in the miRNA pathway in a largely overlapping manner (2,9,19). Therefore, the functional significance of the AGO paralog expansion in the vertebrates is poorly understood.
Recent deep sequencing studies suggest that individual miRNAs often exist as populations of variants (iso-miRs) that differ in length and sometimes nucleotide composition (8,20–24). Interestingly, the 3′ ends of Argonaute-associated miRNAs have recently been shown to be trimmed by the 3′-to-5′ exonuclease Nibbler in Drosophila (25,26). This reaction may potentially contribute to the developmentally regulated iso-miR dynamics detected for some miRNAs in this organism (22). However, it is currently unclear whether mammalian miRNAs also undergo extensive 3′-terminal trimming and if they do what are the molecular mechanisms underlying this process.
Here we demonstrate that a large fraction of mature miRNAs undergo progressive 3′-terminal shortening during mammalian nervous system development. We further show that, of the four Argonautes, only AGO2-loaded miRNAs are efficiently trimmed and that the AGO2 fractional abundance increases at later developmental stages, thus suggesting a mechanism for the widespread shortening of miRNAs in the adult brain. Importantly, we identify unique structural features of the AGO2 PAZ domain promoting the trimming reaction and provide evidence that biological activity of miRNA can be modulated by the nature of the Argonaute paralog.
MATERIALS AND METHODS
Plasmids and primers
Plasmids encoding miR-124 (RIP-miR-124 or pEM208) and the corresponding control vector (RI vector or pEM157), as well as the human AGO2 (pcDNA3-3xMyc-AGO2) and nlCre (pEM784) expression constructs have been described previously (27–29). The pTet-Off plasmid encoding a tetracycline transactivator (tTA) protein was from Clontech. We used standard molecular cloning techniques (30) and enzymes from NEB to generate several new constructs described in the Supplementary Table S1. Site-specific mutations were introduced using modified Quikchange site-directed mutagenesis protocol (Stratagene), in which the Pfu Turbo DNA polymerase was substituted with the KAPA HiFi DNA polymerase (KAPA Biosystems). Plasmid maps and sequences are available on request. All primers used in this study are listed in the Supplementary Table S2.
Cells and animals
HEK293T cells were obtained from ATCC and cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Hyclone) containing 10% Fetal Bovine Serum (FBS; Hyclone, characterized grade), 100 IU/ml penicillin and 100 µg/ml streptomycin (Invitrogen) and 1 mM sodium pyruvate (Invitrogen) at 37°C and 5% CO2. Transfections were done using Lipofectamine 2000 (Invitrogen) as recommended. Transgenic N2a cell populations were generated using the HILO-RMCE procedure described elsewhere (29). Tissue samples were prepared from C57BL/6J mice using protocols approved by the Institutional Animal Care and Use Committee.
miRNA Northern-blot analysis
Total RNAs were extracted with Trizol (Invitrogen) as recommended and resolved by 15% PAGE containing 7.5 M urea and 1 × TBE. The RNAs were then electrotransferred to Hybond N+ membranes (GE Healthcare) in 0.5 × TBE for 1 h at 2.5 mA/cm2 and UV cross-linked at 0.15 J/cm2. The membranes were blocked with ExpressHyb (Clontech) for 1 h at 37°C and hybridized for 2 h at 37°C with complimentary DNA oligonucleotide probes labeled with T4 polynucleotide kinase and [γ-32P]ATP (Perkin Elmer, 600 Ci/mmol). The membranes briefly washed with 2 × SSC/0.05% SDS and 0.1 × SSC/0.1% SDS were imaged with a Typhoon Trio Variable Mode Imager (GE Healthcare) and the band intensities were quantified using ImageQuant TL (GE Healthcare). In some Northern experiments, we used the following synthetic 22-mer RNAs as markers:
miR-124(wt) (5′pUAAGGCACGCGGUGAAUGCCAA),
miR-124(3′C) (5′pUAAGGCACGCGGUGAAUGCCAC),
miR-124(3′G) (5′pUAAGGCACGCGGUGAAUG CCAG) and
miR-124(3′U) (5′UAAGGCACGCGGUGAAUGCCAU).
RT-qPCR analyses
Total RNAs were isolated from cultured cells and tissues using Trizol (Invitrogen) as recommended and treated with 50 U/ml of RQ1 DNase (Promega) at 37°C for 60 min to remove traces of genomic DNA. Reverse transcription (RT) was carried out using SuperScript III (Invitrogen) and random decamer primers (N10) at 50°C for 60 min as described (29). The reverse transcriptase was inactivated at 70°C for 10 min, the reactions were diluted with sterile DEPC-treated water (Invitrogen) to 200 µl. cDNA samples were analyzed by quantitative real-time PCR (qPCR) using a StepOnePlus real-time PCR system (Applied Biosystems) and Fast SYBR Green Master Mix (Applied Biosystems) with 0.15 µM of each forward and reverse primers. The following cycling program was used: 95°C for 20 s, followed by 45 cycles of a three-step reaction, denaturation at 95°C for 5 s, annealing and extension at 60°C for 45 s. Data were normalized to the expression levels of the GAPDH mRNA.
miRNA duplexes
miRNA duplexes were prepared by annealing complementary synthetic RNAs (Dharmacon). Mixtures containing 20 µM each of the two strands in the 1× siRNA buffer (Dharmacon) were incubated at 90°C for 1 min and slowly cooled down to the room temperature. 22-mer duplexes were prepared by annealing the 5′GGUGUUCACAGCGGACCUUGAU3′ antisense strand and either the 5′pUAAGGCACGCGGUGAAUGCCAA3′ or the 5′pUAAGGCACGCGGUGAAUGCCAmA3′ miRNA strands (5′p indicates a 5′ phosphate and mA3′ is a 2′-OMe-modified 3′ adenosine). The 21-mer duplex was prepared by annealing the 5′GUGUUCACAGCGGACCUUGAU3′ antisense strand and the 5′pUAAGGCACGCGGUGAAUGCCA3′ miRNA strand.
Co-immunoprecipitation analysis
Co-immunoprecipitation of mature miR-124 and Argonaute proteins was done in principle as described (27). Dishes (10 cm) containing ∼60% confluent HEK293T cell cultures were typically co-transfected with a 3xMyc-tagged Argonaute expression plasmid and a miR-124 expression plasmid and incubated for 48 h. Alternatively, cells were first transfected with a 3xMyc-tagged Argonaute expression plasmid for 24 h followed by transfection with a synthetic miRNA duplex. Cells were then washed with ice-cold 1 × PBS followed by a 15-min incubation in a 1-ml hypotonic lysis buffer containing 10 mM Tris–HCl pH 7.5, 10 mM KCl, 2 mM MgCl2, 5 mM DTT and 1 × Complete EDTA-free protease inhibitor cocktail (Roche). The cells were scraped off the plate and the suspensions were supplemented with the 5 × ATP depletion mix [450 mM KCl, 100 mM glucose, 1 mg/ml yeast tRNA (Invitrogen), 0.4 U/µl recombinant RNasin (rRNasin, Promega) and 0.5 U/ml hexokinase (Sigma)] added to the final concentration of 1×. The cells were then homogenized in a Dounce homogenizer by 15 strokes of pestle B. The lysates were cleared by centrifugation for 30 min at 16 000g, 4°C. To immunoprecipitate the 3xMyc-tagged Argonaute complexes, anti-c-Myc agarose beads (Sigma) were pre-blocked for 2 h at 4°C in W1 buffer (0.5% Nonidet P-40, 150 mM NaCl, 2 mM MgCl2, 2 mM CaCl2, 20 mM Tris–HCl, pH 7.5, 5 mM DTT and 1 × Complete EDTA-free protease inhibitor cocktail) additionally containing 1 mg/ml yeast tRNA and 1 mg/ml BSA. The beads (∼35 µl resin) were then incubated with the cleared lysates for 4 h at 4°C, washed once with 1 ml of ice-cold W1 buffer and twice with 1 ml of ice-cold W2 buffer (0.5% Nonidet P-40, 650 mM NaCl, 2 mM MgCl2, 2 mM CaCl2, 20 mM Tris, pH 7.5, 5 mM DTT and 1 × Complete EDTA-free protease inhibitor cocktail), 5 min each wash. The co-immunoprecipitated RNAs were eluted from the beads using Trizol and analyzed by Northern blotting and phosphorimaging. ‘Input’ lanes contained RNAs Trizol-extracted from 5% aliquots of the cleared lysates.
Pulse-chase experiments
HEK293T cells were co-transfected with the pTet-Off plasmid and an appropriate miR-124 expression plasmid and incubated for 24 h. Doxycycline (Dox) was then added to the cultures to the final concentration of 2 µg/ml to repress the tTA interaction with the TET promoter and the incubation was continued for another 48 h. Total RNA samples isolated at different points of this time course were analyzed by Northern blotting and phosphorimaging. pEM142 (Supplementary Table S1) was used as a control plasmid.
In vitro dicing
Total RNA (20 µg) isolated from pEM208-transfected HEK293T cells 24-h post-transfection was incubated for 1 h at 37°C in 20 µl reactions with 0.17–0.5 U of the purified human Dicer enzyme (Roche) in 1× reaction buffer containing 30 mM Tris–HCl pH 6.8, 50 mM NaCl, 3 mM MgCl2, 0.1% Triton X-100, 15% glycerol, 1 mM DTT and 2 U/µl rRNasin (Promega). Reaction products were extracted with a 1:1 mixture of acidic phenol/chloroform, precipitated with ethanol and analyzed by Northern blotting and phosphorimaging.
Primer extension
Total RNA (10 µg) from the E12.5 or the adult mouse brain was annealed with 200 fmol of 5′ [32P]-labeled miR-124_p_ext primer complementary to the 3′ end of the mature miR-124 in 10 µl reaction mixtures containing 10 mM Tris pH 7.4, 0.5 mM EDTA and 50 mM KCl. The reactions were incubated for 5 min at 70°C, 20 min at 37°C and finally for 10 min at 20°C. The annealed RNA–DNA hybrids were supplemented with 10 µl of the 2 × AMV RT mixture containing 4 µl of the 5 × AMV RT buffer (Promega), 2 µl of 10 mM dNTPs, 2 µl of DEPC water (Invitrogen), 1 µl of rRNasin (40 U/µl; Promega) and 1 µl of the AMV reverse transcriptase (10 U/µl; Promega) and incubated at 42°C for 20 min. The reaction products were extracted with phenol–chloroform, separated by 15% PAGE containing 7.5 M urea and the gel was exposed with an X-ray film.
miRNA deep sequencing
Deep sequencing of small RNAs from E12.5 and adult mouse brains was done as recommended by Illumina using a GAIIx sequencing platform. The fastq files were pre-filtered to remove reads containing bases with phred scores <10, followed by clipping the adapter sequences and post-filtering to remove reads containing bases with phred scores <20 and discarding reads <18 nt long using FASTX (http://hannonlab.cshl.edu/fastx_toolkit/). The collapsed reads (31) were parsed with a bowtie (32) index containing mature mouse miRNA sequences (miRBase v17; miRBase) extended by 3 nt at the 5′ ends and by 6 nt at the 3′ ends. No mismatches were allowed. To analyze reads with non-templated 3′-terminal nucleotides, parsing was repeated with reads computationally clipped by 1–3 nt at their 3′ ends. Sequences represented by <100 reads in either of the two brain samples were discarded. This filter was relaxed to <10 reads for the pair-wise analysis of trimming efficiencies of pre-miRNA ‘siblings’. We further discarded miRNA entries that could not be unambiguously distinguished from other miRNAs after removing three 3′-terminal nucleotides from either the experimental reads of the miRBase sequences. Downstream statistical analyses were carried out using in-house Perl and R scripts.
Ligation-mediated RT-PCR
The LM-RT-PCR protocol was modified from (23). Briefly, 500 µg of total RNA from E12.5 or adult mouse brain was resolved by 10% PAGE containing 7.5 M urea and the 14–30 nt RNA fraction was isolated using a miRNA extraction kit (Abnova). One-half of the gel-purified RNA was ligated with 200 pmol of the 3′ RNA adapter (5′P-UCGUAUGCCGUCUUCUGCUUGidT; Dharmacon) at 16°C for 12 h in a 10 -µl reaction containing 5 U of T4 RNA ligase (Ambion). The ligation mixtures were heat inactivated at 68°C for 8 min, resolved by 10% PAGE containing 7.5 M urea and the 36–46 nt RNA fraction was extracted from the gel (Abnova) and subjected to RT with the 3′_RT primer and the SuperScript III RT (Invitrogen). The miR-124-specific sequences were then amplified from the cDNA samples using the miR-124_F1 and 3′_PCR primers and KAPATaq HotStart DNA Polymerase (KAPA Biosystems) as recommended. The PCR products were purified using a Nucleospin Extract II kit (Macherey-Nagel), ligated with the pGEM-T vector (Promega) according to the manufacturer’s instructions and transformed into chemically competent Top10 E. coli cells (Invitrogen). A total of 30 clones for each sample were sequenced using T7 sequencing primer.
Immunoblotting
Cells were washed with ice-cold PBS and the proteins were extracted with 20 mM Tris–HCl, pH 7.5, 150 mM NaCl, 5 mM EDTA, 10% glycerol, 1% Nonidet P-40, 1 mM PMSF and 1× Complete EDTA free protease inhibitor cocktail (Roche). Mouse brain tissues were washed with ice-cold PBS, frozen in liquid nitrogen and homogenized with mortar and pestle prior to adding the extraction buffer. Lysates were cleared by centrifugation for 10 min at 20 000g, 4°C and the protein concentration was determined using a BCA protein assay kit (Thermo Scientific) The samples were separated by 4–20% gradient SDS–PAGE (Thermo Scientific), transferred to nitrocellulose membranes and incubated with the following primary antibodies: rabbit polyclonal anti-Ago1 (Millipore; 1:1000 dilution); mouse monoclonal anti-Ago1 (kind gift of Mikiko Siomi; 1:500 dilution), rabbit monoclonal anti-Ago2 (Cell Signaling Technology; 1:1000), mouse monoclonal anti-β-tubulin (Invitrogen; 1:3000), mouse monoclonal anti-GAPDH (Ambion; 1:10000), mouse monoclonal anti-c-Myc (Sigma; 1:3000). The secondary anti-rabbit and anti-mouse horseradish peroxidase (HRP) conjugates were from GE Healthcare (1:5000). Protein bands were visualized using enhanced chemiluminescence (ECL) reagents from Thermo Scientific as recommended.
In silico protein analyses
Argonaute amino acid sequences were aligned using MUSCLE (http://www.ebi.ac.uk/Tools/msa/muscle/) and the similarity plots were generated in JProfileGrid [(33); http://www.profilegrid.org/] using a 5-amino acid sliding window. The structure of the AGO1-small RNA complex [1SI3; (14)] was downloaded from the Protein Data Bank (http://www.pdb.org) and visualized by PyMOL (http://www.pymol.org/).
RESULTS
Mammalian Argonaute identity affects miRNA length
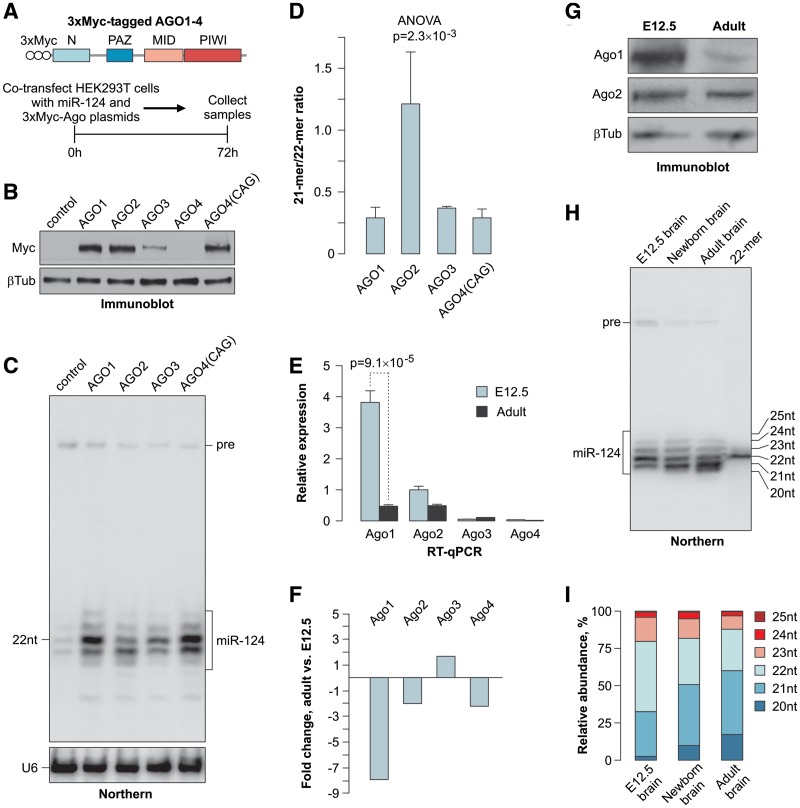
To understand the biological significance of the mammalian Argonaute paralog expansion, we co-transfected human HEK293T cells with expression plasmids encoding 3xMyc-tagged versions of the human AGO1–4 under the control of the CMV or CAG promoters (Figure 1A) together with the RIPmiR-124 construct encoding miR-124, an evolutionarily conserved nervous system-specific miRNA studied in our laboratory (28,34). The AGO1–3 proteins were well expressed from a CMV promoter, whereas a CAG promoter was required to express AGO4 at an optimal level (Figure 1B).
Figure 1.
Mammalian Argonautes modulate the mature miRNA length. (A) Top, diagram of the 3xMyc-AGO constructs and bottom, workflow of the 3xMyc AGO/miR-124 co-expression experiment. (B) Immunoblot analysis of the 3xMyc-tagged AGO1–4 proteins expressed from a CMV promoter and AGO4 expressed from a CAG promoter using a Myc tag-specific antibody. β-tubulin is a lane loading control. (C) Northern-blot analysis of miR-124 co-expressed with the corresponding 3xMyc AGO proteins or a control plasmid. The positions of the mature miR-124 and the pre-miR-124 precursor (pre) are marked on the right. U6 snRNA specific probe is used as a loading control. (D) Quantification of the results in (C) showing a significantly increased ratio between the 21- and 22-mer in cells over-expressing 3xMyc AGO2 as compared to the other three Argonautes. Data are averaged from three independent transfection experiments ±SD. (E) RT-qPCR analysis of the Argonaute expression in E12.5 and adult (P60) mouse brains. Data were averaged from three biological replicates ±SD and analyzed by the t test. (F) Quantification of the data in (E) showing a dramatic decrease in the Ago1 expression in the adult brain. Negative fold changes correspond to down-regulation in the adult brain and positive fold changes correspond to up-regulation in the adult brain. (G) Immunoblot analysis showing a relatively uniform expression of Ago2 in the E12.5 and adult mouse brains and a reduction in the Ago1 protein levels during mouse brain development. β-tubulin is a loading control. (H) Northern-blot analysis of RNAs from E12.5, newborn (P0) and adult (P60) mouse brains showing progressive shortening of mature miR-124 during development. (I) Quantification of the data in (H) showing a change in the percentage of the corresponding miR-124 isoforms (20–25 nt) as a function of development.
A high-resolution Northern-blot analysis of the miR-124 RNA revealed that: (i) Argonaute over-expression led to dramatically increased steady-state levels of the mature miRNA and (ii) the miR-124 isoform population was dominated by 21-mers in the presence of recombinant AGO2 and by 22-mers in the presence of AGO1, AGO3 or AGO4 (Figure 1C and D). Similar result was obtained when we co-expressed miR-124 with the four mouse Argonautes (Ago1–4; Supplementary Figure S1A– S1C).
The excess mature miRNA accumulated in the Argonaute over-expressing cells was likely associated with the corresponding AGO proteins, as discussed previously (35). To test this prediction, we co-expressed miR-124 and the 3xMyc-tagged AGO proteins in HEK293T cells, pulled down the complexes using a Myc tag-specific antibody and analyzed the co-immunoprecipitated miR-124 by Northern blotting (Supplementary Figure S1D). Similar to the distributions observed in Figure 1C and D, the most abundant AGO2-associated form was 21-mer, whereas 22-mer was predominant in the AGO1, AGO3 and AGO4 pull-downs (Supplementary Figure S1D and S1E). Thus, the miR-124 isoform composition of the total RNA samples corresponded to the miRNA patterns physically associated with the over-expressed Argonautes.
Increase in fractional abundance of Ago2 correlates with shortening of mature miR-124 during mouse brain development
Since the above results suggested that the Argonaute identity might affect the length of mature miRNAs, we examined the Ago1–4 expression in developing mouse brain. Quantitative RT-PCR (RT-qPCR) analyses showed that the embryonic day 12.5 (E12.5) brain expressed readily detectable amounts of the Ago1 and Ago2 mRNAs (Figure 1E). Notably, the apparent Ago1 mRNA abundance decreased ∼8-fold in the adult brain, while the Ago2 expression decreased only ∼2-fold (Figure 1E and F). The expression of Ago3 and Ago4 was negligible at both developmental stages (Figure 1E). Immunoblot analyses confirmed that the originally high Ago1 protein levels in the E12.5 brain were dramatically reduced in the adult sample, whereas the Ago2 expression was relatively uniform at both developmental stages (Figure 1G).
To investigate the effect of this increase in the Ago2 fractional abundance, we analyzed the mature miR-124 in the E12.5, newborn and adult mouse brains (Figure 1H). Interestingly, 22-mers were predominant in the E12.5 sample, whereas 21-mers became the most abundant form in the newborn and adult samples (Figure 1H and I). This indicated that the change in the Argonaute expression pattern might underlie the miR-124 isoform dynamics during brain development.
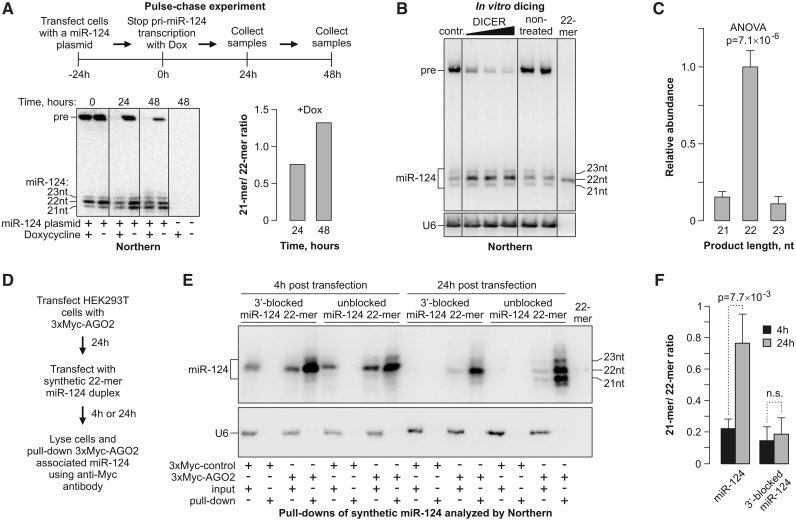
Mature miRNAs are shortened following the Dicer processing step
The Argonaute-dependent change in the miR-124 length could theoretically be due to: (i) altered cleavage preferences of Drosha or Dicer or (ii) a post-Dicer trimming of the Argonaute-loaded mature miRNAs. To distinguish between these possibilities, we carried out a pulse-chase experiment, in which miR-124 was expressed from a Dox-repressible promoter in HEK293T cells containing substantial amounts of endogenous AGO2 (15). The transcription was stopped with Dox 24-h post-transfection and the subsequent changes in the miR-124 RNA were analyzed by Northern blotting (Figure 2A). The originally prominent pre-miR-124 precursor band disappeared within 24 h of the Dox treatment, presumably as a result of the Dicer processing reaction (Figure 2A, bottom). This allowed us to monitor the subsequent changes in the mature miR-124 species. Notably, the 22-mer miR-124 species were chased into 21-mers from 24 to 48 h after adding Dox (Figure 2A).
Figure 2.
miRNA length is reduced following its recruitment to AGO2. (A) Pulse-chase experiment showing that the mature miR-124 length is reduced after the completion of the Drosha- and Dicer-dependent processing steps. Top, the experimental workflow; bottom left, Northern-blot analysis of the corresponding RNA samples; bottom right, quantification of the Northern-blot data showing an increase in the 21- to 22-mer ratio in samples from collected 24 and 48 h after halting the pri-miR-124 transcription by Dox. (B) RNA from miR-124-expressing HEK293T cells in (A) at the time point 0 was incubated in vitro with increasing amounts of purified human Dicer and the reaction products were analyzed by Northern blotting. (C) Quantification of the newly formed products in (B) showing that Dicer generates predominantly 22-mer species. Data are averaged from the three Dicer lanes in (B) ±SD. (D–F) AGO2-loaded synthetic 22-mers are efficiently trimmed unless protected at the 3′ end. (D) Outline of the experiment. (E) HEK293T cells expressing 3xMyc-AGO2 were transfected with synthetic siRNA-like duplexes containing either an unmodified 22-mer miR-124 or 22-mer protected at the 3′ end with a 2′-OMe group. The AGO2/miR-124 complexes were pulled down 4- and 24-h post-transfection and analyzed by Northern blotting. (F) Quantifications of the data in (E) confirming efficient trimming of the unmodified but not 3′-protected 22-mers loaded into AGO2. Data are averaged from three independent experiments ±SD.
This result was consistent with the model that pre-miR-124 was initially processed by Dicer into 22-mers that were subsequently trimmed to 21-mers. To directly determine the Dicer cleavage specificity, we incubated the total RNA from the RIPmiR-124-transfected HEK293T cells harvested at 24-h post-transfection with purified human Dicer in vitro (Figure 2B). Northern-blot quantification of the newly formed mature miR-124 species processed from the pre-miR-124 substrate confirmed that Dicer indeed generated predominantly 22-mer products (Figure 2C).
The 3′ termini of some Argonaute-loaded miRNAs are known to undergo enzymatic trimming in Drosophila (25,26). To test whether miR-124 might be shortened in the mammalian cells through a similar process, we transfected HEK293T cells transiently expressing 3xMyc-tagged AGO2 with synthetic miR-124 22-mer duplexes designed similarly to siRNAs and analyzed the lengths of the AGO2-associated miRNAs by Northern blotting 4- and 24-h post-transfection (Figure 2D–F and Supplementary Figure S2A). The fraction of the AGO2-bound 21-mers increased over time, indicative of an ongoing trimming reaction. On the other hand, essentially no trimming was detected when we repeated this experiment with 22-mer miR-124, in which the 3′-terminal nucleotide was protected by a 2′-OMe group (Figure 2D–F). This suggested that the AGO2-associated 21-mers were derived from the 22-mers as a result of the 3′ terminal nucleotide removal.
Since it was formally possible that different Argonaute proteins might have distinct miRNA loading preferences, we transfected HEK293T cells expressing 3xMyc-tagged AGO1 or AGO2 with an equimolar mixture of the 22- and 21-mer miR-124 duplexes and analyzed the isoform distributions in the Myc-tag pull-down fractions 4-h post-transfection (Supplementary Figure S2B and S2C). Both AGO1 and AGO2 recruited 22- and 21-mers with comparable efficiencies thus supporting the model that the length of mature miRNAs is altered following their loading into AGO2.
Mammalian miRNAs undergo large-scale 3′-terminal trimming during nervous system development
To ensure that the 3′-terminal trimming also accounted for the miR-124 isoform dynamics in the developing mouse brain, we analyzed the 3′-terminal sequences of miR-124 at the E12.5 and the adult stages using ligation-mediated RT-PCR followed by cloning and sequencing of the miR-124-specific PCR products (Supplementary Figure S3A). All 30 clones derived from the E12.5 sample terminated in … CCAA3′, which corresponded to the 22-mer sequence 5′UAAGGCACGCGGUGAAUGCCAA3′. On the other hand, … CCA3′ (21-mer) was the most frequently encountered terminus in the adult clones followed by … CCAA3′ (22-mer), … CC3′ (20-mer) and … C3′ (19-mer) (Supplementary Figure S3A). Primer extension analysis of the E12.5 and adult miR-124 showed that the predominant 5′ end position was 5′UAAGGC … in both samples, thus confirming that the reduced length of miR-124 in the adult brain was due to changes at the 3′ end (Supplementary Figure S3B).
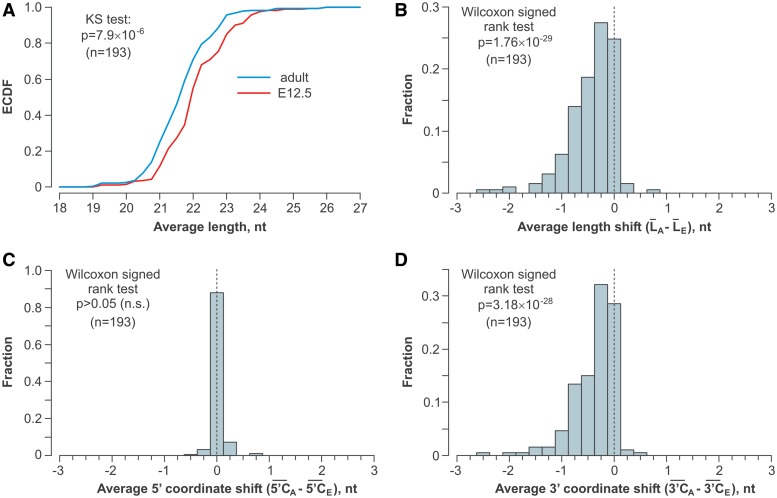
To find out if the 3′ ends of other miRNAs were shortened during brain development, we analyzed the small RNA fraction from the E12.5 and the adult brain by deep sequencing and focused on 193 miRNAs that were detectably expressed in both samples (>100 reads), could be unambiguously distinguished from other miRNAs and had no non-templated nucleotides at the 3′ end. Notably, the cumulative functions (ECDFs) of the overall miRNA length distributions in the two samples differed significantly (P = 7.9 × 10−6, KS test), with the adult miRNAs being shorter than the E12.5 ones (Figure 3A). This conclusion was further supported by pair-wise comparisons of the average lengths of specific miRNAs in the adult and E12.5 samples (Figure 3B; P = 1.76 × 10−29, Wilcoxon signed rank test; also see Supplementary Tables S3 and S4). The differences in the miRNA lengths were clearly due to shortening of the 3′ but not the 5′ end in the adult sample (Figure 3C and D). Notably, Northern-blot analyses of select miRNAs confirmed our deep sequencing data (Supplementary Figure S4).
Figure 3.
Widespread shortening of the miRNA 3′ ends during brain development. Statistical analyses of deep sequencing data for 193 microRNAs expressed in the E12.5 and the adult mouse brain and containing no non-templated nucleotides. (A) The overall length distribution of miRNAs expressed in the adult brain is shifted toward shorter species as compared to E12.5. (B) Average lengths of specific miRNAs tend to decrease in the adult brain as compared to E12.5. (C) The miRNA 5′ end coordinates do not differ significantly between the two brain samples. (D) The 3′ end coordinates of the adult miRNAs are diminished significantly as compared to E12.5.
To ensure that the above result was not due to a sample preparation artifact, we repeated the statistical analyses with 80 unambiguous miRNAs that were detectably expressed in both the E12.5 and adult samples (>100 reads) and contained one to three non-templated nucleotides at their 3′ ends (Supplementary Figure S5). These modified miRNA isoforms represented ∼10% of the total reads in both samples and the non-templated extensions consisted predominantly of A’s and U’s (Supplementary Figure S5A), as described earlier (21,23,24). Importantly, when we computationally clipped the non-templated extensions and analyzed the lengths and the 5′ and 3′ coordinates of the remaining genome-encoded ‘stumps’, a large-scale 3′-terminal trimming was again detected for the adult miRNAs (Supplementary Figure S5B–S5D). We concluded that the miRNA 3′ ends receded in the adult brain but the probability of their subsequent extension by terminal transferases was comparable between the E12.5 and the adult samples.
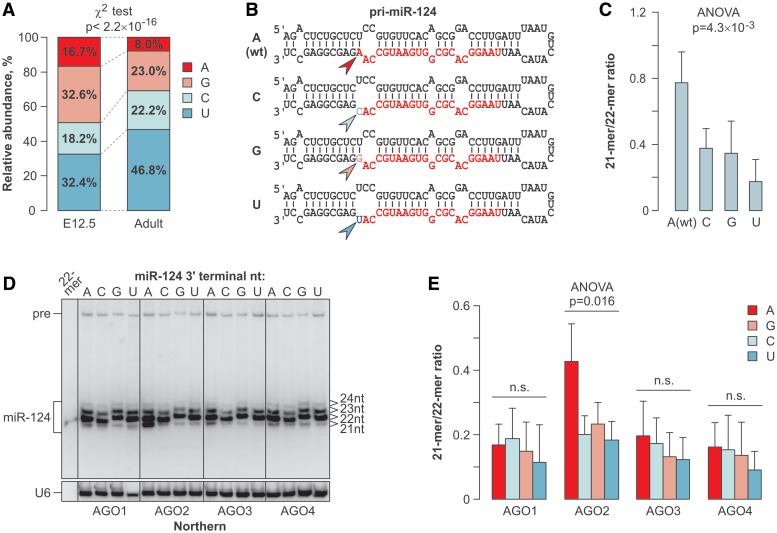
miRNA trimming efficiency depends on the nature of the 3′-terminal nucleotide
We then returned to the 193 miRNAs devoid of the non-templated extensions and analyzed their 3′-terminal nucleotide usage. All 4 nt occurred fairly randomly at the 3′ ends of the E12.5 miRNAs (Figure 4A). However, the 3′ termini of the adult miRNAs were noticeably depleted in A’s and enriched in U’s (Figure 4A; P < 2.2 × 10−16, χ2 test).
Figure 4.
3′-Terminal nucleotide preferences of the trimming reaction. (A) The usage of the 3′-terminal genome-encoded nucleotides is relatively uniform in the E12.5 miRNAs, whereas it becomes significantly depleted in A’s and enriched in U’s in the adult sample. (B) Diagram of the miR-124 expression constructs genetically modified to introduce all 4 nt (arrowheads) at the 3′-terminal position of the mature miRNA (colored sequence). (C) Of the four miR-124 variants described in (B), A3′ is trimmed most efficiently and U3′ is trimmed least efficiently in the HEK293T cells. (D) The four 3′-terminal miR-124 variants were co-expressed in HEK293T cells with the AGO1–4 and the trimming efficiency was analyzed by Northern blotting 48-h post-transfection. (E) Quantification of the data in (D).
To test whether the corresponding nucleotide preferences of the trimming reaction could explain this bias, we prepared a set of miR-124 constructs encoding all 4 nt at the 3′-terminal position of the mature 22-mer and analyzed the trimming behavior of the corresponding miRNAs in the HEK293T cells (Figure 4B and C; Supplementary Figure S6). Notably, the A3′-terminated (wild-type) miR-124 was trimmed most efficiently and the U3′ miR-124 was trimmed least efficiently (P = 4.3 × 10−3; ANOVA test; Figure 4B and C; Supplementary Figure S6).
Co-expression of the four 3′-terminal miR-124 variants with the 3xMyc-tagged Argonautes showed that AGO2-overexpressing cells trimmed A3′ most efficiently and U3′ least efficiently (Figure 4D and E), similar to the preferences of endogenous HEK293T Argonaute blend. Trimming of all four miR-124 variants was rather inefficient in the presence of over-expressed AGO1, AGO3 or AGO4 (Figure 4D and E). These results suggested that the nature of the 3′-terminal nucleotide modulates trimming of AGO2-loaded miRNAs.
Specific structural feature of the AGO2 PAZ domain enables efficient miRNA trimming
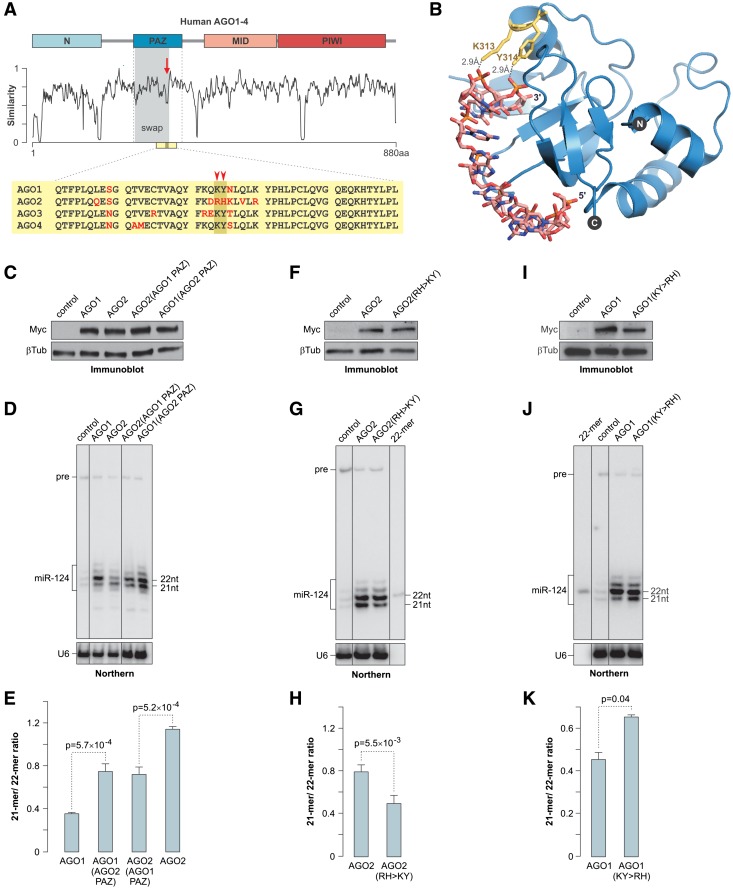
To elucidate the mechanism for the efficient shortening of AGO2-associated miRNAs, we aligned the amino acid sequences of human Argonautes and focused on the regions showing the highest divergence (Figure 5A). The least conserved element within the PAZ domain sequence corresponded to the AGO1 300–319 amino acid segment known to fold into a partially α-helical loop interacting with the small RNA 3′ end [Figure 5A; (14)]. The K313 and Y314 residues were of particular interest since they formed hydrogen bonds with the phosphate groups of the 3′ penultimate and ultimate nucleoside monophosphates, respectively (Figure 5B). Notably, the KY motif was conserved in AGO1, AGO3 and AGO4 across tetrapod vertebrates, but it was substituted with RH in AGO2 (Figure 5A and Supplementary Figure S7).
Figure 5.
Structure of the Argonaute PAZ domain modulates miRNA trimming efficiency. (A) Human Argonaute sequence similarity plot. The diagram on the top indicates the positions of the N, PAZ, MID and PIWI domains. The divergent sequence element within the PAZ domain is indicated by the arrow and the sequence swapped between AGO1 and AGO2 in (C–E) is highlighted in gray. A fragment of the PAZ sequence alignment is presented at the bottom with non-conserved amino acid residues shown in red. The position of the AGO1 K313 and Y314 residues is marked by the arrowheads. (B) Crystal structure of the AGO1 PAZ domain associated with a small RNA [1SI3; (14)] showing hydrogen bonds between the K313 and Y314 side chains and the 3′ penultimate and ultimate nucleoside triphosphates. (C, F and I) Immunoblot analyses showing comparable expression of the wild-type AGO1 and AGO2, (C) their reciprocal PAZ domain-swapped mutants, (F) AGO2(RH > KY) mutant and (I) AGO1(KY > RH) mutant in HEK293T cells. (D, G and J) Northern-blot analyses of miR-124 co-expressed with the corresponding Argonaute proteins in HEK293T cells. Cells were harvested at (D) 72 hours or (G and J) 48-h post-transfection. (E, H and K) Quantification of the data in (D, G and J) showing the effect of the PAZ domain structure on the miRNA trimming efficiency. Data were averaged from three independent transfection experiments ±SD and compared using the t test.
To address the role of the PAZ domain in the miRNA trimming, we generated reciprocal swap mutants of AGO1 and AGO2 that contained heterologous PAZ domain segments comprising the KY/RH motif (see Figure 5A). The engineered AGO1(AGO2 PAZ) and AGO2(AGO1 PAZ) proteins were expressed in HEK293T cells at levels similar to their wild-type counterparts (Figure 5C). Strikingly, miR-124 was trimmed more efficiently in the presence of AGO1(AGO2 PAZ) as compared to the wild-type AGO1 protein (Figure 5D and E). On the other hand, trimming of AGO2-loaded miR-124 was suppressed by grafting the AGO1 PAZ domain (Figure 5D and E).
We hypothesized that the KY residues might stabilize the 3′ end of AGO1-, AGO3- and AGO4-bound miRNAs within the PAZ domain and restrict its availability to yet-to-be identified mammalian miRNA trimming enzyme(s). To test this prediction, we mutated the RH element in AGO2 to KY and co-expressed the mutant protein with miR-124 in HEK293T cells (Figure 5F). Satisfyingly, the RH > KY mutation significantly suppressed trimming of the AGO2-loaded miR-124 (Figure 5G and H). On the other hand, the reciprocal KY > RH mutation of AGO1 stimulated miR-124 trimming (Figure 5I–K). The control mutation (D597A) inactivating the AGO2 slicer activity (9) had no effect on the miRNA trimming efficiency (Supplementary Figure S8). Therefore, the unusually high trimming propensity of AGO2-loaded miRNAs requires specific structural features of the AGO2 PAZ domain.
AGO1-loaded miR-124 is more potent in inducing neuron-like differentiation of neuroblastoma cells than AGO2-loaded miR-124
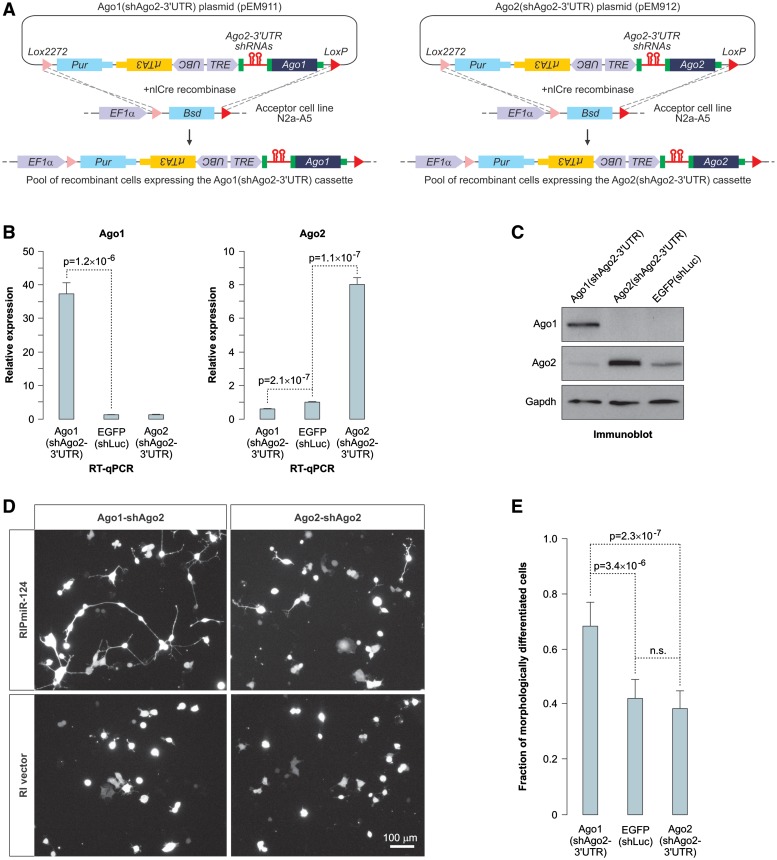
To address the role of the dramatic decrease in the Ago1/Ago2 ratio during nervous system development, we took advantage of the high-efficiency and low-background recombination-mediated cassette exchange (HILO-RMCE) system (29). Using this approach we generated two recombinant populations of mouse Neuro2a (N2a) neuroblastoma cells expressing Dox-inducible cassettes with two intronic shRNAs against the endogenous Ago2-3′-UTR followed by a transgenic open reading frame (ORF) of either Ago1 or Ago2 (Figure 6A). The sequences recognized by the Ago2-3′-UTR-specific shRNAs were absent from the transgenic Ago2 sequence.
Figure 6.
AGO1 stimulates miR-124-induced neuronal differentiation of neuroblastoma cells. (A) Diagram of the HILO-RMCE procedure used to obtain pools of transgenic N2a cells expressing Dox-inducible Ago1(shAgo2-3′-UTR) and Ago2(shAgo2-3′-UTR) cassettes. (B and C) Transgenic N2a cells generated as described in (A) or cells containing a Dox-inducible EGFP(shLuc) control transgene were treated with Dox for 72 h and the Ago1 and Ago2 expression was analyzed by (B) RT-qPCR or (C) immunoblotting. Data in (B) were averaged from six independent amplification experiments ±SD and compared using t test. (D) Ago1(shAgo2-3′-UTR) and Ago2(shAgo2-3′-UTR) cells pre-treated with Dox for 72 h were transiently transfected with the miR-124 expression plasmid RIP-miR-124 additionally encoding the dsRed2 fluorescent marker or the corresponding control vector (RI vector) and the transfected cell morphology was examined 36-h post-transfection using dsRed2 fluorescence. Note that the expression of RIP-miR-124 leads to the appearance of a larger number of neuron-like cells containing long processes in the Ago1(shAgo2-3′-UTR) cells as compared to the Ago2(shAgo2-3′-UTR) cells. No differentiated cells are obvious in the RI vector-transfected cultures. (E) Quantification of the RIPmiR-124-induced morphological differentiation in Ago1(shAgo2-3′-UTR), Ago2(shAgo2-3′-UTR) and EGFP(shLuc) cells. Shown are fractions of cells containing at least one primary process longer than the average diameter of the cell soma. Ten randomly selected fields were imaged for each sample and the data were averaged ±SD and compared using the t test.
The Argonaute expression patterns in the two transgenic N2a populations changed dramatically upon Dox treatment (Figure 6B and C). Ago1 was over-expressed and Ago2 was down-regulated in the Ago1(shAgo2-3′-UTR) cells, whereas the overall Ago2 levels increased in the Ago2(shAgo2-3′-UTR) cells as compared to the control cell population expressing a Dox-inducible EGFP ORF and a firefly specific shRNA [EGFP(shLuc); Figure 6B and C]. As expected, transiently transfected miR-124 was trimmed more efficiently in the Ago2(shAgo2-3′-UTR) as compared to the Ago1(shAgo2-3′-UTR) cells (Supplementary Figure S9A and S9B).
Since miR-124 expression is known to induce neuron-like differentiation of N2a cells (28), we examined the efficiency of this process in the two Argonaute backgrounds. Strikingly, miR-124 expressed in the Dox-treated Ago1(shAgo2-3′-UTR) cells was significantly more potent in inducing the neurite outgrowth phenotype as compared to miR-124 expressed in the Dox-treated Ago2(shAgo2-3′-UTR) cells (Figure 6D and E). This effect was not due to an overall higher activity of miR-124 in the Ago1(shAgo2-3′-UTR) cells since miR-124 repressed three of its previously published targets (Ptbp1, Vamp3 and Ctdsp1) equally well in both backgrounds (Supplementary Figure S9C and S9D). We concluded that the Ago1- and Ago2-loaded miR-124 may have distinct biological activities in the context of mammalian brain development possibly through differential regulation of a subset of targets.
DISCUSSION
Multiple post-transcriptional mechanisms have been implicated in the regulation of miRNA activity in a range of biological contexts (8). Our study provides an important update to the mammalian catalog of post-transcriptional modifications by showing that a large subset of miRNAs undergo 3′-terminal miRNA trimming during mouse brain development.
Several lines of evidence strongly suggest that, similar to the situation in Drosophila (25,26), mammalian trimming occurs after the pre-miRNA dicing step and the recruitment of mature miRNAs into Argonaute-containing complexes. (i) miRNA was trimmed in the absence of detectable amounts of the pre-mRNA precursor (Figure 2A). (ii) Similarly, AGO2-loaded synthetic miRNAs were efficiently trimmed (Figure 2D–F). (iii) Notably, we failed to detect a significant correlation between the trimming efficiencies of mature miRNA ‘siblings’ encoded by the same pre-miRNA in our deep sequencing data (Supplementary Table S5), which is consistent with the model that trimming occurs after the two strands of the precursor miRNA duplex are physically separated.
Further work will be required to establish the identity of the enzyme(s) trimming the mammalian miRNA 3′ ends. An obvious starting point of these studies would be characterization of the mammalian Nibbler homologs (25,26). Our observation that the nature of the miRNA 3′ terminal nucleotide affects the efficiency of the trimming reaction (Figure 4 and Supplementary Figure S6) might reflect biochemical preferences and possibly facilitate the identification of this hypothetical 3′-to-5′ exonuclease.
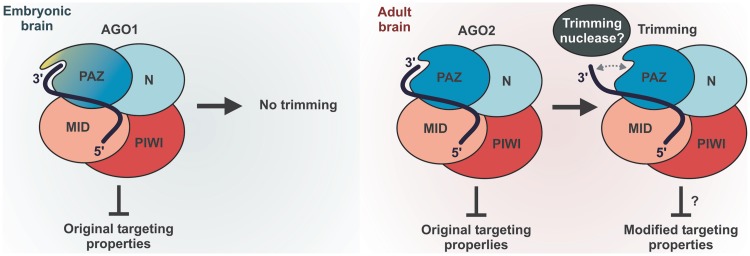
A surprising conclusion from our work is that, of the four mammalian Argonaute paralogs, only Ago2-loaded miRNAs undergo efficient 3′-terminal trimming (Figure 1, Supplementary Figure S1 and Figure 5). Given the increase in the fractional abundance of Ago2 during brain development due to dramatic down-regulation of Ago1 (Figure 1E–G), this provides a molecular mechanism for the large-scale resection of the miRNA 3′ ends in adult mouse brain (Figure 7).
Figure 7.
Model outlining the role of mammalian Argonaute identity in miRNA trimming.
We link the efficient trimming of Ago2-loaded miRNAs to the unusual structure of the Ago2 PAZ domain featuring the RH residues in its miRNA 3′-interaction loop instead of the KY motif conserved across other miRNA-specific Argonautes (Supplementary Figure S7). A likely outcome of the KY > RH substitution could be a loss of at least one of the two hydrogen bonds between the KY residues and the miRNA 3′ end due to the relatively small size and the lack of hydroxyl in the histidine side chain. This might theoretically weaken the interaction of the miRNA 3′ end with the PAZ domain thus making it more accessible to a trimming exonuclease (Figure 7). Based on our phylogenetic analyses (Supplementary Figure S7), it is tempting to speculate that the Ago2 RH motif might have emerged in an ancestor of the tetrapod vertebrates as an adaptation enabling developmental control of the miRNA trimming.
What could be the biological function of miRNA trimming? A growing body of evidence suggests that the 3′-terminus of a miRNA may modulate its interaction with cognate mRNAs in addition to the 5′-proximal ‘seed’sequence (3,36,37). It is therefore conceivable that the removal of 1 or 2 nt from the 3′ terminus could diminish the miRNA binding affinity to a subset of targets requiring 3′ compensatory interactions. The higher biological activity of Ago1-associated miR-124 in neuroblastoma cells observed in our study (Figure 6) would generally be consistent with this model. Indeed, trimming of Ago2-loaded miR-124 might diminish its target repertoire thus potentially leading to a loss of important differentiation-specific targets (Figure 7). We are currently investigating the effect of Argonaute identity on the miR-124 targeting properties and potential significance of the stronger differentiation phenotype of Ago1-loaded miR-124 for mouse brain development.
In conclusion, our data uncover pervasive 3′-terminal trimming of mammalian miRNAs and suggest that the efficiency of this process might be developmentally regulated by changes in fractional abundance of Ago2, the only mammalian Argonaute compatible with efficient trimming reaction. Identification of the mammalian miRNA trimming enzyme(s) and understanding the possible functions of trimmed miRNAs in mammals and Drosophila will be important questions for future studies.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–5 and Supplementary Figures 1–9.
FUNDING
Singapore National Research Foundation [NRF-RF2008-06 to E.V.M.]. Funding for open access charge: Singapore National Research Foundation [NRF-RF2008-06 to E.V.M.].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank G. Hannon, A. McMahon, G. Meister, M. Siomi and T. Tuschl for reagents and S. Oliferenko for comments on the manuscript.
REFERENCES
- 1.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 2.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell. Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ivey KN, Srivastava D. MicroRNAs as regulators of differentiation and cell fate decisions. Cell Stem Cell. 2010;7:36–41. doi: 10.1016/j.stem.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 5.Fineberg SK, Kosik KS, Davidson BL. MicroRNAs potentiate neural development. Neuron. 2009;64:303–309. doi: 10.1016/j.neuron.2009.10.020. [DOI] [PubMed] [Google Scholar]
- 6.Bushati N, Cohen SM. microRNA functions. Annu. Rev. Cell Dev. Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 7.Leung AK, Sharp PA. MicroRNA functions in stress responses. Mol. Cell. 2010;40:205–215. doi: 10.1016/j.molcel.2010.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 9.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 10.Murphy D, Dancis B, Brown JR. The evolution of core proteins involved in microRNA biogenesis. BMC Evol. Biol. 2008;8:92. doi: 10.1186/1471-2148-8-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nat. Rev. Mol. Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- 12.Höck J, Meister G. The Argonaute protein family. Genome Biol. 2008;9:210. doi: 10.1186/gb-2008-9-2-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boland A, Huntzinger E, Schmidt S, Izaurralde E, Weichenrieder O. Crystal structure of the MID-PIWI lobe of a eukaryotic Argonaute protein. Proc. Natl Acad. Sci. USA. 2011;108:10466–10471. doi: 10.1073/pnas.1103946108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma JB, Ye K, Patel DJ. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature. 2004;429:318–322. doi: 10.1038/nature02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 16.Yang JS, Maurin T, Robine N, Rasmussen KD, Jeffrey KL, Chandwani R, Papapetrou EP, Sadelain M, O’Carroll D, Lai EC. Conserved vertebrate mir-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis. Proc. Natl Acad. Sci. USA. 2010;107:15163–15168. doi: 10.1073/pnas.1006432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cifuentes D, Xue H, Taylor DW, Patnode H, Mishima Y, Cheloufi S, Ma E, Mane S, Hannon GJ, Lawson ND, et al. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010;328:1694–1698. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Azuma-Mukai A, Oguri H, Mituyama T, Qian ZR, Asai K, Siomi H, Siomi MC. Characterization of endogenous human Argonautes and their miRNA partners in RNA silencing. Proc. Natl Acad. Sci. USA. 2008;105:7964–7969. doi: 10.1073/pnas.0800334105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berezikov E, Robine N, Samsonova A, Westholm JO, Naqvi A, Hung JH, Okamura K, Dai Q, Bortolamiol-Becet D, Martin R, et al. Deep annotation of Drosophila melanogaster microRNAs yields insights into their processing, modification, and emergence. Genome Res. 2011;21:203–215. doi: 10.1101/gr.116657.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wyman SK, Knouf EC, Parkin RK, Fritz BR, Lin DW, Dennis LM, Krouse MA, Webster PJ, Tewari M. Post-transcriptional generation of miRNA variants by multiple nucleotidyl transferases contributes to miRNA transcriptome complexity. Genome Res. 2011;21:1450–1461. doi: 10.1101/gr.118059.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fernandez-Valverde SL, Taft RJ, Mattick JS. Dynamic isomiR regulation in Drosophila development. RNA. 2010;16:1881–1888. doi: 10.1261/rna.2379610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morin RD, O’Connor MD, Griffith M, Kuchenbauer F, Delaney A, Prabhu AL, Zhao Y, McDonald H, Zeng T, Hirst M, et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008;18:610–621. doi: 10.1101/gr.7179508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu N, Abe M, Sabin LR, Hendriks GJ, Naqvi AS, Yu Z, Cherry S, Bonini NM. The Exoribonuclease Nibbler Controls 3′ End Processing of MicroRNAs in Drosophila. Curr. Biol. 2011;21:1888–1893. doi: 10.1016/j.cub.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han BW, Hung JH, Weng Z, Zamore PD, Ameres SL. The 3′-to-5′ exoribonuclease nibbler shapes the 3′ ends of micrornas bound to Drosophila Argonaute1. Curr. Biol. 2011;21:1878–1887. doi: 10.1016/j.cub.2011.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karginov FV, Conaco C, Xuan Z, Schmidt BH, Parker JS, Mandel G, Hannon GJ. A biochemical approach to identifying microRNA targets. Proc. Natl Acad. Sci. USA. 2007;104:19291–19296. doi: 10.1073/pnas.0709971104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol. Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khandelia P, Yap K, Makeyev EV. Streamlined platform for short hairpin RNA interference and transgenesis in cultured mammalian cells. Proc. Natl Acad. Sci. USA. 2011;108:12799–12804. doi: 10.1073/pnas.1103532108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook J, Russell RW. Molecular Cloning: A Laboratory Manual. 3rd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 31.Friedländer MR, Chen W, Adamidi C, Maaskola J, Einspanier R, Knespel S, Rajewsky N. Discovering microRNAs from deep sequencing data using miRDeep. Nat. Biotechnol. 2008;26:407–415. doi: 10.1038/nbt1394. [DOI] [PubMed] [Google Scholar]
- 32.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roca AI, Almada AE, Abajian AC. ProfileGrids as a new visual representation of large multiple sequence alignments: a case study of the RecA protein family. BMC Bioinformatics. 2008;9:554. doi: 10.1186/1471-2105-9-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Makeyev EV, Maniatis T. Multilevel regulation of gene expression by microRNAs. Science. 2008;319:1789–1790. doi: 10.1126/science.1152326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Diederichs S, Haber DA. Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell. 2007;131:1097–1108. doi: 10.1016/j.cell.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 36.Broderick JA, Salomon WE, Ryder SP, Aronin N, Zamore PD. Argonaute protein identity and pairing geometry determine cooperativity in mammalian RNA silencing. RNA. 2011;17:1858–1869. doi: 10.1261/rna.2778911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.