Abstract
The recent discovery that the human and other mammalian genomes produce thousands of long non-coding RNAs (lncRNAs) raises many fascinating questions. These mRNA-like molecules, which lack significant protein-coding capacity, have been implicated in a wide range of biological functions through diverse and as yet poorly understood molecular mechanisms. Despite some recent insights into how lncRNAs function in such diverse cellular processes as regulation of gene expression and assembly of cellular structures, by and large, the key questions regarding lncRNA mechanisms remain to be answered. In this review, we discuss recent advances in understanding the biology of lncRNAs and propose avenues of investigation that may lead to fundamental new insights into their functions and mechanisms of action. Finally, as numerous lncRNAs are dysregulated in human diseases and disorders, we also discuss potential roles for these molecules in human health.
INTRODUCTION
Recent advances in technologies, such as tiling arrays and RNA deep sequencing (RNA-seq), have made it possible to survey the transcriptomes of many organisms to an unprecedented degree. Several studies utilizing these technologies have unequivocally demonstrated that the genomes of mammals, as well as other organisms, produce thousands of long transcripts that have no significant protein-coding capacity and thus are referred to as long (or large) non-coding RNAs (lncRNAs) (1–6). LncRNAs are strikingly similar to mRNAs: they are RNA polymerase II transcripts that are capped, spliced and polyadenylated, yet do not function as templates for protein synthesis (7).
Although a functional lncRNA known as Xist was discovered and characterized in the early 1990s (8–10), the prevailing view until recently was that such transcripts are rare and only a handful of functional lncRNAs are represented in the genome. However, numerous publications in the past several years have now documented important functions for lncRNAs, affecting many biological processes, including regulation of gene expression, dosage compensation, genomic imprinting, nuclear organization and compartmentalization, and nuclear-cytoplasmic trafficking (7,11–14). It is very likely that additional functions for lncRNAs will be discovered, as only a small percentage of lncRNAs have been studied in detail to date. Furthermore, there are a number of studies that have shown many lncRNAs are dysregulated in various human diseases and disorders, although it is not yet clear if these lncRNAs are causal or symptomatic of the disease state (15,16).
In this review, we will discuss a number of important topics regarding lncRNAs: (i) How many functional lncRNAs are transcribed in mammals?; (ii) Known biological functions of lncRNAs; (iii) How do lncRNAs exert their effects? And finally, (iv) What are the potential roles of lncRNAs in human disease? Although in this review we will focus on mammalian lncRNAs, it is important to point out that lncRNAs are being actively investigated in many other organisms (17,18).
HOW MANY FUNCTIONAL lncRNAs ARE PRESENT IN MAMMALS?
Prior to advances in technologies that made it possible to survey transcriptomes in an unbiased manner and to a much greater depth than previously possible, lncRNAs were discovered and characterized using traditional gene cloning methods. Initially, these transcripts were thought to code for proteins, but subsequent experimental and bioinformatic data indicated that these transcripts lack long open reading frames (ORFs). The few lncRNAs known prior to the past decade were thought to arise sporadically in the genome (10,19–21). This picture changed radically when, in the early 2000s, the FANTOM consortium examined over 60 000 full-length cDNAs and identified over 11 000 lncRNAs in mouse (22). A significant proportion of these transcripts is overlapping with, and is transcribed in the antisense direction, to protein-coding genes. They are thus referred to as natural antisense transcripts (NATs). Another independent study found that ∼40% of protein-coding genes in human cells also express NATs (23). To date, numerous studies demonstrated that NATs regulate their overlapping protein-coding partners in cis, either concordantly or discordantly (6,24,25). Furthermore, a number of studies have reported lncRNAs that are expressed solely from the introns of protein-coding genes (26,27).
More recently, it was reported that ‘intergenic’ regions of the genome, which were previously thought to be gene ‘deserts’ or ‘junk’ DNA, also express thousands of long non-coding RNAs, termed large intervening non-coding RNAs (lincRNAs) (1,3–5). Prior to advances in RNA sequencing (RNA-seq) technologies, lincRNAs were discovered by using a chromatin signature of actively transcribed genes (4,5). Essentially, actively transcribed protein-coding genes typically display a specific histone modification pattern: H3K4 trimethylation in the promoter region and H3K36 trimethylation in the body of the gene (28,29). By examining these chromatin marks genome-wide and eliminating those corresponding to protein-coding genes and microRNAs, it was shown that the human and mouse genomes produce over 3300 lincRNAs (4,5). Subsequently, RNA-seq experiments confirmed these observations and revealed an additional 5000 lincRNAs (1,30). It is now estimated that the human genome produces over 8000 lincRNAs, with 4500 of these considered to be high-confidence lincRNAs. These lincRNAs are multi-exonic, capped, polyadenylated and localized to the nucleus, cytoplasm or both. Intriguingly, many of these lincRNAs show tissue-specific expression patterns suggesting potential roles in cell identity (1,4,5).
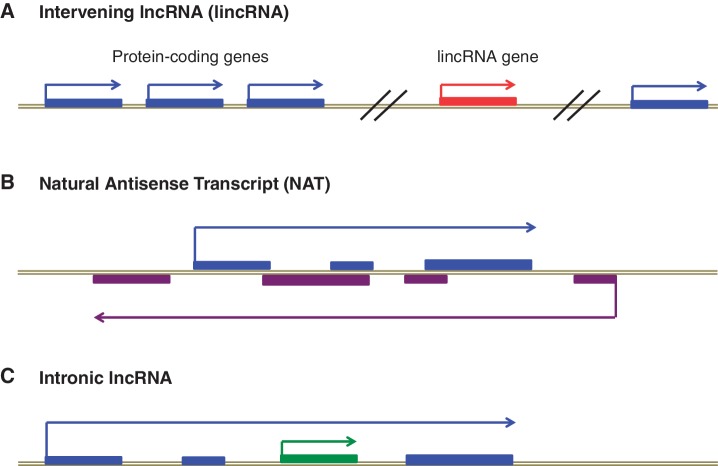
The exact number of distinct human lncRNAs, which consist of NATs, lincRNAs and intronic lncRNAs (Figure 1), is still a matter of intense debate. However, based on recent publications utilizing the most advanced sequencing platforms and algorithms to assemble transcripts from deep RNA-sequencing reads, the number of total lncRNAs (lincRNAs + NATs + intronic lncRNAs) is in the range of ∼20 000 transcripts. Nonetheless, whether the final number will be larger or smaller, it is now clear that the human and other mammalian genomes encode thousands of lncRNAs, yet neither the biological processes in which most of these molecules function, nor their mechanisms of action, have been determined (7).
Figure 1.
LncRNAs fall into one of three categories. (A) Long intervening non-coding RNAs (lincRNAs) are transcribed from regions far away from protein-coding genes. (B) Natural Antisense Transcripts (NATs) are transcribed from the opposite strand of a protein-coding gene. (C) Intronic lncRNAs (shown in green) are transcribed from within introns of protein-coding genes.
A crucial outstanding question is whether all lncRNAs are functional. Some have argued that the mere transcription of lncRNAs does not indicate functional significance, especially given that many of these transcripts do not appear to be conserved even between closely related species (1). In this view, establishing the functional significance of any individual lncRNA would require experimental evidence. To date, over 200 lncRNAs have been studied functionally and/or mechanistically. Although the depth of analysis varies considerably, many of these lncRNAs show evidence of functionality, at least in vitro. In contrast, only a few lncRNAs have been studied in animal models, with results suggesting that these few lncRNAs are not essential to the viability of the organism (31). One intriguing hypothesis is that many lncRNAs are genetically redundant and therefore loss of one lncRNA is easily compensated for by one or more other lncRNAs. It is also critical to point out that mouse knockouts of many protein-coding genes are also viable and display no obvious phenotypes (32).
In summary, it is now clear that the human and other mammalian genomes produce thousands of lncRNAs (33). Due to the complexity of the lncRNA population, it will be many years before we can elucidate the functions of all lncRNAs, particularly as many have multiple alternatively spliced forms. In an effort to expedite progress toward this important goal, some researchers are utilizing bioinformatic strategies such as ‘guilt by association’ or high throughput approaches such as siRNA/shRNA screens to home in on candidate lncRNAs for functional and mechanistic studies (4,5,34–36). As elaborated in the next section, it is very likely that lncRNAs are involved in a wide range of biological functions as they have been implicated in key biological processes thus far.
KNOWN BIOLOGICAL FUNCTIONS OF lncRNAs
Although only a very small percentage of all lncRNAs identified to date have been examined experimentally, an emerging paradigm suggests that lncRNAs function in many biological contexts. Thus far, lncRNAs have been implicated in such diverse processes as regulation of gene expression both in cis and in trans, guidance of chromatin-modifying complexes, X chromosome inactivation (Xi) and genomic imprinting, nuclear compartmentalization, nuclear-cytoplasmic trafficking, RNA splicing and translational control (11–14). In the following sections, we will discuss the evidence supporting the established roles in which lncRNAs function.
Regulation of gene expression by lncRNAs both in cis and in trans
Regulation of gene expression is a complex process that typically requires many factors and co-factors that either open up chromatin and make DNA accessible to RNA polymerases, or conversely, lead to chromatin condensation and DNA inaccessibility. Recent studies have clearly shown that a number of lncRNAs also contribute to gene regulation by various mechanisms (11).
One of the best-studied lncRNAs to date is Xist, which is responsible for the initiation and spreading of X chromosome inactivation (Xi) in female somatic cells. Xist was discovered in 1991 and, despite having been studied extensively, the exact mechanism of Xist-mediated X chromosome inactivation is yet to be fully elucidated (37,38). Nonetheless, it is widely accepted that Xist is required for the silencing of hundreds of genes on the inactive X chromosome. It is thought that a small repeat region within Xist, which is referred to as RepA, is initially transcribed from both X chromosomes along with Xist’s antisense partner Tsix (39). Tsix, also a lncRNA, prevents RepA from binding to either X chromosome. However, post-cellular differentiation, RepA, in association with the chromatin-modifying complex PRC2 (polycomb repressive complex 2), binds to one of the two X chromosomes at the X inactivation center to initiate X inactivation (39). Subsequently, production of full-length Xist from the X chromosome destined for inactivation, which also binds to PRC2, leads to the spreading of X inactivation from the X inactivation center to the entire X chromosome in cis (39). On the active X chromosome, Tsix is believed to prevent Xist transcription and thus maintains an active chromatin state. It is currently not known what prevents Xist from ‘escaping’ the inactive X and acting on the active X or other chromosomes, in trans, or how Xist is prevented from silencing genes that escape X inactivation; as many as 20% of genes escape Xi in human females (40,41). Finally, it is worth noting that, in addition to Xist and Tsix, several other lncRNAs are transcribed from the X inactivation center and these also appear to play roles in the counting and choice in Xi (38,42). Once Xi is established, the inactive X chromosome is condensed into facultative heterochromatin and appears as a condensed round body (Barr Body), usually at the periphery of nuclei (43). The inactive X chromosome, in contrast to the active X and the autosomes, is marked with repressive chromatin marks and DNA methylation at CpG islands, with the exception of regions that escape Xi (41,44,45).
Another epigenetic phenomenon that, similar to Xi, also utilizes lncRNAs to regulate gene expression is known as genomic imprinting (46). The expression of imprinted genes depends on their parental origin, and the level of differential expression of the two alleles of an imprinted gene can vary from one imprinted gene to another. Since imprinted genes play critical roles in mammalian development, their expression must be tightly regulated (47). Intriguingly, many imprinted gene loci express, in addition to mRNAs, a significant number of lncRNAs that appear to play major roles in regulating the expression of neighboring imprinted protein-coding genes in cis (48). One such lncRNA is Air, which is monoallelically expressed from the paternal allele, associates with the histone methyltransferase G9a and localizes to chromatin to silence three imprinted genes known as Slc22a3, Slc22a2 and Igf2r in cis (49). Ablation of Air results in the biallelic expression of Slc22a3 and loss of G9a recruitment to the Slc22a3 promoter, suggesting that Air plays a role in guiding the methyltransferase G9a to chromatin at the Slc22a3 promoter (49). In a similar mechanism to Air, the lncRNA Kcnq1ot1 also regulates the expression of imprinted genes in a lineage-specific manner by directing G9a, PRC2 and Dnmt1 to the Kcnq1 locus in cis (50,51). Collectively, these observations and others suggest that lncRNAs are intimately involved in regulating the expression of imprinted genes, potentially in a complex and layered manner.
In contrast to Air and Kcnq1ot1, which regulate gene expression in cis, an elegant study from Rinn et al. (52) led to the discovery of an intervening lncRNA (lincRNA), HOTAIR, that regulates human HOXD genes expression in trans. Subsequent studies demonstrated that HOTAIR regulates gene expression in trans on a genome-wide scale by associating with the chromatin-modifying complexes PRC2, LSD1 and CoREST/REST (4,52–54). HOTAIR guides as well as serves as a scaffold for PRC2 and LSD1/CoREST complexes at their endogenous target genes (54). Subsequently, the LSD1/CoREST complex demethylates histone H3 at lysine 4 (H3K4), while PRC2 methylates histone H3 at lysine 27 (H3K27) leading to the loss of an activating histone mark (i.e. H3K4 dimethylation) and the gain of a repressive histone mark (H3K27 trimethylation) at genes targeted by HOTAIR (54). In contrast to the human HOTAIR, mouse Hotair does not appear to regulate Hoxd genes suggesting that HOTAIR may have distinct functions in human and mouse (31).
In addition to the lncRNAs discussed earlier, several other human and mouse lncRNAs (e.g. Tug1, linc-P21, PANDA, Evf-2 and others) have also been shown to regulate gene expression by guiding their protein partners to specific genomic loci (4,34,36,55,56). Collectively, these studies demonstrate that lncRNAs regulate gene expression in trans as well as in cis. However, the molecular mechanisms that determine a lncRNA’s ability to regulate gene expression in cis versus in trans are not currently known. It is likely that the sequences and/or the secondary structures of lncRNAs inherently affect their mechanisms of action, an area of lncRNA research that remains in its infancy due to lack of appropriate tools.
A subset of lncRNAs is required for maintaining pluripotency
The ability of a stem cell to give rise to all three germ layers (endoderm, mesoderm and ectoderm) is referred to as pluripotency. Previous studies have identified key transcription and chromatin remodeling factors that are required for maintaining pluripotency (57,58). Intriguingly, some of these transcription factors were found to bind to the promoters of over 100 lincRNAs in mouse ES cells (5). Subsequently, another study found that two lncRNAs, which are regulated by the transcription factors Oct4 and Nanog, to be essential for maintaining pluripotency (59). Furthermore, the authors found that knockdown and overexpression of these two lncRNAs results in dramatic changes in Oct4 and Nanog mRNA levels, pointing to the involvement of a feedback loop in the regulatory mechanism (59).
Recently, the role of lincRNAs in pluripotency was examined in a high throughput manner using shRNAs in mES cells (34). The authors demonstrated that 26 lincRNAs are required for the maintenance of pluripotency, as knockdown of each lincRNA led to either an exit from the pluripotent state or activation of lineage commitment programs. Since some of these lincRNAs interact with chromatin-modifying complexes, they could potentially guide and/or serve as scaffolds for such complexes at specific gene loci that are critical for maintaining the pluripotent state. It is also possible that some lincRNAs function as ‘environmental sensors’ that alert stem cells to maintain pluripotency or to differentiate depending on changes in the environment. Indeed, a recent study identified a lncRNA termed ANCR (Anti-differentiation non-coding RNA) that is required for maintaining cells in an undifferentiated state in the epidermis (60). Future studies are needed to dissect the exact roles of lncRNAs in maintaining pluripotency and promoting cellular differentiation programs.
Nuclear organization: Paraspeckle formation requires the lncRNA NEAT1
Regulation of gene expression is a complex process and can take place at the transcriptional, co-transcriptional, post-transcriptional and translational levels. Recently it was suggested that nuclear structures known as paraspeckles may regulate gene expression post-transcriptionally by the retention of hyperedited mRNAs in the nucleus (61–64). Paraspeckles are identified cytologically based on the presence of a few protein components, which include PSP1, PSP2 and p54 (63). Although the function of PSP1 is not known, PSP2 is involved in splicing, while P54 is involved in splicing as well as several other processes including transcriptional regulation, DNA unwinding and nuclear retention of hyperedited dsRNA (61–63,65). Typically, each nucleus contains several paraspeckles, which are dynamic structures and require ongoing RNA polymerase II transcription (65). During telophase, when transcription is shut down, and in cells treated with drugs that inhibit RNA polymerase II, paraspeckles disappear (61–63).
Intriguingly, the formation and maintenance of paraspeckles require the lncRNA NEAT1 (nuclear-enriched autosomal transcript 1), which localizes exclusively to paraspeckles (61,65–67). Paraspeckles are dynamic structures that are absent in embryonic stem cells but appear after differentiation coincident with NEAT1 activation (61–63). Moreover, depletion of NEAT1 is sufficient to cause loss of paraspeckles in the nucleus, and overexpressing NEAT1, but not paraspeckle-associated proteins, leads to an increase in the number of paraspeckles suggesting an essential role for NEAT1 in paraspeckle formation and/or integrity (61,62,65). Although the mechanism of NEAT1 mediated paraspeckle formation is not completely clear, it is possible that NEAT1 serves as a scaffold for proteins involved in paraspeckle formation, and therefore loss of NEAT1 prevents the ability of these proteins to co-localize. It would not be surprising if future research uncovers other lncRNAs that are involved in forming or maintaining the integrity of other nuclear and/or cytoplasmic structures.
Regulation of alternative splicing by lncRNAs
Alternative splicing of pre-mRNAs increases the proteomic complexity of cells by resulting, in many cases, in several protein products with non-overlapping functions from a single mRNA. The lncRNA MALAT1 (metastasis-associated lung adenocarcinoma transcript 1), which was initially identified in a screen for genes associated with metastasis, plays a critical role in pre-mRNA alternative splicing (68). MALAT1 localizes to nuclear speckles, which contain several proteins that are known to be involved in alternative splicing (68). Furthermore, depletion of MALAT1 is sufficient to alter the alternative splicing patterns of a subset of mRNAs (68). It appears that MALAT1 forms a molecular scaffold for several proteins present in nuclear speckles, as well as modulates the phosphorylation of SR proteins (68). SR proteins are serine/arginine-rich proteins which are involved in the regulation and selection of the splice sites in pre-mRNAs. By regulating the phosphorylation of SR proteins, MALAT1 may thus regulate the cellular levels of active SR proteins and subsequently the splicing of many pre-mRNAs (68). Finally, it remains to be determined if other lncRNAs may also play a role in alternative splicing. Performing biochemical assays such as RNA co-immunoprecipitation followed by deep sequencing of RNAs (RIP-seq) on proteins that are known to be involved in alternative splicing may lead to the identification of other lncRNAs with functions similar to MALAT1.
HOW DO lncRNAs EXERT THEIR EFFECTS?
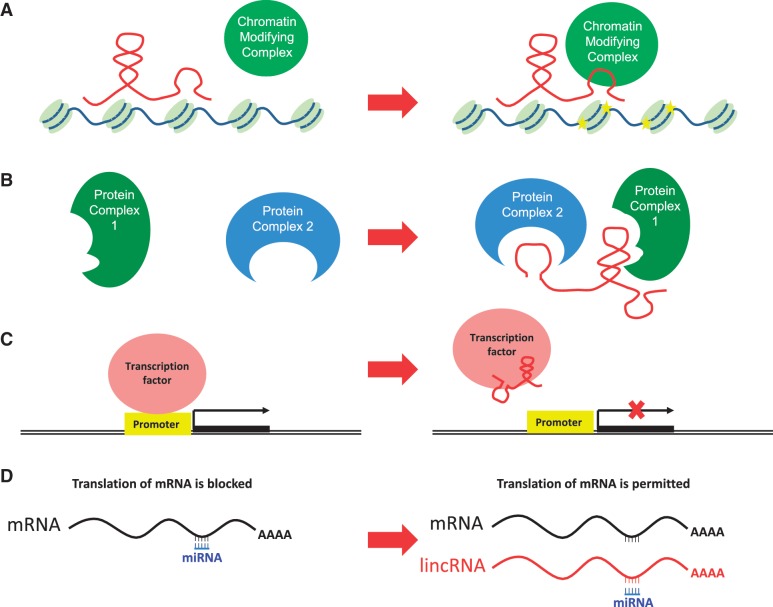
Thus far lncRNAs have been implicated in numerous biological functions and pathways, and their mechanisms of actions are very diverse (Figure 2) (7,11). Here we will discuss several mechanisms by which lncRNAs exert their effects, although it is worth noting that in every case much remains to be learned about the detailed mechanism of action.
Figure 2.
LncRNAs exert their effects by diverse mechanisms. (A) lncRNAs can act as guides and tethers for chromatin-modifying complexes, and thus contribute to tissue-specific gene expression. (B) lncRNAs can act as molecular scaffolds for protein complexes that lack protein–protein interaction domains. (C) lncRNAs can bind to transcription factors and prevent them from binding to their target DNA sequence. (D) lncRNAs can interact directly with microRNAs (miRNAs) and prevent them from binding to mRNA, thus regulating protein synthesis.
lncRNAs ‘guide’ chromatin-modifying complexes to specific genomic loci in cis and in trans
Cellular identity in an organism is determined by epigenetic factors that modulate specific gene expression programs (69). These epigenetic factors, such as chromatin-modifying complexes and DNA methyltransferases, activate and repress specific genes by enzymatically modifying chromatin and DNA (70). One of the most puzzling questions in biology is how do these ubiquitous enzymes, which lack DNA binding capacity, recognize their target genes in the various cell types. Emerging evidence suggest that some lncRNAs ‘guide’ chromatin-modifying complexes as well as other nuclear proteins to specific genomic loci to exert their effects (14,71,72). In essence, some lncRNAs may function as ‘GPS’ devices to target other cellular components to their sites of action. Below, we will highlight several examples of lncRNAs that have been shown to possess such activity.
As discussed previously, the lncRNA HOTAIR directs the chromatin-modifying complexes PRC2 and LSD1 to numerous gene loci on a genome-wide scale in trans (4,52–54). By contrast, other lncRNAs, such as Air, Kcnq1ot1 and Evf-2, target chromatin-modifying complexes to their target genes in cis (39,49,50,56). Currently, the sequence of events that leads to a lncRNA-mediated guidance of a protein complex to chromatin have not been fully elucidated. However, recent evidence suggests that lncRNAs may bind to chromatin first, and then serve as docking stations for chromatin-modifying complexes. In support of this model, a recent study, which utilized a novel technology that allows the genomic occupancy of a lncRNA to be determined, found that HOTAIR localizes to chromatin independent of its protein-binding partner PRC2 (73).
Since numerous lncRNAs are known to bind to chromatin-modifying complexes (4,74), it is conceivable that many of these lncRNAs also function by ‘guiding’ their protein partners to chromatin. To fully understand this mechanism of action, a number of key questions must be addressed: (i) Are there specific motifs in lncRNAs that are responsible for the targeting mechanism to specific genomic regions? (ii) How do proteins recognize and specifically bind to certain lncRNA(s) but not others? (iii) Do these lncRNAs directly interact with DNA to form lncRNA:DNA hybrids or triplexes or (iv) do DNA binding proteins serve as intermediates between a lncRNA and DNA? These key questions will be critical to answer in order to fully understand the mechanisms through which chromatin-associated lncRNAs function.
lncRNAs serve as structural links in ribonucleoprotein complexes (RNPs)
While it is clear that the majority of RNA does not float around in the cell naked but rather is complexed with protein, the precise molecular composition of most ribonucleoprotein complexes (RNPs) has not been determined. Recent studies have shown that many lncRNAs exist in the cell as part of RNPs (71). Thus far, lncRNAs have been shown to form RNPs with chromatin-modifying complexes, transcription factors, splicing factors, as well as other classes of proteins (71). For example, as discussed earlier, HOTAIR forms a RNP with the chromatin-modifying complexes PRC2 and LSD1/CoREST (54). Mutational analysis of HOTAIR revealed that HOTAIR integrity is required for PRC2 interaction with LSD1/CoREST (54). This suggests that, in addition to HOTAIR’s role in guiding PRC2 and LSD1 to chromatin, HOTAIR also serves as a structural bridge between PRC2 and LSD1/CoREST at endogenous target genes. Also, Xist has recently been shown to interact with the transcription factor YY1, which helps tether Xist, and consequently PRC2, to the inactive X chromosome (75). In essence, Xist appears to be forming a molecular bridge between YY1 and PRC2 to repress genes on the inactive X chromosome.
The lncRNAs NEAT1 and MALAT1 form molecular scaffolds for several proteins that are core components of paraspeckles and speckles, respectively (61–63,68). It is not currently known how lncRNAs recognize their protein partners and initiate the formation of these nuclear compartments. It is possible that secondary structures of lncRNAs contribute to the specificity of lncRNAs interactions with their protein partners. Mutation analysis of structural lncRNAs may lead to the identification of lncRNA ‘domains’ that are critical for their functions as molecular scaffolds. In summary, lncRNAs appear to provide an extensive infrastructure within the nucleus, and potentially in the cytoplasm, that makes it possible for various proteins to co-localize and coordinate their functions to accomplish a specific biological function (71). Disruption of such lncRNAs may lead to undesired biological consequences (76).
LncRNAs regulate distinct transcriptional programs
A few lncRNAs have been shown to be activated in response to specific stimuli, and subsequently activate specific transcriptional programs that allow the cell to respond to these stimuli. For example, lncRNAs, such as linc-P21, PANDA, Tug1 and others, are transcriptionally activated in response to DNA damage by the direct binding of the tumor-suppressor protein p53 to their promoters (4,5,36,55). Subsequently, these lncRNAs regulate gene expression by distinct pathways. Linc-P21, which represses numerous genes in the p53 pathway, requires the RNA/DNA binding protein hnRNP K and other as yet unidentified factors (36). By contrast, the lncRNA PANDA, which is also activated by p53, requires the downstream effector NF-YA to regulate the expression of pro-apoptotic genes (55), while Tug1 functions via its interaction with the chromatin-modifying complex PRC2 (4).
The lncRNA Gas5 (growth arrest specific 5), which is highly expressed in cells that have arrested growth, serves as a negative regulator of glucocorticoid receptors (GR), a specific class of nuclear receptors (77). Gas5 interacts directly with the DNA binding domain of GRs, preventing them from binding to their DNA response elements, thereby in effect acting as a molecular decoy (77). The ability of a lncRNA to modulate the effects of a transcription factor can lead, in some cases, to significant changes in gene expression and subsequently profound effects on the cells ability to respond to external stimuli. Further studies are needed to determine the underlying mechanisms of how a lncRNA, once activated, modulates the activity of transcription factor(s) to allow the cells to respond to their environment.
Regulation of microRNAs by lncRNAs
In 2007, a study in Arabidopsis thaliana found the non-coding RNA IPS1 to bind to the microRNA miR-399 and block its ability to regulate PHO2 mRNA (78). Recently, evidence came to light suggesting that some mammalian lncRNAs may also regulate gene expression post-transcriptionally by binding to miRNAs, and consequently preventing specific miRNAs from binding to their target mRNAs. In a 2011 published study, it was demonstrated that a lincRNA, linc-MD1, serves as a ‘sponge’ for two miRNAs, which regulate transcription factors involved in muscle differentiation (79). These findings are very intriguing since they demonstrate that distinct classes of non-coding RNAs cooperate to regulate gene expression. It is conceivable that other lncRNAs can also serve as ‘sponges’ for miRNAs in a tissue and developmental stage-specific manner. However, it is not clear how cells regulate the expression levels of lncRNAs and miRNAs, and how lncRNAs receive a signal to bind or not to bind a miRNA. Potentially, novel protein partners of lncRNAs may be critical for regulating lncRNAs interactions with miRNAs in a spatial and temporal manner. There are many key questions, which are yet to be determined regarding this mechanism of lncRNA-mediated regulation, that will require tremendous effort to answer but are key to understanding this mode of action.
WHAT ARE THE POTENTIAL ROLES OF lncRNAs IN HUMAN DISEASE?
In contrast to the extensive evidence that links dysregulation of protein-coding genes to disease etiology, to date only a few lncRNAs have been implicated in human disease (76). However, we are beginning to observe, in some cases, strong associations between lncRNAs and human disease, and it is reasonable to expect that a concrete, mechanistic understanding of these connections will emerge in the coming years. Since both the role of lncRNAs dysregulation in human disease and their molecular mechanisms remain unclear, it is timely to ask important questions such as: (i) How many lncRNAs are differentially regulated in a given human disease compared to healthy counterparts? (ii) What are the molecular and biological functions of lncRNAs that are dysregulated in human disease? (iii) Do disease-specific lncRNAs change their subcellular localization? (iv) How stable are lncRNAs and is their stability altered in various disease states? Thus, an immediate goal of lncRNA research is to determine whether lncRNAs are useful signatures for early disease detection, or can be used as candidate drug targets for disease intervention (80).
LncRNAs have been found to be dysregulated in a wide range of human diseases and disorders, including various types of cancers. This includes breast cancer (53), colorectal cancer (81), prostate cancer (82), hepatocellular carcinoma (83–85), leukemia (86,87), melanoma (88) and possibly others (76,89). On a more mechanistic level, recent studies have revealed the contribution of lncRNAs as proto-oncogenes, e.g. GAGE6 (90), as tumor suppressor genes, e.g. ‘p15 antisense RNA and lincP21’ (36,91), as drivers of metastatic transformation, e.g. HOTAIR in breast cancer (53), and as regulators of alternative splicing, e.g. MALAT1 (68).
Although the entire mechanisms of action of most lncRNAs that are dysregulated in cancer have not been fully elucidated, a number of studies have provided some insights into the mechanisms of few such lncRNAs. For example, the lncRNA linc-P21 functions as a repressor in the p53 pathway by directing the RNA/DNA binding protein hnRNPK to chromatin (36). The lncRNA, SPRY4-IT1, which is up-regulated in human melanomas compared to melanocytes and keratinocytes, affects cell dynamics, including increased rate of wound closure upon ectopic expression. This suggests that the higher expression of SPRY4-IT1 may have an important role in the molecular etiology of human melanomas (88).
Aside from their roles in cancer, lncRNAs are known to be dysregulated in several other diseases, including heart disease (92,93), Alzheimer’s disease (94), psoriasis (95), spinocerebellar ataxia type 8 (96) and fragile X syndrome (97). Finally, we refer the reader to an excellent review article that covers the role of lncRNAs in human disease in depth (76).
CONCLUDING REMARKS
The discovery of thousands of lncRNAs has certainly changed our view of the complexity of mammalian genomes and transcriptomes, as well as many other aspects of biology including transcriptional and post-transcriptional regulation of gene expression. To date, we have only scratched the surface in terms of elucidating the functions and mechanisms of lncRNAs; nonetheless, recent studies suggest that lncRNAs are likely to exert their effects by diverse mechanisms (7,11,13,14,16,71,72,76). In many cases lncRNAs have been shown to work cooperatively with proteins by forming ribonucleoprotein complexes (RNPs) (71). These RNPs appear to depend on their lncRNA constituent(s) for proper localization to specific regions within the cell, as well as to coordinate the interactions between protein complexes that do not have interacting domains (4,52–54,68,98,99). Furthermore, lncRNAs appear to be intimately associated with both chromatin and chromatin-modifying complexes suggesting that lncRNAs are critical for genome organization and regulation of gene expression at the transcriptional levels (11,14,72,73). Lastly, since lncRNAs show strong tissue-specific expression, they are likely to play a major role in cell identity and spatial organization in multicellular organisms (1,4). We envision that lncRNAs work within complex networks involving proteins and other types of ncRNAs (e.g. miRNAs) to achieve a system level of cellular organization.
The studies discussed earlier and others clearly demonstrate that lncRNAs, or at least the ones that have been examined to date, are functional, despite lack of conservation in many cases even among closely related species. This conundrum could be explained by the hypothesis that lncRNAs rely on ‘conserved’ secondary structures and not their primary sequence to perform their functions. This hypothesis would help explain how many lncRNAs with distinct sequences bind the same protein complex (4,34,71); as well as how lncRNAs have functional orthologs that are not conserved at the sequence level (100). Currently, there are no reliable methods to determine the secondary structures of lncRNAs, and thus, the above hypothesis remains to be experimentally tested. However, an emerging technology that is referred to as parallel analysis of RNA structure (PARS), which is based on deep sequencing of RNA fragments that have been treated with structure-specific enzymes, has shown promise in predicting secondary structure of RNAs in yeast (101). This technology can potentially be modified to perform similar analysis in higher eukaryotes. Finally, it can not be excluded that a small percentage of lncRNAs encodes short peptides (102).
Dysregulation of lncRNAs, which has been observed in numerous human diseases, suggest that lncRNAs can be utilized in medicine as biomarkers and/or drug targets (76). Such therapies would be useful in cases where drugs designed to target proteins have failed, or even in conjunction with available drugs in order to enhance their effects. For example, a previous study found that knocking down a lncRNA enhances the effects of chemotherapeutic drugs in vitro (103).
In conclusion, the potential roles of lncRNAs in biology and medicine could be tremendous, and will require many years of intensive research before they can be fully deciphered and applied. However, if recent publications are an indication of the progress in this area of scientific research, then this field is certainly moving at a supersonic speed.
FUNDING
Funding for open access charge: Waived by Oxford University Press.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to thank Dr Jo Ann Wise for her insightful and constructive critical comments. Also, we would like to thank Callie Merry, Maya Ratnam and Jill Marinis for discussion and constructive feedback.
REFERENCES
- 1.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 4.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl Acad. Sci. USA. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 7.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 8.Brown CJ, Hendrich BD, Rupert JL, Lafreniere RG, Xing Y, Lawrence J, Willard HF. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71:527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- 9.Brockdorff N, Ashworth A, Kay GF, McCabe VM, Norris DP, Cooper PJ, Swift S, Rastan S. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992;71:515–526. doi: 10.1016/0092-8674(92)90519-i. [DOI] [PubMed] [Google Scholar]
- 10.Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, Willard HF. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349:38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- 11.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol. Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagano T, Fraser P. No-nonsense functions for long noncoding RNAs. Cell. 2011;145:178–181. doi: 10.1016/j.cell.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 13.Clark MB, Mattick JS. Long noncoding RNAs in cell biology. Semin. Cell Dev. Biol. 2011;22:366–376. doi: 10.1016/j.semcdb.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 14.Mattick JS, Amaral PP, Dinger ME, Mercer TR, Mehler MF. RNA regulation of epigenetic processes. Bioessays. 2009;31:51–59. doi: 10.1002/bies.080099. [DOI] [PubMed] [Google Scholar]
- 15.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 16.Qureshi IA, Mattick JS, Mehler MF. Long non-coding RNAs in nervous system function and disease. Brain Res. 2010;1338:20–35. doi: 10.1016/j.brainres.2010.03.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geisler S, Lojek L, Khalil AM, Baker KE, Coller J. Decapping of long noncoding RNAs regulates inducible genes. Mol. Cell. 2012;45:279–291. doi: 10.1016/j.molcel.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pauli A, Valen E, Lin MF, Garber M, Vastenhouw NL, Levin JZ, Fan L, Sandelin A, Rinn JL, Regev A, et al. Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res. 2012;22:577–591. doi: 10.1101/gr.133009.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee JT, Davidow LS, Warshawsky D. Tsix, a gene antisense to Xist at the X-inactivation centre. Nat. Genet. 1999;21:400–404. doi: 10.1038/7734. [DOI] [PubMed] [Google Scholar]
- 20.Brown CJ, Lafreniere RG, Powers VE, Sebastio G, Ballabio A, Pettigrew AL, Ledbetter DH, Levy E, Craig IW, Willard HF. Localization of the X inactivation centre on the human X chromosome in Xq13. Nature. 1991;349:82–84. doi: 10.1038/349082a0. [DOI] [PubMed] [Google Scholar]
- 21.Brannan CI, Dees EC, Ingram RS, Tilghman SM. The product of the H19 gene may function as an RNA. Mol. Cell. Biol. 1990;10:28–36. doi: 10.1128/mcb.10.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okazaki Y, Furuno M, Kasukawa T, Adachi J, Bono H, Kondo S, Nikaido I, Osato N, Saito R, Suzuki H, et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- 23.He Y, Vogelstein B, Velculescu VE, Papadopoulos N, Kinzler KW. The antisense transcriptomes of human cells. Science. 2008;322:1855–1857. doi: 10.1126/science.1163853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 25.Werner A, Carlile M, Swan D. What do natural antisense transcripts regulate? RNA Biol. 2009;6:43–48. doi: 10.4161/rna.6.1.7568. [DOI] [PubMed] [Google Scholar]
- 26.Kampa D, Cheng J, Kapranov P, Yamanaka M, Brubaker S, Cawley S, Drenkow J, Piccolboni A, Bekiranov S, Helt G, et al. Novel RNAs identified from an in-depth analysis of the transcriptome of human chromosomes 21 and 22. Genome Res. 2004;14:331–342. doi: 10.1101/gr.2094104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rinn JL, Euskirchen G, Bertone P, Martone R, Luscombe NM, Hartman S, Harrison PM, Nelson FK, Miller P, Gerstein M, et al. The transcriptional activity of human Chromosome 22. Genes Dev. 2003;17:529–540. doi: 10.1101/gad.1055203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 30.Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X, Fan L, Koziol MJ, Gnirke A, Nusbaum C, et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat. Biotechnol. 2010;28:503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schorderet P, Duboule D. Structural and functional differences in the long non-coding RNA hotair in mouse and human. PLoS Genet. 2011;7:e1002071. doi: 10.1371/journal.pgen.1002071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Austin CP, Battey JF, Bradley A, Bucan M, Capecchi M, Collins FS, Dove WF, Duyk G, Dymecki S, Eppig JT, et al. The knockout mouse project. Nat. Genet. 2004;36:921–924. doi: 10.1038/ng0904-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Amaral PP, Dinger ME, Mercer TR, Mattick JS. The eukaryotic genome as an RNA machine. Science. 2008;319:1787–1789. doi: 10.1126/science.1155472. [DOI] [PubMed] [Google Scholar]
- 34.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loewer S, Cabili MN, Guttman M, Loh YH, Thomas K, Park IH, Garber M, Curran M, Onder T, Agarwal S, et al. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat. Genet. 2010;42:1113–1117. doi: 10.1038/ng.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pontier DB, Gribnau J. Xist regulation and function explored. Hum. Genet. 2011;130:223–236. doi: 10.1007/s00439-011-1008-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Augui S, Nora EP, Heard E. Regulation of X-chromosome inactivation by the X-inactivation centre. Nat. Rev. Genet. 2011;12:429–442. doi: 10.1038/nrg2987. [DOI] [PubMed] [Google Scholar]
- 39.Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carrel L, Willard HF. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434:400–404. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- 41.Khalil AM, Driscoll DJ. Trimethylation of histone H3 lysine 4 is an epigenetic mark at regions escaping mammalian X inactivation. Epigenetics. 2007;2:114–118. doi: 10.4161/epi.2.2.4612. [DOI] [PubMed] [Google Scholar]
- 42.Tian D, Sun S, Lee JT. The long noncoding RNA, Jpx, is a molecular switch for X chromosome inactivation. Cell. 2010;143:390–403. doi: 10.1016/j.cell.2010.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barr ML, Bertram EG. A morphological distinction between neurones of the male and female, and the behaviour of the nucleolar satellite during accelerated nucleoprotein synthesis. Nature. 1949;163:676. doi: 10.1038/163676a0. [DOI] [PubMed] [Google Scholar]
- 44.Panning B. X inactivation in mouse ES cells: histone modifications and FISH. Methods Enzymol. 2004;376:419–428. doi: 10.1016/S0076-6879(03)76028-5. [DOI] [PubMed] [Google Scholar]
- 45.Heard E, Rougeulle C, Arnaud D, Avner P, Allis CD, Spector DL. Methylation of histone H3 at Lys-9 is an early mark on the X chromosome during X inactivation. Cell. 2001;107:727–738. doi: 10.1016/s0092-8674(01)00598-0. [DOI] [PubMed] [Google Scholar]
- 46.Reik W, Murrell A. Genomic imprinting. Silence across the border. Nature. 2000;405:408–409. doi: 10.1038/35013178. [DOI] [PubMed] [Google Scholar]
- 47.Li Y, Sasaki H. Genomic imprinting in mammals: its life cycle, molecular mechanisms and reprogramming. Cell Res. 2011;21:466–473. doi: 10.1038/cr.2011.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mohammad F, Mondal T, Kanduri C. Epigenetics of imprinted long noncoding RNAs. Epigenetics. 2009;4:277–286. [PubMed] [Google Scholar]
- 49.Nagano T, Mitchell JA, Sanz LA, Pauler FM, Ferguson-Smith AC, Feil R, Fraser P. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322:1717–1720. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
- 50.Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, Nagano T, Mancini-Dinardo D, Kanduri C. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol. Cell. 2008;32:232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 51.Mohammad F, Mondal T, Guseva N, Pandey GK, Kanduri C. Kcnq1ot1 noncoding RNA mediates transcriptional gene silencing by interacting with Dnmt1. Development. 2010;137:2493–2499. doi: 10.1242/dev.048181. [DOI] [PubMed] [Google Scholar]
- 52.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, Horlings HM, Shah N, Umbricht C, Wang P, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat. Genet. 2011;43:621–629. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Feng J, Bi C, Clark BS, Mady R, Shah P, Kohtz JD. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 2006;20:1470–1484. doi: 10.1101/gad.1416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 58.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 59.Sheik Mohamed J, Gaughwin PM, Lim B, Robson P, Lipovich L. Conserved long noncoding RNAs transcriptionally regulated by Oct4 and Nanog modulate pluripotency in mouse embryonic stem cells. RNA. 2010;16:324–337. doi: 10.1261/rna.1441510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kretz M, Webster DE, Flockhart RJ, Lee CS, Zehnder A, Lopez-Pajares V, Qu K, Zheng GX, Chow J, Kim GE, et al. Suppression of progenitor differentiation requires the long noncoding RNA ANCR. Genes Dev. 2012;26:338–343. doi: 10.1101/gad.182121.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33:717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen LL, Carmichael GG. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol. Cell. 2009;35:467–478. doi: 10.1016/j.molcel.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bond CS, Fox AH. Paraspeckles: nuclear bodies built on long noncoding RNA. J. Cell Biol. 2009;186:637–644. doi: 10.1083/jcb.200906113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Prasanth KV, Prasanth SG, Xuan Z, Hearn S, Freier SM, Bennett CF, Zhang MQ, Spector DL. Regulating gene expression through RNA nuclear retention. Cell. 2005;123:249–263. doi: 10.1016/j.cell.2005.08.033. [DOI] [PubMed] [Google Scholar]
- 65.Sunwoo H, Dinger ME, Wilusz JE, Amaral PP, Mattick JS, Spector DL. MEN varepsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009;19:347–359. doi: 10.1101/gr.087775.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mao YS, Sunwoo H, Zhang B, Spector DL. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat. Cell Biol. 2011;13:95–101. doi: 10.1038/ncb2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shevtsov SP, Dundr M. Nucleation of nuclear bodies by RNA. Nat. Cell Biol. 2011;13:167–173. doi: 10.1038/ncb2157. [DOI] [PubMed] [Google Scholar]
- 68.Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 70.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 71.Khalil AM, Rinn JL. RNA-protein interactions in human health and disease. Semin. Cell Dev. Biol. 2011;22:359–365. doi: 10.1016/j.semcdb.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Koziol MJ, Rinn JL. RNA traffic control of chromatin complexes. Curr. Opin. Genet. Dev. 2010;20:142–148. doi: 10.1016/j.gde.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol. Cell. 2011;44:667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, Song JJ, Kingston RE, Borowsky M, Lee JT. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol. Cell. 2010;40:939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jeon Y, Lee JT. YY1 tethers Xist RNA to the inactive X nucleation center. Cell. 2011;146:119–133. doi: 10.1016/j.cell.2011.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS. Non-coding RNAs: regulators of disease. J. Pathol. 2010;220:126–139. doi: 10.1002/path.2638. [DOI] [PubMed] [Google Scholar]
- 77.Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci. Signal. 2010;3:ra8. doi: 10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, Garcia JA, Paz-Ares J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 2007;39:1033–1037. doi: 10.1038/ng2079. [DOI] [PubMed] [Google Scholar]
- 79.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mattick JS. The genetic signatures of noncoding RNAs. PLoS Genet. 2009;5:e1000459. doi: 10.1371/journal.pgen.1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S, et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71:6320–6326. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- 82.Chung S, Nakagawa H, Uemura M, Piao L, Ashikawa K, Hosono N, Takata R, Akamatsu S, Kawaguchi T, Morizono T, et al. Association of a novel long non-coding RNA in 8q24 with prostate cancer susceptibility. Cancer Sci. 2011;102:245–252. doi: 10.1111/j.1349-7006.2010.01737.x. [DOI] [PubMed] [Google Scholar]
- 83.Yang Z, Zhou L, Wu LM, Lai MC, Xie HY, Zhang F, Zheng SS. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann. Surg. Oncol. 2011;18:1243–1250. doi: 10.1245/s10434-011-1581-y. [DOI] [PubMed] [Google Scholar]
- 84.Yang F, Zhang L, Huo XS, Yuan JH, Xu D, Yuan SX, Zhu N, Zhou WP, Yang GS, Wang YZ, et al. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology. 2011;54:1679–1689. doi: 10.1002/hep.24563. [DOI] [PubMed] [Google Scholar]
- 85.Lai MC, Yang Z, Zhou L, Zhu QQ, Xie HY, Zhang F, Wu LM, Chen LM, Zheng SS. Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med. Oncol. 2011 doi: 10.1007/s12032-011-0004-z. June 16 (doi:10.1007/s12032-011-0004-z; epub ahead of print). [DOI] [PubMed] [Google Scholar]
- 86.Calin GA, Pekarsky Y, Croce CM. The role of microRNA and other non-coding RNA in the pathogenesis of chronic lymphocytic leukemia. Best Pract. Res. Clin. Haematol. 2007;20:425–437. doi: 10.1016/j.beha.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 87.Calin GA, Liu CG, Ferracin M, Hyslop T, Spizzo R, Sevignani C, Fabbri M, Cimmino A, Lee EJ, Wojcik SE, et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell. 2007;12:215–229. doi: 10.1016/j.ccr.2007.07.027. [DOI] [PubMed] [Google Scholar]
- 88.Khaitan D, Dinger ME, Mazar J, Crawford J, Smith MA, Mattick JS, Perera RJ. The melanoma-upregulated long noncoding RNA SPRY4-IT1 modulates apoptosis and invasion. Cancer Res. 2011;71:3852–3862. doi: 10.1158/0008-5472.CAN-10-4460. [DOI] [PubMed] [Google Scholar]
- 89.Huarte M, Rinn JL. Large non-coding RNAs: missing links in cancer? Hum. Mol. Genet. 2010;19:R152–R161. doi: 10.1093/hmg/ddq353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li L, Feng T, Lian Y, Zhang G, Garen A, Song X. Role of human noncoding RNAs in the control of tumorigenesis. Proc. Natl Acad. Sci. USA. 2009;106:12956–12961. doi: 10.1073/pnas.0906005106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP, Cui H. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ishii N, Ozaki K, Sato H, Mizuno H, Saito S, Takahashi A, Miyamoto Y, Ikegawa S, Kamatani N, Hori M, et al. Identification of a novel non-coding RNA, MIAT, that confers risk of myocardial infarction. J. Hum. Genet. 2006;51:1087–1099. doi: 10.1007/s10038-006-0070-9. [DOI] [PubMed] [Google Scholar]
- 93.Pasmant E, Laurendeau I, Heron D, Vidaud M, Vidaud D, Bieche I. Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res. 2007;67:3963–3969. doi: 10.1158/0008-5472.CAN-06-2004. [DOI] [PubMed] [Google Scholar]
- 94.Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, Finch CE, St Laurent G, 3rd, Kenny PJ, Wahlestedt C. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nat. Med. 2008;14:723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sonkoly E, Bata-Csorgo Z, Pivarcsi A, Polyanka H, Kenderessy-Szabo A, Molnar G, Szentpali K, Bari L, Megyeri K, Mandi Y, et al. Identification and characterization of a novel, psoriasis susceptibility-related noncoding RNA gene, PRINS. J. Biol. Chem. 2005;280:24159–24167. doi: 10.1074/jbc.M501704200. [DOI] [PubMed] [Google Scholar]
- 96.Daughters RS, Tuttle DL, Gao W, Ikeda Y, Moseley ML, Ebner TJ, Swanson MS, Ranum LP. RNA gain-of-function in spinocerebellar ataxia type 8. PLoS Genet. 2009;5:e1000600. doi: 10.1371/journal.pgen.1000600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Khalil AM, Faghihi MA, Modarresi F, Brothers SP, Wahlestedt C. A novel RNA transcript with antiapoptotic function is silenced in fragile X syndrome. PLoS One. 2008;3:e1486. doi: 10.1371/journal.pone.0001486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yang L, Lin C, Liu W, Zhang J, Ohgi KA, Grinstein JD, Dorrestein PC, Rosenfeld MG. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell. 2011;147:773–788. doi: 10.1016/j.cell.2011.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011;147:1537–1550. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kertesz M, Wan Y, Mazor E, Rinn JL, Nutter RC, Chang HY, Segal E. Genome-wide measurement of RNA secondary structure in yeast. Nature. 2010;467:103–107. doi: 10.1038/nature09322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhu Y, Yu M, Li Z, Kong C, Bi J, Li J, Gao Z, Li Z. ncRAN, a newly identified long noncoding RNA, enhances human bladder tumor growth, invasion, and survival. Urology. 2011;77:510 e511–e515. doi: 10.1016/j.urology.2010.09.022. [DOI] [PubMed] [Google Scholar]