Abstract
Restriction endonuclease Bse634I recognizes and cleaves the degenerate DNA sequence 5′-R/CCGGY-3′ (R stands for A or G; Y for T or C, ‘/’ indicates a cleavage position). Here, we report the crystal structures of the Bse634I R226A mutant complexed with cognate oligoduplexes containing ACCGGT and GCCGGC sites, respectively. In the crystal, all potential H-bond donor and acceptor atoms on the base edges of the conserved CCGG core are engaged in the interactions with Bse634I amino acid residues located on the α6 helix. In contrast, direct contacts between the protein and outer base pairs are limited to van der Waals contact between the purine nucleobase and Pro203 residue in the major groove and a single H-bond between the O2 atom of the outer pyrimidine and the side chain of the Asn73 residue in the minor groove. Structural data coupled with biochemical experiments suggest that both van der Waals interactions and indirect readout contribute to the discrimination of the degenerate base pair by Bse634I. Structure comparison between related enzymes Bse634I (R/CCGGY), NgoMIV (G/CCGGC) and SgrAI (CR/CCGGYG) reveals how different specificities are achieved within a conserved structural core.
INTRODUCTION
Type II restriction endonucleases (REases) recognize specific DNA sequences (usually 4–8 bp in length) and cleave them at specific positions within or close to their recognition site. The majority of restriction enzymes faithfully recognize a unique target site in DNA (e.g. EcoRI binds to GAATTC) and discriminate it against other sequences with an extreme specificity [discrimination factor is up to 1010 (1,2)]. However, a significant number of REases is able to interact with degenerate recognition sequences meaning that more than one base is permitted at a particular position of the target site. For example, the thermophilic Bse634I restriction enzyme recognizes sequence R/CCGGY, where Y (pYrimidine) stands for ‘C or T’, R (puRine) stands for ‘A or G’, and ‘/’ indicates the cleavage position (3). In fact, among 314 recognition sites of Type II REases categorized in the REBASE (4), 83 represent degenerate sequences, including 53 containing an R:Y degenerate base pair at various positions. Molecular mechanisms that enable REase cleavage at several alternative target sites, but prevent cutting at non-cognate DNA sequences, are not yet fully understood. Deciphering of these mechanisms may reveal novel strategies for the engineering of highly specific nucleases which target unique sites in genomes but are refractory for the off-side cleavage (5).
In order to understand molecular mechanisms of the degenerate sequence recognition, we have focused on the REases that contain an R:Y base pair in their target sites. Currently, there are 32 structures of TypeII REases in the DNA-bound form and 5 of them, namely BsoBI (C/YCGRG) (6), EcoO109I (RG/GNCCY) (7), HincII (GTY/RAC) (8), SgrAI (CR/CCGGYG) (9,10) and BstYI (R/GATCY) (11) represent enzymes which recognize R:Y degenerate target sites. In most cases, the crystal structures are solved with one version of the degenerate target site and only for HincII, the structures were obtained with two cognate oligonucleotide variants providing us with the first structural mechanism of the alternative sequence recognition. Crystallographic studies supported by biochemical experiments (12,13) demonstrate that HincII specificity for the degenerate GTYRAC sequence arises from the indirect readout of the conformational preferences at the central pyrimidine–purine step (8). However, it remains to be established whether the same or different mechanisms are responsible for the degenerate sequence recognition by other restriction enzymes.
Bse634I restriction enzyme is specific for the degenerate hexanucleotide sequence R/CCGGY and belongs to a large family of evolutionarily related REases that interact with closely related DNA sequences containing the conserved CCGG or CCNGG core and cut their target sites before the first C (14,15) (Table 1). The crystal structure of Bse634I in the DNA free form revealed a tetramer that had two DNA-binding sites (16). Structural comparison of Bse634I and NgoMIV–DNA (17) complex suggested that both enzymes presumably share a conserved mechanism for the recognition of the conserved CCGG core (18), however, it did not provide an answer how Bse634I discriminates the degenerate outer base pairs.
Table 1.
Structurally characterized REases that interact with /CCGG or /CCNGG target sequencesa
| REase | Subtype | Recognition sequence | Oligomeric state | PDB ID | References |
|---|---|---|---|---|---|
| Bse634I | IIF | R/CCGGY | Tetramer | 1KNV | (18) |
| Cfr10I | IIF | R/CCGGY | Tetramer | 1CFR | (19) |
| SgrAI | IIF | CR/CCGGYG | dimer/tetramer | 3DVO, 3MQ6, 3DPG, 3DW9 | (9,10) |
| NgoMIV | IIF | G/CCGGC | Tetramer | 1FIU, 4ABT | (17) |
| Ecl18kI | IIF | /CCNGG | dimer/tetramer | 2FQZ | (20) |
| PspGI | IIP | /CCWGG | dimer | 3BM3 | (21) |
| EcoRII | IIE | /CCWGG | dimer | 1NA6, 3HQG | (22,23) |
a‘/’ indicates the cleavage position
Here, we present the crystal structures of the Bse634I (R226A mutant) in the complex with the oligoduplexes representing both palindromic variants (GCCGGC and ACCGGT) of the target site (Table 2). The structures of Bse634I with the AT-1 and AT-2 oligoduplexes containing the ACCGGT site were solved at 2.3 and 2.7 Å resolution, respectively. The crystal structure of Bse634I with the GC-1 oligoduplex, containing the GCCGGC site, was solved at 2.3 Å resolution. The crystal structures reveal structural principles for an alternative target site recognition by Bse634I. The structural data coupled with the biochemical experiments suggest that both van der Waals interactions and indirect readout contribute to the discrimination of the degenerate base pair by Bse634I. Furthermore, a direct structural comparison of Bse634I (R/CCGGY), NgoMIV (G/CCGGC) and SgrAI (CR/CCGGYG) in the DNA-bound form demonstrates how different specificities are achieved within the group of related enzymes.
Table 2.
DNA oligoduplexes used in this study
| Name | Sequencea | Comment |
|---|---|---|
| NS | 5′-AGCGTAGCACTGGGCTGCTAGTC-3′ | Non-cognate, unmodified |
| 3′-TCGCATCGTGACCCGACGATCAG-5′ | ||
| AT | 5′-CGCACGATCACCGGTGATGCACGC-3′ | Cognate, unmodified |
| 3′-GCGTGCTAGTGGCCACTACGTGCG-5′ | ||
| GC | 5′-CGCACGATCGCCGGCGATGCACGC-3′ | Cognate, unmodified |
| 3′-GCGTGCTAGCGGCCGCTACGTGCG-5′ | ||
| TA | 5′-CGCACGATCTCCGGAGATGCACGC-3′ | Mis-cognate, unmodified |
| 3′-GCGTGCTAGAGGCCTCTACGTGCG-5′ | ||
| CG | 5′-CGCACGATCCCCGGGGATGCACGC-3′ | Mis-cognate, unmodified |
| 3′-GCGTGCTAGGGGCCCCTACGTGCG-5′ | ||
| AU | 5′-CGCACGATCACCGGUGATGCACGC-3′ | Cognate, modified |
| 3′-GCGTGCTAGUGGCCACTACGTGCG-5′ | ||
| IC | 5′-CGCACGATCICCGGCGATGCACGC-3′ | Cognate, modified |
| 3′-GCGTGCTAGCGGCCICTACGTGCG-5′ | ||
| UA | 5′-CGCACGATCUCCGGAGATGCACGC-3′ | Mis-cognate, modified |
| 3′-GCGTGCTAGAGGCCUCTACGTGCG-5′ | ||
| CI | 5′-CGCACGATCCCCGGIGATGCACGC-3′ | Mis-cognate, modified |
| 3′-GCGTGCTAGIGGCCCCTACGTGCG-5′ | ||
| GC-1 | 5′-TCGCGCCGGCGCG-3′ | Crystallization |
| 3′-GCGCGGCCGCGCT-5′ | ||
| AT-1 | 5′-TCGCACCGGTGCG-3′ | Crystallization |
| 3′-GCGTGGCCACGCT-5′ | ||
| AT-2 | 5′-TTCGACCGGTCGA-3′ | Crystallization |
| 3′-AGCTGGCCAGCTT-5′ |
aThe central tetranucleotide CCGG of the Bse634I recognition sequence is underlined. Modified bases are in bold face.
MATERIALS AND METHODS
Oligonucleotides
All oligonucleotides used in this study were synthesized and HPLC purified by Metabion (Martinsried, Germany). For the DNA-binding and kinetic experiments, the upper strand of each DNA duplex was labeled at the 5′-end with [γ-33P]ATP (Hartmann Analytic, Braunschweig, Germany) using T4 DNA polynucleotide kinase (ThermoFisher, Vilnius, Lithuania). Oligoduplexes were assembled by slow annealing from 95°C to room temperature in a buffer [8 mM Tris-OAc (pH 7.9 at 25°C), 16.5 mM KOAc] or in water (for crystallization).
Protein purification and crystallization
Mutants P203G and P203S were constructed by two-step megaprimer method (24). The protein coding region of the resulting plasmids was sequenced in order to ensure that the only desired mutations were introduced. The wt Bse634I and R226A (25), P203G and P203S (this study) mutants were expressed and purified as described (18). The identity of the purified P203G and P203S mutant proteins was checked by mass spectrometry. The concentration of the protein monomers was estimated spectrophotometrically using extinction coefficient 34 280 M−1 cm−1. R226A–DNA complex was crystallized by the sitting drop vapor diffusion technique. The protein in a buffer containing 5 mM CaCl2, 20 mM Tris–HCl, 50 mM NaCl, pH 7.5 was mixed with an equimolar amount of DNA and an equal volume of a crystallization solution was added. Final concentration of the protein was in the range from 4 to 8 mg/ml. The crystals grew in a week, and after several months degraded severely. Solutions used as reservoirs for crystallization of the R226A mutant complexes with GC-1 and AT-1 oligonucleotides contained 100 mM NaOAc pH 4.25–5.5, 10 mM CaCl2 or Ca(OAc)2, 4–8% (w/v) of PEG8000. The crystals of the R226A complex with the AT-2 oligoduplex were obtained with the reservoir solution which contained 0.1 M Bis–Tris pH 5.5, 0.5% polyvinylpyrrolidone and 16% of PEG400.
Data collection and structure determination
Crystallographic data for R226A-GC-1 were collected at EMBL Hamburg outstation DESY X12 beamline, and data sets from crystals with AT-1 and AT-2 oligoduplexes were collected at X11 DESY beamline. MOSFLM (26,27), SCALA (28) and TRUNCATE (29) were used for data processing. Initial phases were obtained by molecular replacement using MOLREP (30) and PDB entry 1KNV as an initial model. Coordinates for B-form DNA oligonucleotides were generated using NAB software http://structure.usc.edu/make-na/server.html. The model was rebuilt and refined using COOT (31), CNS (32) and REFMAC (33) programs. The crystal structure of the Bse634I complex with AT-2 was refined using non-crystallographic symmetry (NCS) restraints between all eight subunits present in the asymmetric unit. Data collection and refinement statistics are shown in Supplementary Table S1. All molecular scale representations were prepared using MOLSCRIPT (34) and RASTER3D (35) software.
DNA-binding experiments
Binding of the modified and unmodified DNA oligoduplexes by wt and mutant Bse634I was studied by the gel mobility shift assay. The radiolabeled oligoduplexes (0.1 nM) were mixed in 20 μl samples with different concentrations of Bse634I (varying from 0.1 nM to 5 μM in terms of monomer). The reaction was carried out in a buffer containing 40 mM Tris-OAc (pH 8.3 at 25°C), 5 mM Ca(OAc)2, 0.1 mg/ml BSA and 10% (v/v) glycerol. Before loading on the gel the samples were incubated for 10 min at room temperature. The protein–DNA complexes were resolved by electrophoresis in 6% non-denaturing PAGE [acrylamide/N,N′-methylenebisacrylamide 29:1 (w/w)] in 40 mM Tris–OAc running buffer containing 5 mM Ca(OAc)2 at 6 V/cm for 3 h. The gels were visualized using a Cyclone Phosphor-Imager (Perkin-Elmer, Wellesley, MA, USA).
DNA-cleavage experiments
The DNA cleavage reactions were performed by mixing the radiolabeled oligoduplexes (100 nM) with Bse634I (100 nM in terms of tetramer) in the reaction buffer [10 mM Tris-HCl (pH 8.5 at 37°C), 100 mM KCl, 10 mM MgCl2 and 0.1 mg/ml BSA] at 25 and 50°C (reactions which required incubation time >10 min were carried out under mineral oil). The samples were collected at timed intervals and quenched with a loading dye solution [95% (v/v) formamide, 25 mM EDTA, 0.01% (w/v) bromphenol blue]. Separation of the DNA hydrolysis products was performed by denaturing PAGE: the 20% polyacrylamide gel [acrylamide/N,N′-methylenebisacrylamide 19:1 (w/w)] in Tris–borate buffer containing 8.5 M of urea was run at 30 V/cm. The radiolabeled DNA was detected as described above. The rate constants of the DNA cleavage were determined by fitting single exponentials to the time-courses of the substrate depletion in the experiments with Gnuplot software.
RESULTS
Crystals of Bse634I R226A mutant
Our multiple attempts to obtain crystals of the wild type Bse634I–DNA complex were unsuccessful, therefore, we switched to the crystallization trials of the mutational variants. We succeeded in the co-crystallization of the Bse634I R226A mutant with three different oligoduplexes GC-1, AT-1 and AT-2 (Table 2). Bse634I restriction enzyme is arranged as a tetramer comprised of two primary dimers and requires two DNA copies for its optimal activity (18). The Arg226 residue is located at the tetramerization interface of Bse634I and makes an intricate network of interactions with the amino acid residues of two neighboring subunits. Its replacement by alanine disrupts the Bse634I tetramer into monomers in solution and impairs the Bse634I cleavage at low protein concentrations [(25) and M.Z., unpublished results].
Two different crystal forms of the R226A mutant in complex with DNA were obtained. Data collection and refinement statistics is presented in the Supplementary Table S1. The crystals in the P21212 space group were produced with the cognate oligoduplexes GC-1 and AT-1 (Table 2) representing both palindromic variants of the Bse634I recognition sequence GCCGGC and ACCGGT, respectively. The crystallographic asymmetric unit in both crystals contains two protein monomers and two DNA chains. The primary dimer is generated by rotation of the protein and DNA monomer around the crystallographic 2-fold axis.
The second crystal form in the P21 space group was obtained with the AT-2 oligoduplex, which differs from the AT-1 oligoduplex by the flanking sequence (Table 2). The asymmetric unit in the crystal consists of eight protein subunits that build up two tetramers (Supplementary Figure S1A). Two oligoduplexes are bound within the DNA-binding clefts of each Bse634I tetramer. Interestingly, three additional ‘out-of-site’ duplexes fill up the space between the protein tetramers and form the continuous quasi-infinite DNA strands, each built by the repetition of three crystallographically independent oligoduplexes. The unpaired 5′-terminal thymine of the AT-2 oligoduplex makes H-bonds to the last A:T pair of the next DNA molecule, forming a nearly perfect Hoogsteen pair (Supplementary Figure S1B).
Overall complex architecture
In both crystal forms, two primary dimers are stacked back-to-back to each other and form a tetramer (Figure 1A and Supplementary Figure S1A). Each primary dimer binds one DNA duplex, therefore the tetramer binds two DNA molecules simultaneously. In all three R226A–DNA complex structures, the tetramer arrangement is very similar to that of the Bse634I in the DNA-free form (18). Thus, although R226A substitution disrupts the Bse634I tetramer into monomers in solution at low protein concentrations, in the crystal, the mutant protein is assembled into the tetramer similar to the wt enzyme.
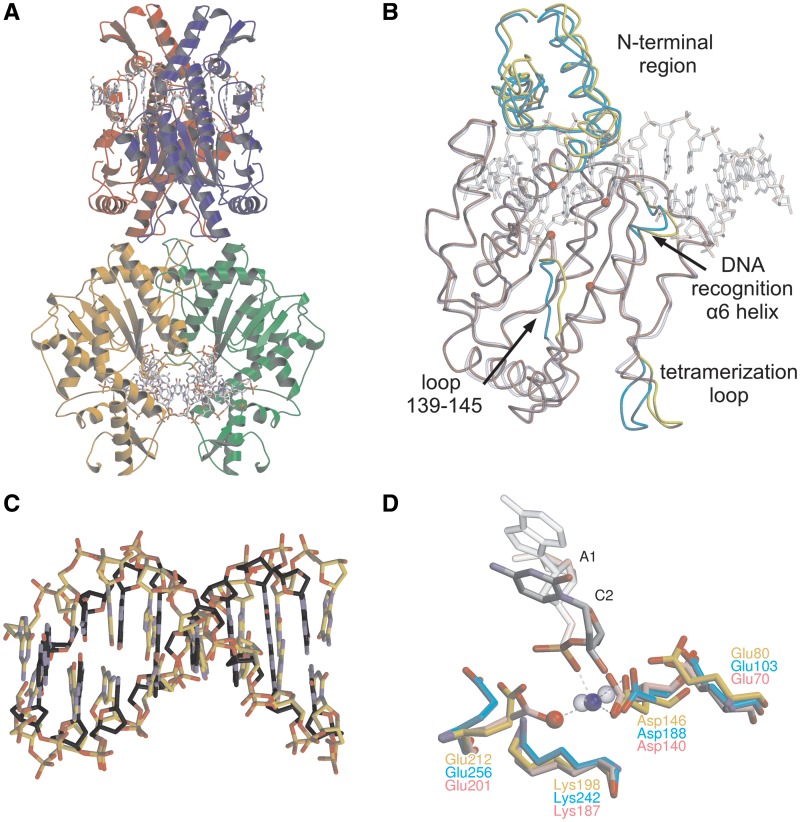
Figure 1.
The crystal structure of Bse634I (R226A)–DNA complex. (A) General architecture of the tetramer bound to the GC-1 oligoduplex. Individual monomers are shown in different colors. Two DNA molecules bound by Bse634I are in a ball-and-stick representation. (B) Superposition of DNA-free Bse634I (PDB ID 1KNV) monomer A (transparent blue) with the monomer A (red) in the Bse634I–GC-1 complex. Protein chains are traced by Cα, DNA is shown in a transparent ball-and-stick representation and CPK colors. Protein chain fragments that differ in both structures are cyan and yellow for apo- and DNA bound monomers, respectively. Cα atoms of the catalytic motif (Glu80, Asp146, Lys198 and Glu212) are shown as red spheres. (C) Superposition of the eight central base pairs (recognition site and adjacent flanking base pairs) of the GC-1 oligoduplex complexed with Bse634I (gold) and B-form DNA of the same sequence generated by NAB software (grey). (D) Superposition of the catalytic center residues of the Bse634I and AT-1 complex (gold) and corresponding residues in NgoMIV–DNA (PDB ID 4ABT, pink) and SgrAI–DNA (PDB ID 3DVO, cyan) complexes. Ca2+ is shown as a blue sphere, a water molecule as a red sphere. Ca2+ in the active sites of NgoMIV and SgrAI are shown as blue transparent spheres.
Bse634I undergoes relatively small changes upon DNA binding (Figure 1B). First, the N-terminal part (residues 23–89) of the Bse634I monomer which is flexible in the apo-structure (18), moves closer to the DNA and becomes fixed by the DNA backbone contacts. The DNA binding also induces a slight shift of the protein segment (residues 139–145) in the vicinity of the active site residue Asp146 and upstream the α6 ‘recognition’ helix (residues 202–205) which bears the residues involved in the central CCGG core recognition. The tetramerization loop (residues 258–264), which makes major contacts between the primary dimers within the tetramer, also shows slightly different conformations in the DNA bound- and apo-structures.
The DNA duplex bound to Bse634I in all three structures is in the B-form conformation (Figure 1C). Unpaired thymines at the 5′-end of the oligoduplex are highly flexible and poorly resolved. According to CURVES (36) analysis, DNA in the complex with the R226A mutant shows a slight 11–18° bending towards the minor groove (the bending angle value varies depending on the individual structure). The most profound kink occurs at the central dinucleotide step of the site RCCGGY coinciding with the dyad axis of oligonucleotide.
Active center organization
The active center of Bse634I (Figure 1D) is formed by Glu80, Asp146, Lys198 and Glu212 residues which (except of Glu80) follow a sequence motif PDX46–53KX13E characteristic for the REase family presented in Table 1. The Bse634I active site is structurally similar to those of NgoMIV (17), and SgrAI enzymes (9). Indeed, Cα atoms of the catalytic residues corresponding to Glu212 and Asp146 of Bse634I nearly coincide between all three enzymes. However, the side chain residues of Glu212 are not well-resolved in the individual Bse634I subunits. This fact, probably, reflects an incompleteness of the active center assembly, whether due to the mutation, or due to a relatively low pH of the crystallization mixture.
Calcium ion was included in all crystallization mixtures, because it is a necessary pre-requisite for the DNA binding by Bse634I (16). Nevertheless, only in the crystal structure of Bse634I with AT-1 oligoduplex, an electron density that could be asserted to calcium was found in a close vicinity of the scissile phosphate. The octahedral coordination sphere of the metal ion is incomplete and only four ligands could be identified. The long axis of octahedron is built up by a phosphate oxygen of C2 nucleotide and a carbonyl oxygen of Leu197. Other two ligands are carboxyl oxygens of Asp146 and a water molecule (Figure 1D).
Contacts to the conserved CCGG core
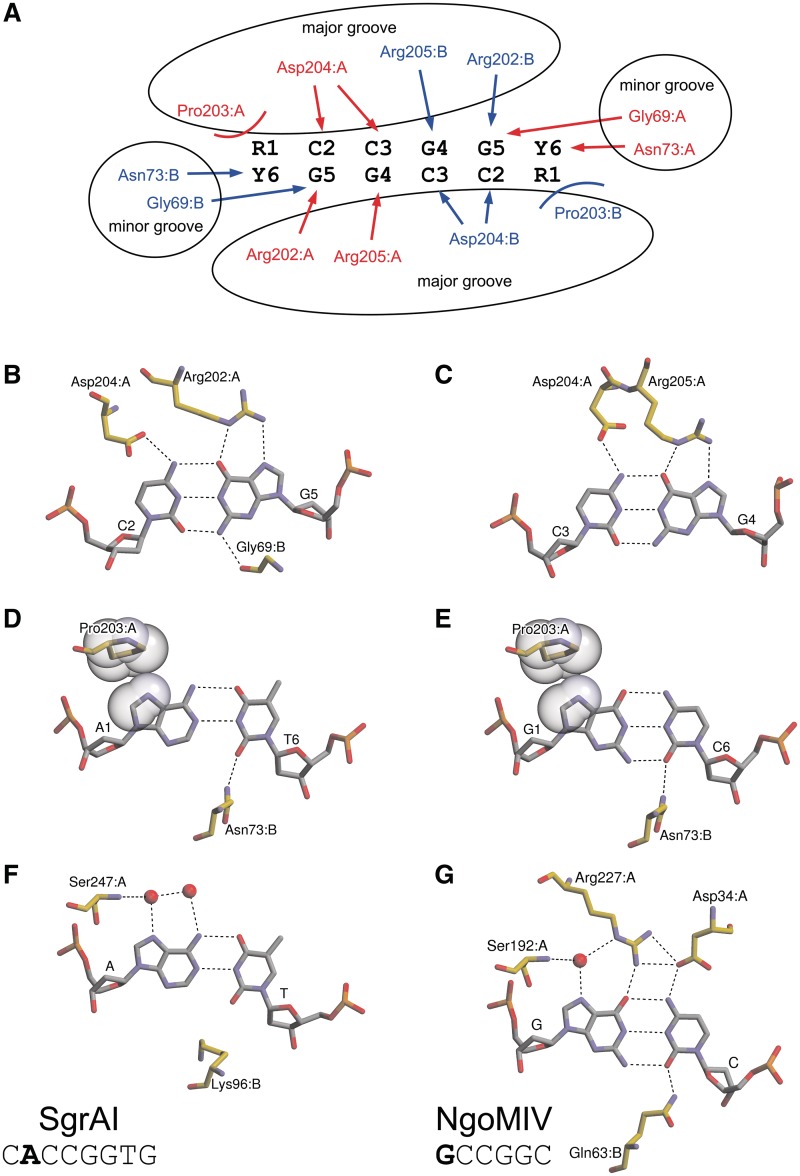
Two symmetry-related α6 helices, dubbed recognition helices (18), protrude into the major groove of DNA, as it was predicted analyzing the apo-structure. The side chain atoms of the amino acid residues Arg202, Asp204 and Arg205 located at the N-terminus of the α6 helix and on the upstream loop are engaged in the sequence-specific hydrogen bond interactions with the donor and acceptor atoms of the CCGG bases. One protein subunit makes contacts in the major groove with one half-site of the symmetric CCGG sequence (Figure 2A). More specifically, the Nε and Nη2 atoms of the Arg202 and Arg205 residues make bidentate hydrogen bonds to the O6 and N7 atoms of G5 and G4 bases, respectively, while carboxyl oxygens of Asp204 bridge N4 atoms of the neighboring cytosines C2 and C3 (Figure 2B and C). The Nη1 atom of Arg205 also makes a direct hydrogen bond to the main chain oxygen atom of Ser200 and through the bridging water molecule interacts with the catalytic Lys198 residue (not shown). In the minor groove, the main chain oxygen atom of Gly69 is within a hydrogen bond distance (2.9–3.1 Å depending on the particular subunit) to the N2 amino group of the G5 base (Figure 2B).
Figure 2.
Specific interactions of Bse634I with the recognition sequence. (A) Schematic representation of the degenerate target site and interactions between the Bse634I and its target site. Residues belonging to monomers A and B are shown in red and blue, respectively. H-bonds are indicated by arrows, van der Waals interactions to purines–by curved lines. (B) Bse634I interactions with the C2:G5 base pair. (C) Bse634I interactions with the C3:G4 base pair. (D) Bse634I contacts to the outer A1:T6 base pair in the AT-1 complex. (E) Bse634I contacts to the outer G1:C6 base pair in the GC-1 complex. C8 and N7 atoms of R1 base and side chain atoms of Pro203 residue are shown in CPK representation. (F) SgrAI interactions with the A:T base pair in complex with the oligoduplex containing the cognate 8 bp site CACCGGTG (PDB ID 3DVO). (G) NgoMIV interactions with the G:C base pair in the 6 bp target site GCCGGC (PDB ID 4ABT).
Interactions with an outer R1:Y6 base pair
The recognition pattern of the conserved CCGG sequence by Bse634I is nearly identical to that of NgoMIV (17). However, in contrast to NgoMIV which strictly prefers the G:C base pair outside the CCGG core, Bse634I tolerates both G:C and A:T within its target site. The crystal structures of Bse634I in the complex with the oligoduplexes containing either GCCGGC (GC-1; Table 2) or ACCGGT sites (AT-1 and AT-2; Table 2) reveal the protein interaction networks with two alternative target sequences. In both structures, A1:T6 and G1:C6 base pairs are contacted by the Bse634I dimer from both minor and major groove sides (Figure 2D and E) and both monomers contribute the amino acid residues for the outer base pair recognition (Figure 2A). In the major groove, Pro203 is in the van der Waals contact distance both to A1 and G1 bases (Figure 2D and E). In the minor groove, the Nδ atom of Asn73 residue coming from the neighboring monomer makes a hydrogen bond to the O2 atom of T6 and C6 pyrimidines.
The outer base pair R1:Y6 is distorted in the Bse634I–DNA complex. The local base pair step parameters as calculated by x3dna (37,38) are represented in the Supplementary Figure S2. The Y6 pyrimidine base is displaced from the minor groove by Asn73 and shifted towards the major groove relatively to the first flanking base pair (Supplementary Figure S2C and S2D). This results in the distortion of stacking between Y6 base and the first base outside of the recognition sequence.
Thus, Bse634I seems to combine both direct and indirect mechanisms to discriminate the RCCGGY sites from mis-cognate YCCGGR sequences. The direct interactions are limited by van der Waals contact to the purine base made by Pro203 in the major groove and a H-bond made to the complementary pyrimidine by the Asn73 in the minor groove. In silico mutagenesis suggests that the R1 purine replacement by the T base should introduce a steric clash of the C5 methyl group with the Pro203 residue and interfere with the Bse634I interaction with the TCCGGA site.
Bse634I binding and cleavage studies
In order to test this hypothesis, we analyzed the DNA binding and cleavage by Bse634I using a set of the oligoduplexes containing cognate (ACCGGT or GCCGGC), mis-cognate (TCCGGA or CCCGGG) and modified cognate and mis-cognate sites (Table 2). In the modified substrates, the outer T is replaced by U (uracil), which lacks the 5-methyl group in the pyrimidine ring, and G is replaced by I (inosine), which lacks 2-amino group in the purine ring. Flanking sequences in all oligoduplexes used in these experiments were identical (Table 2).
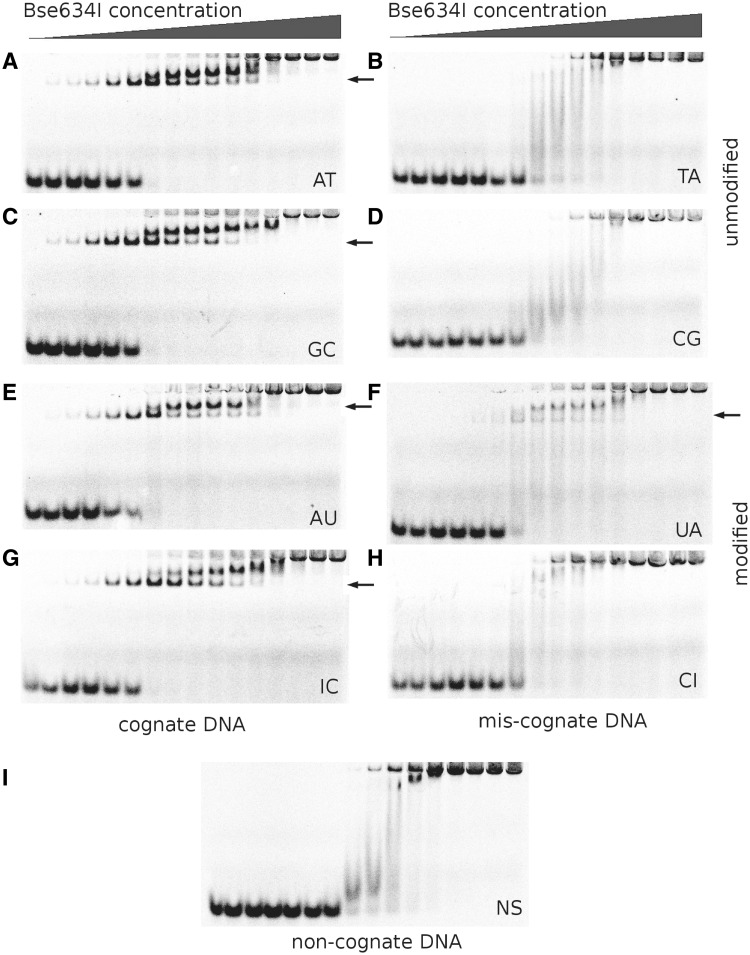
The electrophoretic mobility shift assay revealed that Bse634I binds the cognate oligoduplexes AT and GC (Figure 3A and C) at the low protein concentrations. Further increase of the Bse634I concentration gives rise to the second slowly migrating protein–DNA complex. In the case of the non-cognate oligoduplex NS (Figure 3I), only this slowly migrating complex is observed at the increased protein concentrations. Based on the DNA-binding patterns presented here and in the previous studies (16), we conclude that the complexes with the cognate DNA observed at low protein concentrations represent the specific complexes where Bse634I is bound to the recognition site while lower mobility complexes at increased protein concentrations correspond to non-specific complexes. The cognate oligoduplexes containing U instead of T (AU) and I instead of G (IC) showed the DNA-binding pattern similar to that for the specific oligoduplexes GC and AT indicating that the elimination of the 5-methyl group in thymine or 2-amino group in guanine in the outer base pair has no effect on the Bse634I binding (Figure 3E and G). The crystal structures are consistent with this observation since there are no direct or indirect contacts with methyl or amino groups in the Bse634I complexes with cognate DNA.
Figure 3.
Binding of specific (A, C, E and G), mis-cognate (B, D, F and H), and non-cognate (I) DNA oligoduplexes by the wt Bse634I. Labels shown on the picture indicate oligoduplex (Table 2) used in the gel-shift experiment. I stands for Inosine and U for Uridine. Concentration of Bse634I in samples is increasing from 0.1 nM to 5 μM (in terms of monomer; see ‘Materials and Methods’ section). The first lane in all gels contains no protein. Specific or specific-like Bse634I–DNA complexes are designated by arrows.
On the other hand, the oligoduplexes TA (Figure 3B) and CG (Figure 3D) containing mis-cognate sites are bound by the protein similarly to the non-cognate DNA (Figure 3I). However, T to U replacement in the mis-cognate oligoduplex UA results in the different binding pattern (compare panels F and B in Figure 3). Indeed, the band corresponding to a specific protein–DNA complex appears in the gel, albeit at the higher protein concentrations in comparison to the cognate AT site. The modified CI oligoduplex forms only a non-specific protein–DNA complex, similarly to the mis-cognate CG duplex (Figure 3H and D).
On the next step, we analyzed Bse634I cleavage of cognate, mis-cognate and modified oligoduplexes. The cleavage reaction was carried out under single turnover conditions using an optimal enzyme concentration that yields a maximal cleavage rate. Reaction rate constants were determined as described in the ‘Materials and Methods’ section.
The cleavage rates for the cognate and the modified cognate oligoduplexes AT, AU, GC and IC are within the range 4–13 min−1 at 25°C (Table 3) indicating that modifications in the cognate AU and IC duplexes have no significant effect on the cleavage activity. However, in the case of the unmodified and the modified mis-cognate duplexes (TA, CG, UA and CI), the cleavage rates at 25°C were too slow to be reliably measured. Therefore, the Bse634I cleavage of TA, CG, UA and CI oligoduplexes was studied at 50°C taking an advantage of the fact that Bse634I is a thermophilic enzyme (3). Under these conditions, the cleavage rate of the cognate oligoduplex was too high to be measured by the conventional techniques. On the other hand, the mis-cognate TA and CG duplexes even at 50°C were hydrolyzed very slowly (∼20% of the oligoduplex was hydrolyzed after 48 h incubation). However, under these conditions, the Bse634I cleavage rate of the modified mis-cognate UA duplex was increased nearly by three orders of magnitude compared to the TA oligoduplex. The cleavage rate of the CI duplex was very close to that of the unmodified CG and TA oligonucleotides (Table 3).
Table 3.
Rate constants of oligoduplex cleavage by Bse634I
| Oligoduplex |
k (min−1) |
|||||
|---|---|---|---|---|---|---|
| wt |
P203S |
P203G |
||||
| 25°C | 50°C | 25°C | 50°C | 25°C | 50°C | |
| AT | 10 ± 1.3 | – | 15 ± 1.8 | – | 2.9 ± 0.2 | – |
| GC | 4.1 ± 0.9 | – | 7.8 ± 1.2 | – | 12 ± 4.0 | – |
| AU | 5.4 ± 1.3 | – | 11 ± 2.4 | – | 1.5 ± 0.4 | – |
| IC | 13 ± 2.9 | – | 17 ± 1.5 | – | 14 ± 2.1 | – |
| TA | nca | 5.7 ± 0.7 × 10−5 | nc | 5.7 ± 1.6 × 10−4 | nc | 1.2 ± 0.3 × 10−4 |
| CG | nc | 8.2 ± 0.8 × 10−5 | 1.3 ± 0.1 × 10−4 | 9.4 ± 1 × 10−2 | nc | 3.9 ± 0.5 × 10−4 |
| UA | nc | 6.9 ± 0.8 × 10−2 | 1.1 ± 0.1 × 10−2 | 5.8 ± 1 | 1.7 ± 2.6 × 10−3 | 3.8 ± 0.3 × 10−1 |
| CI | nc | 3.8 ± 0.9 × 10−5 | 1.4 ± 0.1 × 10−4 | 2.4 ± 0.8 × 10−1 | nc | 1.5 ± 0.9 × 10−3 |
| NS | nc | nc | nc | 5.4 ± 1.6 × 10−5 | nc | 3.7 ± 2 × 10−5 |
aNo cleavage products were observed after 48 h of incubation.
To further probe the importance of the van der Waals interactions in the discrimination of the degenerate R:Y base pair, we engineered P203G and P203S mutants replacing the proline with glycine (no side chain) or serine (equivalent to Pro in SgrAI) and performed DNA binding and cleavage experiments (Table 3, Supplementary Figures S3 and S4). Functional analysis revealed that the Pro203 replacement resulted in the ∼10-fold increase in the oligoduplex TA cleavage rate in comparison to the wt Bse634I (Table 3). This finding would be consistent with our hypothesis that van der Waals interactions between Pro and T methyl group are important for the discrimination of the degenerate base pair by Bse634I. However, the Pro replacement by Gly or Ser seems to be not fully equivalent to the T substitution by U in DNA, since the P203G and P203S mutants display increased cleavage rate for all tested mis-cognate substrates indicating that the Pro mutations compromise Bse634I specificity.
DISCUSSION
Mechanisms of degenerate sequence recognition by Bse634I
The Bse634I restriction enzyme identified in the thermophilic Bacillus stearothermophilus strain cleaves two alternative target sites ACCGGT and GCCGGC (conserved sequences underlined) with nearly equal rates (Table 3). In order to understand structural mechanisms of the degenerate sequence recognition by Bse634I, we have solved the crystal structures of Bse634I in complex with three oligoduplexes representing two alternative sites.
Structural analysis revealed that the recognition of the conserved CCGG sequence is achieved by a direct readout of the unique pattern of the donor and acceptor atoms on the base edges of the CCGG tetranucleotide core (Figure 2A–C). The degenerate base pair outside the conserved core introduces an ambiguity for the base pair discrimination by a direct readout, since A:T and G:C base pairs expose a different set of donor and acceptor atoms in the major and minor grooves. Not surprisingly, there are no direct H-bonds with the G:C and A:T bases in the major groove and only the Pro203 residue makes van der Waals contact to the purine ring of the external base pair. In the minor groove, contacts to the degenerate base pair are limited to a single H-bond between the Nδ atom of Asn73 residue and O2 atom of the pyrimidine base. This raises a question how these interactions can discriminate RCCGGY sites against YCCGGR sites.
In silico experiments indicate that in the case of TCCGGA sequence a steric clash would occur between the C5 methyl group of T base and Pro203 residue. The importance of this van der Waals interaction for the R versus T discrimination is supported by biochemical data. Indeed, the modified UA oligoduplex (Table 2) which contains U instead of T and lacks 5-methyl group forms the specific Bse634I–DNA complex (Figure 3F) and is cleaved nearly three orders of magnitude faster in comparison to the unmodified mis-cognate oligoduplexes TA and CG (Table 3). Taken together, these data indicate that the steric clash between the C5 methyl group of T and Pro203 residue plays an important role in the R versus T discrimination by Bse634I. However, the Bse634I preference for R against T in the first position of the target site cannot be fully explained by a disruption of the favored van der Waals contact between Pro203 and the purine base. Cleavage rate of the UA oligoduplex containing the UCCGGA site is still much lower than of the cognate duplex.
Moreover, the steric expulsion mechanism does not provide an answer why C1:G6 base pair is not accepted by Bse634I. In the crystal structures with ACCGGT and GCCGGC oligoduplexes, the O2 atom of the outer pyrimidine Y6 makes a H-bond to the side chain Nδ2 of Asn73. Interestingly, the Gln63 residue in NgoMIV that is structurally equivalent to the Asn73 of Bse634I also makes a H-bond with O2 of the outer C6 base (Figure 2G). However, it seems unlikely that this single minor groove contact can discriminate between R and Y in the first position of the target site. Theoretically, the N3 nitrogen of A6 base should be able to make a H-bond with Asn73 similarly to Y6 pyrimidine.
Our present hypothesis is that the Bse634I selection against CCCGGG (and to some extent against the TCCGGA site) is achieved by an indirect readout mechanism due to a certain combination of distortions in DNA conformation. In the Bse634I structures with both cognate oligoduplexes, the outer pyrimidine base is displaced towards the major groove resulting in the distortion of the stacking interaction between Y6 base and the first base outside of the recognition sequence (Supplementary Figure S2). Interestingly, SgrAI which recognizes CRCCGGYG sequence (9) displays a similar DNA distortion.
Other REases which interact with the recognition sites containing the R:Y base pair employ different mechanisms for the degenerate base pair recognition. There are five other REases with the known structure that accept the R:Y base pair in their palindromic recognition sites (summarized in the Supplementary Table S2). Three of them are solved in complex with the oligoduplex containing only one variant of the degenerate target site, while the HincII crystal structure was solved with two alternative variants of the recognition sequence. [EcoO109I crystal structure with oligoduplex containing the non-palindromic recognition sequence contains DNA in both overlapping orientations (7).] Crystal structure analysis indicates that the REases achieve specificity for their degenerate target sites by decreasing the number of direct contacts which often become limited to the N7 position of purines or due to the ambivalence of the water-mediated hydrogen bonding (Supplementary Table S2).
How different specificities are generated within a group of related restriction enzymes
It has been reported before that apo-Bse634I (RCCGGY) shows close structural resemblance to the 6 bp cutter NgoMIV (GCCGGC) and 8 bp cutter SgrAI (CRCCGGYG) (conserved CCGG sequence is underlined) (14,15). All three enzymes cut their target sites before the CCGG core and share similar architecture of the active site (Figure 1D). The Ca2+ ion present in the Bse634I–AT-1 complex occupies exactly the same position as the first Ca2+ ion in the SgrAI structure with cognate DNA (9). In Bse634I, Ca2+ ion is coordinated by the oxygen atoms of the scissile phosphate, the side chain oxygen atoms of Asp146 and the main chain carbonyl of the Leu197 preceding the catalytic Lys198. The water molecule which serves as a fourth Ca2+ ion ligand in Bse634I overlaps well with the water molecule coordinated by Ca2+ ion in SgrAI and NgoMIV structures. Furthermore, Glu80 of Bse634I coincides with Glu103 of SgrAI, which coordinates the second Ca2+ ion (Figure 1D). The electron density which could be assigned to the second metal ion, however, is absent in the Bse634I structure.
Structural comparison of the enzymes in the DNA-bound forms confirmed that Bse634I, SgrAI and NgoMIV share a conserved mechanism for the recognition of the CCGG core within their target sites. The key contacts to the CCGG bases (Figure 2B and C) are made by the conserved stretch of RXDR residues located at the N-terminal tip of the recognition helices and are identical between Bse634I, NgoMIV and SgrAI.
In contrast to NgoMIV, which is strictly specific for the GCCGGC site, Bse634I recognizes two alternative palindromic sequences ACCGGT and GCCGGC. Outside the CCGG core, NgoMIV makes an intricate network of H-bonds to the outer G:C base pair (Figure 2G) both in the major and minor grooves. These interactions unambiguously specify the G:C base pair. Unlike NgoMIV, Bse634I makes only two contacts to the base pair outside the CCGG core, namely the hydrogen bond by Asn73 to the O2 of pyrimidine in the minor groove and the van der Waals contact by Pro203 to the purine from the major groove (Figure 2A, D and E). Thus, the direct contacts made by Bse634I to the outer base pair are restrained in comparison to NgoMIV presumably due to an ambiguity introduced by the degenerate base pair.
The target site of Bse634I (RCCGGY) overlaps with the central part of SgrAI site CRCCGGYG (underlined). SgrAI makes no direct contacts with the degenerate R:Y base pair (9) except of the putative water-mediated H-bond between Ser247 and N7 atom of purine (Figure 2F). The Cα atom of Lys96 occupies a structurally equivalent position of Asn73 in Bse634I, however, H-bond to the O2 oxygen of thymine in the minor groove is missing. Thus, the direct contacts to the degenerate base made by SgrAI are restrained similarly to Bse634I. Moreover, in the SgrAI–DNA complex, the DNA is distorted similarly to Bse634I (Supplementary Figure S2). The side chain of Lys96 contacts the outer G (underlined) of the SgrAI recognition sequence CRCCGGYG from the minor groove and unstacks this base from the Y that just precedes it (9). Thus, it seems that both in SgrAI and Bse634I an indirect mechanism plays an important role in the discrimination of the degenerate base pair, however, it remains to be determined to what extent the specific structural features of the RY helical step (39–42) contribute to the discrimination of the degenerate base pair.
Structural comparison suggests that the N-terminal domains of Bse634I and SgrAI exhibit the plasticity which may contribute to the recognition of the 8 bp sequence by SgrAI. The Arg31 residue of SgrAI which makes bidentate H-bonds to the external G residue (underlined) in the CACCGGTG sequence is located in the loop (residues 19–39) at the N-terminal domain. The N-terminal loop (residues 10–26) of Bse634I is structurally equivalent to the loop (residues 19–39) in SgrAI. However, the N-terminal loop in all Bse634I structures is rather flexible and shows a different orientation or is disordered in the individual monomers. The structurally equivalent loop in SgrAI is much more rigid presumably due to the larger number of contacts to the DNA backbone and the protein body. Furthermore, the structural equivalent of the Arg31 residue is missing in the similar loop of Bse634I which is too short to reach the major groove of the DNA substrate outside of the hexanucleotide recognition sequence. Further structural studies of Kpn2I (T/CCGGA), AgeI (A/CCGGT) and BsaWI (W/CCGGW, where W stands for A or T) restriction enzymes ongoing in the lab should contribute to our understanding how Nature engineered different specificities within the conserved structural fold.
ACCESSION NUMBERS
Coordinates and structure factors of Bse634I (R226A) complexes with DNA are deposited under PDB ID 3V1Z (GC-1), 3V20 (AT-1) and 3V21 (AT-2).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2 and Supplementary Figures 1–4.
FUNDING
Research Council of Lithuania (contract Nr. MIP-102/2010, to M.Z.); European Community's Research Infrastructure Action under the Sixth Framework Programme ‘Structuring the European Research Area Specific Programme’ (contract Number RII3-CT-2004-506008); and by Max-Planck Society (contract with Institute of Biotechnology, to S.G.). Funding for open access charge: Research Council of Lithuania/Sixth Framework Programme ‘Structuring the European Research Area Specific Programme’ (contract Number RII3-CT-2004-506008).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr M. Groves, Dr G. Bourenkov and Dr M.S. Weiss at EMBL beamlines X11 and X12 at the DORIS storage ring, Hamburg for the invaluable help with beamline operation. We are also very grateful to Dr Zita Liutkevičiūtė for the mass-spectrometry of Bse634I mutants.
REFERENCES
- 1.Engler LE, Welch KK, Jen-Jacobson L. Specific binding by EcoRV endonuclease to its DNA recognition site GATATC. J. Mol. Biol. 1997;269:82–101. doi: 10.1006/jmbi.1997.1027. [DOI] [PubMed] [Google Scholar]
- 2.Jen-Jacobson L, Engler LE, Jacobson LA. Structural and thermodynamic strategies for site-specific DNA binding proteins. Structure. 2000;8:1015–1023. doi: 10.1016/s0969-2126(00)00501-3. [DOI] [PubMed] [Google Scholar]
- 3.Repin VE, Lebedev LR, Puchkova L, Serov GD, Tereschenko T, Chizikov VE, Andreeva I. New restriction endonucleases from thermophilic soil bacteria. Gene. 1995;157:321–322. doi: 10.1016/0378-1119(94)00781-m. [DOI] [PubMed] [Google Scholar]
- 4.Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE–a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 2010;38:D234–D236. doi: 10.1093/nar/gkp874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Silva G, Poirot L, Galetto R, Smith J, Montoya G, Duchateau P, Pâques F. Meganucleases and other tools for targeted genome engineering: perspectives and challenges for gene therapy. Curr. Gene Ther. 2011;11:11–27. doi: 10.2174/156652311794520111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van der Woerd MJ, Pelletier JJ, Xu S, Friedman AM. Restriction enzyme BsoBI-DNA complex: a tunnel for recognition of degenerate DNA sequences and potential histidine catalysis. Structure. 2001;9:133–144. doi: 10.1016/s0969-2126(01)00564-0. [DOI] [PubMed] [Google Scholar]
- 7.Hashimoto H, Shimizu T, Imasaki T, Kato M, Shichijo N, Kita K, Sato M. Crystal structures of type II restriction endonuclease EcoO109I and its complex with cognate DNA. J. Biol. Chem. 2005;280:5605–5610. doi: 10.1074/jbc.M411684200. [DOI] [PubMed] [Google Scholar]
- 8.Horton NC, Dorner LF, Perona JJ. Sequence selectivity and degeneracy of a restriction endonuclease mediated by DNA intercalation. Nat. Struct. Biol. 2002;9:42–47. doi: 10.1038/nsb741. [DOI] [PubMed] [Google Scholar]
- 9.Dunten PW, Little EJ, Gregory MT, Manohar VM, Dalton M, Hough D, Bitinaite J, Horton NC. The structure of SgrAI bound to DNA; recognition of an 8 base pair target. Nucleic Acids Res. 2008;36:5405–5416. doi: 10.1093/nar/gkn510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park CK, Joshi HK, Agrawal A, Ghare MI, Little EJ, Dunten PW, Bitinaite J, Horton NC. Domain swapping in allosteric modulation of DNA specificity. PLoS Biol. 2010;8:e1000554. doi: 10.1371/journal.pbio.1000554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Townson SA, Samuelson JC, Xu SY, Aggarwal AK. Implications for switching restriction enzyme specificities from the structure of BstYI bound to a BglII DNA sequence. Structure. 2005;13:791–801. doi: 10.1016/j.str.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 12.Joshi HK, Etzkorn C, Chatwell L, Bitinaite J, Horton NC. Alteration of sequence specificity of the type II restriction endonuclease HincII through an indirect readout mechanism. J. Biol. Chem. 2006;281:23852–23869. doi: 10.1074/jbc.M512339200. [DOI] [PubMed] [Google Scholar]
- 13.Babic AC, Little EJ, Manohar VM, Bitinaite J, Horton NC. DNA distortion and specificity in a sequence-specific endonuclease. J. Mol. Biol. 2008;383:186–204. doi: 10.1016/j.jmb.2008.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tamulaitis G, Solonin AS, Siksnys V. Alternative arrangements of catalytic residues at the active sites of restriction enzymes. FEBS Lett. 2002;518:17–22. doi: 10.1016/s0014-5793(02)02621-2. [DOI] [PubMed] [Google Scholar]
- 15.Pingoud V, Conzelmann C, Kinzebach S, Sudina A, Metelev V, Kubareva E, Bujnicki JM, Lurz R, Lüder G, Xu S-Y, et al. PspGI, a type II restriction endonuclease from the extreme thermophile Pyrococcus sp.: structural and functional studies to investigate an evolutionary relationship with several mesophilic restriction enzymes. J. Mol. Biol. 2003;329:913–929. doi: 10.1016/s0022-2836(03)00523-0. [DOI] [PubMed] [Google Scholar]
- 16.Zaremba M, Sasnauskas G, Urbanke C, Siksnys V. Conversion of the tetrameric restriction endonuclease Bse634I into a dimer: oligomeric structure-stability-function correlations. J. Mol. Biol. 2005;348:459–478. doi: 10.1016/j.jmb.2005.02.037. [DOI] [PubMed] [Google Scholar]
- 17.Deibert M, Grazulis S, Sasnauskas G, Siksnys V, Huber R. Structure of the tetrameric restriction endonuclease NgoMIV in complex with cleaved DNA. Nat. Struct. Biol. 2000;7:792–799. doi: 10.1038/79032. [DOI] [PubMed] [Google Scholar]
- 18.Grazulis S, Deibert M, Rimseliene R, Skirgaila R, Sasnauskas G, Lagunavicius A, Repin V, Urbanke C, Huber R, Siksnys V. Crystal structure of the Bse634I restriction endonuclease: comparison of two enzymes recognizing the same DNA sequence. Nucleic Acids Res. 2002;30:876–885. doi: 10.1093/nar/30.4.876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bozic D, Grazulis S, Siksnys V, Huber R. Crystal structure of Citrobacter freundii restriction endonuclease Cfr10I at 2.15 A resolution. J. Mol. Biol. 1996;255:176–186. doi: 10.1006/jmbi.1996.0015. [DOI] [PubMed] [Google Scholar]
- 20.Bochtler M, Szczepanowski RH, Tamulaitis G, Grazulis S, Czapinska H, Manakova E, Siksnys V. Nucleotide flips determine the specificity of the Ecl18kI restriction endonuclease. EMBO J. 2006;25:2219–2229. doi: 10.1038/sj.emboj.7601096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Szczepanowski RH, Carpenter MA, Czapinska H, Zaremba M, Tamulaitis G, Siksnys V, Bhagwat AS, Bochtler M. Central base pair flipping and discrimination by PspGI. Nucleic Acids Res. 2008;36:6109–6117. doi: 10.1093/nar/gkn622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Golovenko D, Manakova E, Tamulaitiene G, Grazulis S, Siksnys V. Structural mechanisms for the 5'-CCWGG sequence recognition by the N- and C-terminal domains of EcoRII. Nucleic Acids Res. 2009;37:6613–6624. doi: 10.1093/nar/gkp699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou XE, Wang Y, Reuter M, Mücke M, Krüger DH, Meehan EJ, Chen L. Crystal structure of type IIE restriction endonuclease EcoRII reveals an autoinhibition mechanism by a novel effector-binding fold. J. Mol. Biol. 2004;335:307–319. doi: 10.1016/j.jmb.2003.10.030. [DOI] [PubMed] [Google Scholar]
- 24.Barik S. Site-directed mutagenesis by double polymerase chain reaction. Mol. Biotechnol. 1995;3:1–7. doi: 10.1007/BF02821329. [DOI] [PubMed] [Google Scholar]
- 25.Zaremba M, Sasnauskas G, Urbanke C, Siksnys V. Allosteric communication network in the tetrameric restriction endonuclease Bse634I. J. Mol. Biol. 2006;363:800–812. doi: 10.1016/j.jmb.2006.08.050. [DOI] [PubMed] [Google Scholar]
- 26.Leslie A. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography. 1992;26 [Google Scholar]
- 27.Leslie AGW. The integration of macromolecular diffraction data. Acta Crystallogr. D Biol. Crystallogr. 2006;62:48–57. doi: 10.1107/S0907444905039107. [DOI] [PubMed] [Google Scholar]
- 28.Evans P. Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 29. CCP4 (Collaborative Computational Project, N. 4). (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D. Biol. Crystallogr., 50, 760–763. [DOI] [PubMed]
- 30.Vagin AA. MOLREP: an automated program for molecular replacement. J. Appl. Cryst. 1997;30:1022–1025. [Google Scholar]
- 31.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 32.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 33.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 34.Kraulis P. Molscript—a program to produce both detailed and schematic plots of protein structures. J. Appl. Cryst. 1991;24:946–950. [Google Scholar]
- 35.Merritt EA, Murphy ME. Raster3D Version 2.0 A program for photorealistic molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 1994;50:869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
- 36.Lavery R, Sklenar H. Defining the structure of irregular nucleic acids: conventions and principles. J. Biomol. Struct. Dyn. 1989;6:655–667. doi: 10.1080/07391102.1989.10507728. [DOI] [PubMed] [Google Scholar]
- 37.Lu X-J, Olson WK. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu X-J, Olson WK. 3DNA: a versatile, integrated software system for the analysis, rebuilding and visualization of three-dimensional nucleic-acid structures. Nat. Protoc. 2008;3:1213–1227. doi: 10.1038/nprot.2008.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ulyanov NB, Zhurkin VB. Sequence-dependent anisotropic flexibility of B-DNA. A conformational study. J. Biomol. Struct. Dyn. 1984;2:361–385. doi: 10.1080/07391102.1984.10507573. [DOI] [PubMed] [Google Scholar]
- 40.Suzuki M, Yagi N, Finch JT. Role of base-backbone and base-base interactions in alternating DNA conformations. FEBS Lett. 1996;379:148–152. doi: 10.1016/0014-5793(95)01506-x. [DOI] [PubMed] [Google Scholar]
- 41.Olson WK, Gorin AA, Lu XJ, Hock LM, Zhurkin VB. DNA sequence-dependent deformability deduced from protein-DNA crystal complexes. Proc. Natl Acad. Sci. USA. 1998;95:11163–11168. doi: 10.1073/pnas.95.19.11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fujii S, Kono H, Takenaka S, Go N, Sarai A. Sequence-dependent DNA deformability studied using molecular dynamics simulations. Nucleic Acids Res. 2007;35:6063–6074. doi: 10.1093/nar/gkm627. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.