Abstract
In most organisms, the primary function of homologous recombination (HR) is to allow genome protection by the faithful repair of DNA double-strand breaks. The vital step of HR is the search for sequence homology, mediated by the RAD51 recombinase, which is stimulated further by proteins mediators such as the tumor suppressor BRCA2. The biochemical interplay between RAD51 and BRCA2 is unknown in Leishmania or Trypanosoma. Here we show that the Leishmania infantum BRCA2 protein possesses several critical features important for the regulation of DNA recombination at the genetic and biochemical level. A BRCA2 null mutant, generated by gene disruption, displayed genomic instability and gene-targeting defects. Furthermore, cytological studies show that LiRAD51 can no longer localize to the nucleus in this mutant. The Leishmania RAD51 and BRCA2 interact together and the purified proteins bind single-strand DNA. Remarkably, LiBRCA2 is a recombination mediator that stimulates the invasion of a resected DNA double-strand break in an undamaged template by LiRAD51 to form a D-loop structure. Collectively, our data show that LiBRCA2 and LiRAD51 promote HR at the genetic and biochemical level in L. infantum, the causative agent of visceral leishmaniasis.
INTRODUCTION
DNA double-strand breaks (DSBs) are one of the most cytotoxic lesions that can occur in the genomic DNA. In human cells, these lesions are repaired by non-homologous end joining (NHEJ) but also homologous recombination (HR). In this context, HR protects the genome from DNA translocations or rearrangements. Several studies have demonstrated a role for RAD51 and BRCA2 in both mitotic and meiotic recombination. BRCA2 plays a pivotal role in controlling the function and localization of RAD51 during HR, a central process for the maintenance of genome stability and cancer prevention (1). The BRC repeats located at the center of BRCA2 confer a very sophisticated mode of regulation. Indeed, the BRC repeats bind RAD51 and control RAD51 nucleoprotein filament formation in conjunction with the BRCA2 DNA-binding domain, whereas a BRC-unrelated RAD51-binding motif located at the C-terminus of BRCA2 appears to stabilize the filament (2–5). BRCA2 contains also a characteristic OB (oligonucleotide/oligosaccharide-binding) pocket, which consists of a highly curved five-stranded β-sheet that closes on itself to form a β-barrel, which confers single-strand DNA (ssDNA)-binding activity (6,7). Orthologs of BRCA2 have been found in eukaryotes such as human, mouse, rat, Xenopus, Caenorhabditis elegans, Trypanosoma and Leishmania but this protein is absent from yeast, archaea and bacteria (8). Saccharomyces cerevisiae does not bear a BRCA2 homolog, a function that might be replaced by RAD52. However, RAD52 orthologs have not been detected in Leishmania infantum, although these enzymes have a central role in recombination and DNA repair (9). Budding yeast or human RAD52 promote single-strand annealing of complementary ssDNA independent of RAD51 (10,11) and enhance RAD51-mediated strand invasion by direct interaction with the recombinase RAD51 (12). Although the NHEJ factors Ku70 and Ku80 have been identified in trypanosomes (13,14), it is not clear whether these human parasites possess a NHEJ activity as no DNA ligase IV and XRCC4 homologs were detected within their genomes (15).
The most common view in the control of the HR process is that HR should be suppressed up to a level where genomic stability is maintained. However, organisms such as Trypanosoma brucei use HR as a beneficial mechanism for antigenic variation (16) or Leishmania for drug resistance (17). First, it has been assumed that HR and DSB formation play an important role in antigenic variation. Variant surface glycoprotein (VSG) switching allows some of the infecting T. brucei to evade the host immune response, therefore allowing survival of the parasite and new transmission to another mammal. Analysis in T. brucei revealed that VSG switching frequency is highly dependent on RAD51 and BRCA2 proteins. Moreover, it was shown that the induction of a DSB adjacent to the 70-bp repeats upstream of the transcribed VSG induced VSG switching. The switching frequency was increased 250-fold compared to control cells without an I-SceI recognition sequence. Interestingly, VSG switching occurred through break-induced replication (16). Hence, understanding the biochemical role of BRCA2 in HR might be beneficial in understanding trypanosomatid diseases. Moreover, L. infantum amplify regions of its genome upon drug selection by HR between homologous repeated sequences (17).
The large size of the human BRCA2 protein (384 kDa) poses a great technical challenge for biochemical analyses. Amongst all BRCA2 proteins, Brh2 of Ustilago maydis, has been extensively characterized because of its smaller size (127 kDa). Brh2 binds D-loops specifically (18) and promotes strand invasion in an homologous duplex (19). Brh2 facilitates the second-end capture step in the DNA DSB model, by enhancing the annealing of a second complementary single-strand to the displaced strand of a D-loop to form a duplexed D-loop (20). It was recently reported that the human BRCA2 protein forms rings on DNA (21), binds to approximately six RAD51 molecules and promotes RAD51 filament formation on RPA-covered ssDNAs (22). In addition, it stimulates RAD51-mediated DNA strand exchange (23). To date, however, D-loop formation was not performed for the human BRCA2 protein (21–23) and evidence of human BRCA2 being able to stimulate RAD51-mediated D-loop formation is lacking.
The Rad51 gene from the parasite Leishmania major has been identified previously (24). Comparative bioinformatic analyses revealed that the size of the L. infantum BRCA2 protein was about three times smaller (125 kDa) than its human counterpart. Moreover, our analyses revealed that LiBRCA2 possesses key features of the BRCA2 family. We exploited the smaller size of the Leishmania BRCA2 protein to better understand its function in HR, along with the LiRAD51 recombinase.
MATERIALS AND METHODS
Nucleic acids
The L. infantum Brca2 and Rad51 genes were amplified by PCR from genomic DNA as protein-encoding genes are intronless in Leishmania. The sequences were identical to the NCBI reference sequences A4HYJ9 and A4I3C9, respectively. The LiBrca2 PCR fragment was obtained with the combination of primers JYM1599 and JYM1600 (Supplementary Table S1) and the purified PCR fragment was cloned in a modified pFASTBAC1 plasmid (Invitrogen) encoding GST and His tags to yield the GST–LiBRCA2–HIS construct. Similarly, LiRad51 was amplified with primers JYM1669 and JYM1670 (Supplementary Table S1) then cloned in a modified pFASTBAC1 plasmid (Invitrogen) encoding GST to generate the LiRAD51–GST construct. LiBRCA2–GFP was obtained following the amplification of LiBrca2 with primers JYM1894 and JYM1896 (Supplementary Table S1) and the resulting PCR product was cloned in the Leishmania expression vector pSPαHYGαGFP. Finally, the LiRAD51–DsRed version was generated with primers JYM1899 and JYM1900 (Supplementary Table S1), and the purified PCR product was cloned in the pSP72αNEOα-DsRed vector (constructed by inserting DsRed in pSP72αNEOα (25)).
Generation of the LiBrca2 null mutant
To generate a single knockout of LiBrca2 (e.g. LinJ.20.0070 in geneDB see http://www.genedb.org/), the entire ORF of LinJ.20.0070 was substituted by a neomycin phosphotransferase cassette flanked by 5′- and 3′-regions of LinJ.20.0070. The upstream LiBrca2 region was amplified with primers MOU1001 and MOU1002 (Supplementary Table S1), and the LiBrca2 downstream region was obtained with primers combination MOU1003 and MOU1004 (Supplementary Table S1). Targeting flanks were amplified from L. infantum genomic DNA and ligated to the NEO marker gene as previously reported (26). An insertional inactivation strategy was performed to target the second LiBrca2 allele. A polypyrimidine stretch (Y)-hygromycin-a-tubulin fragment (YHYGa) was obtained by PCR with the primers MOU1005 and MOU1006 of the Leishmania expression vector psp72Yhygroa (27). The YHYGa purified fragment was then fused with the 5′- and 3′-flanking regions of LiBrca2 after their respective PCR amplification with primers MOU1007 and MOU1008 (upstream region) and MOU1009 and MOU1010 (downstream region) (Supplementary Table S1). The final targeting cassette was inserted into the ORF of LinJ.20.0070 by HR. For episomal complementation of LinJ.20.0070, the LiBrca2 gene was amplified using 1 ng of L. infantum genomic DNA, with two primers containing either XbaI (MOU 1011) or SalI (MOU1012) (Supplementary Table S1) restriction sites. The PCR product was first cloned in the pGEM-T Easy vector then the construct was digested with both restriction enzymes. The LiBrca2 fragment was subcloned into the Leishmania expression vector pSP72αZEOα (28) within the XbaI and SalI cloning sites. All constructs were confirmed by DNA sequencing. The empty vector pSP72αZEOα was transfected in L. infantum wild-type cells (LiWT) and in the LinJ.20.0070 null mutant (LinJ.20.0070−/−) and these cells were used as controls.
Recombinants and transfectants were selected as following: 5 µg of both linear inactivation cassettes (neo or hyg) were obtained by PCR amplification, purified and transfected into promastigotes by electroporation as previously reported (25). For plasmid transfection (episomal complementation or empty vector), 10–20 µg was used. Recombinants and transfectants were selected in the presence of 300 µg/ml hygromycin B, 40 µg/ml G418 (Geneticin, Gibco-BRL) and 800 µg/ml zeocin (Invitrogen) depending on the marker used. After 2–3 passages, cells resistant to the antibiotic selection were cloned on SDM–agar plates (1%) in the presence of 20 µg/ml of G418 or 160 µg/ml of hygromycin B. Clones were picked up after 7–10 days and used for analysis. PCR analysis of three different clones after integration of the HYG and NEO cassettes was done using primers pairs a + b, c + d, e + f respectively MOU1007 + MOU1110, MOU1113 + MOU1114, MOU1115 + MOU1116 (Supplementary Table S1).
Southern blot analysis
Genomic DNA of the selected clones was isolated using DNAzol as recommended by the manufacturer (Invitrogen). Digested genomic DNAs with ApaI and ClaI were subjected to Southern blot hybridization with [α-32P]dCTP-labeled DNA probes according to standard protocols (29). All probes were obtained by PCR from Leishmania genomic DNA or from plasmids containing markers then purified appropriately before being labeled.
Transformation efficiency
We used the construct 5′-GSH1-BLA-3′GSH1 (26) to target the single copy gene GSH1 on chromosome 18 and confer blasticidin resistance in order to monitor the integration efficiency. Briefly, 4 × 107 cells of WT pSP72αZEOα, LinJ.20.0070−/− pSP72αZEOα, and LinJ.20.0070−/− pSP72αZEOα encoding LiBRCA2 were transfected with 5 µg of the linear 5′GSH1-BLA-3′GSH1 construct and after 24 h following electroporation, cells were preselected with 40 µg/ml blasticidin. After another 24 h cells were plated on SDM–agar plates (1% agar and 100 µg/ml blasticidin). LiWT cells and complemented cells were plated at a density of 5 × 106 cells/plate and the null mutant LinJ.20.0070 −/− at 7 × 106 cells. All these strains were also transfected with an empty plasmid (pSPαBLAαGSH1) as a control. Colonies were counted after 10–15 days of plating. Genomic DNA was extracted from selected clones using the DNAzol method (Invitrogen) and integration of the BLAST marker gene (BLA) into the GSH1 chromosomal locus was confirmed by PCR (primers MOU1117 and MOU1118) (Supplementary Table S1).
Western blotting of LiRAD51 and LiBRCA2 in Leishmania protein extracts
Leishmania infantum promastigotes were grown in SDM–79 medium supplemented with 10% heat-inactivated fetal bovine serum, 5 μg/ml hemin and 5 μM biopterin at pH 7.0, 28°C up to the logarithmic phase (OD600 nm ∼ 0.5). Cultures containing 5 × 106 cells resuspended in 5 ml medium were prepared with 12 μg/ml of phleomycin and incubated at 28°C for 0, 24, 48, 72 and 96 h. At each time point, the cells were centrifuged at 3000 rpm for 5 min, washed once with HEPES/NaCl buffer (21 mM HEPES, pH 7.2, 137 mM NaCl, 5 mM KCl, 0.7 mM Na2HPO4, 6 mM glucose) then resuspended in 30 µl of 2× Laemmli buffer. The protein samples (20 µl) were loaded onto a 8% SDS–PAGE and analyzed by western blot using the indicated antibodies. LiBRCA2 and hRAD51 antibodies were generated in the laboratory and were used at 1:5000 concentration whereas the α-tubulin antibody was purchased from Abcam and used at 1: 2500 dilution.
Protein sequence analysis
The domains presented in the Supplementary Figure S1 were identified using pfam with an E-value cutoff of 10 (30). The phylogenetic tree was generated by first gathering BRC repeat sequences for each different species. The sequences corresponding to the BRC repeat domain in each protein was extracted and numbered in function of their location relative to the N-terminal of the protein so the closest domain in N-terminal is assigned number 1. The alignment used in Supplementary Figure S1B is the alignment reported by hmmsearch when aligning sequence to the model. The phylogenetic tree of Supplementary Figure S1B was generated within Jalview using Neighbor-joining with the Blosum 62 scoring matrix (31). The tree reported in Supplementary Figure S1C was generated on a selected portion of global multiple alignment produced using the software MAFFT in FFT-NS-i mode (32,33). We selected the portion located just before OB1 (human starting amino acid S2590). The tree building option on the MAFFT website was used to produce the tree of Supplementary Figure S1C using the Neighbor-joining method with JTT substitution model without heterogeneity among sites and 1000 bootstraps.
Fluorescence microscopy
Promastigote cells expressing the green fluorescence protein (GFP)-fusion proteins or DsRed-fusion proteins were first washed with HEPES/NaCl buffer then incubated 10 min with 0.5 µg/ml DAPI (Sigma) in the dark. The cells were washed twice with HEPES/NaCl buffer and immobilized in HEPES/NaCl buffer containing 10% glycerol. Live parasites were mounted on microscope slides with coverslips sealed with nail varnish and air-dried for 15 min. The slides were visualized by epifluorescence microscopy. Bright field and fluorescence images were taken using a Nikon Eclipse TE300 inverted microscope equipped with a Photometrics coolSNAPfx camera. Visualization of the DAPI, GFP and DsRed fluorophores were achieved using respectively 340/380 nm, 460/500 nm, 532/588 nm excitation filters and 435/485 nm, 510/560 nm, 607/683 nm emission filters with a 100× objective. The images were processed using ImageJ version 1.42 (National Institutes of Health, USA; http://rsb.info.nih.gov/ij).
Purification of human RPA, human RAD51 and L. infantum RAD51 and BRCA2
The human RPA protein was purified according to (34), and human RAD51 was purified as described in (35). GST–LiBRCA2–His was purified from baculovirus-infected Sf9 cells. Sf9 insect cells (1 l culture) were infected with the BRCA2 baculovirus for 3 days at 27°C. The cell pellet (600 ml) was resuspended in 18 ml of PBS washing buffer [300 mM NaCl, 1 mM EDTA, 0.05% Triton, 1 mM DTT in 1× PBS (GIBCO)] containing protease inhibitors PMSF (1 mM), aprotinin (0.019 UIT/ml) and leupeptin (1 µg/ml). The suspension was lysed using a Dounce homogenizer (15 strokes), sonicated three times for 30 s on ice. Insoluble material was removed by centrifugation in small aliquots of 1 ml (10 min, 13 000 rpm, 4°C). An amount of 1 ml of glutathione sepharose beads (GE Healthcare) was added to the combined supernatant and incubated during 1 h at 4°C. The beads were washed two times with 1× PBS then incubated for 30 min in HSP buffer (5 mM ATP, 15 mM MgCl2 in 1× PBS) at 4°C. The beads were further washed two times with 1× PBS containing 500 mM NaCl and one more time in the Talon buffer (50 mM NaHPO4 pH 7.0, 500 mM NaCl, 10% glycerol, 0.05% Triton, 5 mM imidazole). The LiBRCA2 protein was eluted by cleavage with PreScission protease (60 U/ml, GE Healthcare) during 3 h at 4°C in the Talon buffer. The supernatant was added on 200 µl of Talon resin (Clontech) and incubated for 30 min at 4°C. The resin was washed two times with 4 ml of Talon buffer containing 30 mM imidazole and one time with 80 mM imidazole. LiBRCA2 protein was eluted in buffer containing 500 mM imidazole and dialyzed in storage buffer (20 mM Tris–Acetate, pH 8.0, 200 mM KOAc, 10% glycerol, 1 mM EDTA, 0.5 mM DTT). The GST–LiRAD51 protein was purified as for GST–LiBRCA2–His but excluding the Talon purification step. The beads were washed in PreScission washing buffer (50 mM Tris–HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 1 mM DTT, 0.05% Tween 20) before cleavage. The LiRAD51 protein was eluted following an overnight cleavage at 4°C with 60 U/ml of PreScission protease (GE Healthcare) in PreScission washing buffer.
Gel-filtration analysis
The molecular mass of LiRAD51 (20 µg) and LiBRCA2 (5 µg) were determined by gel filtration using 250 µg of a series of protein standards; bovine thyroglobulin (670 kDa), bovine gamma globulin (158 kDa), chicken ovalbumin (44 kDa), horse myoglobin (17 kDa) and vitamin B-12 (1.35 kDa). Purified proteins were analyzed on a FPLC Explorer 10 system fitted with a 24 ml Superdex 200 PC 3.2/30 column (Pharmacia) equilibrated in gel-filtration buffer (25 mM Tris–HCl pH 7.5, 150 mM KCl, 5% glycerol, 1 mM DTT). Fractions (500 µl) were collected and analyzed by SDS–PAGE followed by western blotting with a monoclonal anti-Histidine antibody (BD Biosciences) for LiBRCA2 or a polyclonal anti-human RAD51 to detect LiRAD51.
D-loop assays
The D-loop assay with 3′-tail DNA was conducted essentially as described (36). A linear duplex DNA was prepared by restriction digestion of pPB4.3 DNA (4.3 kb) (37) with SmaI. Linear DNA with 3′-tails (∼200 nt in length) were generated by treatment of linearized pPB4.3 DNA (100 µg) with T7 gene 6 exonuclease (830 units, 1 min at 25°C) in 800 μl reaction volume containing 20 mM Tris–Acetate pH 7.9, 50 mM KOAc pH 7.9, 10 mM Mg(OAc)2 and 1 mM DTT. Exonuclease digestion was stopped by extraction with phenol/chloroform. Finally, the resected DNA was purified by gel electrophoresis through a 0.8% (w/v) agarose gel and recovered by electroelution. 32P-labeled 3′-tailed DNA substrate (1 µM nucleotides) was incubated for 5 min at 37°C with the indicated concentration of LiRAD51 and/or LiBRCA2 in 25 mM Tris–Acetate pH 7.5, 1 mM DTT, 20 mM creatine phosphate, 5 U/ml phosphocreatine kinase, 1 mM ATP and 0.5 mM MgCl2 in 10 µl. CsCl-purified pPB4.3 replicative form I DNA (300 µM) was added and the reaction was incubated for 1 h 30 min. The reaction was quenched with the addition of one-fifth volume of stop buffer (20 mM Tris–Cl pH 7.5 and 2 mg/ml proteinase K) followed by a 30 min incubation period at 37°C. Labeled DNA products were analyzed by electrophoresis through a 0.8% TAE 1× agarose gel, run at 65 V, dried onto DE81 filter paper and visualized by autoradiography.
DNA-binding assays
Reactions (10 µl) contained 100 nM of 32P-labeled DNA oligonucleotides (e.g. primer JYM925 for the ssDNA assay and primers JYM925/JYM945 for the double-strand DNA (dsDNA) assay, see Supplementary Table S1 for oligonucleotide sequences) and LiBRCA2 and/or LiRAD51, at the indicated concentrations, in binding buffer (20 mM Triethanolamine–HCl pH 7.5, 50 mM NaCl, 2 mM MgCl2, 2 mM ATP and 1 mM DTT). Proteins were added to reactions and incubated at 37°C for 10 min, followed by 15 min of fixation with 0.2% glutaraldehyde. Reactions were loaded onto a 0.8% TBE 1× agarose gel.
Single-strand annealing reactions
Reactions (10 µl) contained denatured 32P-labeled dsDNA (50 nM) with LiRAD51 or/and LiBRCA2, at the indicated concentrations, in HEPES buffer (20 mM HEPES pH 7.5, 5 mM Mg(CH3COO)2, 2 mM ATP, 5 mM DTT and 100 mM NaCl). Incubation was performed at 20°C for 10 min. The reaction products were deproteinized by the addition of one-fifth volume of stop buffer (20 mM Tris–Cl pH 7.5 and 2 mg/ml proteinase K) followed by a 10 min incubation period at 20°C. Labeled DNA products were analyzed by electrophoresis (8% PAGE gel with TBE1X buffer, run at 150 V for 3 h), the gel was dried onto DE81 filter paper and visualized by autoradiography.
Immunoprecipitation
Immunoprecipitation from purified proteins were performed as follows. Purified LiBRCA2 (750 ng) and hRPA (1 µg) were incubated in 100 μl of IPR buffer (20 mM Tris–acetate pH 8.0, 125 mM KOAc, 0.5 mM EDTA, 0.5% NP40, 10% glycerol, 0.1 mg/ml BSA) for 30 min at 37°C. An amount of 1 μg anti-Histidine (Clontech) was added for 30 min at 4°C followed by the addition of 15 μl protein A/G sepharose beads (Pierce) for 30 min. Immunoprecipitates were washed four times in IPR buffer containing 300 mM KOAc and visualized by western blotting using the indicated antibodies.
Sf9 insect cells (20 ml) were coinfected with the LiBRCA2 and LiRAD51 baculoviruses (dilution 1/150) for 2 days at 27°C. The cell pellet was resuspended in 5 ml of IP100 buffer (20 mM Tris–Acetate pH 8.0, 100 mM KOAc, 0.5 mM EDTA, 0.5% NP40, 10% glycerol, 0.1 mg/ml BSA, 1 mM DTT) containing protease inhibitors PMSF (1 mM), aprotinin (0.019 UIT/ml) and leupeptin (1 µg/ml). The suspension was sonicated two times for 10 s. Insoluble material was removed by centrifugation in small aliquots of 1 ml (10 min, 13 000 rpm, 4°C). 2 µg anti-Histidine (Clontech) was added to 1 mg of protein and incubated during 2 h at 4°C followed by the addition of 15 μl protein A/G sepharose beads (Pierce) for 1 h at 4°C. Immunoprecipitates were washed four times in the IP100 buffer with the indicated concentration of KOAc and visualized by western blotting using the indicated antibodies.
Biotin pull-down assay
Magnetic beads (Roche) containing 5′-biotinylated ssDNA polydT 83-mer (corresponding to the final concentration of 1 μM nucleotide) were pre-incubated with a blocking buffer (25 mM Tris–acetate pH 7.5, 40 mM KOAc, 1 mM DTT, 0.02% Igepal and 10 mg/ml BSA) for 20 min at 37°C. The beads were captured with the magnetic particle separator, washed once with DC buffer (25 mM Tris–Acetate pH 7.5, 40 mM KOAc, 1 mM DTT, 0.02% Igepal, 1 mg/ml BSA, 5 mM ATP and 0.5 mM MgCl2) and resuspended in the same buffer. hRPA was added (17 μl) and the reaction was incubated for 5 min at 37°C. Then LiRAD51 (200 nM) was added and the reaction was incubated for 5 min at 37°C. Finally, the indicating concentration of LiBRCA2 protein was added (in 20 μl) and the reaction was incubated further for 5 min at 37°C. The beads were captured and were washed twice with 50 μl of washing buffer (25 mM Tris–Acetate pH 7.5, 40 mM KOAc, 1 mM DTT, 0.02% Igepal, 5 mM ATP and 0.5 mM MgCl2). Finally, 15 μl of 2× Laemmli buffer was added and beads were heated 5 min at 95°C. The beads were captured again and the supernatants were loaded onto a 8% SDS–PAGE Ruby (Invitrogen) and visualized by UV.
RESULTS
Bioinformatic analyses of LiBRCA2
Analysis of the Leishmania genome suggested that LinJ20.0070 is a BRCA2 ortholog. We scrutinized whether LiBRCA2 possessed features reminiscent of the BRCA2 family of enzymes. We started by first detecting all the significant protein domains within the selected list of BRCA2 proteins using pfam (30) with a permissive E-value. We detected two BRC repeats, one OB-fold and also at least one putative zf–RanBP (Zn-finger in Ran-binding protein) domain for every Leishmania species analyzed (Supplementary Figure S1B). We also detected 15, 2, 7, 8 and 4 BRC repeats for T. brucei, Trypanosoma cruzi, mouse, human and Arabidopsis, respectively. The OB-fold domain could be detected in all proteins analyzed. The helical domain could only be detected in mouse, human and Arabidopsis and the Tower domain is only present in mouse and human.
Since the BRC repeats are present in all the BRCA2 proteins, we were interested to study the evolutionary link between the BRC repeats of all the different species. For this purpose we extracted the BRC repeats detected by pfam and aligned them to produce a phylogenetic tree (Supplementary Figure S1A). All the first BRC repeats of Leishmania are clustering together with all the BRC repeats of T. brucei or cruzi. The second BRC repeats of Leishmania are forming a cluster with the two first BRC repeats of Arabidopsis. The two BRC repeats of Leishmania are within a cluster also composed of mouse and human BRC 3, 4, 7 and 8. We hypothesized that the two BRC repeats of all Leishmania species analyzed (Supplementary Figure S1A and S1B) are related to the mouse and human BRC repeats 3, 4, 7 and 8. Furthermore, we analyzed globally the evolution of the BRCA2 proteins with a phylogenetic tree produced from an alignment of all the BRCA2 proteins. We can observe from Supplementary Figure S1C that the L. major and L. donovani are the most similar form of BRCA2 followed by L. infantum and L. braziliensis. Collectively, these bioinformatic analyses strongly suggested that L. infantum possesses a BRCA2 homolog at amino acid and phylogenic levels. A schematic representation of LiBRCA2 in comparison to human BRCA2 is depicted in Figure 1A.
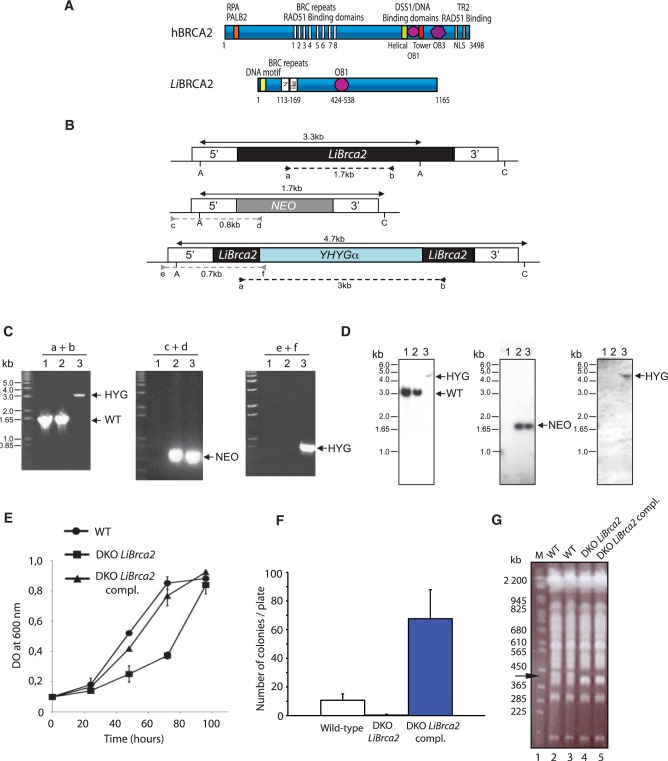
Figure 1.
Inactivation of LiBrca2 and phenotypic analyses. (A) Schematic representation of LiBRCA2 in comparison to human BRCA2. BRC repeats, OB-folds, and RAD51-binding regions are shown. (B) Schematic representation of the LiBrca2 chromosomal locus (top) and inactivation cassettes with the neomycin phosphotransferase (Neo, middle) and hygromycin phosphotransferase (Hyg, bottom) markers. The position of the primers pairs (a + b, c + d, e + f) used in (C) is depicted. (C) PCR analysis of three different clones after integration of the HYG and NEO cassettes. Clone in lane 1 is identical to L. infantum WT (e.g. no integration), clone in lane 2 has a NEO insertion in one allele, and clone in lane 3 has both NEO/HYG insertions. (D) Southern blots of three different clones digested with ApaI (A) and ClaI (C) after hybridizing with a 700 bp specific LiBrca2 (left), NEO (middle) and HYG probes (right) on the DNA isolated from the three clones described in (C). (E) Growth retardation of LiBrca2 null mutants grown as promastigotes. Growth of L. infantum WT (closed circles), DKO LiBrca2 (closed squares), and DKO LiBrca2 complemented with a plasmid expressing LiBRCA2 (closed triangles), was measured over time. (F) Inactivation of LiBrca2 impairs gene targeting. Gene targeting was measured in L. infantum WT, DKO LiBrca2 and DKO LiBrca2 complemented with a plasmid expressing LiBRCA2. (G) DKO LiBrca2 cells display genomic instability. Pulsed field gel electrophoresis was performed on L. infantum WT cells (lanes 2 and 3), DKO LiBrca2 (lane 4) and DKO LiBrca2 complemented with a plasmid expressing LiBRCA2 (lane 5). The arrow shows a chromosomal band present in WT cells but absent in the other strains. Molecular weight markers (BioRad, S. cerevisiae DNA size standard) were used in lane 1.
Inactivation of LinJ.20.0070, the Leishmania BRCA2 gene
Having established that L. infantum contains an homolog of BRCA2, we generated a LiBrca2 null mutant. The BRCA2 ortholog LinJ.20.0070 is a single copy gene. In an attempt to inactivate LinJ.20.0070, one gene inactivation cassette with neomycin phosphotransferase (NEO) and one gene disruption cassette with hygromycin phosphotransferase (HYG) marker (Figure 1B) were prepared. Linear NEO or HYG fragments were transfected into L. infantum and recombination events of clones selected on agar plates were followed by PCR reactions and Southern blot hybridizations (Figure 1C and D). Digestion with ApaI and ClaI should yield a 3.3-kb band in WT, while replacing one allele with NEO should yield a 1.7-kb fragment hybridizing to a 3′-UTR probe. Insertional inactivation with YHYGα into the second LinJ.20.0070 allele of LinJ.20.0070/NEO cells and hybridizing with the same probe should yield a single 4.7-kb band. Five clones from each independent transfection were selected. The DNA of one such representative LinJ.20.0070/NEO (LinJ.20.0070+/−) and NEO/HYG (LinJ.20.0070−/−) clone was isolated, analyzed by PCR using primers (a–f) (Figure 1B). The use of primers a and b should lead to a fragment of 1.7-kb in the WT and the single knockout LinJ.20.0070 should lead to a 3-kb fragment upon insertion of HYG (Figure 1B and C). The integration of NEO and HYG at their homologous locus were confirmed by primer pairs c + d and e + f, respectively. The DNA of these representative LinJ.20.0070/NEO and NEO/HYG clones were digested with ApaI and ClaI and analyzed by Southern blots after hybridizing with a 3′-UTR LinJ.20.0070 probe, NEO and HYG probes. Bands of the expected size hybridizing with all the probes were observed (Figure 1D, lanes 1–3), confirming the generation of a BRCA2 L. infantum null mutant, termed DKO LiBrca2.
Phenotypes of LiBrca2 null mutants
Although deletion of BRCA2 in mouse leads to an embryonic lethality phenotype (38), the LiBrca2 mutants were viable. LiBrca2 null mutants, adapted to in vitro culture, had a specific growth retardation even when grown as promastigotes in SDM medium when compared to wild-type cells (WT) (Figure 1E). This growth retardation was rescued upon the re-introduction of an episomal construct encoding BRCA2 (DKO LiBrca2 complemented). We have succeeded in generating an independent BRCA2 null mutant and observed the same growth defect (data not shown).
Since gene disruption in Leishmania is mediated by double crossed-over HR, we sought to determine whether the loss of BRCA2 would decrease the efficiency of HR. This was assayed by measuring the transformation efficiencies of promastigotes transfected with the linear construct 5′-GSH1-BLA-GSH1-3′ (26) which integrates a blasticidin deaminase cassette into the GSH1 locus on chromosome 18. There was at least a 10-fold decrease in the transformation frequency, as determined by colony numbering, when compared to WT (Figure 1F). Adding back an episomal BRCA2 increased the number of colonies at least to the wild-type levels and in one set of experiments to 6-fold increase in colony numbers. The number of episomal copies per cell can be variable, which might explain the differences between each experiment. The correct integration of the GSH1/BLAS cassette was tested by PCR in selected colonies (data not shown).
BRCA2 inactivation in mammalian cells causes accumulation of aberrant chromosomes, including gross chromosomal rearrangements occurring from chromosomal loss, breakage or translocations (39). The karyotype of the L. infantum BRCA2 null mutant was analyzed by pulsed field gel electrophoresis. Size variations were observed in chromosomes between 365 and 450 kb in DKO LiBrca2 (Figure 1G, lane 4) compared to WT cells (Figure 1G, lanes 2 and 3). Reintroduction of BRCA2 in the null mutant did not restore chromosome integrity (Figure 1G, lane 5).
LiRAD51 expression is modulated by DNA damage
To date, it is unclear to what extent HR proteins are expressed in L. infantum. The expression of LiBRCA2 and LiRAD51 was monitored over time using western blotting in mock- or phleomycin-treated cells (Figure 2A). To do this, we used antibodies raised against LiBRCA2 and human RAD51 that recognized LiBRCA2 and LiRAD51, respectively. LiRAD51 and hRAD51 are 74.5% similar and 63% identical at the amino acid level (Supplementary Figure S2). The proteins recognized by the antibodies were at the predicted molecular weight, 41 kDa for LiRAD51 and 125 kDa for LiBRCA2. The levels of LiBRCA2 did not fluctuate over time in cells treated with the DNA damaging agent phleomycin. However, the levels of LiRAD51 were vastly induced 96 h after DNA damage. This observation is in accordance to the induction of LmRAD51 expression in L. major promastigotes after phleomycin treatment (24).
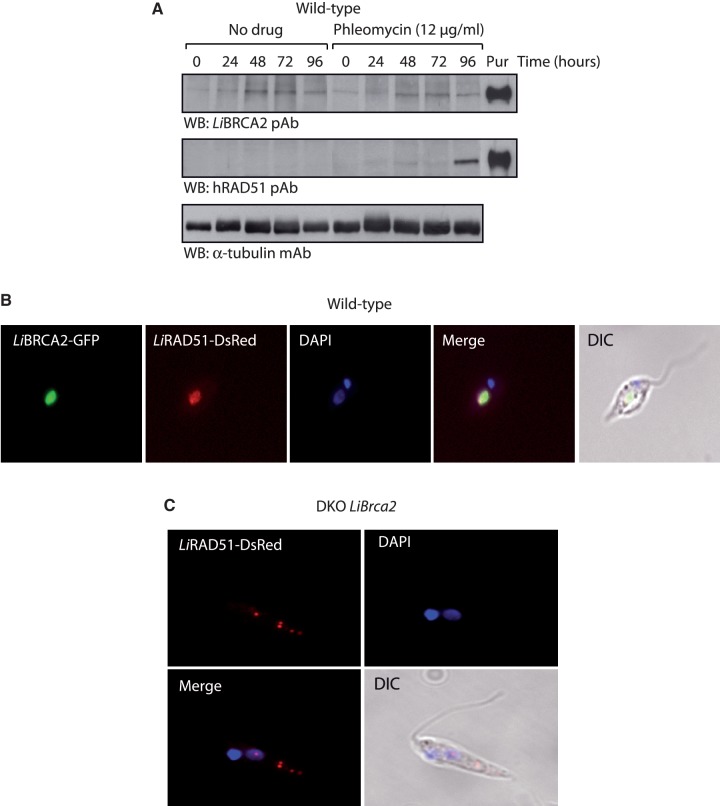
Figure 2.
Expression and cytological analysis of LiRAD51 and LiBRCA2. (A) Phleomycin treatment induces LiRAD51 expression in L. infantum WT cells. Lysates from either untreated (no drug) or phleomycin-treated L. infantum WT promastigotes were subjected to western blotting with anti-LiBRCA2, anti-hRAD51 or α-tubulin antibodies. Purified (Pur) LiBRCA2 or LiRAD51 (30 ng) are also shown. pAb, polyclonal antibody; mAb, monoclonal antibody. (B) Cytological analysis of LiBRCA2–GFP, LiRAD51–DsRed, in WT L. infantum in the absence of exogenous DNA damage. (C) Localization of LiRAD51–DsRed in DKO LiBrca2 in the absence of exogenous DNA damage. The kinetoplast (darker stained disk-shaped structure) and nucleus (lighter stained disk-shaped structure) are stained with DAPI.
In vivo localization of LiBRCA2
In human cells, RAD51 and BRCA2 are known to accumulate as foci at DNA damage sites. The formation of RAD51 foci is impaired in BRCA2-deficient cells. Furthermore, cell fractionation assays using CAPAN-1 cells (which contains BRCA2 6174delT, a truncated version), revealed that RAD51 is present in majority in the cytoplasm, rather than the nuclear fraction (2). We therefore investigated whether these functions are conserved in Leishmania species. To do this, we expressed LiRAD51 fused to DsRed and LiBRCA2 fused to GFP. When both LiBRCA2–GFP and LiRAD51–DsRed were expressed in WT Leishmania cells, both LiRAD51 and LiBRCA2 colocalized in the nucleus (Figure 2B and Supplementary Figure S3A). The expression of GFP or DsRed alone in L. infantum lead to a much different pattern of staining than LiBRCA2–GFP and LiRAD51–DsRed (Supplementary Figure S3B). Interestingly, LiRAD51 was no longer found in the nucleus in the absence of LiBRCA2, but in the cytosol as part of several punctuated stained regions (Figure 2C and Supplementary Figure S3C). These results show that LiBRCA2, like human BRCA2, control the nuclear accumulation of LiRAD51.
Expression and purification of L. infantum RAD51 and BRCA2
To open the door to biochemical studies of L. infantum RAD51 and BRCA2, we purified these proteins to homogeneity. First, the LiRAD51 and LiBRCA2 genes were amplified by PCR from L. infantum genomic DNA and cloned in pET16b vector. Following expression of LiBRCA2 in Escherichia coli, very low expression was achieved, and LiBRCA2 was highly degraded after Talon purification (data not shown). Therefore, we developed a two-step affinity purification procedure to purify full length LiBRCA2 without any degradation products. LiBrca2 gene was cloned in a modified pFASTBAC vector to add, respectively, a glutathione-S-transferase tag at the N-terminus and a 10-histidine tag at the C-terminus of the proteins. Recombinant baculoviruses were used to infect SF9 insect cells, followed by the purification of the recombinant BRCA2 protein. A cell lysate expressing GST–LiBRCA2–His was subjected to GST affinity purification followed by GST cleavage with PreScission protease and purification on Talon beads. Opposite to LiBRCA2, LiRAD51 was well expressed in insect cells and purified by GST affinity purification followed by PreScission cleavage of the GST tag. This led to >95% pure preparations of both LiBRCA2 and LiRAD51 (Figure 3A).
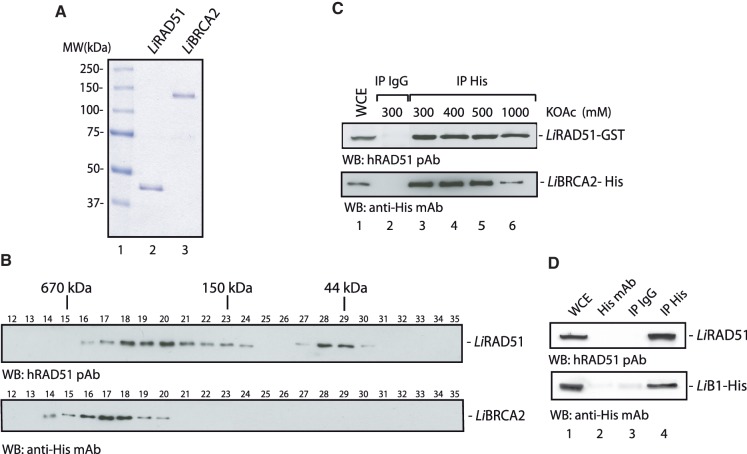
Figure 3.
Purification of L. infantum RAD51 and BRCA2. (A) SDS–PAGE of purified LiRAD51 and LiBRCA2. Purified proteins (400 ng) were loaded on a 8% SDS–PAGE, run then stained with Coomassie blue. Lane 1, molecular weight markers (Bio-Rad); lane 2, purified LiRAD51; lane 3, purified LiBRCA2. (B) Native molecular masses of purified LiRAD51 and LiBRCA2 as determined by gel filtration through Superdex 200 columns. Eluted fractions 12–35 are presented. Size standards (670, 150 and 44 kDa) are indicated. The upper panel was reacted with an anti-hRAD51 antibody and the bottom panel was revealed with an anti-His antibody. (C) LiBRCA2 interacts with LiRAD51. Sf9 cells were coinfected with LiRAD51–GST and LiBRCA2–His baculoviruses, followed by immunoprecipitation with IgG alone (lane 2) or anti-His antibody (lanes 3–6). The protein complexes were washed with buffer containing increasing salt (KOAc) concentration, as indicated. Eluted proteins were run onto an SDS–PAGE along with the whole cell extract (lane 1). Blots were reacted with an anti-hRAD51 (upper panel) or anti-His (bottom panel) antibodies. (D) The N-terminus of LiBRCA2 (LiBRCA2–B1) interacts with LiRAD51. Immunoprecipitations were conducted as described in (C) from Sf9 cells coinfected with LiRAD51–GST and LiBRCA2–B1-His baculoviruses. Lane 1, whole cell extract; lane 2, immunoprecipitations with an anti-His antibody alone; lane 3, immunoprecipitations with IgG alone; lane 4, immunoprecipitations with an anti-His antibody. Blots were reacted with anti-hRAD51 polyclonal antibody (upper panel) or with an anti-His monoclonal antibody (bottom panel).
The native molecular weight of purified LiBRCA2 and LiRAD51 were determined by gel filtration through Superdex 200 (Figure 3B). Similar to human RAD51 (2), two peaks were observed, in fractions 18–20 (corresponding to an averaged molecular weight of ∼350 kDa) and in fractions 28–29 (corresponding to monomeric LiRAD51). LiBRCA2 eluted in fractions 14–20, with a peak observed at fractions 17–18 indicative of multimeric forms. This is in agreement with the observation that human BRCA2 forms multimeric rings (21).
As LiBRCA2 mediates the recruitment of the recombinase LiRAD51 to the nucleus, we therefore tested the possibility that both proteins interact. Using coimmunoprecipitation assays, we observed a co-complex between LiRAD51 and LiBRCA2 (Figure 3C). The interaction between LiBRCA2 and LiRAD51 was highly resistant to salt washes (Figure 3C, lanes 3–6). It has been shown that BRC repeats are involved in the interaction between BRCA2 and RAD51. Leishmania infantum possesses two BRC repeats located at its N-terminus (Figure 1A and Supplementary Figure S1). To test whether LiRAD51 interacts with the N-terminus of LiBRCA2, the LiBRCA2 N-terminal domain (LiBRCA2–B1–His, amino acids 1–122) was coexpressed with LiRAD51 in insect cells, followed by immunoprecipitation analysis (Figure 3D). A co-complex between LiBRCA2–B1–His and LiRAD51 was observed, suggesting that the interaction between LiBRCA2 and LiRAD51 is mediated through the BRC repeats.
LiRAD51 binds DNA, promotes single-strand annealing and strand invasion
As a prerequisite for HR activities, RAD51 binds DNA. Escherichia coli RecA protein shows a high affinity for ssDNA compared with duplex DNA, while RAD51 protein binds both ssDNA and dsDNA (40). Using electromobility shift assays, we found that LiRAD51 showed similar DNA binding to hRAD51, binding 5'-32P-end-labeled single- and dsDNA with a better affinity for ssDNA (Figure 4A and B, compare lanes 3 and 4). Having established conditions under which LiRAD51 bound DNA, we next investigated their ability to promote single-strand annealing. As shown with Schizosaccharomyces pombe RAD51 (41), LiRAD51 promoted single-strand annealing of complementary ssDNAs of 60 nt. While low levels of spontaneous strand annealing was observed in the control alone (18.2%), LiRAD51 formed efficiently renatured DNA products (57.3%) (Figure 4C).
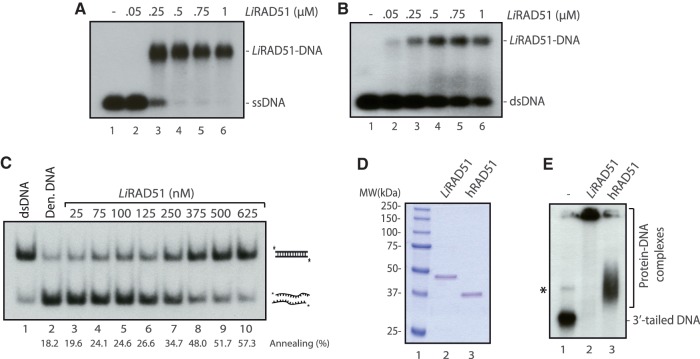
Figure 4.
DNA-binding activity of purified LiRAD51. (A) LiRAD51 binds ssDNA. Lanes 1–6, a P32-labeled-60-mer ssDNA was incubated with increasing concentrations of LiRAD51 (0–1000 nM). (B) LiRAD51 binds dsDNA. Lanes 1–6, a P32-labeled-60-mer dsDNA was incubated with increasing concentration of LiRAD51 (0–1000 nM). (C) LiRAD51 is proficient in single-strand annealing. Lane 1, purified 60-bp duplex DNA. Reactions contained denatured 60-bp duplex DNA in buffer with no protein (lane 2) or with purified LiRAD51 (25–625 nM), lanes 3–10. The percentage of annealing is indicated at the bottom. (D) SDS–PAGE of purified LiRAD51 (lane 2) and human RAD51 (lane 3). Molecular weight markers (Bio-Rad) were run in lane 1. Proteins (300 ng) were loaded on a 10% SDS–PAGE and stained with Coomassie blue. (E) LiRAD51 and human RAD51 show different 3′-tailed DNA-binding profiles. Reactions contained labeled 3′-tailed DNA with no protein (lane 1); with LiRAD51 (lane 2); or human RAD51 (lane 3). The asterisk represents annealed 3′-tailed molecules.
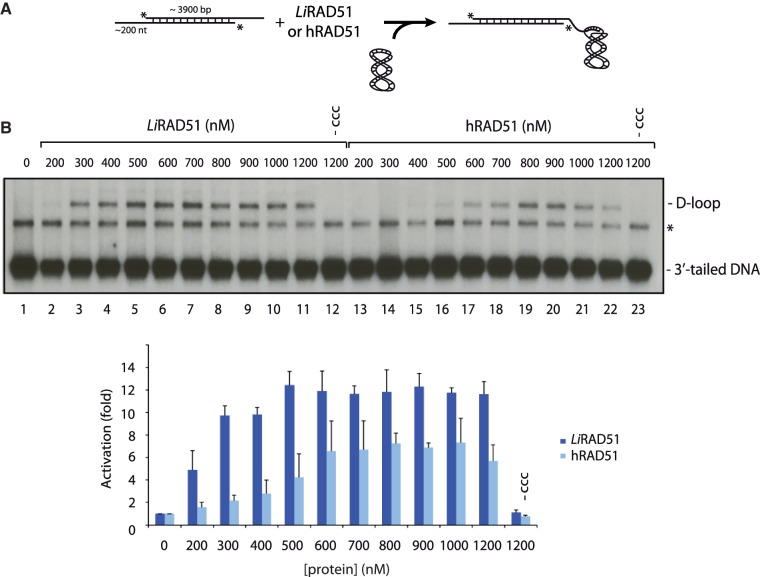
As it is the first time that LiRAD51 is analyzed biochemically, it was important to compare its relative activity to human RAD51. First, both proteins were quantified to insure that equal amounts of proteins were used in the following experiments (Figure 4D). Since recombination is thought to be initiated by DNA that contains single-stranded tails and these substrates would be subsequently used in D-loop assays, we analyzed the binding of both LiRAD51 and hRAD51 to these substrates. While hRAD51 formed complexes of relatively homogeneous size, we observed the formation of LiRAD51–DNA networks that tended to smear up the gel (Figure 4E). This result is characteristic of the formation of networks containing a variable number of protein and DNA molecules. A key step in HR is the formation of a D-loop structure, characterized by the invasion of an ssDNA substrate into a homologous duplex DNA (Figure 5A). D-loop assays were performed with long 3′-tailed DNA substrate and homologous supercoiled DNA in magnesium buffer. Adding purified LiRAD51 and human RAD51 (200–1200 nM) to these substrates led to an increase of D-loop products in a concentration dependent manner. Quantification of the reaction revealed that LiRAD51 was more efficient than human RAD51. At lower concentration (Figure 5B, compare lanes 3–5 and 14–16), LiRAD51 was at least 3-fold more efficient than hRAD51. These results show that LiRAD51 is a functional recombinase, homologous to hRAD51.
Figure 5.
Comparison between LiRAD51 and hRAD51 D-loop activities. (A) Schematic representation of the D-loop assay using a 4.3-kb resected dsDNA and supercoiled DNA templates. (B) Upper panel: D-loop reactions contained the 4.3-kb resected dsDNA with no protein (lane 1), or with purified LiRAD51 (200–1200 nM, lanes 2–11). Lane 12 contains LiRAD51 (1200 nM) without supercoiled DNA (-ccc). Reactions with human RAD51 (200–1200 nM, lanes 13–22); or human RAD51 (1200 nM) without supercoiled DNA (lane 23) are shown. Lower panel: quantification of results presented in fold activation compared to the control at each protein concentration (nM). The asterisk represents annealed 3′-tailed molecules.
LiBRCA2 controls LiRAD51 DNA binding
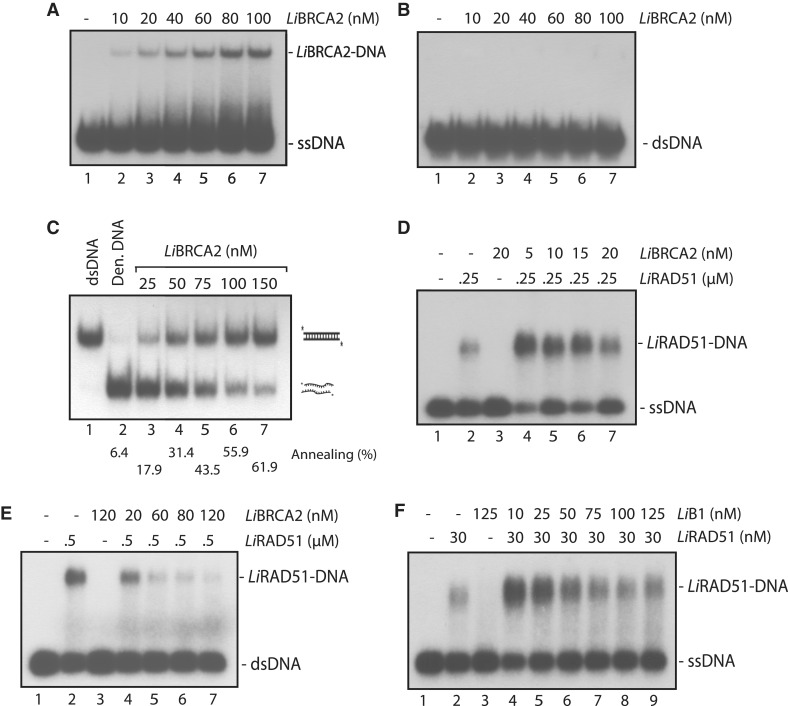
Next, we investigated the DNA-binding properties of LiBRCA2. Similar to hBRCA2 (21), LiBRCA2 bound ssDNA in a concentration-dependent manner, whereas it did not form stable complexes with dsDNA (Figure 6A and B). Interestingly, LiBRCA2 catalyzed the annealing of complementary ssDNAs (Figure 6C). Having established LiBRCA2 DNA-binding properties, we next tested whether LiBRCA2 affects the DNA-binding affinities of LiRAD51. Using conditions where LiBRCA2 binding to DNA was minimal (5–20 nM) we observed a significant increase in LiRAD51 binding on ssDNA upon the addition of LiBRCA2 (Figure 6D). In comparison, LiBRCA2 inhibited the loading of LiRAD51 on dsDNA (Figure 6E).
Figure 6.
DNA binding, annealing and interacting properties of LiBRCA2. (A) LiBRCA2 binds ssDNA preferentially over dsDNA. A 60-mer ssDNA was incubated with increasing concentration (0–100 nM) of LiBRCA2 (lanes 1–7). (B) LiBRCA2 does not bind 60-mer dsDNA. A 60-mer dsDNA was incubated with increasing concentration (0–100 nM) of LiBRCA2 (lanes 1–7) (C) LiBRCA2 promotes single-strand annealing. Lane 1, purified 60-bp duplex DNA. Reactions (lanes 2–7) contained denatured 60-bp duplex DNA in buffer with an increasing concentration of LiBRCA2 (0–150 nM), lanes 2–7. (D–F) LiBRCA2 stimulates and disrupts LiRAD51–dsDNA complexes. (D) LiBRCA2 (0–20 nM) was added to pre-assembled LiRAD51–ssDNA complexes (with LiRAD51 at a concentration of either 0 or 250 nM). (E) LiBRCA2 (0–120 nM) was added to pre-assembled LiRAD51–dsDNA complexes (with LiRAD51 at a concentration of either 0 or 500 nM). (F) The N-terminus of LiBRCA2 affects LiRAD51 DNA-binding activity. Reactions (lanes 1–9) were conducted as in (D), but using none or 30 nM of LiRAD51 and LiBRCA2–B1 at concentrations varying between 0 and 125 nM.
In order to better understand how LiBRCA2 stimulates LiRAD51 DNA-binding ability, we purified a N-terminal fragment of LiBRCA2 encompassing amino acids 1–122. This truncation comprises the BRC repeats, but not the OB-fold ssDNA-binding domain. It was recently shown that the BRC5, 6, 7, 8 repeats of human BRCA2 stimulates RAD51 to bind ssDNA (42). At a concentration of 30 nM, 12.1% of ssDNA was bound by LiRAD51 (Figure 6F, lane 2). However, when this reaction was supplemented by the LiBRCA2 fragment B1 (10–125 nM), LiRAD51 bound ssDNA more strongly at all LiBRCA2–B1 concentrations (Figure 6F, lanes 4–9), although the stimulation of LiRAD51 was more effective at low concentration. Since LiBRCA2–B1 does not bind DNA by itself (Figure 6F, lane 3), this result suggests that LiBRCA2–B1 stimulates LiRAD51–DNA binding through its BRC motifs.
Stimulation of LiRAD51 by LiBRCA2 and mediator function
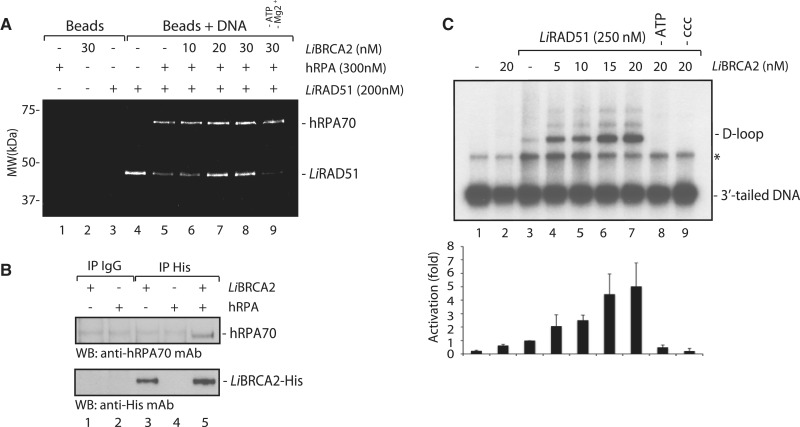
By definition, a recombination mediator assists RAD51 to displace the replicative protein A (RPA) to allow RAD51 to polymerize as nucleoprotein filaments (43,44). It was previously shown that the Ustilago maydis BRCA2 homolog acts as a recombination mediator (45). We therefore tested whether LiBRCA2 assisted RAD51 to displace hRPA. In the absence of hRPA, LiRAD51 bound beads coated with ssDNA (Figure 7A, lane 4). The addition of hRPA prevented the binding of LiRAD51 on ssDNA beads (Figure 7A, lane 5). The addition of LiBRCA2 alleviated the inhibitory effect of hRPA, and led to the accumulation of LiRAD51 on ssDNA (Figure 7A, lanes 7–8). These results demonstrate that LiBRCA2 is a typical recombination mediator. This might be accomplished through interaction with hRPA (Figure 7B).
Figure 7.
LiBRCA2 is a mediator molecule. (A) RPA (300 nM) bound to an ssDNA oligonucleotide prevents LiRAD51 (200 nM) assembly (lane 5), whereas the addition of LiBRCA2 (0–30 nM) stimulates LiRAD51 DNA-binding activity in the presence of 300 nM hRPA (lanes 6–8). Background controls demonstrating the interaction of hRPA (lane 1), LiBRCA2 (lane 2) and LiRAD51 (lane 3) on magnetic beads alone are also shown. The reaction was conducted without ATP and Mg2+ in the presence of LiBRCA2, hRPA and LiRAD51 (lane 9). (B) Interaction between LiBRCA2 and hRPA. LiBRCA2 (750 ng) and/or hRPA (1 µg) were pooled together and coimmunoprecipitation experiments were performed using either IgG (lanes 1 and 2) or His (lanes 3–5) antibodies. Blots were reacted with the indicated antibodies. (C) LiBRCA2 stimulates LiRAD51-catalyzed D-loop formation. Reactions contained DNA alone (lane 1); LiBRCA2 (20 nM, lane 2); LiRAD51 (250 nM, lanes 3–9); or a combination of both proteins (lanes 4–9) with linear DNA molecule containing 3′-single-strand tails (1 µM). Reactions 8 and 9, do not contain ATP (-ATP) or supercoiled DNA (-ccc), respectively. The asterisk represents annealed 3′-tailed molecules. At the bottom, the intensities of bands obtained for each D-loop reaction were quantified in terms of fold activation compared to the control.
As LiBRCA2 is a recombination mediator, we therefore tested whether it could also stimulate LiRAD51 strand invasion ability. When limiting concentrations of LiRAD51 were used, only limited D-loop products were produced (Figure 7C, lane 3). However, when LiRAD51 was supplemented with LiBRCA2 (5–20 nM), strand invasion occurred with a higher level than either protein alone (Figure 7C, compare lanes 4–7 to lanes 2 and 3). No D-loop was observed in the absence of ATP or supercoiled DNA (Figure 7C, lanes 8 and 9). These results show that LiBRCA2 promotes LiRAD51 activation in D-loop formation, a key step in HR.
DISCUSSION
Mutations in BRCA1 or BRCA2 (BReast CAncer) encoding genes lead to ∼10% of familial breast cancers as well as increasing the risk of ovarian cancer. An hallmark of these cancers is genomic instability, characterized by an increase in chromosomal breaks and translocations. In normal cells, this is suppressed by HR which allows the faithful repair of DNA DSBs. The characterization of RAD51 and BRCA2 in a simpler organism such as L. infantum, now opens the door to biochemical studies of several aspects of HR, genomic stability and genetic variability.
We showed that LiBRCA2 possesses several critical features important for the regulation of DNA recombination. First, using cytological analysis, we observed that LiBRCA2 regulates the nuclear accumulation of LiRAD51, suggesting a conserved mechanism of nuclear transport by LiBRCA2 on LiRAD51. The large size of the human BRCA2 protein (384 kDa) poses a great technical challenge for biochemical analyses. To circumvent this problem, we purified a BRCA2 homolog form L. infantum which is three times smaller (125 kDa). One key signature of human BRCA2, is eight conserved motifs of approximately 35 amino acids, known as the BRC repeats. We have analyzed the BRCA2 protein sequence of Leishmania for the presence of these repeats and generated a BRC repeats tree. Using this analysis we found that L. infantum, T. cruzi, L. donovani, L. major, all possessed two BRC repeats, very close to human BRC repeats 7 and 8. This is in contrast with T. brucei BRCA2 which contains an expansion of 15 BRC repeats, which is important for RAD51 localization and the efficiency of T. brucei HR (46). LiBRCA2 also contains one OB-fold domain and a putative Zn-Finger that is unique to the Leishmania BRCA2. Recently, the function of the human BRCA2 BRC repeats has been categorized based on their functional mechanisms. BRC1,2,3,4 domains bind to free RAD51 with high avidity, while BRC5, 6, 7, 8 bind poorly free RAD51, but bind the RAD51–ssDNA filament with high affinity (42). Our results show that a fragment of LiBRCA2 containing two BRC repeats (related to human BRC repeats 3, 4, 7 and 8, respectively) stimulates LiRAD51 binding to ssDNA, especially at low LiBRCA2/LiRAD51 ratio. At higher LiBRCA2/LiRAD51 ratios, the enhancement of LiRAD51 DNA binding was reduced. This dual mode of regulation has been reported where BRC3 peptide disrupted hRAD51 nucleoprotein filaments at high concentration, but formed stable complexes at lower concentration (47). As we observed that LiRAD51 levels are very low compared to LiBRCA2 without DNA damage, this suggests that LiBRCA2 exerts a suppressive role on LiRAD51 in normal conditions. However, when Leishmania is challenged with DNA damage, LiRAD51 levels are increased substantially, suggesting that LiBRCA2 might enhance LiRAD51-dependent recombination for DNA repair.
During the course of these studies, three independent groups have purified full-length human BRCA2 (21–23). Similar to LiBRCA2, it was shown that hBRCA2 is an ssDNA-binding protein (21). However, D-loop formation was not performed for the human BRCA2 protein. In addition, only strand exchange reactions with oligonucleotides, not full-length substrates, were reported. We provide the first evidence of BRCA2 being able to stimulate RAD51-mediated HR by D-loop formation using long DNA substrates. Our results suggest that the minimal domain requirement for a BRCA2 protein to function in such setting is two BRC repeats and an OB-fold.
LiBRCA2 is a functional homolog of RAD52
Interestingly, RAD52 is absent in Leishmania and Saccharomyces cerevisiae does not bear a BRCA2 homolog. In human cells, BRCA2 stimulates the assembly of RAD51 nucleoprotein filament. Saccharomyces cerevisiae does not have a BRCA2 homolog and RAD52 is taking charge of this function. Both proteins interact with RPA and RAD51 and their dual inactivation lead to a synthetic lethality phenotype in human cells, demonstrating that both protein mediate pathways leading to RAD51 activation in mammalian cells (48). In Leishmania, RAD52 is absent, and BRCA2 might also sustain the function of RAD52 shown in lower organisms. We observed that LiBRCA2 stimulated LiRAD51 for HR, and interacted with LiRAD51 and human RPA. We have not used Leishmania RPA or SSB, as the orthologs need to be fully characterized. However, the functional interchangeability between RPA and SSB from different species is supported by results indicating that SSB can substitute for budding yeast and human RPA in strand exchange mediated by S. cerevisiae and human RAD51, respectively (49,50). Consistent with the observation that RAD52 deficient cells accumulate genomic instability, we observed that BRCA2 deficient-cells undergo chromosome loss. We also found that LiBRCA2 is proficient in single-strand annealing, a function normally attributed to RAD52 (10,51). Thus, our results add significant strength to the idea that BRCA2 is a functional homolog of RAD52 in Leishmania.
In order to characterize the function of LiBRCA2 in vivo, we inactivated BRCA2, and assessed HR and cell survival. Gene targeting was severely affected in the absence of LiBRCA2. RNAi approaches are not functional in Leishmania (52), leading to the use of conventional genetic inactivation. DNA integration in Leishmania occurs predominantly by HR, whose efficiency can be an obstacle to insertional mutagenesis strategies. Collectively, as well as defining the functions of LiBRCA2 and LiRAD51, our work also shows that upregulation of HR through overexpression of a recombinase gene might be possible in this unicellular eukaryote.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1, Supplementary Figures 1–3, Supplementary Reference [53].
FUNDING
CIHR grants (to M.O. and J.Y.M.). M.M.G. is a FQRNT and CIHR Vanier scholar; R.B. is a FQRNT scholar; M.O. holds a Canada Research Chair in Antimicrobial Resistance; J.Y.M. is an FRSQ senior investigator. Funding for open access charge: CIHR.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank members of the Masson lab and Danielle Légaré for critical reviewing of the manuscript and members of the Bioimaging Platform of the Centre de Recherche en Infectiologie at the CHUL-CHUQ for their precious help in microscopy-based studies.
REFERENCES
- 1.Moynahan ME, Jasin M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat. Rev. Mol. Cell. Biol. 2010;11:196–207. doi: 10.1038/nrm2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davies AA, Masson JY, McIlwraith MJ, Stasiak AZ, Stasiak A, Venkitaraman AR, West SC. Role of BRCA2 in control of the RAD51 recombination and DNA repair protein. Mol. Cell. 2001;7:273–282. doi: 10.1016/s1097-2765(01)00175-7. [DOI] [PubMed] [Google Scholar]
- 3.Esashi F, Christ N, Gannon J, Liu Y, Hunt T, Jasin M, West SC. CDK-dependent phosphorylation of BRCA2 as a regulatory mechanism for recombinational repair. Nature. 2005;434:598–604. doi: 10.1038/nature03404. [DOI] [PubMed] [Google Scholar]
- 4.Esashi F, Galkin VE, Yu X, Egelman EH, West SC. Stabilization of RAD51 nucleoprotein filaments by the C-terminal region of BRCA2. Nat. Struct. Mol. Biol. 2007;14:468–474. doi: 10.1038/nsmb1245. [DOI] [PubMed] [Google Scholar]
- 5.Carreira A, Hilario J, Amitani I, Baskin RJ, Shivji MK, Venkitaraman AR, Kowalczykowski SC. The BRC repeats of BRCA2 modulate the DNA-binding selectivity of RAD51. Cell. 2009;136:1032–1043. doi: 10.1016/j.cell.2009.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang H, Jeffrey PD, Miller J, Kinnucan E, Sun Y, Thoma NH, Zheng N, Chen PL, Lee WH, Pavletich NP. BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science. 2002;297:1837–1848. doi: 10.1126/science.297.5588.1837. [DOI] [PubMed] [Google Scholar]
- 7.Bochkarev A, Bochkareva E. From RPA to BRCA2: lessons from single-stranded DNA binding by the OB-fold. Curr. Opin. Struct. Biol. 2004;14:36–42. doi: 10.1016/j.sbi.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 8.Lo T, Pellegrini L, Venkitaraman AR, Blundell TL. Sequence fingerprints in BRCA2 and RAD51: implications for DNA repair and cancer. DNA Repair. 2003;2:1015–1028. doi: 10.1016/s1568-7864(03)00097-1. [DOI] [PubMed] [Google Scholar]
- 9.Passos-Silva DG, Rajão MA, Nascimento de Aguiar PH, Vieira-da-Rocha JP, Machado CR, Furtado C. Overview of DNA repair in Trypanosoma cruzi, Trypanosoma brucei, and Leishmania major. J. Nucleic Acids. 2010;2010:840768. doi: 10.4061/2010/840768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mortensen UH, Bendixen C, Sunjevaric I, Rothstein R. DNA strand annealing is promoted by the yeast Rad52 protein. Proc. Natl Acad. Sci. USA. 1996;93:10729–10734. doi: 10.1073/pnas.93.20.10729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shinohara A, Shinohara M, Ohta T, Matsuda S, Ogawa T. Rad52 forms ring structures and co-operates with RPA in single-strand DNA annealing. Genes Cells. 1998;3:145–156. doi: 10.1046/j.1365-2443.1998.00176.x. [DOI] [PubMed] [Google Scholar]
- 12.Shen Z, Cloud KG, Chen DJ, Park MS. Specific interactions between the human Rad51 and Rad52 proteins. J. Biol. Chem. 1996;271:148–152. doi: 10.1074/jbc.271.1.148. [DOI] [PubMed] [Google Scholar]
- 13.Conway C, Proudfoot C, Burton P, Barry JD, McCulloch R. Two pathways of homologous recombination in Trypanosoma brucei. Mol. Microbiol. 2002;45:1687–1700. doi: 10.1046/j.1365-2958.2002.03122.x. [DOI] [PubMed] [Google Scholar]
- 14.Janzen CJ, Lander F, Dreesen O, Cross GA. Telomere length regulation and transcriptional silencing in KU80-deficient Trypanosoma brucei. Nucleic Acids Res. 2004;32:6575–6584. doi: 10.1093/nar/gkh991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burton P, McBride DJ, Wilkes JM, Barry JD, McCulloch R. Ku heterodimer-independent end joining in Trypanosoma brucei cell extracts relies upon sequence microhomology. Eukaryotic Cell. 2007;6:1773–1781. doi: 10.1128/EC.00212-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boothroyd CE, Dreesen O, Leonova T, Ly KI, Figueiredo LM, Cross GA, Papavasiliou FN. A yeast-endonuclease-generated DNA break induces antigenic switching in Trypanosoma brucei. Nature. 2009;459:278–281. doi: 10.1038/nature07982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ubeda JM, Légaré D, Raymond F, Ouameur AA, Boisvert S, Rigault P, Corbeil J, Tremblay MJ, Olivier M, Papadopoulou B, et al. Modulation of gene expression in drug resistant Leishmania is associated with gene amplification, gene deletion and chromosome aneuploidy. Genome Biol. 2008;9:R115. doi: 10.1186/gb-2008-9-7-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mazloum N, Zhou Q, Holloman WK. DNA binding, annealing, and strand exchange activities of Brh2 protein from Ustilago maydis. Biochemistry. 2007;46:7163–7173. doi: 10.1021/bi700399m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mazloum N, Zhou Q, Holloman WK. D-loop formation by Brh2 protein of Ustilago maydis. Proc. Natl Acad. Sci. USA. 2008;105:524–529. doi: 10.1073/pnas.0707031105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mazloum N, Holloman WK. Second-end capture in DNA double-strand break repair promoted by Brh2 protein of Ustilago maydis. Mol. Cell. 2009;33:160–170. doi: 10.1016/j.molcel.2008.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thorslund T, McIlwraith MJ, Compton SA, Lekomtsev S, Petronczki M, Griffith JD, West SC. The breast cancer tumor suppressor BRCA2 promotes the specific targeting of RAD51 to single-stranded DNA. Nat. Struct. Mol. Biol. 2010;17:1263–1265. doi: 10.1038/nsmb.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu J, Doty T, Gibson B, Heyer WD. Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat. Struct. Mol. Biol. 2010;17:1260–1262. doi: 10.1038/nsmb.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jensen RB, Carreira A, Kowalczykowski SC. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature. 2010;467:678–683. doi: 10.1038/nature09399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McKean PG, Keen JK, Smith DF, Benson FE. Identification and characterisation of a RAD51 gene from Leishmania major. Mol. Biochem. Parasitol. 2001;115:209–216. doi: 10.1016/s0166-6851(01)00288-2. [DOI] [PubMed] [Google Scholar]
- 25.Papadopoulou B, Roy G, Ouellette M. A novel antifolate resistance gene on the amplified H circle of Leishmania. EMBO J. 1992;11:3601–3608. doi: 10.1002/j.1460-2075.1992.tb05444.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mukherjee A, Roy G, Guimond C, Ouellette M. The gamma-glutamylcysteine synthetase gene of Leishmania is essential and involved in response to oxidants. Mol. Microbiol. 2009;74:914–927. doi: 10.1111/j.1365-2958.2009.06907.x. [DOI] [PubMed] [Google Scholar]
- 27.El Fadili A, Kündig C, Ouellette M. Characterization of the folylpolyglutamate synthetase gene and polyglutamylation of folates in the protozoan parasite Leishmania. Mol. Biochem. Parasitol. 2002;124:63–71. doi: 10.1016/s0166-6851(02)00163-9. [DOI] [PubMed] [Google Scholar]
- 28.Richard D, Leprohon P, Drummelsmith J, Ouellette M. Growth phase regulation of the main folate transporter of Leishmania infantum and its role in methotrexate resistance. J. Biol. Chem. 2004;279:54494–54501. doi: 10.1074/jbc.M409264200. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook EF, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. New York: Cold Spring Harbor Laboratory Press, Cold Spring Harbor; 1989. [Google Scholar]
- 30.Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 31.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Henricksen LA, Umbricht CB, Wold MS. Recombinant replication protein A: expression, complex formation, and functional characterization. J. Biol. Chem. 1994;269:11121–11132. [PubMed] [Google Scholar]
- 35.Baumann P, Benson FE, Hajibagheri N, West SC. Purification of human Rad51 protein by selective spermidine precipitation. Mutat. Res. 1997;384:65–72. doi: 10.1016/s0921-8777(97)00028-1. [DOI] [PubMed] [Google Scholar]
- 36.Buisson R, Dion-Côté AM, Coulombe Y, Launay H, Cai H, Stasiak AZ, Stasiak A, Xia B, Masson JY. Cooperation of breast cancer proteins PALB2 and piccolo BRCA2 in stimulating homologous recombination. Nat. Struct. Mol. Biol. 2010;17:1247–1254. doi: 10.1038/nsmb.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Masson JY, Davies AA, Hajibagheri N, Van Dyck E, Benson FE, Stasiak AZ, Stasiak A, West SC. The meiosis-specific recombinase hDmc1 forms rings structures and interacts with hRad51. EMBO J. 1999;18:6552–6560. doi: 10.1093/emboj/18.22.6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suzuki A, de la Pompa JL, Hakem R, Elia A, Yoshida R, Mo R, Nishina H, Chuang T, Wakeham A, Itie A, et al. Brca2 is required for embryonic cellular proliferation in the mouse. Genes Dev. 1997;11:1242–1252. doi: 10.1101/gad.11.10.1242. [DOI] [PubMed] [Google Scholar]
- 39.Yu VP, Koehler M, Steinlein C, Schmid M, Hanakahi LA, van Gool AJ, West SC, Venkitaraman AR. Gross chromosomal rearrangements and genetic exchange between nonhomologous chromosomes following BRCA2 inactivation. Genes Dev. 2000;14:1400–1406. [PMC free article] [PubMed] [Google Scholar]
- 40.Benson FE, Stasiak A, West SC. Purification and characterization of the human Rad51 protein, an analogue of E. coli RecA. EMBO J. 1994;13:5764–5771. doi: 10.1002/j.1460-2075.1994.tb06914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sauvageau S, Stasiak AZ, Banville I, Ploquin M, Stasiak A, Masson JY. Fission yeast rad51 and dmc1, two efficient DNA recombinases forming helical nucleoprotein filaments. Mol. Cell. Biol. 2005;25:4377–4387. doi: 10.1128/MCB.25.11.4377-4387.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carreira A, Kowalczykowski SC. Two classes of BRC repeats in BRCA2 promote RAD51 nucleoprotein filament function by distinct mechanisms. Proc. Natl Acad. Sci. USA. 2011;108:10448–10453. doi: 10.1073/pnas.1106971108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sung P. Mediating repair. Nat. Struct. Mol. Biol. 2005;12:213–214. doi: 10.1038/nsmb0305-213. [DOI] [PubMed] [Google Scholar]
- 44.Schild D, Wiese C. Overexpression of RAD51 suppresses recombination defects: a possible mechanism to reverse genomic instability. Nucleic Acids Res. 2010;38:1061–1070. doi: 10.1093/nar/gkp1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang H, Li Q, Fan J, Holloman WK, Pavletich NP. The BRCA2 homologue Brh2 nucleates RAD51 filament formation at a dsDNA-ssDNA junction. Nature. 2005;433:653–657. doi: 10.1038/nature03234. [DOI] [PubMed] [Google Scholar]
- 46.Hartley CL, McCulloch R. Trypanosoma brucei BRCA2 acts in antigenic variation and has undergone a recent expansion in BRC repeat number that is important during homologous recombination. Mol. Microbiol. 2008;68:1237–1251. doi: 10.1111/j.1365-2958.2008.06230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Galkin VE, Esashi F, Yu X, Yang S, West SC, Egelman EH. BRCA2 BRC motifs bind RAD51-DNA filaments. Proc. Natl Acad. Sci. USA. 2005;102:8537–8542. doi: 10.1073/pnas.0407266102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Feng Z, Scott SP, Bussen W, Sharma GG, Guo G, Pandita TK, Powell SN. Rad52 inactivation is synthetically lethal with BRCA2 deficiency. Proc. Natl Acad. Sci. USA. 2011;108:686–691. doi: 10.1073/pnas.1010959107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sugiyama T, Zaitseva EM, Kowalczykowski SC. A single-stranded DNA-binding protein is needed for efficient presynaptic complex formation by the Saccharomyces cerevisiae Rad51 protein. J. Biol. Chem. 1997;272:7940–7945. doi: 10.1074/jbc.272.12.7940. [DOI] [PubMed] [Google Scholar]
- 50.Baumann P, West SC. Heteroduplex formation by human Rad51 protein: effects of DNA end-structure, hRP-A and hRad52. J. Mol. Biol. 1999;291:363–374. doi: 10.1006/jmbi.1999.2954. [DOI] [PubMed] [Google Scholar]
- 51.Ploquin M, Bransi A, Paquet ER, Stasiak AZ, Stasiak A, Yu X, Cieslinska AM, Egelman EH, Moineau S, Masson JY. Functional and structural basis for a bacteriophage homolog of human RAD52. Curr. Biol. 2008;18:1142–1146. doi: 10.1016/j.cub.2008.06.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robinson KA, Beverley SM. Improvements in transfection efficiency and tests of RNA interference (RNAi) approaches in the protozoan parasite Leishmania. Mol. Biochem. Parasitol. 2003;128:217–228. doi: 10.1016/s0166-6851(03)00079-3. [DOI] [PubMed] [Google Scholar]
- 53.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.