Abstract
miR-21 is the most commonly over-expressed microRNA (miRNA) in cancer and a proven oncogene. Hsa-miR-21 is located on chromosome 17q23.2, immediately downstream of the vacuole membrane protein-1 (VMP1) gene, also known as TMEM49. VMP1 transcripts initiate ∼130 kb upstream of miR-21, are spliced, and polyadenylated only a few hundred base pairs upstream of the miR-21 hairpin. On the other hand, primary miR-21 transcripts (pri-miR-21) originate within the last introns of VMP1, but bypass VMP1 polyadenylation signals to include the miR-21 hairpin. Here, we report that VMP1 transcripts can also bypass these polyadenylation signals to include miR-21, thus providing a novel and independently regulated source of miR-21, termed VMP1–miR-21. Northern blotting, gene-specific RT-PCR, RNA pull-down and DNA branching assays support that VMP1–miR-21 is expressed at significant levels in a number of cancer cell lines and that it is processed by the Microprocessor complex to produce mature miR-21. VMP1 and pri-miR-21 are induced by common stimuli, such as phorbol-12-myristate-13-acetate (PMA) and androgens, but show differential responses to some stimuli such as epigenetic modifying agents. Collectively, these results indicate that miR-21 is a unique miRNA capable of being regulated by alternative polyadenylation and two independent gene promoters.
INTRODUCTION
Mature microRNAs (miRNAs) are short endogenous non-coding RNAs that post-transcriptionally inhibit the expression of target genes (1,2). miRNAs are initially expressed within long transcripts known as primary microRNAs (pri-miRNAs). Many pri-miRNAs are also protein-coding (3), thus they are processed both as pri-miRNAs and as pre-mRNAs, including 5′-capping, splicing and polyadenylation. For miRNA processing the stem–loop intermediate, or pre-miRNA, is released from the nascent transcript by the Microprocessor complex (4,5), which minimally consists of DGCR8 and Drosha. DGCR8 acts as a molecular anchor to measure the distance from the dsRNA–ssRNA junction and guide the RNase III enzyme, Drosha, for cleavage to release the pre-miRNA (6). Current knowledge supports that release of pre-miRNAs occurs co-transcriptionally, along with pre-mRNA processing events, before the transcript is discharged from the DNA template (7). The unleashed pre-miRNA is subsequently transported to the cytoplasm by Exportin-5 and further cleaved by Dicer to generate the mature miRNA. Once incorporated into the RNA-induced silencing complex, the single-stranded mature miRNA will target mRNAs by complete or partial sequence complementary, resulting in message destabilization and decreased translation (8).
The current estimate for human miRNAs hairpin precursors is 1527 (9,10). Their rate of discovery has increased significantly with second and third-generation sequencing technologies (11); however, the pri-miRNAs which encode them and their transcription start sites (TSSs) remain poorly characterized. One of the first characterized pri-miRNAs was miR-21 (12). Interestingly miR-21 belongs to a rare class of miRNAs by being located near the 3′-untranslated region (3′-UTR) of a coding gene. Specifically, miR-21 is located immediately downstream of the vacuole membrane protein-1 (VMP1) gene, also known as TMEM49. However, the VMP1 transcript is polyadenylated prior to the miR-21 hairpin and thus was not thought to contribute to its expression (13,14). Instead, the primary miR-21 transcript was found to initiate within intron 11 of VMP1, resulting in a 3.4 kb capped, polyadenylated and unspliced transcript (12). Subsequent alternative isoforms and TSSs were detected upstream of this initial location, namely within intron 10 of VMP1, to generate an unspliced 4.3 kb pri-miR-21 transcript (15). This larger transcript was confirmed by northern blotting and a putative miR-21 promoter, miPPR-21, was characterized and verified (15,16). Additional miR-21 promoters and primary transcripts have been characterized from within the terminal intronic regions of VMP1 (17,18). Therefore, the full complement of miR-21-encoding transcripts is complex and has yet to be fully characterized.
miR-21, as measured by the mature miRNA, is the most commonly elevated miRNA in cancer (19). There are many mechanisms associated with elevated miR-21 levels. The encoding genetic locus, 17q23, is amplified in many solid tumors (20–22). In addition, miR-21 expression is stimulated by a variety of cancer associated pathways such as hypoxia, inflammation, AP-1 and steroid hormones (15–17,23,24). Elevated miR-21 expression results in enhanced growth and reduced apoptosis in a number of cell culture and animal models. Importantly, elevated miR-21 alone was shown to be sufficient to induce a pre-B malignant lymphoid-like phenotype in animal models and continuous miR-21 expression was necessary to maintain the tumor phenotype, indicating that miR-21 is a true oncogene (25). Similarly, miR-21 deletion in mouse cancer models reduced tumor formation (26,27). Thus, there is significant evidence that miR-21 is an important gene in cancer biology.
Detailed analysis of the miPPR-21 promoter identified specific transcription factor binding sites, such as AP-1, Ets/PU.1, C/EBP-α, STAT3, and others (28). Phorbol-12-myristate-13-acetate (PMA), a known stimulant of AP-1 transcription factors, was found to induce the miPPR-21 promoter and the resulting 4.3 kb pri-miR-21 transcript, but not the overlapping VMP1 transcript (15). Similar studies have reported disparate responses of VMP1 and miR-21 following stimulation, indicating that these two overlapping genes are independently regulated (12,15,17,29). Expressed sequence taq (EST) database analysis further suggests that these two genes are independent, and that VMP1 mRNA does not extend into the miR-21 hairpin. However, the overlapping nature of VMP1 and miR-21 and their processing by polyadenylation and Microprocessor cleavage has made it difficult to confirm this autonomy. Here, we have applied precise detection, sizing and capture techniques to identify and characterize an alternatively polyadenylated isoform of VMP1, VMP1–miR-21, that initiates from the coding VMP1 gene and extends the 3′-UTR to include the miR-21 hairpin. VMP1–miR-21 is commonly expressed in many cell lines and is processed by Drosha, suggesting that it is a true primary miR-21 transcript. These new data indicate that miR-21 is a unique miRNA that is regulated both by alternative polyadenylation and multiple independent promoters.
MATERIALS AND METHODS
Cell culture and siRNA transfection
Cells were grown using the American type culture collection (ATCC) recommended media in the presence of 10 µg/ml ciprofloxacin hydrochloride (US Biological) and 10% fetal bovine serum. Drosha siRNA (23) designed to anneal to Drosha mRNA at 5′-AAGGACCAAGUAUUCAGCAAG-3′ was synthesized by Thermo Scientific. Control non-target siRNA #1 (Dharmacon) was also applied. Hiperfect reagent (Qiagen) was used to transfect 20 nM of siRNAs according to the manufacturer protocol.
RNA extraction and northern blotting
Total RNA was extracted from cells at 80% confluency with TRIzol reagent (Invitrogen). About 10–20 µg of RNA was separated by 1.2% agarose denaturing gel and northern blotting completed as previously described (30). Transcript size was estimated relative to High Range ssRNA Ladder (New England Biolabs). Primers and northern blotting probes (Supplementary Table SI) were generated from Michigan cancer foundation-7 (MCF-7) cDNA PCR amplified using the GoTaq® qPCR Master Mix (Promega), single bands isolated by QIAquick PCR Purification Kit (Qiagen) and probes labeling by α-32P-dCTP (Perkin Elmer) and ChromaSpin + TE-10 Columns (Clontech). After an ON incubation in Ultrahyb Oligo Buffer (Ambion), blots were washed in serial dilutions of hot 2 × SSC buffer and 2 × SSC buffer + 0.5% SDS. Radioactive signals were directly quantified using an Instant Image.
Reverse transcription and RT-PCR
One microgram of total RNA was subjected to random reverse transcription using the QuantiTect Reverse Transcription Kit (Qiagen). For gene-specific RT-PCR, gene-specific primers (GSP) for reverse transcription (Supplementary Table SII) were applied, with DNase I Amplification Grade (Invitrogen) digestion per 5 µg total RNA. Two microgram GSP was applied with ThermoScritp™ Reverse Transcriptase (Invitrogen) according to the manufacturer protocol. As a control for this step, a sample without reverse transcriptase was processed in parallel. Following RNase H (Invitrogen) digestion, cDNA was amplified by PCR using Power SYBR Green Mix (Applied Biosystems) and the specific primers (Supplementary Table SI).
RNA pull-down
Fifty microgram of TRIzol-extracted total RNA was applied to the µMACS Streptavidin Kit (Miltenyi Biotec Inc.) following the manufacturer protocol. Heat-denatured RNA and 0.5 µg of biotinylated capture DNA probe (Supplementary Table SII) was annealed and mixed with 100 µl of µMACS Streptavidin Microbead, vortexed and incubated for 2 min. Beads were magnetically captured, washed and specific transcripts eluted in pre-heated RNAse-free water. Ten microliters was applied for RT-PCR.
Mature miR-21 quantification
TaqMan MicroRNA RT Kit (Applied Biosystems) was used in combination with the miR-21-specific reverse transcription primers followed by PCR using miR-21 TaqMan specific primers and TaqMan 2X Universal PCR Master Mix, No AmpErase UNG (Applied Biosystems). RNU6B was used as normalization control.
3′-Rapid amplification of cDNA ends
GeneRacer Kit (Invitrogen) was applied to 5 µg of total RNA from Drosha depleted MCF-7 following the manufacturer protocol. Oligo-ligated RNA was reverse transcribed using the SuperScript III module and oligo-dT primers. The 3′-rapid amplification of cDNA ends (RACE)-ready cDNA was PCR amplified using the Phusion® High-Fidelity PCR Master Mix (Finnzymes). Single products were PCR purified with QIAquick Gel Extraction Kit (Qiagen) and cloned using the TOPO TA Cloning® Kit (Invitrogen). Plasmid DNA from insert-positive colonies (white–blue screening) was extracted with QIAPrep Spin Miniprep Kit (Qiagen) and sequenced.
Drosha QuantiGene branched-chain DNA assay
VMP1–miR-21 and pri-miR-21 levels were measured using the branched-chain DNA (bDNA) assay described elsewhere (31). DU-145 cells were transfected for 48 h with Drosha siRNA or non-target siRNA #1 and Drosha knock-down verified by RT-PCR. The QuantiGene® Screen Assay Kit 2.0 (Panomics) was used to specifically capture the miR-21 containing transcripts (chr17:55 273 238–55 273 669) to a 96-well plate during a 55°C overnight incubation. Unbound material was washed for three times, blocked and labeling probes targeting RNA regions in intron 11 (chr17:55 271 009–55 271 910, specific for pri-miR-21) or alternatively exons 2–5 of VMP1 (bases 125–538 of VMP1–miR-21) were added. Signal amplifier molecules were added, washed and luminescent substrate added for quantification in a microplate luminometer. A ‘no-template’ background provided the background values for each set of probes. Values were normalized to an identical QuantiGene assay for the housekeeping gene cyclophylin B.
Transcriptional induction
For transcription initiation studies, HL-60 cultures were treated for 6 h with 160 nM of PMA (Sigma) or vehicle (DMSO). HCT116 and HEK293 cells were seeded at 20% confluence in p35 mm plates. After 24 h, 5-aza-2′-deoxycytidine (Fluka) or MS-275 (Syndax) were added to the media at a final concentration of 10 and 1 µM, respectively. As a control, equivalent amounts of vehicle were used. Media and treatment were refreshed every 48 h.
Additional Supplementary Methods can be found at Nucleic Acids Research online.
RESULTS
Identification of a VMP1–miR-21 fusion transcript
The precursor miR-21 hairpin is located immediately downstream of the VMP1 coding gene on chromosome 17q23 (Figure 1). Despite their close proximity, previous studies indicate that VMP1 and primary miR-21 transcripts are regulated by independent promoters and that VMP1 transcripts are polyadenylated prior to miR-21 (12–15,18). We hypothesized that alternative polyadenylation of VMP1 may occur at some rate to produce a read-through VMP1–miR-21 fusion transcript. To investigate this, an androgen receptor (AR) induction model was applied (16,23). Cells were treated with the synthetic androgen, R1881, and changes in VMP1 transcript levels were measured by quantitative RT-PCR. In all four AR-positive prostate cancer cell lines studied, androgen treatment induced VMP1 gene expression (Supplementary Figure S1A). Using these cells, an RT-PCR strategy was applied to detect the putative VMP1–miR-21 fusion transcript by amplifying a region starting upstream of the reported pri-miR-21 TSSs, within VMP1 exon 9, to a region downstream of the miR-21 hairpin. Cells were first pre-treated with Drosha siRNA, to reduce hairpin processing. Indeed, a VMP1–miR-21 amplicon was detected in LNCaP and LAPC-4 cells and verified by sequencing (Supplementary Figure S1B). VMP1 expression levels were then quantified in a panel of prostate cancer cell lines and compared to a mixture of healthy prostate from young men. MCF-7 cells, which are known to express high levels of miR-21, were included as a positive control. All tumor cell lines exhibited greater levels of VMP1 than the normal prostate of young men (Supplementary Figure S2). These results support that VMP1 is expressed in cancer cell lines and that alternative polyadenylation events can lead to the inclusion of the miR-21 hairpin within the VMP1 transcript.
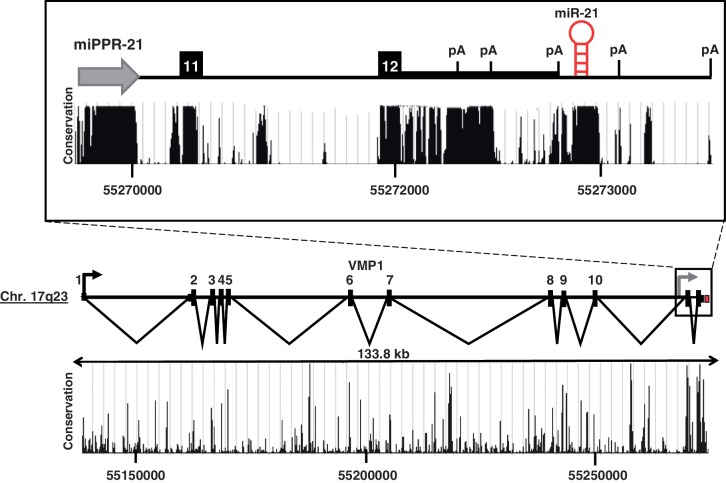
Figure 1.
VMP1 and miR-21. Structure of the VMP1 and miR-21 loci at chromosome 17q23. Scale is indicated. VMP1 exon numbers are noted. Inset highlights the pri-miR-21 gene region. White numbers identify exons 11 and 12 of VMP1. The line of intermediate thickness indicates the 3′-UTR of VMP1. A gray arrow is situated at the miR-21 promoter, miPPR-21. The red hairpin indicates the location of the pre-miR-21 hairpin. The black arrow represents the TSS of VMP1. Evolutionary conservation (UCSC Genome Browser 28 species conservation track, NCBI36/hg18 assembly) is plotted below.
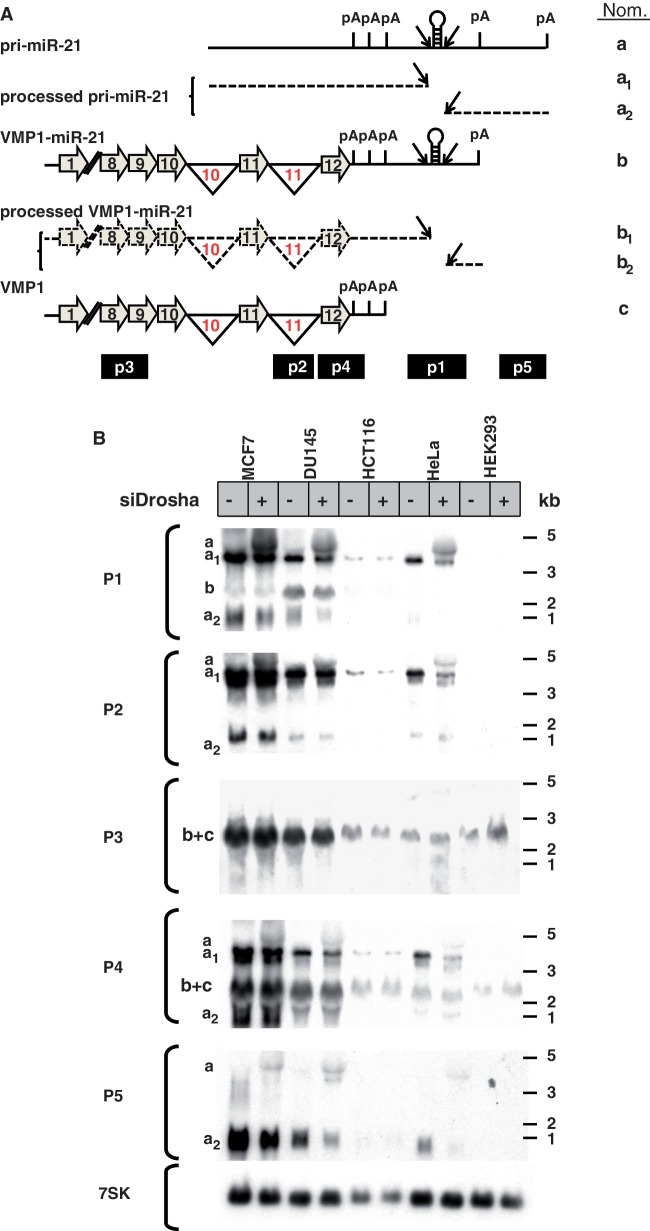
Using these and other cancer cells lines, northern blotting was applied to characterize pri-miR-21 and VMP1 transcripts. Drosha knockdown was performed as above to diminish miR-21 transcript processing. A series of probes were designed to differentially detect the VMP1, pri-miR-21, VMP1–miR-21 transcripts and their cleavage products (Figure 2A). Because of the overlapping nature of these transcripts, some probes detect multiple products. Probe P1 binds to a region spanning from the last poly(A) of VMP1 to ∼180 bp downstream of the miR-21 hairpin. Therefore, it should detect the non-spliced 4.3 kb pri-miR-21 transcript (band ‘a’) as well as the putative spliced, 2.5 kb VMP1–miR-21 transcript (band ‘b’), but not the polyadenylated VMP1 (band ‘c’). Results show that the highest miR-21 expressing cell lines, MCF-7 and DU-145, express both pri-miR-21 and VMP1–miR-21 transcripts (Figure 2B, bands ‘a’ and ‘b’). Of note, HEK293 cells are known to express low levels of miR-21 and accordingly exhibit undetectable levels of pri-miR-21 and VMP1–miR-21 by probe P1 in Figure 2B. However, the VMP1 transcripts (band ‘b+c’) were visible in HEK293 cells with probes P3 and P4. The combination of an absent ‘band b’ with probe P1 in HEK293 and the presence of a ‘band b+c’ with probes P3 and P4 demonstrate the specificity of the northern blots and confirm the common expression of an alternatively polyadenylated VMP1 transcript, VMP1–miR-21. In all of the other cancer cell lines tested, pri-miR-21 was detected by probes P1 (miR-21 hairpin region), P2 (VMP1 intron 11) and P4 (VMP1 exon 12). Similarly, in all cell lines, VMP1 (band ‘c’) was detected by probes P3 and P4; however, the observed size of VMP1–miR-21 (probe P1, band ‘b’) was indistinguishable from VMP1 (probes P3 and P4, band ‘c’). The comparison of the bands detected with these probes suggests that not all VMP1 encompassed miR-21. This is particularly clear when observing HCT116, HeLa and HEK293 cells which express VMP1 (probes P3 and P4) with undetectable levels of VMP1–miR-21 (probe P1). It is possible that cell-specific alternative polyadenylation determines VMP1–miR-21 expression rates.
Figure 2.
VMP1 and pri-miR-21 transcripts by northern blotting. (A) Schematic of the transcripts VMP1, VMP1–miR-21 and pri-miR-21 (not to scale). Horizontal gray arrows represent exons and black lines represent introns (numbered in red) and non-coding sequences. Break symbols (double slash mark) are placed between exons 1 and 8 of VMP1 and VMP1–miR-21. Splicing is indicated by inverted triangles and Drosha cleavage by diagonal arrows. Pri-miR-21 is not spliced and therefore does not include exon or splicing symbols. Processed transcripts are marked by dashed lines. Polyadenylation signals are indicated as pA. Northern probes P1, P2, P3, P4 and P5 are drawn at the site of hybridization along the bottom of the transcripts. (B) Northern blot analysis of total RNA from cells transfected for 48 h with Drosha siRNA (siDrosha, +) or a non-target control siRNA (−). On the left, probes applied for each northern blot and corresponding bands (a–c) are indicated. Molecular size ladder is represented on the right of the panels.
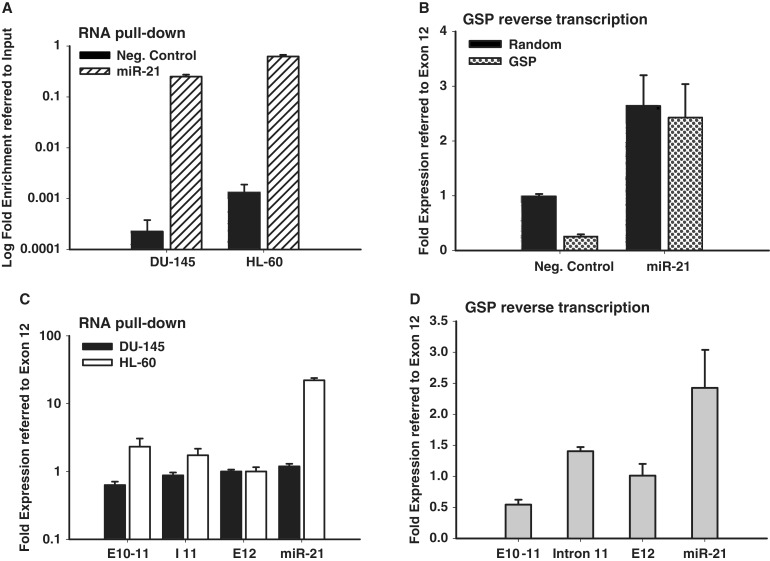
To further verify and characterize VMP1–miR-21 transcripts, two separate miR-21 primary RNA enrichment techniques were applied. In one technique, miR-21-specific RNA pull-down was achieved with a biotinylated oligonucleotide complementary to a region between the miR-21 hairpin and VMP1 polyadenylation [poly(A)] signals (Supplementary Figure S3, Oligo #1). Captured RNA was then reverse transcribed with random primers to generate cDNA. The second enrichment technique utilized reverse transcription by a GSP, located immediately downstream of the miR-21 hairpin (Supplementary Figure S3, Oligo #2). The resulting cDNA from both enrichment protocols was then applied to quantitative PCR (qPCR) for VMP1–miR-21 (exons 10-11), pri-miR-21 (intron 11) or both transcripts (exon 12). Both approaches were verified to enrich for miR-21 primary transcripts or deplete non-specific products, respectively (Figure 3A and B). As shown in Figure 3C, miR-21 RNA pull-down captured VMP1–miR-21 transcripts (exons 10-11) in DU-145 and HL-60 cell, as well as pri-miR-21 (intron 11). Similarly, in Figure 3D, miR-21-specific reverse transcription detected VMP1–miR-21 (exons 10-11) and pri-miR-21 (intron 11) in MCF-7 cells. Collectively, these studies confirm the northern blots and support the common expression of VMP1–miR-21 in multiple cell types.
Figure 3.
Relative quantification of pri-miR-21 and VMP1–miR-21 transcripts. (A) Total RNA from DU-145 and HL-60 cells was specifically pulled-down (RNA pull-down) with a biotinylated probe in the miR-21 hairpin region and control or miR-21-specific RNA was quantified by quantitative RT-PCR. The y-axis represents log fold enrichment after miR-21 pull down, relative to input RNA. (B) Total RNA from MCF-7 cells was reverse transcribed using a miR-21 GSP. The resulting cDNA was quantified by qPCR for control or miR-21 transcripts to determine enrichment by GSP reverse transcription. (C) RNA from miR-21 pull down in DU-145 and HL-60 cells was reverse transcribed and amplicons specific for VMP1–miR-21 (exons 10-11), pri-miR-21 (intron 11) or both transcripts (exon 12 and miR-21) were quantified by qPCR. Ct values obtained for captured transcripts were referred to values of exon 12 and plotted in a logarithmic scale. (D) cDNA from MCF-7 miR-21 GSP reverse transcription was quantified by qPCR for VMP1–miR-21 (exons 10-11), pri-miR-21 (intron 11) or both transcripts (exon 12 and miR-21) amplicons. Ct values obtained for captured transcripts were referred to values of exon 12 and plotted in a logarithmic scale. Error bars represent standard deviations derived from one representative experiment measured per triplicate. Error bars represent standard deviations derived from three independent measurements.
Characterization of VMP1–miR-21 and pri-miR-21 3′-termini
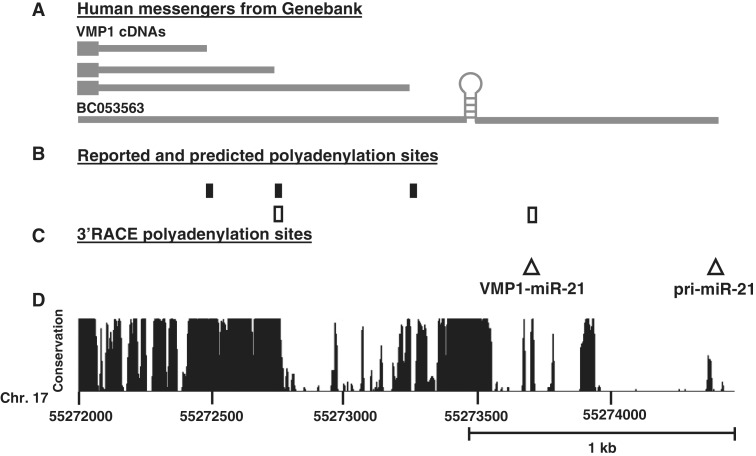
The UCSC Genome Browser poly(A) track identifies four poly(A) signals at the 3′-end of VMP1 (Figure 4B). Three are upstream of the miR-21 hairpin as mapped by cDNA/EST sequences, and another is downstream and predicted using bioinformatics (32). In addition, the 3′-end of the mapped pri-miR-21 transcript (BC053563) terminates several hundred base pairs downstream of the most distal poly(A) signal (Figure 4A). To further characterize the 3′-termini of miR-21 transcripts we performed 3′-RACE. Figure 4C summarizes that two termini were identified, both near canonical AAUAAA poly(A) signals. One is proximal and consistent with the previously predicted poly(A) signal and the second is a novel distal site near the BC053562 terminus. Both poly(A) signals are in highly conserved regions (Figure 4D). A northern probe (P5), which hybridizes to the region between the two poly(A) sites, was generated to identify transcripts which use the most distal poly(A) signal (Figure 2A). Probe P5 detected the 4.3 kb pri-miR-21 (band ‘a’) in Drosha knocked-down cells, and also detected an ∼1 kb band consistent with the 3′-fragment of Drosha-cleaved pri-miR-21 (Figure 2B, band ‘a2’). As would be expected, the intensity of the 3′-cleavage product (fragment ‘a2’), is reduced with Drosha knock-down. If VMP1–miR-21 also utilized the distal poly(A) site, the predicted size would be ∼1 kb greater than VMP1 itself, or 3.6 kb. However no band with a compatible size was observed on northern blotting with probes P1 or P5 (Figure 2B), indicating that VMP1–miR-21 primarily utilizes the proximal signal. This is supported by the small fragment (Figure 2A, band ‘b2’), which is visible on higher percentage gels with probe P1 (Supplementary Figure S4). Collectively, these results indicate that VMP1–miR-21 consistently utilizes the proximal poly(A) signal and pri-miR-21 the distal poly(A) signal (sites 1 and 2, respectively, in Figure 4C).
Figure 4.
Characterization of pri-miR-21 and VMP1–miR-21 3′-termini and polyadenylation sites. (A) Summary of the mRNA sequences identified with the UCSC Genome Browser Human mRNA track, NCBI36/hg18 assembly. (B) Location of poly(A) signals the surrounding miR-21 hairpin according to UCSC Genome Browser poly(A) track,NCBI36/hg18 assembly. Black boxes represent reported polyadenylation signals based on EST/cDNA sequencing and white boxes represent bioinformatic predicted sites. (C) Mapped 3′-termini and polyadenylation sites by 3′-RACE. White triangles (1 and 2) are located at the identified end of the transcripts and confirmed the presence of a canonical AAUAAA site. Northern blotting (Figure 2B and Supplementary Figure S4) supports that VMP1-miR-21 predominantly uses Site 1 while pri-miR-21 uses Site 2. (D) Plot depicting the evolutionary conservation (UCSC Genome Browser 28 species conservation track, NCBI36/hg18 assembly). Scale is indicated below the graph.
Drosha processing of primary miR-21 transcripts
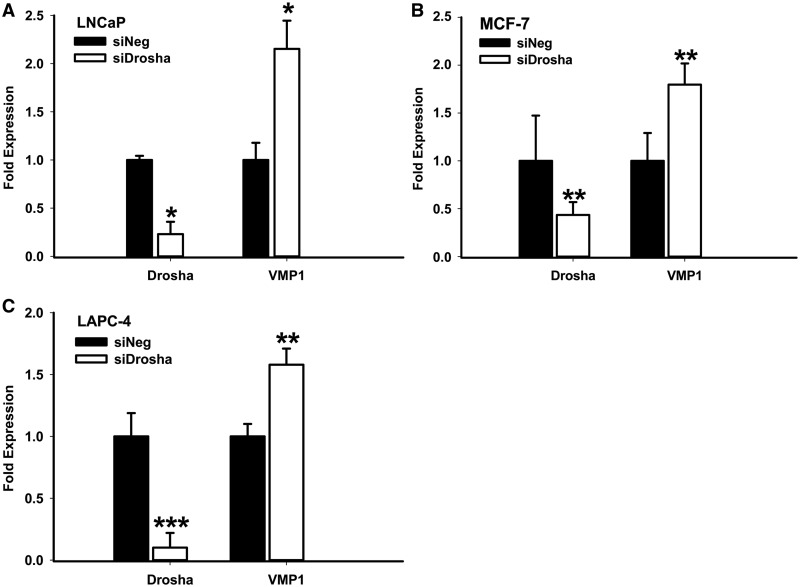
miRNAs are rarely located in the 3′-UTR of coding transcripts. This could be because Microprocessor cleavage would de-stabilize such messages by leaving a free and non-polyadenylated 3′-end (12). VMP1–miR-21 provides a novel endogenous gene to study this phenomenon. To investigate effects of Drosha cleavage on VMP1 mRNA levels, Drosha was knocked-down by siRNA treatment and validated by quantitative RT-PCR (Figure 5, Drosha). In this setting, levels of VMP1 mRNA showed a cumulative increase, irrespective of the cell line (Figure 5, VMP1). We explored whether this increase was due to Drosha-regulated de-stabilization of the mRNA or Drosha-mediated effects on transcriptional processing by repeating this experiment in the presence or absence of the transcriptional inhibitor Actinomycin D (Supplementary Figure S6). In MCF-7 cells, which express high levels of VMP1–miR-21, Drosha knock-down stabilized the existing VMP1 transcripts in the presence and absence of Actinomycin D. On the other hand, in LAPC4 cells, VMP1 levels were not induced in the absence of new transcription. These results suggest that Microprocessor activity reduces the levels of VMP1 transcripts through both mRNA de-stabilization and transcriptional processing.
Figure 5.
Drosha knock-down enhances VMP1 transcript levels. Total RNA was prepared from (A) LNCaP, (B) MCF-7 or (C) LAPC-4 cells, which had been transfected for 72 h with a negative control siRNA (siNeg) or Drosha siRNA (siDrosha). Expression of Drosha and VMP1, relative to actin, was measured by quantitative RT-PCR. Quantification is the result of three independent measurements. *P < 0.05; **P < 0.005; ***P < 0.001 (Student’s t-test).
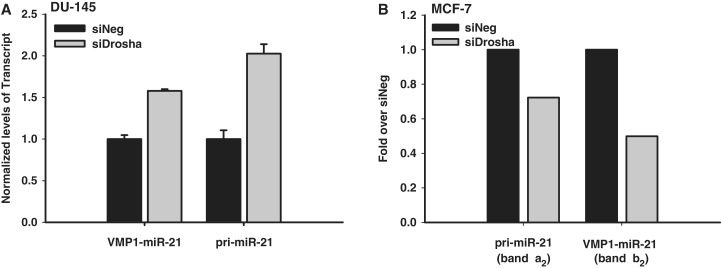
To determine the efficiency of Drosha processing of the two miR-21 primary transcripts, RNA quantification with branched DNA (bDNA) technology was applied (31). Briefly, total RNA was purified from Drosha or control depleted DU-145 cells. Transcripts containing the miR-21 hairpin were then captured to a plate using multiple probes within the vicinity of the miRNA hairpin. Probes targeting VMP1–miR-21 (exons 2–5) or pri-miR-21 (intron 11) were then used to differentiate the amounts of the two miR-21 primary transcripts in the control or Drosha knocked-down states. Because Microprocessor cleavage would separate the captured and probed regions, only unprocessed transcripts should be detected. Drosha knock-down resulted in an increase both VMP1–miR-21 and pri-miR-21 levels, indicating that both transcripts are substrates of the Microprocessor complex (Figure 6A). Interestingly, pri-miR-21 appears to be more significantly affected by Drosha depletion. To further study Microprocessor processing rates, pri-miR21 and VMP1–miR-21 Drosha cleavage products were quantified from MCF-7 cell northern blots (Figure 6B). In these cells, VMP1–miR-21 transcripts appear to be more significantly affected by Drosha knock-down. Both studies support that the novel VMP1–miR-21 transcript is a substrate for Drosha cleavage and that it is processed with comparable efficiency to the pri-miR-21 transcript.
Figure 6.
VMP1-miR-21 processing by Drosha. (A) bDNA assay quantification of VMP1–miR-21 and pri-miR-21 transcripts in DU-145 cells. RNA transcripts were captured with multiple probes in the common miR-21 hairpin region and detected with probes in VMP1 exons 2–5 (VMP1–miR-21) or in the VMP1 intron 11 region (pri-miR-21). Data reported are relative to the control siRNA (siNeg) treatment and normalized to Cyclophylin B. (B) Northern blot quantification of Drosha processing in MCF-7 cells. Northern blots with probe P1 (Figure 2B and Supplementary Figure S4) were quantified for relative pri-miR-21 (band a2) and VMP1–miR-21 (band b2) cleavage products after treatment with control or Drosha siRNA, and normalized to 7SK. The y-axis represents fold band intensity relative to negative control siRNA (siNeg) treatment.
Common and differential transcriptional regulation of pri-miR-21 and VMP1
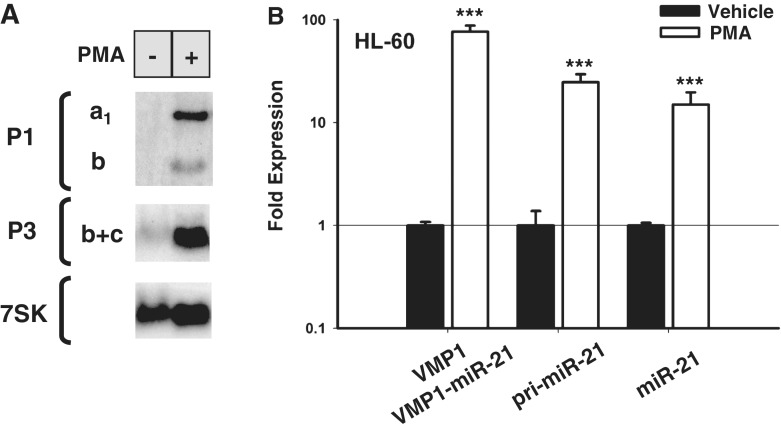
It has been previously reported that mature miR-21 expression is induced by the AP-1 stimulating agent, PMA (15,33). To confirm this, RNA from HL-60 cells was treated with PMA or vehicle for 6 h and analyzed by northern blotting with probes that detect the miR-21 hairpin region (probe P1) as well as VMP1 and VMP1–miR-21 transcripts (probe P3). As expected, bands corresponding to the pri-miR-21 transcript were significantly increased following PMA treatment (Figure 7A, probe P1 and band ‘a1’). Interestingly, VMP1–miR-21 and VMP1 transcripts were similarly stimulated by AP-1 activation (Figure 7A, probes P1 and P3, bands ‘b’ and ‘c’, respectively). This was confirmed by quantitative RT-PCR (Figure 7B). Based on these findings, we anticipate that a portion of PMA-stimulated mature miR-21 is generated through VMP1–miR-21 transcripts. These results slightly differ from a previous study where PMA was not found to stimulate VMP1 expression by northern blotting after 24 h of treatment (15). Differences in these results may be due to the length of PMA treatment schedules or northern blot sensitivity.
Figure 7.
Phorbol ester mediated induction of pri-miR-21 and VMP1–miR-21. (A) Northern blot analysis of total RNA from HL-60 cells treated with PMA (+) or vehicle (−) for 6 h. At the left, probes used for each blot and identifying letters for the transcripts are indicated. (B) Quantitative RT-PCR analysis of HL-60 cells treated with PMA or vehicle. Relative induced expression of VMP1 and VMP1–miR-21 (exons 10-11), pri-miR-21 (intron 11) and an area surrounding miR-21 hairpin (miR-21) are normalized to actin. Note that VMP1 and VMP1–miR-21 are indistinguishable by this quantitative RT-PCR analysis. Error bars represent standard deviations derived from two independent experiments measured per triplicate. ***P < 0.001 (Student’s t-test).
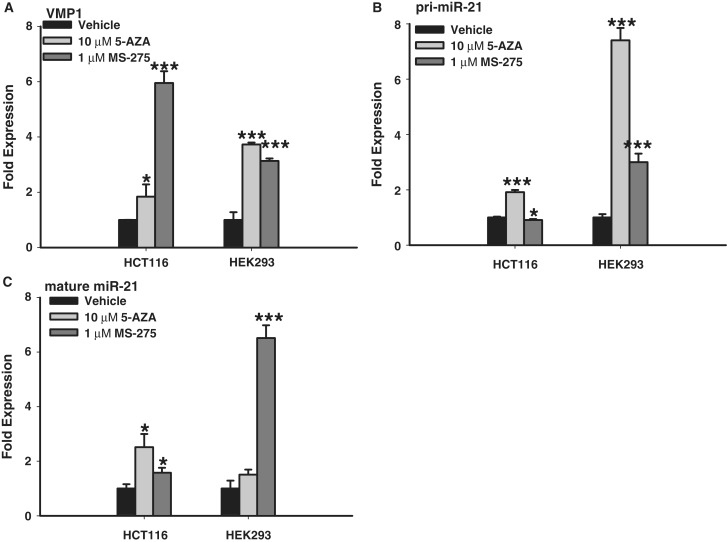
While PMA and androgen stimulation enhanced both VMP1 and miR-21 messages, their genomic organization clearly indicates that VMP1 and pri-miR-21 are regulated by independent promoters. One potential difference could be epigenetic regulation through the presence of CpG islands or chromatin conformation. Previous studies of the pri-miR-21 promoter region have not detected CpG islands (18,34). However, Iorio et al. (35) did identify a short, 300 bp and 50% GC-rich area upstream of the miR-21 hairpin as a putative CpG island. This may account for the elevated expression of miR-21 in divergent models after 5-aza-2′-deoxycitidine (5-AZA-CdR) treatment (36,37). Here, we report that VMP1 also has a potential CpG island surrounding its TSS which is longer at 500 bp and is 55% GC rich (Supplementary Figure S5A). To investigate the methylation potential of these regions we performed bisulfite sequencing of genomic DNA from two cell lines with low basal expression of miR-21, HEK293 and HCT116. The results reveal that the previously identified pri-miR-21 CpG island is commonly methylated in both cell lines, while the larger VMP1 CpG island is not methylated (Supplementary Figure S7). We then separately treated these cells with the epigenetic modifying agents 5-AZA-CdR and MS-275. Quantitative RT-PCR of pri-miR-21 (intron 11) as well as VMP1 (exons 10-11) indicated both similar and differential responses of these two genes following treatment (Figure 8). Specifically, both VMP1 and pri-miR-21 were induced by 5′-AZA-CdR in HEK293 and HCT116 cells. These results suggest that the pri-miR-21 CpG island may directly influence expression of both pri-miR-21 and VMP1 transcripts, although indirect regulation cannot be excluded. On the other hand, the MS-275 histone deacetylase (HDAC) inhibitor showed differential induction of VMP1 and pri-miR-21 in HCT116 cells. In particular, VMP1 levels were strongly induced by MS-275 treatment (Figure 8A), while pri-miR-21 levels were only mildly affected (Figure 8B). Further studies are required to determine if these divergent responses are due to direct epigenetic modification of VMP1 gene regulatory elements in these cells. Finally, we evaluated whether the induced expression of VMP1–miR-21 and pri-miR-21 correlated with elevated levels of mature miR-21 (Figure 8C). Notably, a 3-fold increase in both VMP1 and pri-miR-21 in HEK293 cells following MS-275 treatment correlated with ∼6-fold increase in mature miR-21. However, there are other examples where VMP1 and pri-miR-21 expression did not correlate with mature miR-21 levels, indicating that post-transcriptional processing of VMP1 and pri-miR-21, such as polyadenylation or drosha cleavage, influenced the overall levels of mature miR-21.
Figure 8.
Independent gene regulation of VMP1 and pri-miR-21. (A) Relative VMP1 (amplicon, exons 10-11) expression response to epigenetic modifying agents as quantified by quantitative RT-PCR, when normalized to actin, from HCT116 and HEK293 cells 3 days following treatment with 10 µM 5-azacytidine or 1 µM MS-275. (B) Relative pri-miR-21 (intron 11) expression response as quantified by quantitative RT-PCR, when normalized to actin. (C) Relative mature miR-21 expression response as quantified by quantitative RT-PCR, when normalized to U6. Error bars represent standard deviations from three independent measurements. *P < 0.05; **P < 0.01; ***P < 0.001 (Student’s t-test).
DISCUSSION
Since the discovery of miRNAs, a great amount of effort has been devoted to the characterization of their primary transcripts and promoters. Many studies have applied pre-determined size limits in promoter-prediction studies, for instance by examining a maximum distance of 100 kb upstream the miRNA hairpin. This approach has favored the discovery of more adjacent promoters and shorter pri-miRNAs (15,34,38,39). Similarly, 5′-RACE is biased toward the discovery of shorter products because they are more efficiently reverse transcribed and amplified. In the case of miR-21, several groups have predicted or identified promoters and TSSs within intron 10 or 11 of VMP1 (15,17,18,34). In other studies, where distance from the hairpins has not been as heavily weighed, the miR-21 promoter was hypothesized to be immediately upstream of the VMP1 transcript (40). However, all reported human VMP1 cDNAs and ESTs terminate before the miR-21 hairpin (Figure 4A). Only one human transcript containing the miR-21 hairpin, termed BC053563, is identifiable in Genome Browser analysis. It is not clear why the predominant 4.3 kb pri-miR-21 (15) or the newly discovered 2.5 kb VMP1–miR-21 transcripts were not previously identified in cDNA and EST libraries. It is possible that oligo-dT bias predominantly identified VMP1 transcripts, rather than pri-miR-21 transcripts, from this region. The region between the 3′-terminus of VMP1 and miR-21 is also short and AT-rich, which may have hidden this region from random priming. Cell-specific differences in VMP1 polyadenylation could be also responsible for the lack of VMP1–miR-21 detection. Further, Drosha cleavage would have separated upstream VMP1 and pri-miR-21 regions, keeping them from discovery. Here we demonstrate that VMP1–miR-21 is readily detectable in multiple cancer cell lines, especially when Drosha levels are first knocked down by siRNA treatment.
The overlapping nature of miR-21 and VMP1 has made their transcripts difficult to characterize. Here, we have performed detailed northern blotting and gene-specific quantification studies to distinguish and enumerate the predominant forms of miR-21 primary transcripts in multiple human cell lines. Two predominant miR-21 transcripts are notable, a larger transcript consistent with the 4.3 kb pri-miR-21 identified by Fujita et al. (15), and the novel VMP1–miR-21 transcript. We did not detect any band of compatible size to BC053563 or, alternatively, to the similar transcript described by Cai et al. (12). This may be expressed at levels below the detection sensitivity of our northern blots. VMP1–miR-21 expression was observed in several cell lines, with MCF-7 and DU-145 showing the highest expression in our study (Figure 2B). Interestingly the ratio of pri-miR-21 to VMP1–miR-21 is quite different among the cell lines, suggesting differential regulation or stability. This is consistent with the initial characterization of VMP1 transcripts in rat tissues. Dusetti et al. (13) probed rat tissue northern blots with the full-length VMP1 cDNA and found three bands of 3.5, 2.7 and 1.9 kb. The larger bands are peculiar because the cDNA was only 1.9 kb, but we suggest that this was likely the 5′-Drosha cleavage product of the pri-miR-21 transcript (band ‘a1’ in Figure 2A). In rats, the ratios of these three bands were different in different tissues, which the authors predicted to be due to differential splicing (13). We predict that the differences in expression were likely due to the differential regulation of VMP1 and pri-miR-21 promoters and alternative polyadenylation. VMP1, consisting of 12 exons, has at least 109 predicted splicing variants by the ECgene database (41). However, our northern blot studies with probes P1, P3, and P4 indicate that VMP1 is not significantly alternatively spliced, but rather that it encodes multiple transcripts which are alternatively polyadenylated.
To truly function as a primary miRNA, the miR-21 hairpin must be released by Microprocessor cleavage. In a number of assays we found inhibition of Drosha to stabilize both pri-miR-21 as well as VMP1–miR-21 transcripts, supporting that they are both substrates for the Microprocessor complex. In the case of VMP1–miR-21, which contains the coding region of VMP1, it is logical that Microprocessor cleavage would reduce the level of VMP1 message by leaving an unprotected 3′-terminus. In a model study by Cai et al. (12), the inclusion of miR-30 in an artificial UTR of the firefly luciferase reduced the activity of the enzyme by 2-fold or less. These ratios are similar to the fold of VMP1 mRNA induction observed in our Drosha inhibition study (Figure 5). Our studies in the absence of new transcription indicate that this increase can be due to both VMP1 mRNA transcript stability as well as efficiency of transcription (Supplementary Figure S6). We predict that miR-21 processing would correspondingly reduce VMP1 protein levels, but that the effect would likely be mild because VMP1–miR-21 only accounts for a fraction of the VMP1 coding transcripts. It should also be mentioned that Drosha activity is not restricted to miRNA hairpins. The RNase III enzyme can also negatively regulate mRNAs by cleaving physiological loops contained throughout the transcript. For example Drosha modulates the levels of its partner, DGCR8, by cleaving the highly conserved hairpins in its 5′-UTR (42). This has been confirmed in mouse embryonic cells where several additional Drosha-regulated mRNAs were identified (43). However, our results support a conventional miRNA hairpin cleavage of pri-miR-21 and VMP1–miR-21, rather than internal cleavage, because Drosha siRNA did not have dramatic effects on product sizes in northern blotting (Figure 2B). In addition, RT-PCR detection of the 3′-end of VMP1 was more efficient when Drosha was inhibited. Therefore, we believe that both pri-miR-21 and VMP1–miR-21 are direct sources for pre-miR-21 and mature miR-21 miRNA.
Mounting evidence suggests that, like pre-mRNA, the processing of pri-miRNA is coupled to transcription [reviewed in Ref. (44)]. For example, Pawlicki and Steitz (7) reported that exogenously expressed pri-miRNAs, without poly(A) signals, are retained at the transcription site. Here we found that pri-miR-21 and VMP1–miR-21 utilized different poly(A) signals, despite containing identical overlapping sequences. Specifically, the pri-miR-21 poly(A) is several hundred base pairs downstream of the one used by VMP1–miR-21 (Figure 4). This difference in polyadenylation may be related to pre-mRNA processing, as the VMP1 coding gene is spliced where the 4.3 kb pri-miR-21 is non-coding and is not spliced. There may also be influence from promoter structure and gene context. For instance, it has been reported that transient expression of a pri-miRNA from a miRNA promoter resulted in higher levels of Drosha recruitment, increased retention on the DNA template and enhanced processing of the pri-miRNA compared to the same pri-miRNA driven by promoters such as phosphoglycerate kinase, cytomegalovirus or U1 (45). Therefore, several complex features may contribute to the differential processing of VMP1–miR-21 and pri-miR-21.
VMP1 and pri-miR-21 appear to have several common mechanisms for gene expression. VMP1 was initially identified as a stress-induced gene in acinar cells during acute pancreatitis and in kidney after transient failure (13). According to its observed function in stress paradigms, enforced expression of VMP1 triggered cell death accompanied by vacuolization. More recent studies suggest that VMP1 participates in the autophagy cascade (46–51) and that it is critical for autophagy in metazoan organisms (52). Pri-miR-21 expression is also associated with stress and inflammation, and its regulation has been mapped specifically to interleukin-6 signaling and STAT3 activation at the miPPR-21 promoter region (17). In addition to inflammatory pathways, pri-miR-21 expression is directly induced by the AP-1 transcription factors through three conserved elements within miPPR-21 promoter region (15). Here, we found that VMP1 was also stimulated by PMA (Figure 7A and B). While the promoter of VMP1 has not yet been annotated, visual inspection with the UCSC Genome Browser ‘Human/rat/mouse conserved transcription factor binding sites’ track indicated one AP-1 binding site (TRANSFAC z-score of 2.01) within 1.5 kb of the first intron of VMP-1. Therefore, AP-1 may directly induce VMP1. Finally, we previously reported that pri-miR-21 is induced by androgens in prostate cancer cells through direct AR binding of the miPPR-21 promoter (23). In Supplementary Figure S1A, we also found VMP1 to be induced by androgens in four AR-positive prostate cell lines. Therefore, there appear to be several common and cancer associated regulatory pathways which induce VMP1, VMP1–miR-21 and pri-miR-21 expression.
It has been previously reported that miR-21 can be induced through chromatin modifying agents (36,37). A putative CpG island had been identified near the pri-miR-21 promoter region (37). In addition, we identified a prominent 500 bp CpG island within the predicted VMP1 regulatory region (Supplementary Figure S5). In our studies we found that the previously identified pri-miR-21 CpG island was methylated, while the longer VMP1 CpG island was not (Supplementary Figure S7). 5-Aza-CdR treatment increased both VMP1 and pri-miR-21 expression levels. On the other hand, HDAC inhibitor treatment enhanced VMP1, but not pri-miR-21, expression in HCT116 cells (Figure 8). These results support that VMP1 and pri-miR-21 are differentially regulated by independent promoters and chromatin structure. However, elevated VMP1 expression did not always correlate with elevated mature miR-21 levels. This is likely due to alternative polyadenylation of VMP1 transcripts. For example, in HCT116 and HeLa cells pri-miR-21 and VMP1 are detected in the absence of VMP1–miR-21 (Figure 2B). Recent transcriptome wide RNA sequencing studies indicate that alternative polyadenylation events often differ among different tissue types (53). Polyadenylation site selection is further altered in proliferating and cancerous cells (54,55). Thus, VMP1 may be alternatively polyadenylated in different cell types or in cancer, leading to gains or losses of mature miR-21 levels. Further studies are required to determine influence and mechanisms of alternative polyadenylation on miR-21 expression in various tissues and cancer types.
In summary, we have provided new evidence that miR-21 can be generated through two independently regulated transcripts. One previously reported transcript, pri-miR-21, is non-spliced and preferentially terminates at the most distal poly(A) signal. The second transcript, VMP1–miR-21, is a spliced and coding mRNA generated through alternative polyadenylation of VMP1-initiated transcripts. These two overlapping genes are induced by common and independent transcriptional regulatory pathways. Further characterization of the VMP1 promoter region and mechanisms of alternative polyadenylation should provide new insights into the regulation of the oncogenic miRNA, miR-21, in cancerous tissues.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2 and Supplementary Figures 1–7 and Supplementary Methods.
FUNDING
The ‘National Institutes of Health/National Cancer Institute’ [5R01CA143299 to S.E.L., 5P50CA058236 to S.E.L. (Project 1)]; Department of Defense Prostate Cancer Research Fund [W81XWH-08-13-5 to S.E.L.]; Patrick C. Walsh Prostate Cancer Research Fund (to S.E.L.); Spanish ‘Ministerio de Ciencia e Innovación’ [SAF2011-29730 to J.R.]. Funding for open access charge: National Institutes of Health/NCI R01CA143299.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors wish to thank Michael Sure (Panomics) for assistance with the QuantiGene 2.0 protocol, Oliver Kent, Wasim Chowdhury, Hun-Way Hwang, Erik Wentzel and Tsung-Cheng Chang (Johns Hopkins) for their methodological discussion and advice. We also thank Kaoru Sakabe for MCF-7 cells, Leigh-Ann Cruz for the MS-275, Dr Konstantopoulos for the HL-60 and Michael Haffner for the 5′-AZA-CdR. We also want to acknowledge Luzia Brander for her participation in the project. We finally thank Jacint Boix for his careful reading of the article.
REFERENCES
- 1.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim YK, Kim VN. Processing of intronic microRNAs. EMBO J. 2007;26:775–783. doi: 10.1038/sj.emboj.7601512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 5.Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 6.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pawlicki JM, Steitz JA. Primary microRNA transcript retention at sites of transcription leads to enhanced microRNA production. J. Cell. Biol. 2008;182:61–76. doi: 10.1083/jcb.200803111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Griffiths-Jones S. The microRNA registry. Nucleic Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dusetti NJ, Jiang Y, Vaccaro MI, Tomasini R, Azizi Samir A, Calvo EL, Ropolo A, Fiedler F, Mallo GV, Dagorn JC, et al. Cloning and expression of the rat vacuole membrane protein 1 (VMP1), a new gene activated in pancreas with acute pancreatitis, which promotes vacuole formation. Biochem. Biophys. Res. Commun. 2002;290:641–649. doi: 10.1006/bbrc.2001.6244. [DOI] [PubMed] [Google Scholar]
- 14.Vaccaro MI, Grasso D, Ropolo A, Iovanna JL, Cerquetti MC. VMP1 expression correlates with acinar cell cytoplasmic vacuolization in arginine-induced acute pancreatitis. Pancreatology. 2003;3:69–74. doi: 10.1159/000069150. [DOI] [PubMed] [Google Scholar]
- 15.Fujita S, Ito T, Mizutani T, Minoguchi S, Yamamichi N, Sakurai K, Iba H. miR-21 Gene expression triggered by AP-1 is sustained through a double-negative feedback mechanism. J. Mol. Biol. 2008;378:492–504. doi: 10.1016/j.jmb.2008.03.015. [DOI] [PubMed] [Google Scholar]
- 16.Ribas J, Lupold SE. The transcriptional regulation of miR-21, its multiple transcripts, and their implication in prostate cancer. Cell Cycle. 2010;9:923–929. doi: 10.4161/cc.9.5.10930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Löffler D, Brocke-Heidrich K, Pfeifer G, Stocsits C, Hackermüller J, Kretzschmar AK, Burger R, Gramatzki M, Blumert C, Bauer K, et al. Interleukin-6 dependent survival of multiple myeloma cells involves the Stat3-mediated induction of microRNA-21 through a highly conserved enhancer. Blood. 2007;110:1330–1333. doi: 10.1182/blood-2007-03-081133. [DOI] [PubMed] [Google Scholar]
- 18.Ozsolak F, Poling LL, Wang Z, Liu H, Liu XS, Roeder RG, Zhang X, Song JS, Fisher DE. Chromatin structure analyses identify miRNA promoters. Genes Dev. 2008;22:3172–3183. doi: 10.1101/gad.1706508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl Acad. Sci. USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu G, Sinclair C, Hinson S, Ingle JN, Roche PC, Couch FJ. Structural analysis of the 17q22-23 amplicon identifies several independent targets of amplification in breast cancer cell lines and tumors. Cancer Res. 2001;61:4951–4955. [PubMed] [Google Scholar]
- 21.Kasahara K, Taguchi T, Yamasaki I, Kamada M, Yuri K, Shuin T. Detection of genetic alterations in advanced prostate cancer by comparative genomic hybridization. Cancer Genet. Cytogenet. 2002;137:59–63. doi: 10.1016/s0165-4608(02)00552-6. [DOI] [PubMed] [Google Scholar]
- 22.Griffin CA, Hawkins AL, Packer RJ, Rorke LB, Emanuel BS. Chromosome abnormalities in pediatric brain tumors. Cancer Res. 1988;48:175–180. [PubMed] [Google Scholar]
- 23.Ribas J, Ni X, Haffner M, Wentzel EA, Salmasi AH, Chowdhury WH, Kudrolli TA, Yegnasubramanian S, Luo J, Rodriguez R, et al. miR-21: an androgen receptor-regulated microRNA that promotes hormone-dependent and hormone-independent prostate cancer growth. Cancer Res. 2009;69:7165–7169. doi: 10.1158/0008-5472.CAN-09-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhat-Nakshatri P, Wang G, Collins NR, Thomson MJ, Geistlinger TR, Carroll JS, Brown M, Hammond S, Srour EF, Liu Y, et al. Estradiol-regulated microRNAs control estradiol response in breast cancer cells. Nucleic Acids Res. 2009;37:4850–4861. doi: 10.1093/nar/gkp500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Medina PP, Nolde M, Slack FJ. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010;467:86–90. doi: 10.1038/nature09284. [DOI] [PubMed] [Google Scholar]
- 26.Hatley ME, Patrick DM, Garcia MR, Richardson JA, Bassel-Duby R, van Rooij E, Olson EN. Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21. Cancer Cell. 2010;18:282–293. doi: 10.1016/j.ccr.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma X, Kumar M, Choudhury SN, Becker Buscaglia LE, Barker JR, Kanakamedala K, Liu MF, Li Y. Loss of the miR-21 allele elevates the expression of its target genes and reduces tumorigenesis. Proc. Natl Acad. Sci. USA. 2011;108:10144–10149. doi: 10.1073/pnas.1103735108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krichevsky AM, Gabriely G. miR-21: a small multi-faceted RNA. J. Cell. Mol. Med. 2009;13:39–53. doi: 10.1111/j.1582-4934.2008.00556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wickramasinghe NS, Manavalan TT, Dougherty SM, Riggs KA, Li Y, Klinge CM. Estradiol downregulates miR-21 expression and increases miR-21 target gene expression in MCF-7 breast cancer cells. Nucleic Acids Res. 2009;37:2584–2595. doi: 10.1093/nar/gkp117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol. Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Canales RD, Luo Y, Willey JC, Austermiller B, Barbacioru CC, Boysen C, Hunkapiller K, Jensen RV, Knight CR, Lee KY, et al. Evaluation of DNA microarray results with quantitative gene expression platforms. Nat. Biotechnol. 2006;24:1115–1122. doi: 10.1038/nbt1236. [DOI] [PubMed] [Google Scholar]
- 32.Cheng Y, Miura RM, Tian B. Prediction of mRNA polyadenylation sites by support vector machine. Bioinformatics. 2006;22:2320–2325. doi: 10.1093/bioinformatics/btl394. [DOI] [PubMed] [Google Scholar]
- 33.Kasashima K, Nakamura Y, Kozu T. Altered expression profiles of microRNAs during TPA-induced differentiation of HL-60 cells. Biochem. Biophys. Res. Commun. 2004;322:403–410. doi: 10.1016/j.bbrc.2004.07.130. [DOI] [PubMed] [Google Scholar]
- 34.Corcoran DL, Pandit KV, Gordon B, Bhattacharjee A, Kaminski N, Benos PV. Features of mammalian microRNA promoters emerge from polymerase II chromatin immunoprecipitation data. PLoS One. 2009;4:e5279. doi: 10.1371/journal.pone.0005279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 36.Hulf T, Sibbritt T, Wiklund ED, Bert S, Strbenac D, Statham AL, Robinson MD, Clark SJ. Discovery pipeline for epigenetically deregulated miRNAs in cancer: integration of primary miRNA transcription. BMC Genomics. 2011;12:54. doi: 10.1186/1471-2164-12-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, Casalini P, Taccioli C, Volinia S, Liu CG, Alder H, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699–8707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- 38.Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Monteys AM, Spengler RM, Wan J, Tecedor L, Lennox KA, Xing Y, Davidson BL. Structure and activity of putative intronic miRNA promoters. RNA. 2010;16:495–505. doi: 10.1261/rna.1731910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee Y, Kim B, Shin Y, Nam S, Kim P, Kim N, Chung WH, Kim J, Lee S. ECgene: an alternative splicing database update. Nucleic Acids Res. 2007;35:D99–D103. doi: 10.1093/nar/gkl992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Han J, Pedersen JS, Kwon SC, Belair CD, Kim YK, Yeom KH, Yang WY, Haussler D, Blelloch R, Kim VN. Posttranscriptional crossregulation between Drosha and DGCR8. Cell. 2009;136:75–84. doi: 10.1016/j.cell.2008.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karginov FV, Cheloufi S, Chong MM, Stark A, Smith AD, Hannon GJ. Diverse endonucleolytic cleavage sites in the mammalian transcriptome depend upon microRNAs, Drosha, and additional nucleases. Mol. Cell. 2010;38:781–788. doi: 10.1016/j.molcel.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pawlicki JM, Steitz JA. Nuclear networking fashions pre-messenger RNA and primary microRNA transcripts for function. Trends Cell Biol. 2010;20:52–61. doi: 10.1016/j.tcb.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ballarino M, Pagano F, Girardi E, Morlando M, Cacchiarelli D, Marchioni M, Proudfoot NJ, Bozzoni I. Coupled RNA processing and transcription of intergenic primary microRNAs. Mol. Cell Biol. 2009;29:5632–5638. doi: 10.1128/MCB.00664-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grasso D, Ropolo A, Lo Ré A, Boggio V, Molejón MI, Iovanna JL, Gonzalez CD, Urrutia R, Vaccaro MI. Zymophagy, a novel selective autophagy pathway mediated by VMP1-USP9x-p62, prevents pancreatic cell death. J. Biol. Chem. 2011;286:8308–8324. doi: 10.1074/jbc.M110.197301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Itakura E, Mizushima N. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy. 2010;6:764–776. doi: 10.4161/auto.6.6.12709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grasso D, Sacchetti ML, Bruno L, Lo Ré A, Iovanna JL, Gonzalez CD, Vaccaro MI. Autophagy and VMP1 expression are early cellular events in experimental diabetes. Pancreatology. 2009;9:81–88. doi: 10.1159/000178878. [DOI] [PubMed] [Google Scholar]
- 49.Ropolo A, Grasso D, Pardo R, Sacchetti ML, Archange C, Lo Re A, Seux M, Nowak J, Gonzalez CD, Iovanna JL, et al. The pancreatitis-induced vacuole membrane protein 1 triggers autophagy in mammalian cells. J. Biol. Chem. 2007;282:37124–37133. doi: 10.1074/jbc.M706956200. [DOI] [PubMed] [Google Scholar]
- 50.Vaccaro MI, Ropolo A, Grasso D, Iovanna JL. A novel mammalian trans-membrane protein reveals an alternative initiation pathway for autophagy. Autophagy. 2008;4:388–390. doi: 10.4161/auto.5656. [DOI] [PubMed] [Google Scholar]
- 51.Pardo R, Lo Ré A, Archange C, Ropolo A, Papademetrio DL, Gonzalez CD, Alvarez EM, Iovanna JL, Vaccaro MI. Gemcitabine induces the VMP1-mediated autophagy pathway to promote apoptotic death in human pancreatic cancer cells. Pancreatology. 2010;10:19–26. doi: 10.1159/000264680. [DOI] [PubMed] [Google Scholar]
- 52.Tian Y, Li Z, Hu W, Ren H, Tian E, Zhao Y, Lu Q, Huang X, Yang P, Li X, et al. C. elegans screen identifies autophagy genes specific to multicellular organisms. Cell. 2010;141:1042–1055. doi: 10.1016/j.cell.2010.04.034. [DOI] [PubMed] [Google Scholar]
- 53.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mayr C, Bartel DP. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.