Abstract
Alterations in DNA methylation have been reported to occur during development and aging; however, much remains to be learned regarding post-natal and age-associated epigenome dynamics, and few if any investigations have compared human methylome patterns on a whole genome basis in cells from newborns and adults. The aim of this study was to reveal genomic regions with distinct structure and sequence characteristics that render them subject to dynamic post-natal developmental remodeling or age-related dysregulation of epigenome structure. DNA samples derived from peripheral blood monocytes and in vitro differentiated dendritic cells were analyzed by methylated DNA Immunoprecipitation (MeDIP) or, for selected loci, bisulfite modification, followed by next generation sequencing. Regions of interest that emerged from the analysis included tandem or interspersed-tandem gene sequence repeats (PCDHG, FAM90A, HRNR, ECEL1P2), and genes with strong homology to other family members elsewhere in the genome (FZD1, FZD7 and FGF17). Our results raise the possibility that selected gene sequences with highly homologous copies may serve to facilitate, perhaps even provide a clock-like function for, developmental and age-related epigenome remodeling. If so, this would represent a fundamental feature of genome architecture in higher eukaryotic organisms.
INTRODUCTION
Aging is widely viewed as a complex process, distinct from development and characterized by accumulation of damaged molecules, cells and tissues, ultimately leading to malfunction and degeneration (1,2). Associated with old age is loss of the ability to maintain baseline tissue homeostasis and an increased risk for developing many diseases, including cancer, cardiovascular and neurodegenerative disorders (3,4). The aging phenotype varies among individuals, in part due to genetic factors, but also due to environmental influences and stochastic variation in epigenome structure (5,6).
Characterization of epigenome dysregulation with age is at a very early stage, and an even less explored area concerns the underlying role of epigenetic remodeling during post-natal development, despite numerous lines of evidence linking development and age-related variation, as well as age-related phenotypes (7–10). In mice, similarities have been reported between developmental and adult changes in DNA-methylation patterns (11). Furthermore, cumulative evidence links perinatal events to risk patterns for age-related diseases (12,13). Taken together with increasing focus on stem-cell integrity in the context of aging (14–17), such studies suggest that it will be worthwhile to investigate these links further, as well as to reexamine the widely accepted dichotomy between peri- and post-natal development versus aging.
DNA methylation, a major epigenetic mechanism in mammals, constitutes a post-replication modification at the 5′-position of cytosine, predominantly within CpG dinucleotides, that typically results in transcriptional repression of adjacent genes. DNA-methylation changes during early mammalian development, starting with demethylation during cleavage of the early embryo and followed by genome-wide de novo methylation after the embryonic implantation (18). A substantial fraction of CpG dinucleotides are clustered in domains termed ‘CpG islands’ (19). In normal cells, CpGs located in gene promoter-associated CpG islands tend to be unmethylated (20), whereas methylation predominates in most downstream transcribed gene regions (21–23) and in repetitive regions (satellite DNA, LINEs, SINEs, etc.) (24). Less dense CpG clusters than those in CpG islands, as well as sequences immediately adjacent to islands (‘shores’) are thought to exhibit differentiation-specific—but also highly variable—patterns of DNA methylation (25). Additionally, in differing immune cell types, differential methylation can occur in CpG islands that are not associated with transcription start sites (orphan CpG islands) (26).
In rodents, two early studies, one in mouse tissues and another in germ and liver rat cells, revealed consistent age-related increases in DNA-methylation of ribosomal genes (27,28). These studies suggest that ribosomal DNA repetitiveness constitutes a genomic structure susceptible to age-related hypermethylation. More recently, array-based epigenetic studies in mouse and rat tissues have reported hypomethylation or hypermethylation of specific genomic loci during aging (29,30).
A number of studies have examined selected genes or used array-based approaches to document DNA-methylation drift with age in humans. Hypermethylation occurs in the estrogen receptor gene CpG island in human colon samples with cancer, as well as with age (31). Likewise, genes hypermethylated in human prostate cancer are subject to progressive methylation in normal prostate tissues during aging (32). Given high variability of DNA methylation in many genome regions, including in monozygotic twins (33), several recent reports have presented population-based statistical analyses in support of age-related epigenetic disorder or drift (34–37).
In the present work, we compared methylome structures in DNAs from newborns and adults, with samples derived from cord blood and adult peripheral blood, respectively. Methylated DNA Immunoprecipitation followed by Next Generation Sequencing (MeDIP-Seq) was employed to search for age-related methylation differences on a whole genome scale. A notable advantage of this approach is that it allows a global survey of epigenetic marks without bias toward a specific genomic region, e.g. gene promoters or CpG islands.
Our aim was to reveal key genomic regions with distinct structure and sequence characteristics that are susceptible to developmental remodeling or to age-related dysregulation. As an experimental source we chose primary human peripheral blood monocytes. These cells are attractive for epigenome mapping, since they retain stem-cell-like characteristics (38–41) and can be easily differentiated into dendritic cells (DC). Reproducible changes detected across the span of post-natal development should be of strong inherent interest. Moreover, we reasoned that such changes, if detected, might shed light on more strictly age-related (young versus old adult) epigenome perturbations. To our knowledge, this is the first whole human genome DNA-methylation study to provide gene locus-specific results from comparisons between different age groups.
MATERIALS AND METHODS
Ethics
All human materials used in this study were received under approval of the Institutional Review Board of the National Institute of Child Health and Human Development (NICHD), or of the Clinical Center, National Institutes of Health (NIH).
Human peripheral blood monocyte samples
Cord blood samples were obtained from healthy term newborns. Adult peripheral blood or monocyte-enriched apheresis samples were obtained from healthy volunteers from the NIH Department of Transfusion Medicine. In each of two independent experiments, cord blood and adult blood samples were pooled. For the first experiment, cord blood monocytes were derived from a set of five male and four female newborns; for the second experiment, the pooled set consisted of three male and seven female newborns. The corresponding adult sets included three male and six female donors in the first experiment, versus six male and four female in the second. The age range for adult samples in these experiments was 21 to 73 years old.
Cell purification and culture
Cord blood and adult (monocyte-enriched apheresis or whole blood) samples were diluted 2- to 4-fold in Dulbecco’s PBS (DPBS) with 4% citrate dextrose solution (ACD). Samples were placed on a Ficoll-paque gradient (density 1.077) and centrifuged for 35 min at 900 g at room temperature. The interphase was transferred to a fresh tube, diluted to 40 ml using DPBS and centrifuged twice for 5 min at 200 g to reduce platelet contamination. Samples were resuspended in 20 ml of RPMI 1640, 10% fetal calf serum (FCS) and placed on a lower density Ficoll-paque gradient (20 min, 400 g, room temperature) (42). Interphase cells were collected and washed in DPBS followed by centrifugation for 8 min at 500 g. Pellets were resuspended in 20 ml PBE (PBS, 0.5% BSA, 2 mM EDTA) and centrifuged for 10 min at 500 g. Monocytes were further purified by subtractive magnetic cell sorting using a monocyte isolation kit (Miltenyi, Auburn, CA) following the manufacturer’s protocol. Purified monocytes were washed in DPBS. To obtain DC, cells were plated in RPMI 1640, 10% FCS, 50 ng/ml GM-CSF, 20 ng/ml IL-4. Cultures were refed every other day and DC were harvested on Day 6. Purities of monocyte and DC preparations were assessed by flow cytometry (FACSCalibur;BD Biosciences, San Jose, CA) with the following antibodies: anti-CD14 FITC, anti-biotin PE (Miltenyi); anti-HLA-DR and anti-CD86 (BD Biosciences).
Methylated DNA immunoprecipitation
Methylated DNA immunoprecipitation (MeDIP) was performed as previously described in (43). We used 3–5 µg of fragmented DNA for a standard MeDIP assay. We denatured the DNA for 10 min at 95°C and immunoprecipitated it for 2 h at 4°C with 10 µl monoclonal antibody against 5-methylcytidine (Eurogentec) in a final volume of 500 ml IP buffer [10 mM sodium phosphate (pH 7.0), 40 mM NaCl, 0.05% Triton X-100]. We incubated the mixture with 30 μl of Dynalbeads with M-280 sheep antibody to mouse IgG (Dynal Biotech) for 2 h at 4°C and washed it three times with 700 μl IP buffer. We then treated the beads with proteinase K for 3 h at 50°C and recovered the methylated DNA by phenol–chloroform extraction followed by ethanol precipitation.
Preparation and immunoprecipitation of formaldehydefixed chromatin
Purified monocytes or in vitro differentiated DCs were crosslinked with 1% formaldehyde solution. Nuclei were isolated and micrococcal nuclease (MN) digestion was carried out as described previously (44). The soluble chromatin fragments were diluted in ChIP buffer (50 mM Tris–HCl, pH 7.5, 1% NP-40, 0.25% sodium deoxycholate, 150 mM NaCl, 1 mM EDTA, 5 mM Na–butyrate, protein inhibitors) to a final SDS concentration 0.1% and were used for Chromatin Immunoprecipitation (ChIP). ChIP was performed as described in previous studies (44,45) using polyclonal anti-trimethylated H3K27 antibody (Millipore #07-449).
MeDIP and ChIP sequencing library preparation
Sequencing libraries were prepared following Illumina ChIP Sequencing protocol. The DNA was end-repaired and ligated with Illumina sequencing paired end adaptors as described previously (46). After ligation, the DNA was enriched by 18 cycles of PCR with primers complementary to the paired end adaptor sequences followed by agarose gel size selection and purification.
Targeted bisulfite sequencing
DNA samples were bisulfite converted by using EpiTect Bisulfite Kit (Qiagen). Primer pairs were designed using a custom interface with primer3. Candidate initial and nested primers for upper and lower strands of the bisulfite-converted DNA template overlapped no more than one CpG, at a position ≥10 bases from the primer 3′-end. Where an internal primer base aligned to a potentially methylated CpG template cytidine, the base ‘C’ was designated in the primer, forcing either a C–T or C–C mismatch. Specificities and yields of the PCR reactions (initial followed by nested PCR) were analyzed by DHPLC (44) and real time PCR for 55 and 46 candidate primer sets targeted to CpGs within the ECEL1P2 and FZD1 regions, respectively. After final PCR products were combined based on primer validation and reaction yields, libraries were prepared as described above for next-generation sequencing. Reads were initially aligned using Bowtie (47) to bisulfite-converted template databases with duplicate entries containing either CG or TG at all CpG positions. Alignment parameters (seed length = 18, maximum mismatches within seed = 3, maximum total quality scores at all mismatched read positions = 200) were based on pre-analysis confirming ≤6 CpGs/18 bp and ≤9 CpGs/36 bp read for all possible read positions within the regions of interest. In subsequent summation after string match alignment to score C/T read bases opposite ‘Y’ characters in a second template representation, positions were reported only for C + T calls ≥40. Mismatched internal primer bases returned either a G/Y or CC/YG string match, and so could be discarded. Mean (geometric) C + T call depths for the four data sets reported here were ≥750.
Illumina genome analyzer sequencing
Two lanes per sample of 36-bp single-ended sequencing were performed on the Illumina Genome Analyzer II according to the manufacturer’s protocol. The sequencing was done at the Los Alamos National Laboratory (Los Alamos, NM 87545) and at the NIH Intramural Sequencing Center (Rockville, MD 20852).
Quantitative real time PCR
Selected samples were reverse transcribed in reactions containing 300 ng total RNA [TaqMan Reverse transcription Reagents, Applied Biosystems, Foster City, CA (ABI)]. RNA levels were assayed using a TaqMan Gene-expression Assays (ABI) (FZD1: Hs00268943_s1, FZD7: Hs00275833_s1, TRIP6: Hs01547009_m1, FGF17: Hs00182599_m1) according to the manufacturer’s protocol and standardized against GAPDH levels (TaqMan Human GAPDH).
Genomics analysis
This study utilized the high-performance computational capabilities of the Helix Systems (http://helix.nih.gov) and the Biowulf Linux cluster (http://biowulf.nih.gov) at the NIH, Bethesda, MD. Bioinformatics applications blast, RepeatMasker (Smit, AFA, Hubley, R and Green, P. RepeatMasker Open-3.0.1996–2010 <http://www.repeatmasker.org>) and Bowtie (47) were applied as described in Results. Perl scripts and C programs for additional genomics analyses are provided in the Supplementary Materials or are available on request. Public Database Accession Number [Sequence Read Archive (SRA) division of GenBank[: SRP007666.1.
RESULTS
To investigate structural and dynamic features of the human methylome, we performed MeDIP-Seq. Two independent experiments each involved paired sets of pooled monocyte DNA samples from cord blood samples and from adult blood donors (21–73 years). Taken together, a total of 38 individuals contributed to generate four sample sets. We reasoned that pooling samples would minimize interference due to high inter-individual variation in local DNA-methylation patterns. In support of this strategy, a similar approach has been reported to yield the comparable results to those obtained from individual samples (48).
Genome-wide methylation analysis
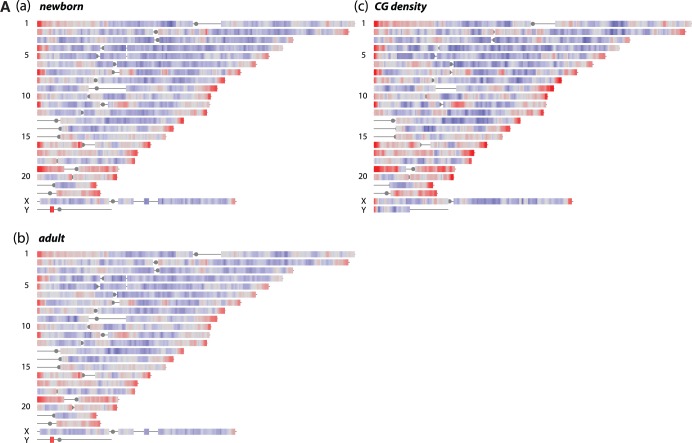
To gain a graphic overview of the results, enrichment patterns were first analyzed at very low (2 Mb) resolution. MeDIP output from reads aligned to single copy (RepeatMasker-excluded) DNA were visualized by a heat map representation of the human chromosomes (Figure 1A, panels a and b). This depiction reveals a striking feature of the organization of the human genome. In many chromosomes, multi-megabase telomere-proximal regions are enriched relative to the input control, whereas under-representation is more common for the internal regions of the chromosome arms. Variation roughly along these lines was noted earlier in fluorescence-based chromosome imaging studies (49). Interestingly, a heat map plot of CG dinucleotide densities, corrected for the single copy (RepeatMasker-excluded) DNA content in 2-Mb windows, generates an almost identical pattern (Figure 1A, panel c). The most straightforward interpretation of these results is that fractional CpG methylation is nearly constant on a 2-Mb scale, so here MeDIP output largely reflects CpG content. Comparison of the two experimental heatmaps reveals only minimal large scale differences between the newborn and adult patterns (Supplementary Figure S1A).
Figure 1.
(A) (a and b) DNA-methylation levels on a whole genome scale. Sequence reads from precipitated MeDIP DNA were analyzed with a 2-Mb genomic window (1-Mb steps) and aligned to single copy (RepeatMasker excluded) regions of the human genome, build 36.1. Red color shows ≥4-fold over-representation, and blue ≤ 4-fold under-representation for MeDIP signals compared to input control DNA. (c) CG dinucleotide densities in single copy regions relative to genome average using same color scale. (B) Over- or under-representation of MeDIP sequence reads flanking transcription start sites (N = 28 939) after normalization for CpG content [values in (a) in Supplementary Figure 1B] × ave/CpG count ratio from (b) in Supplementary Figure 1B). Gray, newborn; black, adult; arrow shows gene orientation. (C) DNA-methylation levels of interspersed repetitive sequences (IRS). The graph represents estimated DNA-methylation levels from MeDIP-Seq for different families of IRS. Enrichment is compared to the control. Columns fully within gray area indicate depletion, i.e. apparent methylation levels lower than in control DNA (≤1, x-axis). Numbers: see key on the right.
Discrimination between non-methylated and methyl-CpG dinucleotides is shown in Figure 1B. This shows over/under-representation versus input for single copy DNA-aligned reads, with positions plotted relative to sense-oriented transcription start sites for all genes. After adjusting for the CpG content of these regions (Supplementary Figure S1B), the estimate for fractional DNA methylation around transcription start sites is clearly low, consistent with the known undermethylation of CpG islands located within promoters (20).
Interspersed repetitive sequences in the genome such as Alus, LINEs, SINEs are generally methylated in normal tissues (24), and can be hypomethylated in cancer cells (50,51). We examined the MeDIP enrichment or depletion relative to input for repetitive sequences grouped into 27 subsets (Figure 1C; shown are averages from the two independent experiments). Apparent DNA methylation levels vary for different subsets of repeated sequences, e.g. high in GA/CT-rich regions and in human satellite DNA, but no significant differences are evident between newborn and adults (it remains possible, of course, that locus-specific age-related differences may occur in such repeats).
Search parameters and variation in detection of methylation differences
Turning to the main focus of the analysis, we next compared MeDIP signals between newborn and adult samples using a sliding 2-kb genomic window. The number of regions sampled in this whole-genome scan is approximately 15 million (window position incremented by 200 bp), but the number of candidate loci for development- or age-related change was found to be strongly dependent on the search parameters selected. In Table 1 are results obtained with very strict parameters: (i) consistent ≥5-fold newborn to adult change in each experiment, and ii) read values elevated in either newborn or adult by ≥6-fold relative to the 2-kb mean. Only two regions were returned from this search, which are listed in the table together with distances to nearby genes. Further details on these regions are described below.
Table 1.
Genes located within 50 kb of age-related change regions
| Symbol | ChrNo | Pos | Fold enrichment | Fold change (nb/ad) | Distance from TS (bp) |
|---|---|---|---|---|---|
| ECEL1P3 | chr2 | 232924316 | 9.58 | 0.14 | 35284 |
| ALPP | chr2 | 232951704 | 9.58 | 0.14 | 7896 |
| ECEL1P2 | chr2 | 232959998 | 9.58 | 0.14 | 398 |
| ALPPL2 | chr2 | 232979797 | 9.58 | 0.14 | 20197 |
| SLC1249 | chr7 | 100288294 | 6.62 | 9.33 | 12906 |
| TRIP6 | chr7 | 100302981 | 6.62 | 9.33 | 1781 |
| ARS2 | chr7 | 100310671 | 6.62 | 9.33 | 9471 |
Threshold criteria: ≥5-fold change and ≥6-fold enrichment in each of three data sets, where fold enrichment is the higher of the newborn(nb)–adult(ad) paired values. ChrNo, chromosome number; Pos, gene position (human genome build 36.1); TS, transcription start.
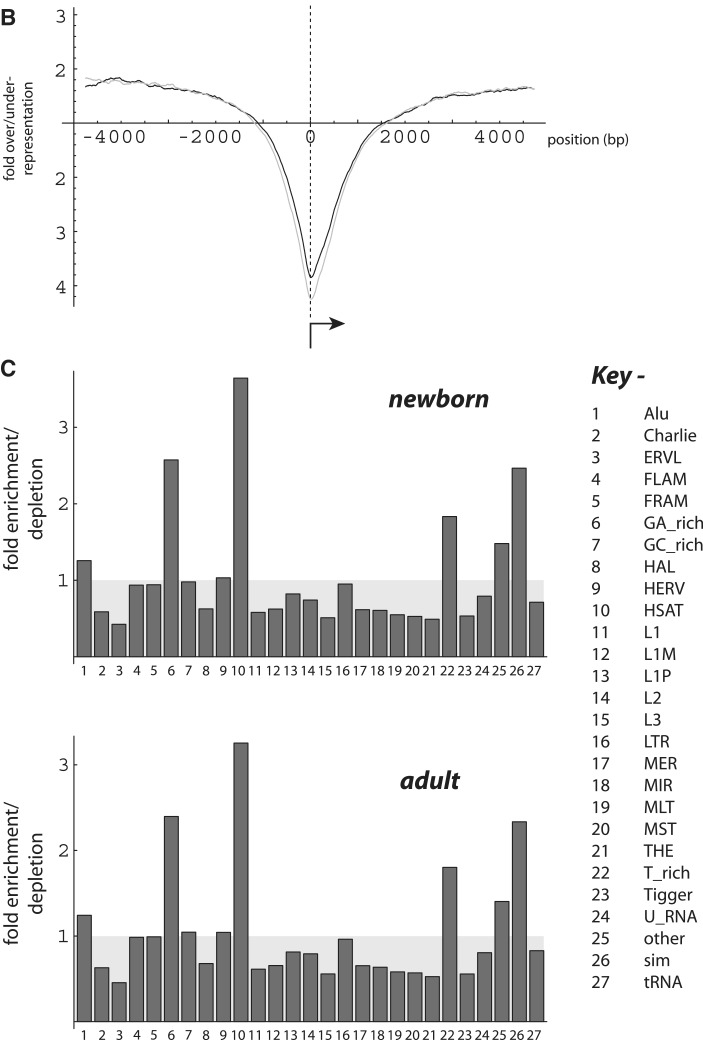
Several strategies were adopted to maximize the number of loci detected while optimizing the signal-to-noise ratio. One option (see following) was to search for changes in MeDIP enrichment extending over larger regions. An alternative option was to include additional data sets. An independent library and sequencing results were available from in vitro-derived DC taken from the same pools of 10 newborns and 10 adults used in the second experiment. During in vitro differentiation, monocytes do not divide at a detectable level (unpublished results) (52,53), so it is unlikely that substantial changes in methylation patterns occur. We reasoned that inclusion of the DC data could be helpful specifically to minimize experimental error associated with library amplification and read sampling. This further provided three biological replicas, which were used to calculate the error bars shown in subsequent figures.
Figure 2 shows several analyses to assess agreement between experiments at 2-kb resolution. With respect to enrichment relative to the mean, the reproducibility with which specific subregions are identified clearly rises as the threshold is increased (Figure 2A). Plotting 173 K paired enrichment values in a binned heatmap format (Figure 2B) provides a measure of the degree of variation observed (correlation coefficient, r = 0.61). As anticipated, high scatter is encountered in calculated post-natal- or age-related change values; however, within a relatively narrow range of parameters, it is possible to detect small numbers of regions that exhibit a high likelihood for consistent change in methylation levels (Figure 2C and D; P < 0.0001 for both r = 0.66 and r = 0.79). A complementary approach to evaluate agreement between experiments is presented in Supplementary Figure S2.
Figure 2.
Experimental agreement between data sets. Enrichment and change values compare data from the first and second independent experiments, where values from the second experiment were derived by averaging the monocyte and dendritic cell data. (A) Abscissa values represent enrichment thresholds (region scored if at least one value ≥ threshold in newborn-adult comparison). For axis values, N(exp) was calculated from the product of the fraction of regions scored in each experiment multiplied by regions sampled (1.5 × 107). N(obs) is the number of regions exceeding threshold in both experiments. (B) Enrichment relative to the mean as in (A), with values in range 2–16 binned and depicted in a heatmap representation. (C) Correlation plot with thresholds for enrichment parameter ≥4.5 and fold-change ≥ 2.5; regions plotted, n = 56. Probability (P) calculated from the Mathematica expression: Abs[r] Sqrt[(n − 2)/(1 − r ^ 2)] > Quantile[StudentTDistribution[n − 2], 0.999]. (D) Correlation plot with thresholds for enrichment parameter ≥4.5 and fold-change ≥3; regions plotted, n = 20.
Methylation differences extending over larger regions
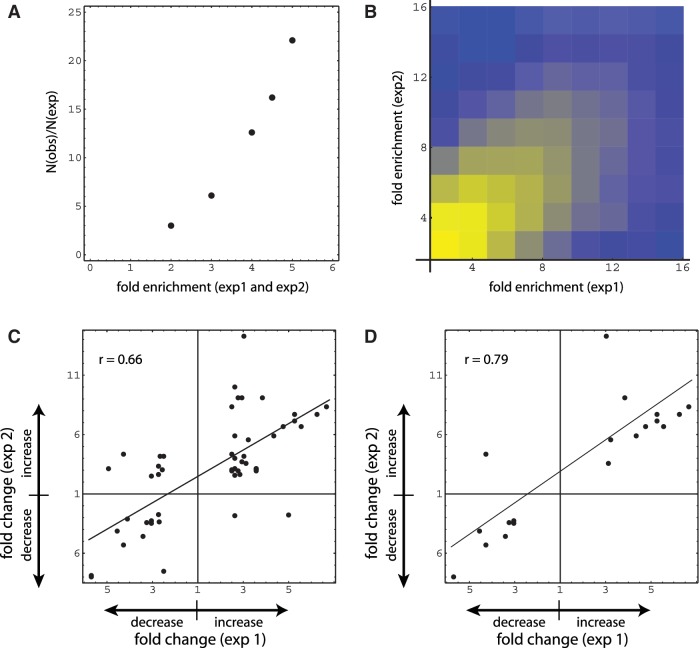
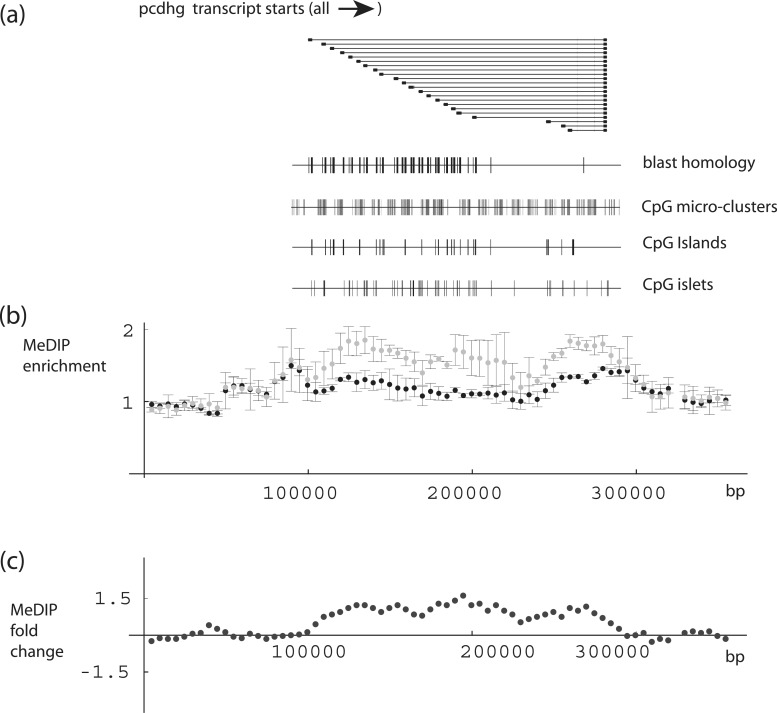
Since epigenome domains are thought to extend in many instances over regions of 10 s of kilobases or more, we examined the sequencing data with the scanning window size varied incrementally ≤200 kb. What emerged from this approach, most strikingly at the 100-kb setting (Figure 3), was detection of a single region that encompasses members of the protocadherin G family (PCDHG). No PCDHG RNA signal could be detected in monocytes (unpublished results); however, PCDHG genes are expressed in the nervous system and encode for cell surface proteins, which play an important role in neuronal connectivity (54). This clustered family displays an unusual organization that is reminiscent of the immunoglobulin and T receptor loci, having multiple variable upstream exons spliced to a constant downstream exon (55,56).
Figure 3.
DNA-methylation levels and structure of the PCDHG gene cluster. (a) Schematic diagram of multiple protocadherin gamma transcripts (human genome build 37.2), together with diagrams showing positions of blast homology domains, CpG micro-clusters, CpG islands and CpG islets within 200-kb PCDHG gene cluster region. CpG islands were calculated according to the original algorithm (57), whereas islets were defined as smaller domains: 200–300 bp versus ≥300 bp for standard islands. CpG micro-clusters were defined by reducing the window size in the standard CpG island algorithm to 50 bp, and requiring the cluster length to fall in the range of 50–200 bp (note that this allows overlap with CpG islets and islands, if the latter contain inhomogeneities at the 50-bp level). Blast search queries for 200-kb region, and corresponding blast database, consisted of 200-bp subregions overlapped by 100-bp steps; search threshold 1 × 10−25. (b) MeDIP enrichment within the PCDHG gene cluster. Each dot represents relative enrichment (read number for specific region (50-kb window; 5-kb step) relative to average reads for given window size). Error bars for each dot calculated for values from three data sets. Black: newborn; gray: adult. (c) DNA-methylation fold change between newborn and adult sample. Each dot represents average change value for the corresponding position in (b).
We posited that the distinct structure of the PCDHG family cluster might reveal features that render certain genome regions susceptible to age-related epigenomic dysregulation. As seen in Figure 3, an increase in MeDIP enrichment with age spans a region containing PCDHG cluster variant coding exons. These coding exons contain numerous CpG islands and islets (overall content ∼38%), and remain undermethylated in both newborn and adult samples. A blast search revealed that there are extensive subregion homologies in the PCDHG cluster, primarily embedded within the variant coding exons (Figure 3). Further examination demonstrated that the regions interspersed between the exons have transition level CpG densities, and that is these latter regions, positioned between or flanking the highly homologous coding exons, which show evidence of methylation changes with age.
Clustered gene families and genes associated with tandem arrays
Motivated by the characteristics of the PCDHG gene cluster, we sought a general means to delineate regions that are normally classified as ‘single copy,’ but in fact have a high degree of homology with one or more regions elsewhere in the human genome. A computationally intensive genome-wide blast cross-homology search was deemed impractical, so virtual ‘sequence reads,’ generated in a uniform tiling pattern across the genome, were aligned using the Bowtie application (47). Such ‘reads’ were retained if: (i) they aligned to more than one genome position (matched at 35/36 positions) and (ii) were devoid of repetitive sequences as classified by RepeatMasker. The subset of sequences so defined (Multipy-Alignable RepeatMasker eXcluded, MAReX) could then be used to annotate the human genome for further analyses. Supplementary Figure S3 compares blast cross-homology output to regions demarcated by the virtual read/Bowtie alignment strategy, showing that similar patterns are produced for the PCDHG gene cluster. At the level of homology required here, MAReX annotated regions comprise 2.7% of the human genome.
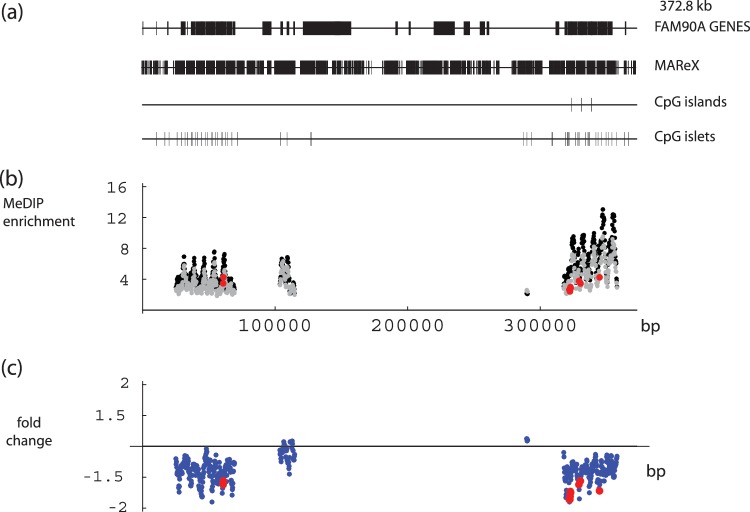
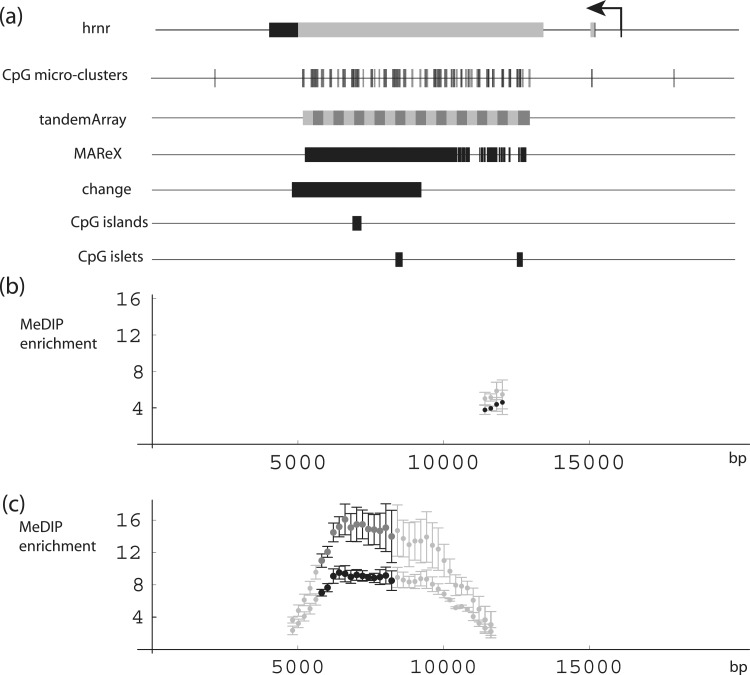
We next analyzed the experimental data using a 2-kb sliding window, but scoring only sequence reads that partially or completely overlap a MAReX domain. Two examples of the results obtained are described. First, Gene family with sequence similarity 90 (FAM90A) (Figure 4) appeared to decrease modestly in DNA methylation with age. This cluster of genes was created by multiple replication and rearrangement events (58), and contains a very high content of multiply-alignable domains within a 375-kb region on chromosome 8. While the magnitude of the decrease is small, the downward drift extends across two relatively large regions. A second interesting example is provided by the hornerin gene (HRNR) (Figure 5). HRNR encodes for a novel profilaggrin-like protein reported to be a differentiation-specific marker in mouse skin (59). In humans, it is expressed in psoriatic and regenerating skin after wounding (60), and it may be present at reduced levels in some cases of atopic dermatitis (61). Within the HRNR-coding region, a 4.4-kb region exhibited consistent increases in MeDIP signals with age in each of three data sets. This change region coincides with a set of tandem repeats that encode repeating HRNR protein domain units. The region of putative methylation instability is rich in CpG micro-clusters (see Figure 3 legend for definition), but less strikingly so in CpG islands or islets. Of note is that neither of the above examples would have been found if the analysis had been limited to uniquely aligned reads. Whether conventional—but lower copy and more complex—repeats, are likewise subject to change, and whether these can be effectively probed with longer or paired end reads, are interesting issues for future study.
Figure 4.
DNA-methylation levels in the FAM90A gene family region. (a) FAM90A gene family structure represented along a region spanning 372.8 kb, including multiply-alignable (MAReX) domains, CpG Islands and CpG islets. (b) MeDIP enrichment across the FAM90A gene family. Values calculated from reads that partially or completely overlap MAReX-annotated genome domains. Data pairs shown only for regions where one or both values ≥ 4-fold enriched. Black: newborn; gray: adult; red: ≥1.5-fold age-related difference in each of three data sets. (c) Apparent fold change in DNA methylation between newborn and adult, values from (b). Red: ≥1.5-fold age-related difference in each of three data sets.
Figure 5.
DNA-methylation levels in hornerin (HRNR) gene region. (a) HRNR gene representation, with coding and non-coding exon regions shown in gray and black, respectively, aligned with CpG micro-clusters, tandem array (repeat elements in alternating gray and black), MAReX domains, region with age-related change in DNA-methylation levels, CpG islands and CpG islets. (b) MeDIP enrichment in HRNR region, with values derived (as conventionally done) from uniquely alignable reads. Gray: adult; black: newborn. Data pairs shown only for regions where one or both values ≥2-fold enriched. Error bars in this and subsequent figures represent three MeDIP data sets. (c) MeDIP enrichment in HRNR region, with values calculated exclusively from reads that partially or completely overlap MAReX-annotated genome domains. Domains with consistent ≥2-fold enrichment and ≥1.5-fold age-related difference in each of three consecutive MeDIP data sets: black. newborn; dark gray, adult. Light gray, domains where one or both values ≥2-fold enriched.
Examples of gene specific age-related methylation differences
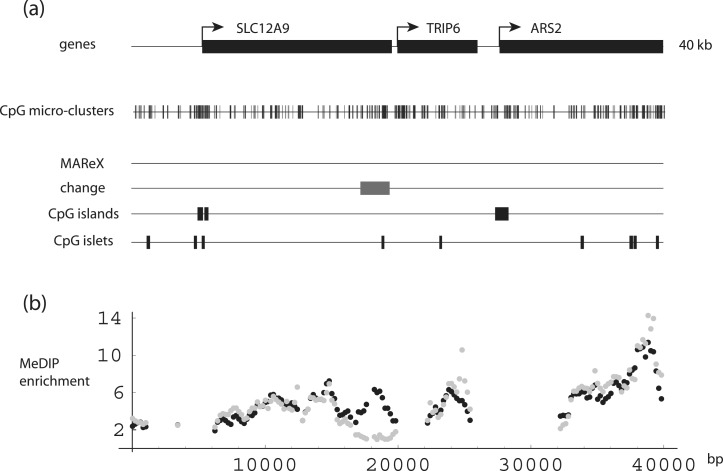
Returning to the seven genes in two regions listed in Table 1, MeDIP enrichment patterns were examined in further detail. On chromosome 7, a region exhibiting a ≥4-fold age-related decrease in each of three data sets is located within the SLC12A9 gene, near (1800-bp upstream from) the TRIP6 transcription start (Figure 6 and Supplementary Figure S4A). This region is devoid of CpG islands, but does contain a single islet. The entire 40-kb region plotted is rich (37%) in CpG micro-clusters (versus 9% for the entire human genome), although these do not coincide noticeably with the change region.
Figure 6.
DNA-methylation levels in the SLC12A9-TRIP6-ARS2 gene region. (a) SLC12A9, TRIP6 and ARS2 gene structures aligned with CpG micro-clusters, MAReX domains, DNA-methylation age-related change region, CpG islands and CpG islets. (b) SLC12A9-TRIP6-ARS2 region MeDIP enrichment. Black: newborn; gray: adult. Data pairs shown only for regions where one or both values ≥ 2-fold enriched (note: Figures 7A and 8 through 10 show both MeDIP and H3K27me3 ChIP data; the latter are omitted in this figure because all values were below the minimal enrichment threshold).
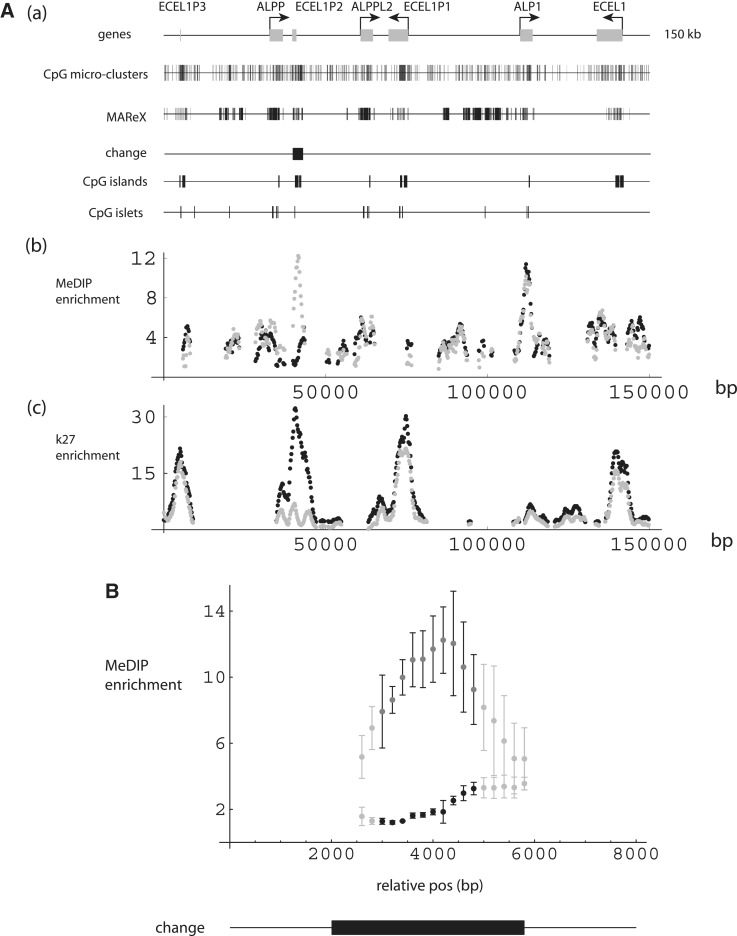
The second region of age-related change in Table 1, ≥5-fold in each data set and 7-fold on average, encompasses a cluster of genes on chromosome 2 generated by multiple genome duplications (62,63). Endothelin converting enzyme-like 1 pseudogene 2 (ECEL1P2) is situated between two alkaline phosphatase genes, the placental (ALPP) and the placental-like 2 (ALPPL2). The latter has also been termed germ-line alkaline phosphatase (64). There is a 7-fold increase in methylation through development across a 3.8-kb region that covers the whole ECEL1P2 gene, including CpG island, islet and micro-clusters (Figure 7A and B). Plotted in this figure are MAReX domains, a subset of which coincides with the ECEL1P2 gene. Examination of Bowtie match patterns revealed, as expected, that the multiple alignments were due to ECEL1 homologs within the chromosome 2 cluster. Additionally plotted are monocyte histone H3–K27 ChIP-seq patterns, which show very substantial enrichment peaks aligned to the ECEL1 gene homologs, with a strong age-related decrease for ECEL1P2. The H3–K27 ChIP-seq data were derived from different individuals (two newborns versus two adults) than those used for MeDIP-seq.
Figure 7.
DNA-methylation levels of the ECEL1–ALP reduplicated region. (A) (a) Schematic diagrams. ECEL1 and ALP gene family structures, with corresponding CpG micro-clusters, MAReX-annotated domains, age-related change region, CpG islands and CpG islets. (b and c) MeDIP and H3K27 trimethylation levels across the ECEL1–ALP region. Black: newborn; gray: adult. Data pairs shown only for regions where one or both values ≥2-fold enriched. (B) ECEL1–ALP subregion centered on area of change (black rectangle). Points plotted in black (newborn) or dark gray (adult) represent regions with ≥6-fold enrichment relative to mean, and ≥2-fold age-related change in the three data sets examined. Remaining data pairs plotted in light gray (newborn and adult) represent regions where one or both values enriched by ≥2-fold.
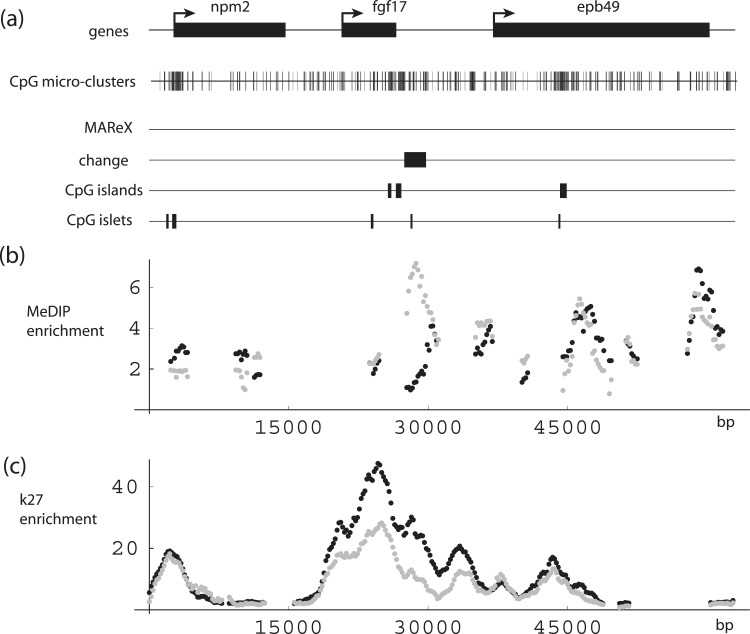
To extend the detection of age-associated methylation alterations associated with nearby genes, we scanned the genome with search parameters set to ≥6-fold MeDIP enrichment and ≥2-fold age-related change in apparent methylation levels in all three data sets. This yielded six additional newborn/adult change regions encompassing 10 genes that have transcription start sites located within a distance of 10 kb (not shown). From this group, fibroblast growth factor 17 (FGF17) ranked just after the ECEL1P2 region in terms of the average fold-change. Downstream of the 3′-end of FGF17 at a distance of 930 bp, there is a 2.2-kb region in which DNA methylation is on average 5-fold higher in the adult samples compared to newborns (Figure 8 and Supplementary Figure S4B). This region contains a CpG islet and multiple CpG micro-clusters, and it also falls within a broad peak of H3–K27 enrichment centered on the FGF17 gene.
Figure 8.
DNA-methylation levels in the FGF17 gene region. (a) Schematic diagrams. NPM2, FGF17 and EPB49 gene structures, together with corresponding CpG micro-clusters, MAReX domains, age-related change region, CpG islands and CpG islets. (b and c) MeDIP and H3K27 trimethylation levels across the FGF17 region. Black: newborn; gray: adult. Data pairs shown only for regions where one or both values ≥2-fold enriched.
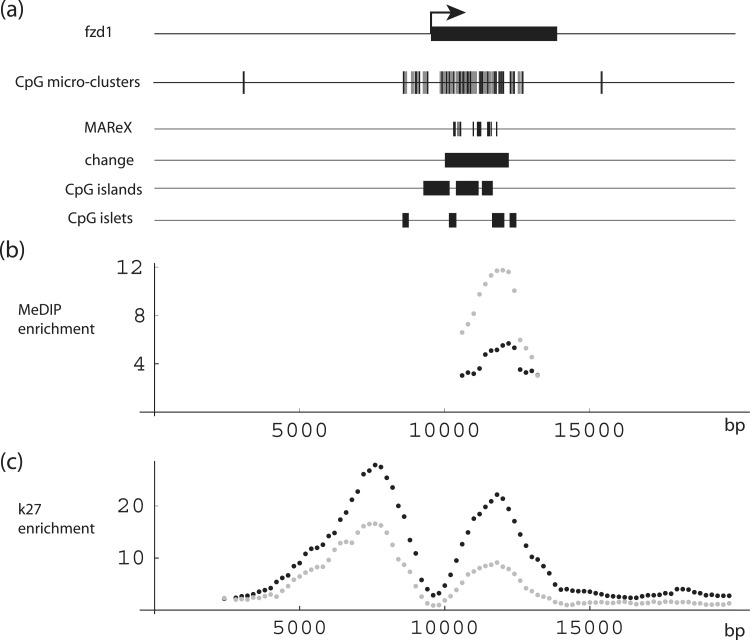
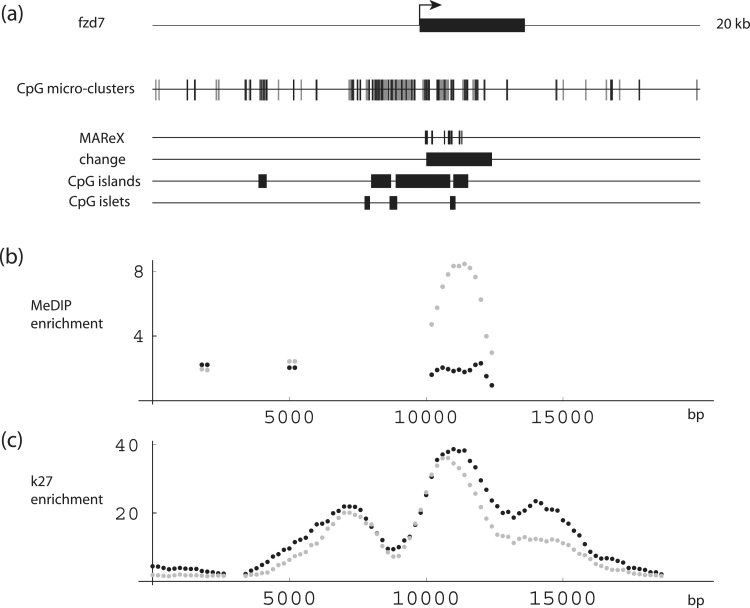
Lastly, Frizzled homolog 1 (FZD1) (Figure 9 and Supplementary Figure S4C) and frizzled homolog 7 (FZD7) (Figure 10 and Supplementary Figure S4D) contained regions with 3- and 5-fold increases in apparent DNA methylation with age, respectively. Frizzled genes encode for transmembrane domain proteins that are receptors for Wnt-signaling proteins. Through the WNT [Wg (wingless) and Int] canonical pathway, FZD products are implicated in cell fate determination, and through the non-canonical pathway they are linked to the early mammalian development (65,66). These regions in both cases include CpG island, islet and micro-cluster features, and striking H3–K27 enrichment peaks are again evident. MAReX domains, which occur within the FZD-coding regions, are due to stretches of high sequence homology shared by FZD1, FZD2 and FZD7.
Figure 9.
DNA-methylation levels in the FZD1 gene region. (a) Schematic diagrams. FZD1 gene structure and corresponding CpG micro-clusters, MAReX domains, age-related MeDIP change region, CpG islands and CpG islets. (b and c) MeDIP and H3K27 trimethylation levels across the FZD1 region. Black: newborn; gray: adult. Data pairs shown only for regions where one or both values ≥2-fold enriched.
Figure 10.
DNA-methylation levels in the FZD7 gene region. (a) Schematic diagrams. FZD7 gene structure and corresponding CpG micro-clusters, MAReX domains, age-related MeDIP change region, CpG islands and CpG islets. (b and c) MeDIP and H3K27 trimethylation levels across the FZD7 region. Black: newborn; gray: adult. Data pairs shown only for regions where one or both values ≥2-fold enriched.
While not the primary focus of this study, RNA measurements were done for several loci. As shown in Supplementary Figure S5A, FZD1 RNA levels increase with age, whereas FZD7 (Supplementary Figure S5B) and TRIP6 RNA (Supplementary Figure S5C) levels exhibit no substantial differences between the two age groups. FGF17 was found to be expressed at such low levels in the monocytes that RNA estimates were considered to be unreliable. Determination of ECEL1P2 and PCDHG gene expression was not attempted, since high homologies between different members of these gene clusters render the development of transcript-specific assays difficult.
Validation of the differentially methylated regions with age by bisulfite sequencing
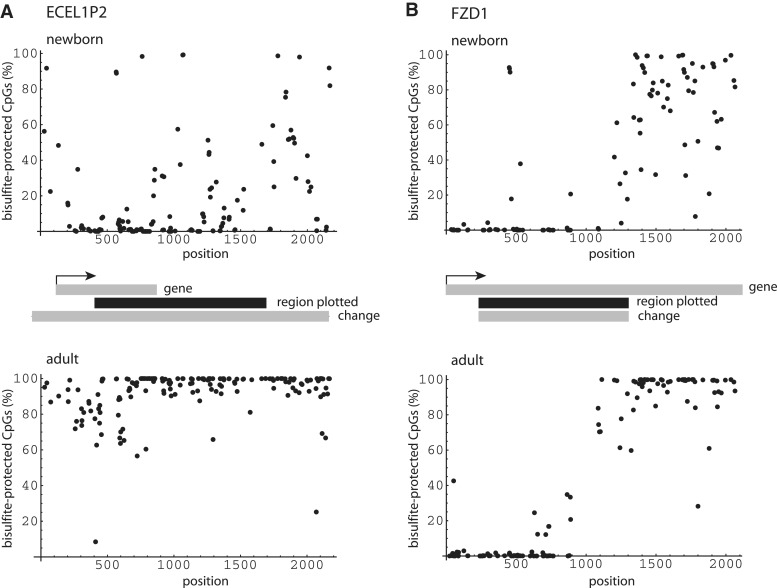
To validate representative MeDIP-Seq results, we employed targeted bisulfite sequencing, which provides single base resolution and is more quantitative than MeDIP. Two 2.2-kb domains were selected, overlapping change regions associated with the ECEL1P2 and FZD1 genes. Pooled monocyte DNA samples from newborn and adult donors were from the same individuals as those used in the initial MeDIP-Seq experiment. To amplify the bisulfite-converted DNA regions of interest, we performed PCR reactions using primers targeted to bracket CpG dinucleotides, followed by a second amplification step with nested primer pairs. Yields of PCR reactions varied widely, so products were normalized before being combined and subjected to next-generation sequencing. As shown in Figure 11, bisulfite analysis confirmed both the ECEL1P2 and FZD1 region MeDIP-Seq results. In the case of ECEL1P2, the percentage of methylated CpGs increased from 19% to 92% (P < 0.0001, t-test for mean difference). Strikingly, the percentage of CpG positions measured to have >90% methylation rose from 5% to 77% (P < 10−40, Fisher’s exact test). For the FZD1 gene, bisulfite analysis revealed a transition at about position 1000, so calculations were based on the downstream subregion, which presumably was the primary determinant for MeDIP enrichment. Here, the percentage of methylated CpGs increased from 69% to 93% (P < 0.0001, t-test for mean difference), while the percentage of CpG positions with >90% methylation rose from 30% to 77% (P < 2 × 10−7, Fisher’s exact test). These results compare to calculated MeDIP change values from the first experiment of 7-fold and 2.6-fold for the ECEL1P2 and FZD1 genes, respectively.
Figure 11.
Methylation in ECEL1P2 and FZD1 by targeted Bisulfite Sequencing (A) Schematic diagram of the percentage of bisulfite protected CpGs (methyl–CpGs) relative to their positions in the region of interest at the ECEL1P2 pseudogene. Coverage of 182 CpGs within the region mapped was 79%. Change (gray rectangle) refers to corresponding region in Figure 7. Upper panel: Newborn. Lower panel: Adult. (B) Percentage of bisulfite protected CpGs relative to the FZD1 gene. Coverage of 79 CpGs within the region spanning positions 1000–2000 was 77%. Change (gray rectangle) refers to corresponding region in Figure 9. Upper panel: Newborn. Lower panel: Adult.
DISCUSSION
In the present study we examined, for the first time on a whole genome scale, age-related differences in human DNA-methylation patterns in cells from newborns and adults. MeDIP followed by next generation sequencing permits the study of DNA methylation without bias towards gene promoters, CpG islands, or other specific regions. This approach was coupled with an experimental design in which pairs of pooled chromatin samples, each derived from 9 or 10 individuals, were derived from cord blood or adult peripheral blood cells. Monocytes were the primary focus of the study, since these cells retain the capacity to differentiate into several different lineages, including DC. In analyzing our data, the primary goal was to identify DNA-methylation domains that undergo age-related epigenome remodeling at sufficient frequencies to emerge from a background of experimental noise and stochastic inter-individual fluctuation (67–72).
Despite the number of samples collected (the data were derived in total from 38 individuals), and that two independent experiments were done, we cannot exclude that factors other than age might have contributed to differences observed. Nevertheless, the findings provide a basis for proposing that a specific class of genome domains is predisposed to progressive post-natal DNA methylation. Once identified, such domains should enable more detailed studies to define how metastable epigenome states may contribute to developmental- and age-related disease.
Several analyses were done to assess MeDIP in this study. Heatmap views at 2-Mb resolution, aligned transcription start sites, and enrichment of interspersed repetitive sequences, all confirmed reproducibility for global and low resolution patterns. At smaller scales, our data support emergence of reproducible signals for developmental- or age-related methylome change primarily for regions that reach high local levels of CpG methylation. Using Bisulfite-Seq, we validated such levels directly for two representative 2-kb regions. These results revealed that transitional, heterogeneous methylation patterns can be present already in newborns. Further experiments will be required to evaluate detailed CpG methylation patterns in individuals, as opposed to pooled samples; however, prior to individual studies, a strategy should be determined, i.e. whether it is adequate to assay single (or small numbers of arbitrarily selected) CpG positions, or instead larger regions should be evaluated, as has been done here.
A striking finding to emerge from scanning the human methylome at a resolution of 100 kb is that the greater part of the protocadherin G (PCDHG) gene cluster is subject to age-related change. PCDHG genes are abundantly expressed in the central nervous system during embryonic development and adulthood (55,73,74), and play a pivotal role in the establishment of specific neuronal connectivity and synapse formation (54–56,74). The unusual genomic organization of this gene cluster is thought to give rise to significant molecular diversity in expression patterns (55,56). Interestingly, the PCDHG cluster is silenced by long- range epigenetic modifications in Wilm’s tumor (75). This provides indirect support to our results, since there is evidence that genes silenced in cancer cells in some instances exhibit a tendency to increased methylation during aging (17,29,31,32,76–78). DNA methylation of another member of PCDH gene family, PCDH10, has been demonstrated to change with post-natal development in mice (29).
In further analysis of the PCDHG gene cluster, we used blast to quantify cross-homologies between variant exons, and to relate these in turn to DNA-methylation patterns. Here it was observed that lower homology regions interspersed between the CpG island-rich exons contribute preferentially to the extended region of age-related change. Nevertheless, considerable evidence exists in the literature that tandemly repeated sequence arrays are prone to progressive DNA methylation and silencing (79–83). An obvious question was whether numerous genome regions might exist where interspersed-tandem arrays of complex ‘single copy’ sequences are predisposed to age-related methylation. This led us to implement a bioinformatic strategy for annotating ‘non-repetitive’ short sequence regions that have one or more near perfect copies elsewhere in the genome, i.e. are multiply-alignable and RepeatMasker excluded (MAReX). Restricting searches to such annotated domains yielded gene regions characterized by multiple duplication and rearrangement events (e.g. FAM90A), as well as genes with internal tandem repeating domains (e.g. HRNR) that would normally have eluded detection.
Strongly supportive of with the idea that selected tandemly arrayed gene clusters can be subject to developmental or age-related DNA methylation are results relating to the ECEL1P2 pseudogene, where we observed a remarkable 7-fold age-related increase in MeDIP enrichment, validated by Bisulfite-Seq analysis. The 150 Kb region encompassing this pseudogene, as well as ECEL1 and alkaline phosphatase gene family copies, has undergone duplication and partial triplication events within the human genome (63). As expected, numerous MAReX annotated domains fall within and between these genes. We note further the strong peaks of H3–K27 enrichment corresponding to the set of ECEL1-related genes. Apparent DNA-methylation levels do not change in ECEL1 and the other ECEL1-related copies, so it seems that a particular combination of CpG micro-cluster density, H3–K27 modification, and other variables not yet identified may be necessary for such age-related epigenome remodeling to occur.
Of particular interest for future study are several genes identified using optimized search parameters, namely FGF17, FZD1 and FZD7, for which there are well established links to differentiation and developmental programs (65,66,84–89). The FZD1 and FZD7 transcripts are unspliced, so these might be considered processed genes that remain functional. In common with ECEL1P2, both FZD genes contain multiply-alignable (MAReX) domains, a consequence of highly homologous regions shared by FZD1, FZD2 and FZD7. FGF17 lacks MAReX domains by the criteria set here, but shares regions with FGF8 and FGF18, which are nearly as homologous as those shared between the FZD1/FZD2/FZD7 set. Change regions for FZD1 and FZD7 fall within transcription units, whereas the corresponding region for FGF17 is located <1 kb from the 3′-end of the gene. Finally, like ECEL1P2, and the PCDHG variable exon region (unpublished data), FGF17, FZD1 and FZD7 are all associated with strong peaks of H3–K27 enrichment.
QPCR analysis revealed an increase in FZD1 expression in adult individuals relative to newborns (2.7-fold; P < 10−5), whereas no significant differences in FZD7 RNA levels were observed (Supplementary Figure S5A and S5B). For FZD1, the direction of expression change is inversely proportional to the H3K27me3 marks, consistent with the generally repressive function of such marks. Conversely, FZD1 expression parallels the direction of DNA-methylation change. The latter result is unexplained, but we note that the CpG island located at the 5′-end of the gene remains unmethylated. Elevated DNA methylation in the gene bodies of expressed genes has been reported, and may play a role in transcription or alternative splicing linked to early stages of cell lineage formation (21–23,26). Bisulfite-Seq analysis confirmed a roughly 1-kb region with high methyl-CpG density within the FZD1 gene body in adults, and it is conceivable that such densities can displace proteins associated with repressive H3K27me3 marks.
Finally, we would like to address intriguing commonalities between the regions reported here to exhibit increasing post-natal DNA methylation. In addition to the expected combinations of CpG islands, islets and micro-clusters, all encompass, or closely flank, domains for which highly homologous copies exist elsewhere in the human genome, and all are associated with H3–K27 enrichment. As already noted, tandemly arrayed transgenes are subject to H3–K27 and DNA methylation (79–83). Yet most tandemly arrayed genes may be silenced too rapidly to exhibit post-natal developmental change (90), whereas small, interspersed or more distant homology regions may interact with appropriate kinetics. Given this background, we postulate that the latter feature will be observed frequently as more details emerge concerning age-related epigenome dynamics. Other investigators have postulated that gene function diversification can explain the high observed content of multi-copy gene families (including conserved pseudogenes), as well as complex reduplicated regions in mammalian genomes (91–95). Our results suggest that such highly homologous multi-copy regions may provide a mechanistic basis for developmental and age-related epigenome remodeling. If so, this fundamental feature of genome architecture in higher eukaryotic organisms merits much more extensive study.
ACCESSION NUMBERS
Public Database Accession Number [Sequence Read Archive (SRA) division of GenBank]: SRP007666.1.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–5.
FUNDING
Funding for open access charge: Intramural Research Program of the National Institutes of Health (National Institute of Child Health and Human Development).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors acknowledge the NIH Blood Bank and all of their staff, especially Cindy Matthews and Amy Melpolder (Department of Transfusion Medicine, National Institutes of Health) for their contribution in the collection of all the adult blood samples used in the present study. The authors are very grateful to Sandy Field (Perinatology Research lab) for her valuable help in the collection of cord blood samples, to Kirin Prasad for contributions to the QPCR analysis, and to the reviewers of the manuscript for helpful suggestions.
REFERENCES
- 1.de Boer J, Andressoo JO, de Wit J, Huijmans J, Beems RB, van Steeg H, Weeda G, van der Horst GT, van Leeuwen W, Themmen AP, et al. Premature aging in mice deficient in DNA repair and transcription. Science. 2002;296:1276–1279. doi: 10.1126/science.1070174. [DOI] [PubMed] [Google Scholar]
- 2.Maslov AY, Vijg J. Genome instability, cancer and aging. Biochim. Biophys. Acta. 2009;1790:963–969. doi: 10.1016/j.bbagen.2009.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Finkel T, Serrano M, Blasco MA. The common biology of cancer and ageing. Nature. 2007;448:767–774. doi: 10.1038/nature05985. [DOI] [PubMed] [Google Scholar]
- 4.Kirkwood TB. Understanding the odd science of aging. Cell. 2005;120:437–447. doi: 10.1016/j.cell.2005.01.027. [DOI] [PubMed] [Google Scholar]
- 5.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 6.Whitelaw NC, Whitelaw E. How lifetimes shape epigenotype within and across generations. Hum. Mol. Genet. 2006;15(Spec No 2):R131–R137. doi: 10.1093/hmg/ddl200. [DOI] [PubMed] [Google Scholar]
- 7.Howard BH. Replicative senescence: considerations relating to the stability of heterochromatin domains. Exp. Gerontol. 1996;31:281–293. doi: 10.1016/0531-5565(95)00022-4. [DOI] [PubMed] [Google Scholar]
- 8.de Magalhaes JP, Church GM. Genomes optimize reproduction: aging as a consequence of the developmental program. Physiology. 2005;20:252–259. doi: 10.1152/physiol.00010.2005. [DOI] [PubMed] [Google Scholar]
- 9.Finch C. Longevity, Senescence, and the Genome. Chicago: University of Chicago Press; 1990. [Google Scholar]
- 10.Martino DJ, Tulic MK, Gordon L, Hodder M, Richman T, Metcalfe J, Prescott SL, Saffery R. Evidence for age-related and individual-specific changes in DNA methylation profile of mononuclear cells during early immune development in humans. Epigenetics. 2011;6:1085–1094. doi: 10.4161/epi.6.9.16401. [DOI] [PubMed] [Google Scholar]
- 11.Takasugi M. Progressive age-dependent DNA methylation changes start before adulthood in mouse tissues. Mech. Ageing Dev. 2011;132:65–71. doi: 10.1016/j.mad.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 12.Godfrey KM, Lillycrop KA, Burdge GC, Gluckman PD, Hanson MA. Epigenetic mechanisms and the mismatch concept of the developmental origins of health and disease. Pediatr. Res. 2007;61:5R–10R. doi: 10.1203/pdr.0b013e318045bedb. [DOI] [PubMed] [Google Scholar]
- 13.Ozanne SE, Constancia M. Mechanisms of disease: the developmental origins of disease and the role of the epigenotype. Nat. Clin. Pract. Endocrinol. Metab. 2007;3:539–546. doi: 10.1038/ncpendmet0531. [DOI] [PubMed] [Google Scholar]
- 14.Bocker MT, Hellwig I, Breiling A, Eckstein V, Ho AD, Lyko F. Genome-wide promoter DNA methylation dynamics of human hematopoietic progenitor cells during differentiation and aging. Blood. 2011;117:e182–e189. doi: 10.1182/blood-2011-01-331926. [DOI] [PubMed] [Google Scholar]
- 15.De Haan G, Gerrits A. Epigenetic control of hematopoietic stem cell aging the case of Ezh2. Ann. N Y Acad. Sci. 2007;1106:233–239. doi: 10.1196/annals.1392.008. [DOI] [PubMed] [Google Scholar]
- 16.Pollina EA, Brunet A. Epigenetic regulation of aging stem cells. Oncogene. 2011;30:3105–3126. doi: 10.1038/onc.2011.45. [DOI] [PubMed] [Google Scholar]
- 17.Teschendorff AE, Menon U, Gentry-Maharaj A, Ramus SJ, Weisenberger DJ, Shen H, Campan M, Noushmehr H, Bell CG, Maxwell AP, et al. Age-dependent DNA methylation of genes that are suppressed in stem cells is a hallmark of cancer. Genome Res. 2010;20:440–446. doi: 10.1101/gr.103606.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jaenisch R. DNA methylation and imprinting: why bother? Trends Genet. 1997;13:323–329. doi: 10.1016/s0168-9525(97)01180-3. [DOI] [PubMed] [Google Scholar]
- 19.Bird A, Taggart M, Frommer M, Miller OJ, Macleod D. A fraction of the mouse genome that is derived from islands of nonmethylated, CpG-rich DNA. Cell. 1985;40:91–99. doi: 10.1016/0092-8674(85)90312-5. [DOI] [PubMed] [Google Scholar]
- 20.Bird AP. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- 21.Ball MP, Li JB, Gao Y, Lee JH, LeProust EM, Park IH, Xie B, Daley GQ, Church GM. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat. Biotechnol. 2009;27:361–368. doi: 10.1038/nbt.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brenet F, Moh M, Funk P, Feierstein E, Viale AJ, Socci ND, Scandura JM. DNA methylation of the first exon is tightly linked to transcriptional silencing. PLoS One. 2011;6:e14524. doi: 10.1371/journal.pone.0014524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laurent L, Wong E, Li G, Huynh T, Tsirigos A, Ong CT, Low HM, Kin Sung KW, Rigoutsos I, Loring J, et al. Dynamic changes in the human methylome during differentiation. Genome Res. 2010;20:320–331. doi: 10.1101/gr.101907.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13:335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- 25.Doi A, Park IH, Wen B, Murakami P, Aryee MJ, Irizarry R, Herb B, Ladd-Acosta C, Rho J, Loewer S, et al. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat. Genet. 2009;41:1350–1353. doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deaton AM, Webb S, Kerr AR, Illingworth RS, Guy J, Andrews R, Bird A. Cell type-specific DNA methylation at intragenic CpG islands in the immune system. Genome Res. 2011;21:1074–1086. doi: 10.1101/gr.118703.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oakes CC, Smiraglia DJ, Plass C, Trasler JM, Robaire B. Aging results in hypermethylation of ribosomal DNA in sperm and liver of male rats. Proc. Natl Acad. Sci. USA. 2003;100:1775–1780. doi: 10.1073/pnas.0437971100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Swisshelm K, Disteche CM, Thorvaldsen J, Nelson A, Salk D. Age-related increase in methylation of ribosomal genes and inactivation of chromosome-specific rRNA gene clusters in mouse. Mutat. Res. 1990;237:131–146. doi: 10.1016/0921-8734(90)90019-n. [DOI] [PubMed] [Google Scholar]
- 29.Maegawa S, Hinkal G, Kim HS, Shen L, Zhang L, Zhang J, Zhang N, Liang S, Donehower LA, Issa JP. Widespread and tissue specific age-related DNA methylation changes in mice. Genome Res. 2010;20:332–340. doi: 10.1101/gr.096826.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thompson RF, Atzmon G, Gheorghe C, Liang HQ, Lowes C, Greally JM, Barzilai N. Tissue-specific dysregulation of DNA methylation in aging. Aging Cell. 2010;9:506–518. doi: 10.1111/j.1474-9726.2010.00577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Issa JP, Ottaviano YL, Celano P, Hamilton SR, Davidson NE, Baylin SB. Methylation of the oestrogen receptor CpG island links ageing and neoplasia in human colon. Nat. Genet. 1994;7:536–540. doi: 10.1038/ng0894-536. [DOI] [PubMed] [Google Scholar]
- 32.Kwabi-Addo B, Chung W, Shen L, Ittmann M, Wheeler T, Jelinek J, Issa JP. Age-related DNA methylation changes in normal human prostate tissues. Clin. Cancer Res. 2007;13:3796–3802. doi: 10.1158/1078-0432.CCR-07-0085. [DOI] [PubMed] [Google Scholar]
- 33.Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, Heine-Suner D, Cigudosa JC, Urioste M, Benitez J, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc. Natl Acad. Sci. USA. 2005;102:10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bjornsson HT, Sigurdsson MI, Fallin MD, Irizarry RA, Aspelund T, Cui H, Yu W, Rongione MA, Ekstrom TJ, Harris TB, et al. Intra-individual change over time in DNA methylation with familial clustering. JAMA. 2008;299:2877–2883. doi: 10.1001/jama.299.24.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boks MP, Derks EM, Weisenberger DJ, Strengman E, Janson E, Sommer IE, Kahn RS, Ophoff RA. The relationship of DNA methylation with age, gender and genotype in twins and healthy controls. PLoS One. 2009;4:e6767. doi: 10.1371/journal.pone.0006767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Christensen BC, Houseman EA, Marsit CJ, Zheng S, Wrensch MR, Wiemels JL, Nelson HH, Karagas MR, Padbury JF, Bueno R, et al. Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS Genet. 2009;5:e1000602. doi: 10.1371/journal.pgen.1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Langevin SM, Houseman EA, Christensen BC, Wiencke JK, Nelson HH, Karagas MR, Marsit CJ, Kelsey KT. The influence of aging, environmental exposures and local sequence features on the variation of DNA methylation in blood. Epigenetics. 2011;6:908–919. doi: 10.4161/epi.6.7.16431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuwana M, Okazaki Y, Kodama H, Izumi K, Yasuoka H, Ogawa Y, Kawakami Y, Ikeda Y. Human circulating CD14+ monocytes as a source of progenitors that exhibit mesenchymal cell differentiation. J. Leukoc. Biol. 2003;74:833–845. doi: 10.1189/jlb.0403170. [DOI] [PubMed] [Google Scholar]
- 39.Randolph GJ, Inaba K, Robbiani DF, Steinman RM, Muller WA. Differentiation of phagocytic monocytes into lymph node dendritic cells in vivo. Immunity. 1999;11:753–761. doi: 10.1016/s1074-7613(00)80149-1. [DOI] [PubMed] [Google Scholar]
- 40.Ruhnke M, Ungefroren H, Nussler A, Martin F, Brulport M, Schormann W, Hengstler JG, Klapper W, Ulrichs K, Hutchinson JA, et al. Differentiation of in vitro-modified human peripheral blood monocytes into hepatocyte-like and pancreatic islet-like cells. Gastroenterology. 2005;128:1774–1786. doi: 10.1053/j.gastro.2005.03.029. [DOI] [PubMed] [Google Scholar]
- 41.Zhou LJ, Tedder TF. CD14+ blood monocytes can differentiate into functionally mature CD83+ dendritic cells. Proc. Natl Acad. Sci. USA. 1996;93:2588–2592. doi: 10.1073/pnas.93.6.2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lehner M, Holter W. Endotoxin-free purification of monocytes for dendritic cell generation via discontinuous density gradient centrifugation based on diluted Ficoll-Paque Plus. Int. Arch Allergy Immunol. 2002;128:73–76. doi: 10.1159/000058006. [DOI] [PubMed] [Google Scholar]
- 43.Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schubeler D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat. Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 44.Russanova VR, Hirai TH, Tchernov AV, Howard BH. Mapping development-related and age-related chromatin remodeling by a high throughput ChIP-HPLC approach. J. Gerontol. A Biol. Sci. Med. Sci. 2004;59:1234–1243. doi: 10.1093/gerona/59.12.1234. [DOI] [PubMed] [Google Scholar]
- 45.Porras A, Kozar S, Russanova V, Salpea P, Hirai T, Sammons N, Mittal P, Kim JY, Ozato K, Romero R, et al. Developmental and epigenetic regulation of the human TLR3 gene. Mol. Immunol. 2008;46:27–36. doi: 10.1016/j.molimm.2008.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Down TA, Rakyan VK, Turner DJ, Flicek P, Li H, Kulesha E, Graf S, Johnson N, Herrero J, Tomazou EM, et al. A Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis. Nat. Biotechnol. 2008;26:779–785. doi: 10.1038/nbt1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Docherty SJ, Davis OS, Haworth CM, Plomin R, Mill J. Bisulfite-based epityping on pooled genomic DNA provides an accurate estimate of average group DNA methylation. Epigenet. Chromatin. 2009;2:3. doi: 10.1186/1756-8935-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brock GJ, Charlton J, Bird A. Densely methylated sequences that are preferentially localized at telomere-proximal regions of human chromosomes. Gene. 1999;240:269–277. doi: 10.1016/s0378-1119(99)00442-4. [DOI] [PubMed] [Google Scholar]
- 50.Issa JP. CpG island methylator phenotype in cancer. Nat. Rev. Cancer. 2004;4:988–993. doi: 10.1038/nrc1507. [DOI] [PubMed] [Google Scholar]
- 51.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 52.van Furth R, Raeburn JA, van Zwet TL. Characteristics of human mononuclear phagocytes. Blood. 1979;54:485–500. [PubMed] [Google Scholar]
- 53.Pickl WF, Majdic O, Kohl P, Stockl J, Riedl E, Scheinecker C, Bello-Fernandez C, Knapp W. Molecular and functional characteristics of dendritic cells generated from highly purified CD14+ peripheral blood monocytes. J. Immunol. 1996;157:3850–3859. [PubMed] [Google Scholar]
- 54.Morishita H, Yagi T. Protocadherin family: diversity, structure, and function. Curr. Opin. Cell Biol. 2007;19:584–592. doi: 10.1016/j.ceb.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 55.Wu Q, Maniatis T. A striking organization of a large family of human neural cadherin-like cell adhesion genes. Cell. 1999;97:779–790. doi: 10.1016/s0092-8674(00)80789-8. [DOI] [PubMed] [Google Scholar]
- 56.Wu Q, Zhang T, Cheng JF, Kim Y, Grimwood J, Schmutz J, Dickson M, Noonan JP, Zhang MQ, Myers RM, et al. Comparative DNA sequence analysis of mouse and human protocadherin gene clusters. Genome Res. 2001;11:389–404. doi: 10.1101/gr.167301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J. Mol. Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 58.Bosch N, Caceres M, Cardone MF, Carreras A, Ballana E, Rocchi M, Armengol L, Estivill X. Characterization and evolution of the novel gene family FAM90A in primates originated by multiple duplication and rearrangement events. Hum. Mol. Genet. 2007;16:2572–2582. doi: 10.1093/hmg/ddm209. [DOI] [PubMed] [Google Scholar]
- 59.Makino T, Takaishi M, Morohashi M, Huh NH. Hornerin, a novel profilaggrin-like protein and differentiation-specific marker isolated from mouse skin. J. Biol. Chem. 2001;276:47445–47452. doi: 10.1074/jbc.M107512200. [DOI] [PubMed] [Google Scholar]
- 60.Takaishi M, Makino T, Morohashi M, Huh NH. Identification of human hornerin and its expression in regenerating and psoriatic skin. J. Biol. Chem. 2005;280:4696–4703. doi: 10.1074/jbc.M409026200. [DOI] [PubMed] [Google Scholar]
- 61.Henry J, Hsu CY, Haftek M, Nachat R, de Koning HD, Gardinal-Galera I, Hitomi K, Balica S, Jean-Decoster C, Schmitt AM, et al. Hornerin is a component of the epidermal cornified cell envelopes. FASEB J. 2011;25:1567–1576. doi: 10.1096/fj.10-168658. [DOI] [PubMed] [Google Scholar]
- 62.Knoll BJ, Rothblum KN, Longley M. Nucleotide sequence of the human placental alkaline phosphatase gene. Evolution of the 5' flanking region by deletion/substitution. J. Biol. Chem. 1988;263:12020–12027. [PubMed] [Google Scholar]
- 63.Rump A, Kasper G, Hayes C, Wen G, Starke H, Liehr T, Lehmann R, Lagemann D, Rosenthal A. Complex arrangement of genes within a 220-kb region of double-duplicated DNA on human 2q37.1. Genomics. 2001;73:50–55. doi: 10.1006/geno.2000.6504. [DOI] [PubMed] [Google Scholar]
- 64.Beckman G, Beckman L, Sikstrom C, Millan JL. DNA polymorphism of alkaline phosphatase isozyme genes: linkage disequilibria between placental and germ-cell alkaline phosphatase alleles. Am. J. Hum. Genet. 1992;51:1066–1070. [PMC free article] [PubMed] [Google Scholar]
- 65.Katoh M. WNT signaling in stem cell biology and regenerative medicine. Curr. Drug Targets. 2008;9:565–570. doi: 10.2174/138945008784911750. [DOI] [PubMed] [Google Scholar]
- 66.Yu H, Smallwood PM, Wang Y, Vidaltamayo R, Reed R, Nathans J. Frizzled 1 and frizzled 2 genes function in palate, ventricular septum and neural tube closure: general implications for tissue fusion processes. Development. 2010;137:3707–3717. doi: 10.1242/dev.052001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Flanagan JM, Popendikyte V, Pozdniakovaite N, Sobolev M, Assadzadeh A, Schumacher A, Zangeneh M, Lau L, Virtanen C, Wang SC, et al. Intra- and interindividual epigenetic variation in human germ cells. Am. J. Hum. Genet. 2006;79:67–84. doi: 10.1086/504729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Heijmans BT, Kremer D, Tobi EW, Boomsma DI, Slagboom PE. Heritable rather than age-related environmental and stochastic factors dominate variation in DNA methylation of the human IGF2/H19 locus. Hum. Mol. Genet. 2007;16:547–554. doi: 10.1093/hmg/ddm010. [DOI] [PubMed] [Google Scholar]
- 69.Illingworth R, Kerr A, Desousa D, Jorgensen H, Ellis P, Stalker J, Jackson D, Clee C, Plumb R, Rogers J, et al. A novel CpG island set identifies tissue-specific methylation at developmental gene loci. PLoS Biol. 2008;6:e22. doi: 10.1371/journal.pbio.0060022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sandovici I, Kassovska-Bratinova S, Loredo-Osti JC, Leppert M, Suarez A, Stewart R, Bautista FD, Schiraldi M, Sapienza C. Interindividual variability and parent of origin DNA methylation differences at specific human Alu elements. Hum. Mol. Genet. 2005;14:2135–2143. doi: 10.1093/hmg/ddi218. [DOI] [PubMed] [Google Scholar]
- 71.Schneider E, Pliushch G, El Hajj N, Galetzka D, Puhl A, Schorsch M, Frauenknecht K, Riepert T, Tresch A, Muller AM, et al. Spatial, temporal and interindividual epigenetic variation of functionally important DNA methylation patterns. Nucleic Acids Res. 2010;38:3880–3890. doi: 10.1093/nar/gkq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Turan N, Katari S, Coutifaris C, Sapienza C. Explaining inter-individual variability in phenotype: is epigenetics up to the challenge? Epigenetics. 2010;5:16–19. doi: 10.4161/epi.5.1.10557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hirano S, Ono T, Yan Q, Wang X, Sonta S, Suzuki ST. Protocadherin 2C: a new member of the protocadherin 2 subfamily expressed in a redundant manner with OL-protocadherin in the developing brain. Biochem. Biophys. Res. Commun. 1999;260:641–645. doi: 10.1006/bbrc.1999.0950. [DOI] [PubMed] [Google Scholar]
- 74.Kohmura N, Senzaki K, Hamada S, Kai N, Yasuda R, Watanabe M, Ishii H, Yasuda M, Mishina M, Yagi T. Diversity revealed by a novel family of cadherins expressed in neurons at a synaptic complex. Neuron. 1998;20:1137–1151. doi: 10.1016/s0896-6273(00)80495-x. [DOI] [PubMed] [Google Scholar]
- 75.Dallosso AR, Hancock AL, Szemes M, Moorwood K, Chilukamarri L, Tsai HH, Sarkar A, Barasch J, Vuononvirta R, Jones C, et al. Frequent long-range epigenetic silencing of protocadherin gene clusters on chromosome 5q31 in Wilms' tumor. PLoS Genet. 2009;5:e1000745. doi: 10.1371/journal.pgen.1000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ahuja N, Li Q, Mohan AL, Baylin SB, Issa JP. Aging and DNA methylation in colorectal mucosa and cancer. Cancer Res. 1998;58:5489–5494. [PubMed] [Google Scholar]
- 77.Issa JP. CpG-island methylation in aging and cancer. Curr. Top Microbiol. Immunol. 2000;249:101–118. doi: 10.1007/978-3-642-59696-4_7. [DOI] [PubMed] [Google Scholar]
- 78.Rakyan VK, Down TA, Maslau S, Andrew T, Yang TP, Beyan H, Whittaker P, McCann OT, Finer S, Valdes AM, et al. Human aging-associated DNA hypermethylation occurs preferentially at bivalent chromatin domains. Genome Res. 2010;20:434–439. doi: 10.1101/gr.103101.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bender J. Cytosine methylation of repeated sequences in eukaryotes: the role of DNA pairing. Trends Biochem. Sci. 1998;23:252–256. doi: 10.1016/s0968-0004(98)01225-0. [DOI] [PubMed] [Google Scholar]
- 80.Bestor T, Tycko B. Creation of genomic methylation patterns. Nat. Genet. 1996;12:363–367. doi: 10.1038/ng0496-363. [DOI] [PubMed] [Google Scholar]
- 81.Dorer DR, Henikoff S. Transgene repeat arrays interact with distant heterochromatin and cause silencing in cis and trans. Genetics. 1997;147:1181–1190. doi: 10.1093/genetics/147.3.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Garrick D, Fiering S, Martin DI, Whitelaw E. Repeat-induced gene silencing in mammals. Nat. Genet. 1998;18:56–59. doi: 10.1038/ng0198-56. [DOI] [PubMed] [Google Scholar]
- 83.Mutskov V, Felsenfeld G. Silencing of transgene transcription precedes methylation of promoter DNA and histone H3 lysine 9. EMBO J. 2004;23:138–149. doi: 10.1038/sj.emboj.7600013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hoshikawa M, Ohbayashi N, Yonamine A, Konishi M, Ozaki K, Fukui S, Itoh N. Structure and expression of a novel fibroblast growth factor, FGF-17, preferentially expressed in the embryonic brain. Biochem. Biophys. Res. Commun. 1998;244:187–191. doi: 10.1006/bbrc.1998.8239. [DOI] [PubMed] [Google Scholar]
- 85.Huang HC, Klein PS. The Frizzled family: receptors for multiple signal transduction pathways. Genome Biol. 2004;5:234. doi: 10.1186/gb-2004-5-7-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mariani FV, Ahn CP, Martin GR. Genetic evidence that FGFs have an instructive role in limb proximal-distal patterning. Nature. 2008;453:401–405. doi: 10.1038/nature06876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Reya T, Duncan AW, Ailles L, Domen J, Scherer DC, Willert K, Hintz L, Nusse R, Weissman IL. A role for Wnt signalling in self-renewal of haematopoietic stem cells. Nature. 2003;423:409–414. doi: 10.1038/nature01593. [DOI] [PubMed] [Google Scholar]
- 88.Sun X, Lewandoski M, Meyers EN, Liu YH, Maxson RE, Jr, Martin GR. Conditional inactivation of Fgf4 reveals complexity of signalling during limb bud development. Nat. Genet. 2000;25:83–86. doi: 10.1038/75644. [DOI] [PubMed] [Google Scholar]
- 89.Winklbauer R, Medina A, Swain RK, Steinbeisser H. Frizzled-7 signalling controls tissue separation during Xenopus gastrulation. Nature. 2001;413:856–860. doi: 10.1038/35101621. [DOI] [PubMed] [Google Scholar]
- 90.Robertson G, Garrick D, Wilson M, Martin DI, Whitelaw E. Age-dependent silencing of globin transgenes in the mouse. Nucleic Acids Res. 1996;24:1465–1471. doi: 10.1093/nar/24.8.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Henikoff S, Greene EA, Pietrokovski S, Bork P, Attwood TK, Hood L. Gene families: the taxonomy of protein paralogs and chimeras. Science. 1997;278:609–614. doi: 10.1126/science.278.5338.609. [DOI] [PubMed] [Google Scholar]
- 92.Itoh N, Ornitz DM. Evolution of the Fgf and Fgfr gene families. Trends Genet. 2004;20:563–569. doi: 10.1016/j.tig.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 93.Marques AC, Vinckenbosch N, Brawand D, Kaessmann H. Functional diversification of duplicate genes through subcellular adaptation of encoded proteins. Genome Biol. 2008;9:R54. doi: 10.1186/gb-2008-9-3-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ohno S. Evolution by Gene Duplication. Berlin: Springer Verlag; 1970. [Google Scholar]
- 95.Young JM, Friedman C, Williams EM, Ross JA, Tonnes-Priddy L, Trask BJ. Different evolutionary processes shaped the mouse and human olfactory receptor gene families. Hum. Mol. Genet. 2002;11:535–546. doi: 10.1093/hmg/11.5.535. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.