Abstract
The imprinted Snurf–Snrpn chromosomal domain contains two large arrays of tandemly repeated, paternally expressed box C/D small-nucleolar RNA (snoRNA) genes: the SNORD115 (H/MBII-52) and SNORD116 (H/MBII-85) gene clusters believed to play key roles in the fine-tuning of serotonin receptor (5-HT2C) pre-mRNA processing and in the etiology of the Prader–Willi Syndrome (PWS), respectively. SNORD115 and SNORD116 were recently proposed to undergo significant conversion into shorter RNA species, the so-called psnoRNAs. Here, we provide evidence that argues against the existence of abundant psnoRNAs in human or mouse brain. Instead, we characterize a previously unsuspected low-abundance, fibrillarin-associated SNORD115-derived smaller RNA species. Based on these findings, we strongly recommend that PWS-encoded SNORD115 and SNORD116 be considered as bona fide box C/D snoRNAs.
INTRODUCTION
Small-nucleolar RNAs (snoRNAs) represent a complex family of small non-coding RNAs (∼65–300 nt in length) that concentrate mostly in the nucleolus where most, but not all, direct site-specific post-transcriptional RNA modifications, particularly onto pre-ribosomal RNAs (rRNAs) or U6 spliceosomal snRNAs (1–3). Except for the RNA component of RNAse MRP, two classes of snoRNAs have been identified so far: the C/D- and H/ACA box snoRNAs that guide, through the formation of specific RNA duplexes with their cellular RNA targets, the biogenesis of 2′-O-ribose methylation and pseudouridines, respectively (4–6). Both C/D and H/ACA snoRNAs function as ribonucleoparticle (snoRNP) complexes consisting of a single snoRNA associated with a specific set of four common proteins, the 2′-O-methyltransferase fibrillarin (Nop1p), Nhpx (Snu13p), NOP56p and NOP58p for C/D snoRNAs and the pseudouridine synthase Dyskerin (Cbf5p), GAR1p, NHP2p and NOP10p for H/ACA snoRNAs (1–3). Importantly, all C/D boxes fold into a highly structured stem–bulge–stem motif—the so-called kink-turn motif (k-turn)—that provides an RNA platform for the biogenesis of snoRNP complexes (3,7).
A growing list of publications has recently demonstrated that C/D and H/ACA snoRNAs are prone to be processed into shorter RNA species, some of which display a microRNA-like silencing function on the mRNA of reporter genes (8–15). Hence, an intriguing possibility is that a subset of tiny regulatory RNAs, particularly microRNAs, may have evolved from snoRNA sequences (16–18). However, our understanding of the functional relevance of these snoRNA-derived small RNAs is still in its infancy, and the possibility that some of them might simply represent metabolically stable, non-functional RNA degradation products cannot be ruled out.
The imprinted Prader–Willi Syndrome (PWS) locus at human 15q11q13—also referred to as the Snurf–Snrpn domain in the mouse (Chr.7C)—expresses numerous C/D snoRNA genes of paternal origin that are organized into two large arrays: the SNORD115 and SNORD116 gene clusters (previously called HBII-52 and HBII-85, respectively). Most PWS-encoded snoRNAs are embedded within, and processed from, repeated introns of huge non-coding transcripts (19–22) (see also Figure 1A). In mouse, both SNORD115 and SNORD116 (previously called MBII-52 and MBII-85, respectively) are mainly, if not exclusively, expressed in the brain (their expression is restricted to neurons) while only SNORD115 is strictly brain-specific in humans (21). Their mode of action remains largely unknown since they lack obvious antisense elements against rRNA or spliceosomal snRNAs, the two classical RNA targets for mammalian C/D snoRNAs. Deciphering their functions is, nevertheless, of primordial importance since SNORD115 is strongly suspected to fine-tune post-transcriptional processing of the serotonin (5-HT2C) receptor pre-mRNA, the first such occurrence for a mammalian snoRNA (23,24), while lack of expression of SNORD116 gene arrays may contribute significantly to the PWS (25–29).
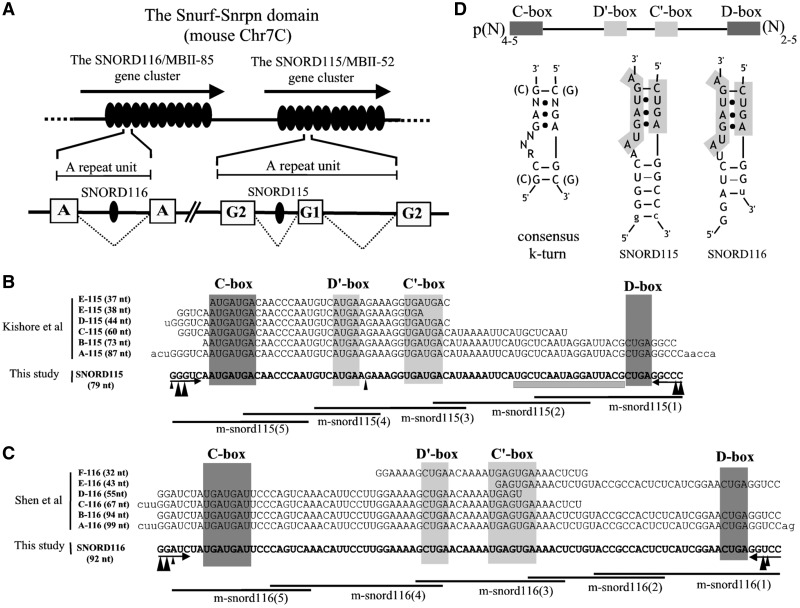
Figure 1.
A complex array of tandemly repeated C/D snoRNA genes mapping at the imprinted Prader–Willi Syndrome (PWS) locus. (A) Schematic representation of the two major arrays of repeated C/D snoRNA genes mapping at the mouse Snurf–Snrpn locus: the SNORD115 and SNORD116 genes [formerly known as MBII-52 and MBII-85, respectively (20)]. C/D snoRNA sequences (symbolized as black ovals) are embedded within repeated introns of long non-protein-coding RNAs comprising repeated exons (white rectangles, G2- and G1-exons for SNORD115 and A-exons for SNORD116). (B) Sequences of full-length mouse SNORD115 (A-115 form) and related psnoRNA species (E-115, D-115, C-115 and B-115 forms) as reported by Kishore et al. (31). The sequence of mouse SNORD115 as originally predicted (20) and used in this study is written in bold characters, with small vertical black arrowheads positioning the 5′ and 3′ extremities determined experimentally. Note that the sequence of the A-form., as well as the D-form, contains additional nucleotides from the surrounding intronic sequences (lower-case letters). The C/D and C′/D′ boxes are highlighted in dark and light grey, respectively, while the canonical (4–5 bp) terminal stem is indicated by inverted small horizontal arrows. The conserved antisense element matching the serotonin receptor (5-HT2C) mRNA is underlined by a horizontal grey bar. The relative positions of DNA oligonucleotides used in this study are also shown below the sequence alignments. (C) Sequences of full-length mouse SNORD116 (A-116 form) and related psnoRNA species (F-116, E-116, D-116, C-116 and B-116 forms) as reported by Shen et al. (30). Legends are as in panel (B). The sequence of the A-116 form considered by Shen et al. (30), as well as the C-116 form, contain additional nucleotides from the surrounding intronic sequences (lower-case letters). Note that the mouse SNORD116 RNA sequence, as originally reported in (20) and written here in bold characters, was edited at 2 nt positions (+7 and +69) to fit with the currently available mouse SNORD116 genomic sequences. (D) Top: Schematic representation of the consensus structure of mammalian box C/D snoRNAs (6). Left: Schematic representation of a consensus K-turn motif (7). SNORD115 (middle) and SNORD116 (right) display canonical 5′–3′-terminal stem structures that bring together the (C) and (D) boxes (highlighted in grey) that form part of the K-turn motif.
SNORD116 and SNORD115 were recently proposed to be massively processed into shorter RNA species, called psnoRNAs (30,31). Remarkably, one SNORD115-derived small RNA appeared to be the predominant cellular form of SNORD115 in mouse brain (31). If correct, such findings would have profound implications for our understanding of the evolution, biogenesis and mode of action of imprinted C/D snoRNAs, and also very likely in terms of their potential impact on the etiology of the Prader–Willi disease (27–29) and also perhaps that of autism (32). In this report, we provide both experimental and conceptual evidence that strongly argues against the notion that PWS-encoded snoRNAs generate psnoRNAs as previously documented. We conclude instead that PWS-encoded snoRNAs represent canonical fibrillarin-associated box C/D snoRNAs whose function remains elusive.
MATERIALS AND METHODS
Unless otherwise noted, all techniques for manipulating nucleic acids were performed according to standard protocols.
Primers
DNA oligonucleotides were purchased from Sigma. m-snord115(1): GGGCCTCAGCGTAATCCTATTGA; m-snord115(2): TCCTATTGAGCATGAATTTTATGT; m-snoRD115(3): TATGTCATCACCTTTCTTCATGAC; m-snoRD115(4): TCTTCATGACATTGGGTTGTC; m-snoRD115(5): TTGGGTTGTCATCATTGACCC; m-snoRD116(1): ACCTCAGTTCCGATGAGAGTGGCGG; m-snoRD116(2): GAGTGGCGGTACAGAGTTTTC; m-snoRD116(3): GTTTTCACTCATTTTGTTCAGC; m-snoRD116(4): CAGCTTTTCCAAGGAATGTTTGACTG; m-snoRD116(5): GACTGGGAATCATCATAGAT; 03: GAAGAAAGGTGATGACATAAAATTCATGC; 01: AATAAAGCGGCCGCGGATCCAA, 02: TTGGATCCGCGGCCGCTTTATT; U3 snoRNA: GCCGGCTTCACGCTCAG; 7SK: GTGTCTGGAGTCTTGGAAGC.
T4-RNA ligase-PCR, primer extension
total RNA (25 µg) extracted from adult mouse brain were ligated to phosphorylated primer 01 (10 pmole). Ligation products were used as a template for cDNA synthesis with primer 02 and AMV-Reverse Transcriptase (Promega). The cDNA product was then used as template for PCR by Taq polymerase (Promega) with primers 03 and 02, and the PCR products were cloned into pGEM T-easy vector (Promega). Individual clones were sequenced. Primer extension was carried out as described in (33).
RNA extraction, Northern-blot analysis
Total RNA was prepared from cells using Tri-Reagent (Euromedex) and treated with RQ1 RNAse free DNAse (Promega) and proteinase K (Sigma) before storage at −20°C in RNAse-free water. Northern-blot analysis with DNA oligoprobes was carried out as described in (33). For hybridization with gel-purified 32P internally labeled riboprobes (200 000 cpm/ml), membranes were incubated overnight at 50°C in 5× SSPE, 50% formamide, 5× Denhardt's, 0.5% SDS and 150 µg/ml tRNA, followed by extensive washes [15 min at 65°C in 2× SSPE/0.1% SDS (×2); 30′ at 65°C in 1× SSPE/0.1% SDS]; 10 min at 65°C in 0.1× SSPE/0.1% SDS and 15′ at room temperature in 0.1× SSPE/0.1% SDS].
Brain extracts, immunoprecipitation
A dissected adult mouse brain was resuspended in 7 ml of NET-150 buffer (50 mM Tris–HCl, pH 7.4, 150 mM NaCl, 0.05% Igepal) and homogenized at 1.53 kbars using the One Shot Model (Constant Systems Ltd.). The brain extract was then clarified by centrifugation [10 min, 10 000 rpm, Beckman–Coulter (JA14) at 4°C] and the supernatant was used for immunoprecipitation as described in (33), using antibodies against fibrillarin (72B9) provided by M. Pollard.
RESULTS AND DISCUSSION
Through poorly understood processing events, SNORD115 was recently shown to generate several smaller RNA species—collectively termed psnoRNAs—that are believed to regulate alternative splicing of several pre-mRNAs (31). The sequences of the psnoRNAs derived from SNORD115 (referred to herein as the B-115, C-115, D-115 and E-115 forms) are given in Figure 1B. Intriguingly, the most abundant B-115 form lacks a few nucleotides at both ends, more especially at its 5′-end, thus rendering this truncated RNA unable to fold into the characteristic 5′–3′ terminal stem as originally proposed (20). Hence, these observations suggest that SNORD115 is massively converted into shorter snoRNA sub-fragments and, as a corollary, that SNORD115 might not be a traditional snoRNA (31). The use of a novel method designed to clone double-stranded RNAs generated by the Ribonuclease Protection Assay (RPA) also led to the conclusion that SNORD116 gives rise to shorter, yet poorly characterized RNA species (30). The sequences of the psnoRNAs derived from SNORD116 (referred to herein as the B-116, C-116, D-116, E-116 and F-116 forms) are given in Figure 1C.
To ascertain the existence of psnoRNAs in adult total mouse brain RNA, we first re-examined directly, using conventional, unbiased methods, the extremities of endogenously expressed SNORD115 species. To delineate their 5′-ends, we carried out primer extensions using the 32P-labeled m-snord115(4) oligonucleotide designed to base pair with all putative psnoRNAs (Figure 1B). As illustrated in Figure 2A, and as expected for C/D snoRNA having a canonical terminal 5′–3′ stem (Figure 1D), we detected two major abundant cDNA products, whose 5′-ends are positioned 4 and 5 nt upstream of the C-Box. Minor stop signals 6 nt upstream of the C-box were also routinely observed. It is important to note that we never observed stop signals mapping 1 nt upstream of the C-box, inconsistent with the existence of the abundant B-115 psnoRNA (Figure 1B). We next determined the 3′-end of SNORD115 species through the use of an RNA ligation-PCR procedure (6). Briefly, total mouse brain RNA was oligonucleotide-tagged at both ends by T4 RNA ligase and amplified by RT-PCR using oligonucleotides as depicted at the top of Figure 2B. Amplified cDNAs were cloned and 40 independent randomly chosen clones were sequenced. As shown at the bottom of Figure 2B, the 3′ termini of 37 out of 40 clones mapped 4–5 nt downstream of the D-box, again in agreement with the presence of a canonical terminal stem structure (Figure 1D). Incidentally, we noted the replacement of genomically encoded Cs by Us (or As) in a subset of sequenced clones, presumably reflecting rebuilding of the 3′-end by post-transcriptional uridylation (or adenylation), as previously reported for other small RNAs (34). We conclude from these results that the bulk of small RNA species produced by the SNORD115 gene array in adult mouse brain corresponds to box C/D-containing small RNAs with canonical 5′–3′-terminal stem structures (Figure 1D).
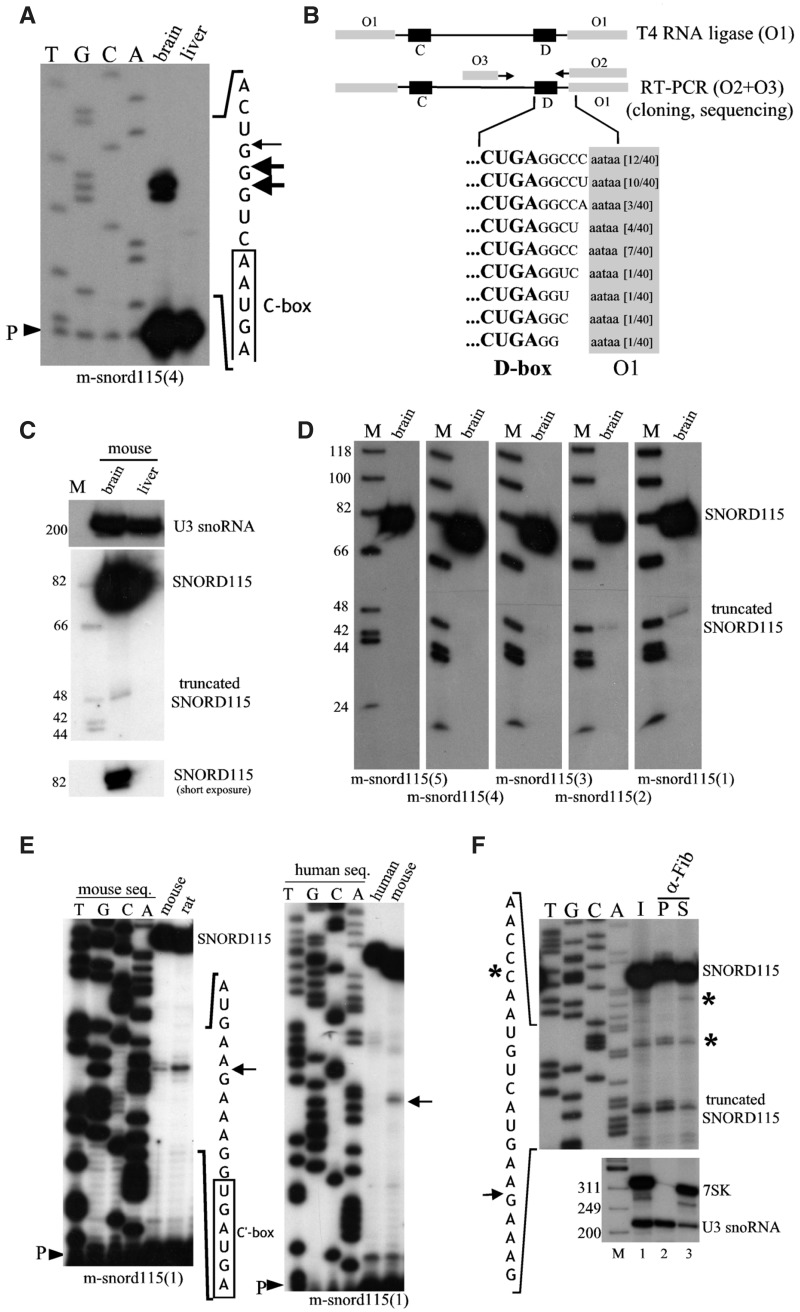
Figure 2.
The major cellular form of SNORD115 possesses a canonical 5′–3′ terminal stem structure and generates a truncated C′–D box small RNA. (A) Identifying the 5′-ends of SNORD115. Primer extension with 32P-labeled oligoprobe m-snord115(4) using 10 µg of total RNA extracted from brain or liver (negative control). P: primer. (B) Identifying the 3′-ends of SNORD115. Mouse brain total RNA was oligonucleotide-tagged at both ends (primer O1) and amplified by RT-PCR (primers O2 and O3). The numbers of sequenced clones are indicated in brackets, sequences of the 3′ arm are highlighted in grey. (C) Northern-blot hybridization to internally 32P-labeled antisense mouse SNORD115 riboprobes using 10 µg of total RNA extracted from brain or liver as indicated above the panels. Two exposures (long and short) are shown for SNORD115. U3 snoRNA was used as a gel loading control. (D) Northern-blot hybridization to 5′-32P end-labeled DNA oligonucleotides (sequences are given in Figure 1B) using 10 µg of total mouse brain RNA. (E) 5′-end identification of the 48-nt-long SNORD115-derived small RNA by primer extension with 32P-labeled snord115(1) using 10 µg of total RNA extracted from rat and mouse brains (left panel) or human and mouse brain (right panel). P: primer. (F) Immunoprecipitation by anti-fibrillarin antibodies (72B9, a generous gift of Dr. M. Pollard). Top panel: full-length SNORD115 and its 5′-truncated form were detected by primer extension as indicated in (E). The asterisks denote signals not reproducibly detected and that most likely correspond to RNA degradation that occurs upon brain extract preparation. I: Input (1/10); S: Supernatant (1/10); P: Pellet. Bottom panel: The same RNA samples were also assayed by Northern-blot analysis with U3- and 7SK-specific oligo-probes used as positive and negative controls, respectively. M: DNA marker (nt).
Spliceosomal small nuclear RNAs (10), transfer RNAs (10,35–39), snoRNAs (8–15,40), vault RNA (41) and also very likely Y RNAs (42) can be sources for smaller RNA species. Indeed, highly sensitive deep-sequencing approaches have revealed that most, if not all, mammalian box C/D snoRNAs appear to be fragmented into smaller RNA pieces (see http://deepbase.sysu.edu.cn/ for examples). However, the functional significance, if any, of such rather low abundance, truncated snoRNA forms remains to be demonstrated. To identify such hypothetical SNORD115-derived small RNA species that may have been overlooked by our 5′- and 3′-end mapping procedures, Northern-blot analysis was performed using a full-length, 32P-internally labeled antisense SNORD115 riboprobe. Interestingly, in addition to the expected full-length, strong SNORD115 signal, we also reproducibly detected the presence of a ∼48- to 49-nt-long RNA with relatively modest steady-state levels relative to full length SNORD115 (Figure 2C). Such truncated RNA species were also detected in rat but not in human brain (see below Figure 2E).
To better characterize these putative novel small RNAs in mouse brain, Northern-blot analyses were performed using various 32P-labeled DNA oligoprobes that systematically overlapped different RNA segments along the SNORD115 sequence (Figure 1B). As shown in Figure 2D, two oligoprobes—m-snord115(1) and m-snord115(2)—detected these shorter 48- to 49-nt-long RNAs, indicating that they comprise the 3′ half of SNORD115. Primer extension using 32P-labeled m-snord115(1) further confirmed the existence of these 5′-truncated snoRNA fragments in rat and mouse brain, by positioning their 5′-termini ∼5–6 nt upstream of the C′ box (Figure 2E, left panel). Interestingly, these short RNA species are not detected in human brain (Figure 2E, right panel). These low-abundance endogenously expressed SNORD115-derived small RNAs—corresponding to C′/D box containing snoRNAs—are very likely to be metabolically stable in adult mouse brain since their association with fibrillarin is in the same order of magnitude as that observed for full-length, 79- to 80-nt-long SNORD115 (Figure 2F).
We next assayed for the presence of psnoRNA species produced by the flanking SNORD116 gene arrays in mouse, rat and human brains. As shown in Figure 3A, Northern-blot analysis carried out with a full-length 32P-internally labeled antisense SNORD116 riboprobe revealed strong signals restricted mainly to a single band. As could be expected, the 5′ and 3′ termini of full-length mouse SNORD116—as determined by primer extension and T4 RNA-ligase based PCR methods (Figure 3B and C, respectively)—were consistent with the observed electrophoretic mobility of the main SNORD116 signal (∼92 nt long). We conclude from these results that human, rat and mouse SNORD116 genes generate mainly, if not exclusively, classical C/D box-containing snoRNAs (Figure 1D). This conclusion is further reinforced by results from Northern blots with various overlapping oligo-probes (Figure 1C) that do not reveal any obvious shorter RNA species, despite a strong signal for full length SNORD115 (Figure 3D).
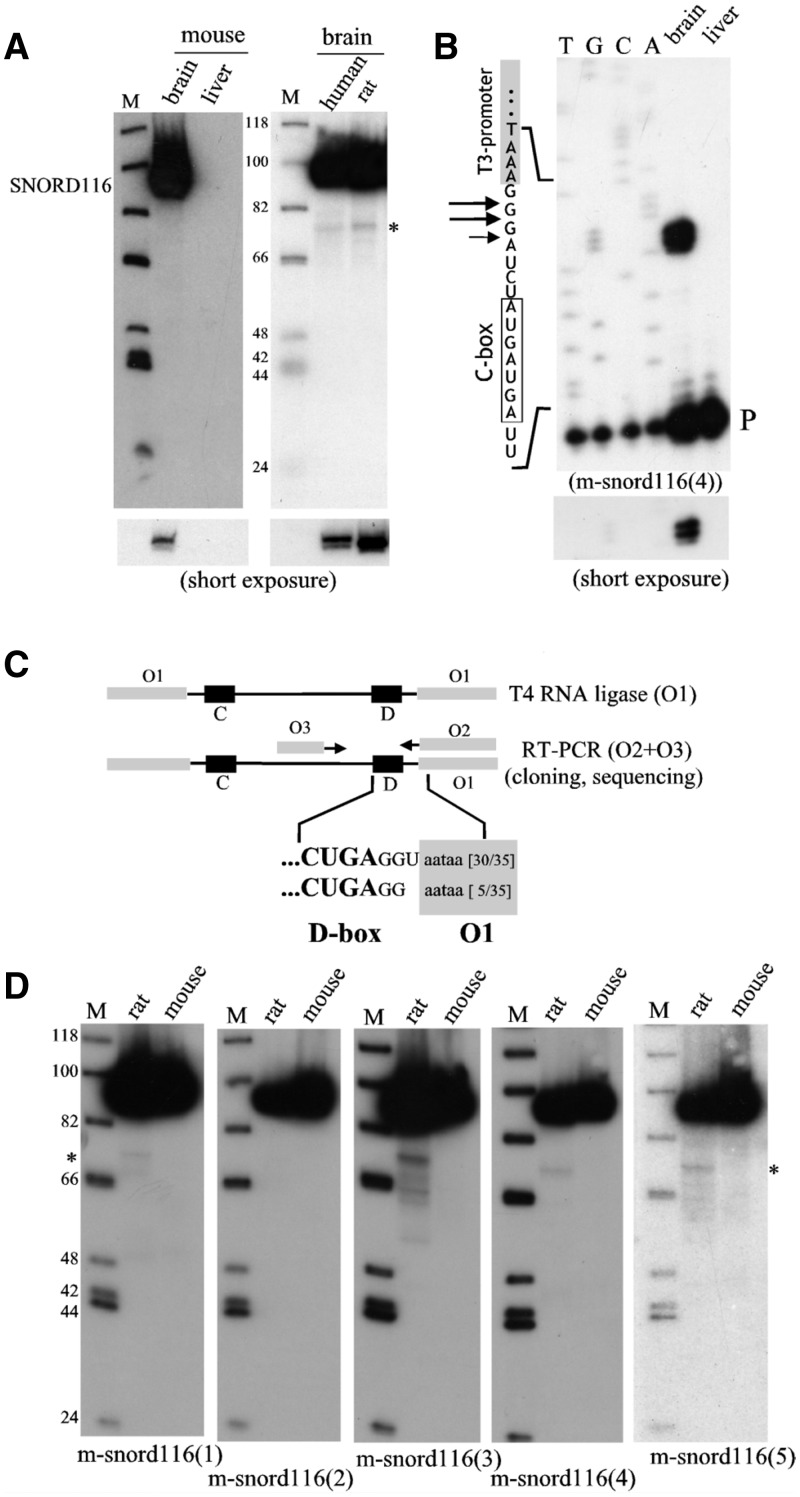
Figure 3.
The major cellular form of SNORD116 possesses a canonical 5′–3′ terminal stem structure. (A) Northern-blot hybridization with internally 32P-labeled antisense mouse (left panel) or human (right panel) SNORD116 riboprobes using 10 µg of total RNA extracted from tissues as indicated above the gels. Two exposures (long and short) are shown for SNORD116. (B and C) Legends are as in Figure 2A and B. Note that the DNA sequencing ladder used to map precisely the 5′ end of SNORD116 was generated from a plasmid harboring a SNORD116 gene copy inserted downstream of T3 promoter, as indicated to the left of the gel. (D) Northern-blot hybridization to 5′-32P end-labeled DNA oligonucleotides (sequences are given in Figure 1C) using 10 µg of total mouse or rat brain RNA. Only a very faint band was observed in rat but not in mouse brains (denoted by an asterisk, see also Figure 3A). Its relevance remains, however, questionable since both the 5′- and 3′-distal oligo-probes revealed it, an observation inconsistent with its apparent electrophoretic mobility. Note that the membrane first hybridized to m-snoRd116(4) was stripped and then re-used for hybridization to m-snoRD116(5).
The conclusions according to which SNORD115 is processed into shorter snoRNA fragments were mostly drawn from a ribonuclease protection assay (RPA). Importantly, the structure of the abundant B115-form, as well in the case of other SNORD115-derived psnoRNAs, was deduced after cloning and sequencing of RNA fragments generated in vitro by RPA, rather than being determined directly from undigested mouse brain RNA preparations. Moreover, we noted that the SNORD115 riboprobe used in RPA does not strictly encompass the SNORD115 sequence as originally published (20): it includes at both ends a few additional nucleotides from surrounding intronic sequences (Figure 1B and its legend). As a consequence, such a riboprobe will reveal fully processed SNORD115 (and possibly any shorter RNA species) as well as any longer SNORD115-containing intronic RNA species.
Keeping these important technical issues in mind, we propose to re-interpret the pattern of protected RNA fragments [Figure 4A in (31)] as follows: (i) the longest 87-nt-long A115-form does not represent full-length SNORD115 but corresponds to a protected RNA fragment generated from unspliced SNORD115-containing precursors and/or partially trimmed, spliced-out SNORD115-containing intermediates (hence their inherent low abundance). This conclusion also explains why the A115-form is apparently not detected by Northern blot using 15% acrylamide gels [see Figure 4B in (31)] since such SNORD115-containing transcripts (potentially several kb in length) are unlikely to be transferred efficiently onto the nylon membrane. (ii) The B115-form simply corresponds to the bona fide 79- to 80-nt-long SNORD115 as originally published (20) and described here (hence its high abundance), and not to any truncated, smaller 73-nt-long RNA fragment lacking the terminal stem. This interpretation also reconciles the discrepancies, between the apparent electrophoretic mobility of the B115-form and its experimentally determined sequence: B-115 is assumed to be a 73-nt-long RNA fragment but migrates just below the 80-nt RNA marker. Finally, we suggest that other lower-abundance psnoRNAs (the C-115, D-115 and E-115 forms) may represent endogenous RNA degradation intermediates and/or internally cleaved RNA species derived from imperfect RNA duplexes formed between the riboprobe (corresponding to a single SNORD115 sequence) and numerous SNORD115 copies with partial sequence similarity.
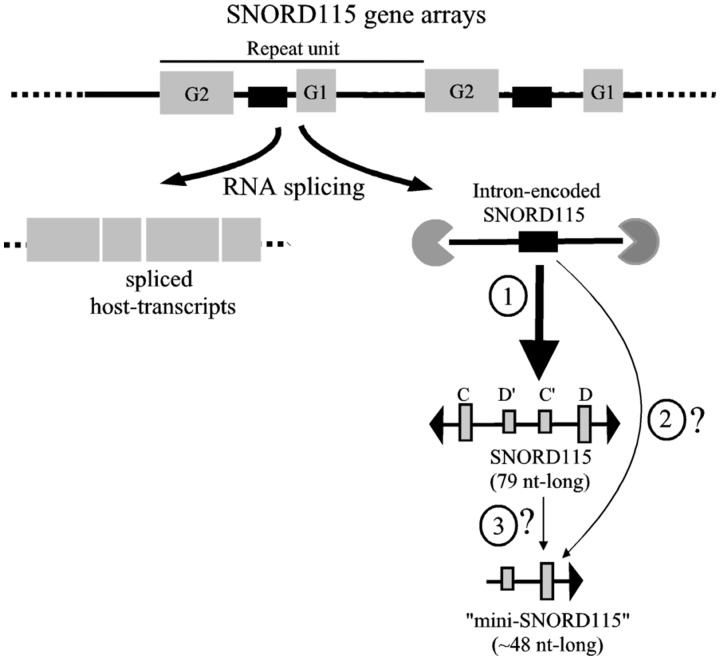
Figure 4.
Revisiting snoRNP biogenesis at the Snurf–Snrpn domain. The major cellular RNA form generated at the SNORD115 gene arrays from the Snurf–Snrpn domain corresponds to a canonical box C/D snoRNAs, most likely produced from exonucleolytic trimming of the debranched, spliced-out intron, as assumed for other intron-encoded snoRNAs (47) (pathway 1). The same holds true for SNORD116 gene arrays (data not shown). SNORD115 also generates low-abundance 5′-truncated C′–D snoRNPs. This may occur during the nucleoplasmic snoRNP assembly events before, or concomitantly with, RNA splicing of the host intron, via a minor (or aberrant) alternative snoRNA-processing pathway (pathway 2). Alternatively, C′/D snoRNPs, whose functional relevance—if any—remains to be demonstrated, may also derive from functional, fully processed snoRNPs, in the course of snoRNP remodeling and/or snoRNP decay (pathway 3).
Cleavage of SNORD116 into smaller RNA species was also inferred from RNA band patterns obtained by RPA experiments (30). As already pointed out for SNORD115, the sequence of the full-length SNORD116 considered in this study does not correspond to the one originally published (20). Indeed, this SNORD116 sequence (referred to herein as the A-116 form) contains a few surrounding intronic nucleotides [Figure 1C; see also Figure 3c and d in (30)], giving rise to a 7- to 8-bp terminal 5′–3′ stem that neither fits the mature 5′ and 3′ termini we have experimentally determined here (Figure 3B and C) nor our current understanding of box C/D-snoRNA structures (Figure 1D). In other words, C/D snoRNAs whose ends have been carefully mapped at the RNA level invariably contain 4–5 nt upstream of the C-box and 2–5 nt downstream of the D-box (Figure 1D), regardless the presence (or absence) of extended base-pairing potentials at their 5′ and 3′ extremities (6,33,43–46). We conclude that the most abundant A-116 form (theoretically 99 nt long) corresponds instead to the 92-nt-long canonical SNORD116 described here. Indeed, this new analysis explains why A-116 migrates just above the 90-nt RNA marker, instead of just below the 100-nt marker, as would be expected for a 99-nt-long RNA having 7–8 bp terminal 5′–3′ stem structures [see Figure 3d in (30)]. The origin of the other shorter psnoRNA species (namely B-116, C-116, D-116, E-116 and F-116) remains unknown since, despite long exposure times and many attempts, we failed to detect them by highly sensitive Northern-blot analysis (Figure 3A–D). As already proposed for SNORD115, they may simply arise from RNA degradation and/or from imperfect RNA duplexes formed between the SNORD116 riboprobe and numerous SNORD116 copies with similar, but not identical sequence similarity.
CONCLUSION
We failed to obtain convincing experimental evidence supporting a model whereby rodent (or human) box C/D SNORD115 and SNORD116 undergo significant conversion into abundant psnoRNAs (30,31). Of note, however, is the observation made in the course of this study of a previously unrecognized 5′-truncated SNORD115 form in the mouse brain. Intriguingly, similar 5′-truncated C′/D fibrillarin-associated snoRNA forms were previously reported at another array of tandemly repeated, brain-specific C/D snoRNA genes mapping at the imprinted rat Dlk1-Dio3 domain: the maternally expressed RBII-36/Bsr locus (33). Hence, production of such ‘mini C′/D snoRNAs’ could be more prevalent than generally assumed (Figure 4). Their biogenesis remains, however, poorly understood since SNORD115 derivatives are not detected upon transient transfection into HeLa cells of a single SNORD115 gene copy embedded within its natural intron and transcribed under the control of a CMV promoter (data not shown). Accordingly, we surmise that their genesis is restricted to neuronal tissues and/or requires large amount of nuclear-retained transcripts generated in their natural chromatin context (21).
Although we do not exclude the possibility that, under some circumstances, such as stressful conditions or at specific developmental stages, some copies of SNORD115 or SNORD116 could be fragmented into tiny amounts of snoRNA sub-fragments with regulatory roles (including perhaps microRNA-like species), we conclude that the major cellular RNA forms of rodent and human SNORD115 and SNORD116 genes do correspond, to classical box C/D-containing snoRNAs. Indeed, they exhibit canonical 5′–3′ terminal stem structures [Figures 1D, 2A and B and 3B and C (6,20,43–46)], associate with fibrillarin, a common C/D snoRNA-binding protein [(24), Figure 2F] and localize predominantly in the nucleolar compartment (21,24). More detailed studies carried out in physiologically relevant contexts should now explore the biological roles of these intriguing PWS-encoded snoRNAs, including the novel low-abundance ∼48-nt-long C′/D SNORD115 derivative reported here, in the development of placental mammals as well as in some human diseases such as PWS or autism (25–29,32,48).
FUNDING
European Union (STREP Prader–Willi Syndrome); Centre National de la Recherche Scientifique (CNRS); Université Paul Sabatier (UPS). Funding for open access charge: Agence Nationale de la Recherche (ANR blanche, snosca).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank E. Käs for careful reading of the manuscript and we are indebted to T. Kiss, B. Clouet d’Orval, H. Seitz, A. Henras and G. Canal for helpful discussions.
REFERENCES
- 1.Henras AK, Dez C, Henry Y. RNA structure and function in C/D and H/ACA s(no)RNPs. Curr. Opin. Struct. Biol. 2004;14:335–343. doi: 10.1016/j.sbi.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 2.Kiss T. Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J. 2001;20:3617–3622. doi: 10.1093/emboj/20.14.3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reichow SL, Hamma T, Ferre-D’Amare AR, Varani G. The structure and function of small nucleolar ribonucleoproteins. Nucleic Acids Res. 2007;35:1452–1464. doi: 10.1093/nar/gkl1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cavaille J, Nicoloso M, Bachellerie JP. Targeted ribose methylation of RNA in vivo directed by tailored antisense RNA guides. Nature. 1996;383:732–735. doi: 10.1038/383732a0. [DOI] [PubMed] [Google Scholar]
- 5.Ganot P, Bortolin ML, Kiss T. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell. 1997;89:799–809. doi: 10.1016/s0092-8674(00)80263-9. [DOI] [PubMed] [Google Scholar]
- 6.Kiss-Laszlo Z, Henry Y, Bachellerie JP, Caizergues-Ferrer M, Kiss T. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell. 1996;85:1077–1088. doi: 10.1016/s0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]
- 7.Klein DJ, Schmeing TM, Moore PB, Steitz TA. The kink-turn: a new RNA secondary structure motif. EMBO J. 2001;20:4214–4221. doi: 10.1093/emboj/20.15.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hutzinger R, Feederle R, Mrazek J, Schiefermeier N, Balwierz PJ, Zavolan M, Polacek N, Delecluse HJ, Huttenhofer A. Expression and processing of a small nucleolar RNA from the Epstein-Barr virus genome. PLoS Pathog. 2009;5:e1000547. doi: 10.1371/journal.ppat.1000547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ender C, Krek A, Friedlander MR, Beitzinger M, Weinmann L, Chen W, Pfeffer S, Rajewsky N, Meister G. A human snoRNA with microRNA-like functions. Mol. Cell. 2008;32:519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 10.Kawaji H, Nakamura M, Takahashi Y, Sandelin A, Katayama S, Fukuda S, Daub CO, Kai C, Kawai J, Yasuda J, et al. Hidden layers of human small RNAs. BMC Genomics. 2008;9:157. doi: 10.1186/1471-2164-9-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brameier M, Herwig A, Reinhardt R, Walter L, Gruber J. Human box C/D snoRNAs with miRNA like functions: expanding the range of regulatory RNAs. Nucleic Acids Res. 2011;39:675–686. doi: 10.1093/nar/gkq776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taft RJ, Glazov EA, Lassmann T, Hayashizaki Y, Carninci P, Mattick JS. Small RNAs derived from snoRNAs. RNA. 2009;15:1233–1240. doi: 10.1261/rna.1528909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saraiya AA, Wang CC. snoRNA, a novel precursor of microRNA in Giardia lamblia. PLoS Pathog. 2008;4:e1000224. doi: 10.1371/journal.ppat.1000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ono M, Scott MS, Yamada K, Avolio F, Barton GJ, Lamond AI. Identification of human miRNA precursors that resemble box C/D snoRNAs. Nucleic Acids Res. 2011;39:3879–3891. doi: 10.1093/nar/gkq1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scott MS, Avolio F, Ono M, Lamond AI, Barton GJ. Human miRNA precursors with box H/ACA snoRNA features. PLoS Comput. Biol. 2009;5:e1000507. doi: 10.1371/journal.pcbi.1000507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rother S, Meister G. Small RNAs derived from longer non-coding RNAs. Biochimie. 2011;93:1905–1915. doi: 10.1016/j.biochi.2011.07.032. [DOI] [PubMed] [Google Scholar]
- 17.Tuck AC, Tollervey D. RNA in pieces. Trends Genet. 2011;27:422–432. doi: 10.1016/j.tig.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 18.Scott MS, Ono M. From snoRNA to miRNA: dual function regulatory non-coding RNAs. Biochimie. 2011;93:1987–1992. doi: 10.1016/j.biochi.2011.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Landers M, Bancescu DL, Le Meur E, Rougeulle C, Glatt-Deeley H, Brannan C, Muscatelli F, Lalande M. Regulation of the large (approximately 1000 kb) imprinted murine Ube3a antisense transcript by alternative exons upstream of Snurf/Snrpn. Nucleic Acids Res. 2004;32:3480–3492. doi: 10.1093/nar/gkh670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cavaille J, Buiting K, Kiefmann M, Lalande M, Brannan CI, Horsthemke B, Bachellerie JP, Brosius J, Huttenhofer A. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc. Natl Acad. Sci. USA. 2000;97:14311–14316. doi: 10.1073/pnas.250426397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vitali P, Royo H, Marty V, Bortolin-Cavaille ML, Cavaille J. Long nuclear-retained non-coding RNAs and allele-specific higher-order chromatin organization at imprinted snoRNA gene arrays. J. Cell Sci. 2010;123:70–83. doi: 10.1242/jcs.054957. [DOI] [PubMed] [Google Scholar]
- 22.Runte M, Huttenhofer A, Gross S, Kiefmann M, Horsthemke B, Buiting K. The IC-SNURF-SNRPN transcript serves as a host for multiple small nucleolar RNA species and as an antisense RNA for UBE3A. Hum. Mol. Genet. 2001;10:2687–2700. doi: 10.1093/hmg/10.23.2687. [DOI] [PubMed] [Google Scholar]
- 23.Kishore S, Stamm S. The snoRNA HBII-52 regulates alternative splicing of the serotonin receptor 2C. Science. 2006;311:230–232. doi: 10.1126/science.1118265. [DOI] [PubMed] [Google Scholar]
- 24.Vitali P, Basyuk E, Le Meur E, Bertrand E, Muscatelli F, Cavaille J, Huttenhofer A. ADAR2-mediated editing of RNA substrates in the nucleolus is inhibited by C/D small nucleolar RNAs. J. Cell Biol. 2005;169:745–753. doi: 10.1083/jcb.200411129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ding F, Li HH, Zhang S, Solomon NM, Camper SA, Cohen P, Francke U. SnoRNA Snord116 (Pwcr1/MBII-85) deletion causes growth deficiency and hyperphagia in mice. PLoS One. 2008;3:e1709. doi: 10.1371/journal.pone.0001709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Skryabin BV, Gubar LV, Seeger B, Pfeiffer J, Handel S, Robeck T, Karpova E, Rozhdestvensky TS, Brosius J. Deletion of the MBII-85 snoRNA gene cluster in mice results in postnatal growth retardation. PLoS Genet. 2007;3:e235. doi: 10.1371/journal.pgen.0030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sahoo T, del Gaudio D, German JR, Shinawi M, Peters SU, Person RE, Garnica A, Cheung SW, Beaudet AL. Prader-Willi phenotype caused by paternal deficiency for the HBII-85 C/D box small nucleolar RNA cluster. Nat. Genet. 2008;40:719–721. doi: 10.1038/ng.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duker AL, Ballif BC, Bawle EV, Person RE, Mahadevan S, Alliman S, Thompson R, Traylor R, Bejjani BA, Shaffer LG, et al. Paternally inherited microdeletion at 15q11.2 confirms a significant role for the SNORD116 C/D box snoRNA cluster in Prader-Willi syndrome. Eur. J. Hum. Genet. 18:1196–1201. doi: 10.1038/ejhg.2010.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Smith AJ, Purmann C, Walters RG, Ellis RJ, Holder SE, Van Haelst MM, Brady AF, Fairbrother UL, Dattani M, Keogh JM, et al. A deletion of the HBII-85 class of small nucleolar RNAs (snoRNAs) is associated with hyperphagia, obesity and hypogonadism. Hum. Mol. Genet. 2009;18:3257–3265. doi: 10.1093/hmg/ddp263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen M, Eyras E, Wu J, Khanna A, Josiah S, Rederstorff M, Zhang MQ, Stamm S. Direct cloning of double-stranded RNAs from RNase protection analysis reveals processing patterns of C/D box snoRNAs and provides evidence for widespread antisense transcript expression. Nucleic Acids Res. 2011;39:9720–9730. doi: 10.1093/nar/gkr684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kishore S, Khanna A, Zhang Z, Hui J, Balwierz PJ, Stefan M, Beach C, Nicholls RD, Zavolan M, Stamm S. The snoRNA MBII-52 (SNORD 115) is processed into smaller RNAs and regulates alternative splicing (2010) Hum. Mol. Genet. 19:1153–1164. doi: 10.1093/hmg/ddp585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakatani J, Tamada K, Hatanaka F, Ise S, Ohta H, Inoue K, Tomonaga S, Watanabe Y, Chung YJ, Banerjee R, et al. Abnormal behavior in a chromosome-engineered mouse model for human 15q11-13 duplication seen in autism. Cell. 2009;137:1235–1246. doi: 10.1016/j.cell.2009.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cavaille J, Vitali P, Basyuk E, Huttenhofer A, Bachellerie JP. A novel brain-specific box C/D small nucleolar RNA processed from tandemly repeated introns of a noncoding RNA gene in rats. J. Biol. Chem. 2001;276:26374–26383. doi: 10.1074/jbc.M103544200. [DOI] [PubMed] [Google Scholar]
- 34.Perumal K, Reddy R. The 3’ end formation in small RNAs. Gene Expr. 2002;10:59–78. [PMC free article] [PubMed] [Google Scholar]
- 35.Li Y, Luo J, Zhou H, Liao JY, Ma LM, Chen YQ, Qu LH. Stress-induced tRNA-derived RNAs: a novel class of small RNAs in the primitive eukaryote Giardia lamblia. Nucleic Acids Res. 2008;36:6048–6055. doi: 10.1093/nar/gkn596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee YS, Shibata Y, Malhotra A, Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs) Genes Dev. 2009;23:2639–2649. doi: 10.1101/gad.1837609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haussecker D, Huang Y, Lau A, Parameswaran P, Fire AZ, Kay MA. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA. 2010;16:673–695. doi: 10.1261/rna.2000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hsieh LC, Lin SI, Kuo HF, Chiou TJ. Abundance of tRNA-derived small RNAs in phosphate-starved Arabidopsis roots. Plant Signal. Behav. 2010;5:537–539. doi: 10.4161/psb.11029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cole C, Sobala A, Lu C, Thatcher SR, Bowman A, Brown JW, Green PJ, Barton GJ, Hutvagner G. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA. 2009;15:2147–2160. doi: 10.1261/rna.1738409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taft RJ, Glazov EA, Cloonan N, Simons C, Stephen S, Faulkner GJ, Lassmann T, Forrest AR, Grimmond SM, Schroder K, et al. Tiny RNAs associated with transcription start sites in animals. Nat. Genet. 2009;41:572–578. doi: 10.1038/ng.312. [DOI] [PubMed] [Google Scholar]
- 41.Persson H, Kvist A, Vallon-Christersson J, Medstrand P, Borg A, Rovira C. The non-coding RNA of the multidrug resistance-linked vault particle encodes multiple regulatory small RNAs. Nat. Cell Biol. 2009;11:1268–1271. doi: 10.1038/ncb1972. [DOI] [PubMed] [Google Scholar]
- 42.Verhagen AP, Pruijn GJ. Are the Ro RNP-associated Y RNAs concealing microRNAs? Y RNA-derived miRNAs may be involved in autoimmunity. Bioessays. 2011;33:674–682. doi: 10.1002/bies.201100048. [DOI] [PubMed] [Google Scholar]
- 43.Darzacq X, Kiss T. Processing of intron-encoded box C/D small nucleolar RNAs lacking a 5’,3’-terminal stem structure. Mol. Cell Biol. 2000;20:4522–4531. doi: 10.1128/mcb.20.13.4522-4531.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qu LH, Henry Y, Nicoloso M, Michot B, Azum MC, Renalier MH, Caizergues-Ferrer M, Bachellerie JP. U24, a novel intron-encoded small nucleolar RNA with two 12 nt long, phylogenetically conserved complementarities to 28S rRNA. Nucleic Acids Res. 1995;23:2669–2676. doi: 10.1093/nar/23.14.2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qu LH, Nicoloso M, Michot B, Azum MC, Caizergues-Ferrer M, Renalier MH, Bachellerie JP. U21, a novel small nucleolar RNA with a 13 nt. complementarity to 28S rRNA, is encoded in an intron of ribosomal protein L5 gene in chicken and mammals. Nucleic Acids Res. 1994;22:4073–4081. doi: 10.1093/nar/22.20.4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nicoloso M, Caizergues-Ferrer M, Michot B, Azum MC, Bachellerie JP. U20, a novel small nucleolar RNA, is encoded in an intron of the nucleolin gene in mammals. Mol. Cell Biol. 1994;14:5766–5776. doi: 10.1128/mcb.14.9.5766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kiss T, Filipowicz W. Exonucleolytic processing of small nucleolar RNAs from pre-mRNA introns. Genes Dev. 1995;9:1411–1424. doi: 10.1101/gad.9.11.1411. [DOI] [PubMed] [Google Scholar]
- 48.Ding F, Prints Y, Dhar MS, Johnson DK, Garnacho-Montero C, Nicholls RD, Francke U. Lack of Pwcr1/MBII-85 snoRNA is critical for neonatal lethality in Prader-Willi syndrome mouse models. Mamm. Genome. 2005;16:424–431. doi: 10.1007/s00335-005-2460-2. [DOI] [PubMed] [Google Scholar]