Abstract
The Escherichia coli AlkB protein (EcAlkB) is a DNA repair enzyme which reverses methylation damage such as 1-methyladenine (1-meA) and 3-methylcytosine (3-meC). The mammalian AlkB homologues ALKBH2 and ALKBH3 display EcAlkB-like repair activity in vitro, but their substrate specificities are different, and ALKBH2 is the main DNA repair enzyme for 1-meA in vivo. The genome of the model plant Arabidopsis thaliana encodes several AlkB homologues, including the yet uncharacterized protein AT2G22260, which displays sequence similarity to both ALKBH2 and ALKBH3. We have here characterized protein AT2G22260, by us denoted ALKBH2, as both our functional studies and bioinformatics analysis suggest it to be an orthologue of mammalian ALKBH2. The Arabidopsis ALKBH2 protein displayed in vitro repair activities towards methyl and etheno adducts in DNA, and was able to complement corresponding repair deficiencies of the E. coli alkB mutant. Interestingly, alkbh2 knock-out plants were sensitive to the methylating agent methylmethanesulphonate (MMS), and seedlings from these plants developed abnormally when grown in the presence of MMS. The present study establishes ALKBH2 as an important enzyme for protecting Arabidopsis against methylation damage in DNA, and suggests its homologues in other plants to have a similar function.
INTRODUCTION
Living organisms are exposed to a variety of environmental DNA-damaging agents, such as chemical mutagens, toxins from fungi and bacteria, as well as ultraviolet and ionizing radiation. In addition, various intracellular metabolites can cause DNA damage. Plants possess various mechanisms for repairing DNA, and genes encoding the components of major DNA repair pathways are found in plant genomes (1,2). However, only a few studies have focused on characterizing DNA repair enzymes from plants, and our understanding of how plants handle the deleterious effects of DNA damage remains vague.
Methylating agents introduce a number of lesions into cellular DNA and RNA. Such agents are present in the environment, e.g. as methyl halides (3,4), or they may be generated intracellularly by normal metabolism. For example, S-adenosylmethionine, which serves as a methyl donor in enzymatic methylation reactions, is also able to induce a low level of aberrant methylations (5). Several different repair mechanisms protect the genome against the harmful effects of methylating and other alkylating agents, including base excision repair (BER) initiated by alkylpurine DNA glycosylases, direct reversal by DNA alkylbase methyltransferases, and oxidative demethylation by AlkB-like dioxygenases (6). The model plant Arabidopsis appears to lack DNA alkylbase methyltransferases, but bioinformatics analyses indicate the existence of a large number of alkylpurine DNA glycosylases, representing all the three families of such glycosylases (7). Moreover, one of these enzymes, termed AMAG, has been shown to be an enzymatically active 3-methyladenine (3-meA) DNA glycosylase (8), and Arabidopsis cell extracts have been shown to contain the enzymatic machinery necessary for performing BER (9).
Escherichia coli AlkB (EcAlkB) is a DNA repair protein which belongs to the superfamily of iron(II) and 2-oxoglutarate dependent dioxygenases (10), enzymes which use ferrous iron as cofactor and 2-oxoglutarate as co-substrate to perform various oxidation reactions, usually hydroxylations. EcAlkB was originally shown to demethylate the lesions 1-meA and 3-meC in DNA by hydroxylating the methyl group, leading to the release of the resulting hydroxymethyl moiety as formaldehyde (11,12). In addition to 1-meA and 3-meC, the structurally similar, but less abundant lesions 1-methylguanine (1-meG) and 3-methylthymine (3-meT) have also been shown to be EcAlkB substrates (13–15). EcAlkB is active also on methylated RNA and on bulkier DNA lesions, such as ethyl and propyl groups, as well as on exocyclic etheno and ethano-lesions, but these activities are generally lower than on the canonical 1-meA and 3-meC lesions in DNA (16–19).
Although homologues of EcAlkB can be found in viruses, eubacteria and eukaryotes, many organisms within these groups lack AlkB homologues (20–24). However, the genomes of multicellular eukaryotes typically encode several different AlkB homologues, and mammals have eight such proteins, ALKBH1–8, as well as the somewhat more distantly related FTO (ALKBH9) protein (21,25,26). Despite extensive studies, the function of the majority of these proteins is still unknown. Also plants, such as Arabidopsis, possess several different ALKBH proteins (21,27).
Among the human AlkB homologues, ALKBH2 and ALKBH3 have been characterized biochemically and shown to have robust repair activities on several of the lesions that are EcAlkB substrates (17,28–31). However, these two proteins display interesting differences with respect to their substrate specificities and subcellular localization, suggesting that they perform distinct cellular functions. ALKBH2 is exclusively active on DNA substrates, whereas ALKBH3 shows comparable activities on DNA and RNA. While ALKBH2 is active both on single-stranded (ss) and double-stranded (ds) DNA substrates, with a slight preference for dsDNA, ALKBH3 displays a strong preference for single-stranded substrates (28,29). ALKBH2 is a nuclear protein which accumulates in replication foci during S phase, while ALKBH3 is found both in the nucleus and the cytosol (28). Moreover, studies of ALKBH2 and ALKBH3 knock-out mice demonstrated that ALKBH2 is the main demethylase for removal of 1-meA in DNA in vivo (32). ALKBH3- or EcAlkB-mediated repair leads to functional recovery of methylated tRNA and mRNA in vitro, and studies of viral AlkB proteins have indicated that AlkB-mediated RNA repair is indeed biologically relevant (23,33). This suggested a possible role for ALKBH3 in RNA repair, but a recent study showed that ALKBH3 is associated with the DNA helicase ASCC3, and that RNAi-mediated knock-down of ASCC3 or ALKBH3 leads to increased cellular sensitivity towards methylating agents and to accumulation of genomic 3-meC, thus indicating that DNA repair may be the only biologically relevant function for ALKBH3 (34).
Weak in vitro repair activities have also been reported for two other ALKBH proteins. The human FTO protein showed repair activity towards 3-meT in ssDNA and 3-methyluridine in ssRNA (25,35), and ALKBH1 was shown to demethylate 3-meC in ssDNA and ssRNA (36). However, ALKBH1 has also been implicated in transcriptional regulation, and FTO was recently shown to efficiently demethylate the RNA modification N6-methyladenine (37–39). Indeed, by showing that mammalian and plant ALKBH8 are tRNA modification enzymes, we and others recently demonstrated that ALKBH proteins have functions other than nucleic acid repair (40–44).
We here report the functional characterization of the Arabidopsis thaliana protein AT2G22260, which is an AlkB homologue with comparable sequence similarities to mammalian ALKBH2 and ALKBH3. Studies of its repair activity, as well as bioinformatics analyses, established this protein as a functional ALKBH2 orthologue. Interestingly, an Arabidopsis alkbh2 mutant displayed hypersensitivity towards the alkylating agent methylmethanesulphonate (MMS), but only showed marginally elevated levels of MMS-induced 1-meA lesions in the genome.
MATERIALS AND METHODS
Protein purification and plasmid construction
Constructs for expression of Arabidopsis ALKBH2 in pET-28a(+) (Novagen, Germany) and pJB658 (45) were generated from total cDNA of Arabidopsis flowers using primers 17-AtALKBH2-fwd and 18-AtALKBH2-rev (primer sequences can be found in Supplementary Table S1). The PCR product was cloned into NdeI and BamHI sites of the vectors. His-tagged protein was expressed from the plasmid pET-28a(+) in the E. coli strain BL21(DE3)-CodonPlus-RIPL (Stratagene, USA) in LB medium. The recombinant protein was isolated using TALON beads as previously described (23).
Phage reactivation experiments
The toluic acid-inducible expression plasmid pJB658 (45) expressing the Arabidopsis AlkB homologue ALKBH2 or EcAlkB was transfected into an F-pilus expressing, AlkB-deficient E. coli strain, HK82/F’. Bacterial cultures were grown at 37°C under shaking, and at A600 = 0.1, the expression of recombinant protein was induced by the addition of 2 mM toluic acid (Fluka/Sigma-Aldrich, USA). The bacteria were further grown after induction until they reached A600 = 0.8. The M13 DNA phage, was treated for 30 min at 30°C with different concentrations of MMS (Fluka/Sigma-Aldrich, USA) or chloroacetaldehyde (CAA; Fluka/Sigma-Aldrich, USA), for introduction of methyl or etheno adducts, respectively. One hundred microlitres of several dilutions of the treated phage was mixed with 300 μl of the induced bacteria and 3 ml LB top agar. The mixture was spread onto LB plates and incubated at 37°C overnight. Phage survival was scored by counting plaques. All phage dilutions and treatments were performed in M9 minimum salt medium.
Bacterial survival experiments
Cultures of E. coli strain HK82/F’ transformed with pJB658-derived plasmids encoding bacterial AlkB proteins were induced with toluic acid as described above and grown until A600 = 0.5. Then, 500 µl of bacterial culture was pelleted by centrifugation, re-suspended in 250 µl of M9 minimum salt medium, and then mixed with an equal volume of M9 minimum salt medium containing the appropriate MMS concentration, followed by 30 min incubation at 30°C. Appropriate (to enable colony counting) dilutions of treated bacteria were plated onto LB plates, which were incubated overnight at 37°C, and survival was assessed by colony counting.
Preparation of oligonucleotide substrates and in vitro repair reactions of site-specific lesions
DNA oligonucleotides with the sequence. TAGACATTGCCATTCTCGATAGGATCCGGTCAAACCTAGACGAATTCCG, containing either 1-meA, 3-meC, or εA at the underlined position, were synthesized by ChemGenes (Wilmington, MA; 1-meA and 3-meC) or Midland Certified Reagent Company Inc. (Midland, TX; εA). Corresponding 5′-[32P]-labelled dsDNA substrates were generated, and repair reactions performed essentially as described previously (31,32).
Oxidative demethylation of [3H]methylated oligonucleotides
The [3H]methylated oligonucleotide (sequence: CATGATAACCGCGACTACACTGAC) was prepared by treatment with N-[3H]methyl-N-nitrosourea, and repair assays performed, as previously described (28,29).
Plant growth conditions
Wild-type Arabidopsis (A. thaliana ecotype Col or Ler) were grown in perlite soil at 18°C under 16 h of light at 100 μE/m2s. Seeds were surface sterilized in 7.5% hypochlorite solution containing 0.1% Tween 20, followed by treatment in 70% ethanol and rinsing in distilled water. Sterilized seeds were plated on solidified Murashige–Skoog (MS) medium (46), supplemented with 2% sucrose (MS-2).
RNA extraction and RT–PCR
RNA extraction was performed using Spectrum Total RNA kit (Fluka/Sigma-Aldrich), following the manufacturer’s instruction. RNA concentration and purity was assessed using NanoDrop2000 (Thermo Scientific). An amount of 1 µg RNA was processed to obtain cDNA using polyT-primer and reverse transcriptase (Superscript II, Invitrogen) following the manufacturer’s instructions. The resulting cDNA was used for Reverse Transcription (RT)–PCR, using primers 1414 and 1415.
GUS staining
GUS staining assay of transgenic plants was performed as previously described (47). Prefixing of plant material in ice-cold 90% acetone for 10 min was followed by rinsing for 10 min in staining buffer (50 mM Na3PO4, pH 7.2; 2 mM potassium–ferrocyanide; 2 mM potassium–ferricyanide; 0.1% Triton X-100; 2 mM X-Gluc) with no substrate and incubated in staining buffer at 37°C for 3–5 h. Following a graded ethanol dehydration series to 50% ethanol, the material was post-fixed in FAA [10:7:2:1 ethanol:distilled water:acetic acid:formaldehyde(37%)] on ice for 30 min and hydrated in an ethanol series to 50 mM Na3PO4 buffer and mounted on microscope slides in chloral hydrate (8:2:1 chloral hydrate:water:glycerol). For all GUS lines, the following tissues were stained; seedlings, mature leaves, flowers and siliques. The GUS staining was observed with a Zeiss Axioplan Imaging2 microscope system equipped with Nomarski optics and cooled LCD imaging facilities.
Gene trap Arabidopsis mutant and transgenic lines
A gene trap insertion line (GT_5_41144, alkbh2-1) was obtained from The Nottingham Arabidopsis Stock centre. Homozygous plants were identified by PCR genotyping as previously described (48), using the primers Ds-5 and Ds-3 in combination with genomic primers 598 and 599. PCR products were cloned into the TOPO Blunt vector (Invitrogen, USA), and clones were sequenced to verify the integration site. To analyse the expression of ALKBH2 in the alkbh2-1 mutant, RT–PCR was performed with primers upstream of the integration site (primers 1418 and 1419), downstream of the integration site (primers 1420 and 1421), as well as spanning the integration site (primers 1414 and 1415). The pALKBH2::GUS construct was generated by Gateway technology (Invitrogen, USA). The promoter fragment, which included 1044 bp of putative promoter sequence, was amplified using the primers; attB1–AtALKBH2–promoter and attB2–AtALKBH2–promoter, recombined into pDONR/Zeo (Invitrogen, USA) and further recombined into pMDC162 (49), creating the construct pMDC162 pAtALKBH2::GUS. The construct was transferred to the Agrobacterium tumefaciens strain C58C1 pGV2260 and ecotype Col plants were transformed using the A. tumefaciens-mediated floral dip method (50). Transformants were selected on MS containing 20 mg/ml hygromycin.
Genotoxic stress treatment of plants
For assessment of sensitivity of alkbh2-1 mutants to MMS or CAA, WT and alkbh2-1 seeds were plated on solidified MS-2 medium supplemented with the genotoxic agents. For MMS treatment, a final concentration of 90 ppm MMS was used. For treatment with CAA 50, 100 and 250 μM was used as previously described (51).
Accumulation of genomic 1-meA
For assessment of accumulation of 1-meA in the genome, 6-day-old seedlings germinated and grown on MS-2 plates were individually transferred to six-well plates containing liquid MS medium supplemented with 90 ppm MMS (Sigma-Aldrich, USA) and incubated in growth chamber at 18°C. Tissue was harvested in liquid N2 and genomic DNA isolated using the Ultraprep Plant DNA kit including an RNase treatment step (AHN Biotechnologie, Germany). Before 1-meA measurements, the amount and purity of the DNA were determined by using NanoDrop 2000 (Thermo Scientific, USA), as well as assayed on gel, and all the DNA preparations were genotyped as described above. Two micrograms of DNA was precipitated using glycogen as a carrier in ethanol supplemented with ammonium acetate, before the 1-meA level in the samples was determined by LC–MS/MS.
LC–MS/MS analysis
Genomic DNA was enzymatically hydrolysed to nucleosides as described (52), added three volumes of methanol and centrifuged (16 000g, 30 min). The supernatants were dried and dissolved in 50 µl 5% methanol in water (v/v) for LC-MS/MS analysis of 1-me(dA), and a portion of each sample diluted for the quantitation of the unmodified deoxynucleosides (dA, dC, dG and T). Chromatographic separation was performed on a Shimadzu Prominence HPLC system with a Zorbax SB-C18 2.1 × 150 mm i.d. (3.5 µm) column equipped with an Eclipse XDB-C8 2.1 × 12.5 mm i.d. (5 µm) guard column (Agilent Technologies). The mobile phase consisted of water and methanol (both added 0.1% formic acid), for 1-me(dA) starting with a 5-min gradient of 5–70% methanol, followed by 6.5 min re-equilibration with 5% methanol, and for unmodified nucleosides maintained isocratically with 85% methanol. Mass spectrometry detection was performed using an Applied Biosystems/MDS Sciex 5000 triple quadrupole (Applied Biosystems) operating in positive electrospray ionization mode, monitoring the mass transitions 266.2→150.1 [1-me(dA)], 252.2→136.1 (dA), 228.2→112.1 (dC), 268.2→152.1 (dG), and 243.2→127.1 (T).
RESULTS
The Arabidopsis genome encodes a putative homologue of mammalian ALKBH2
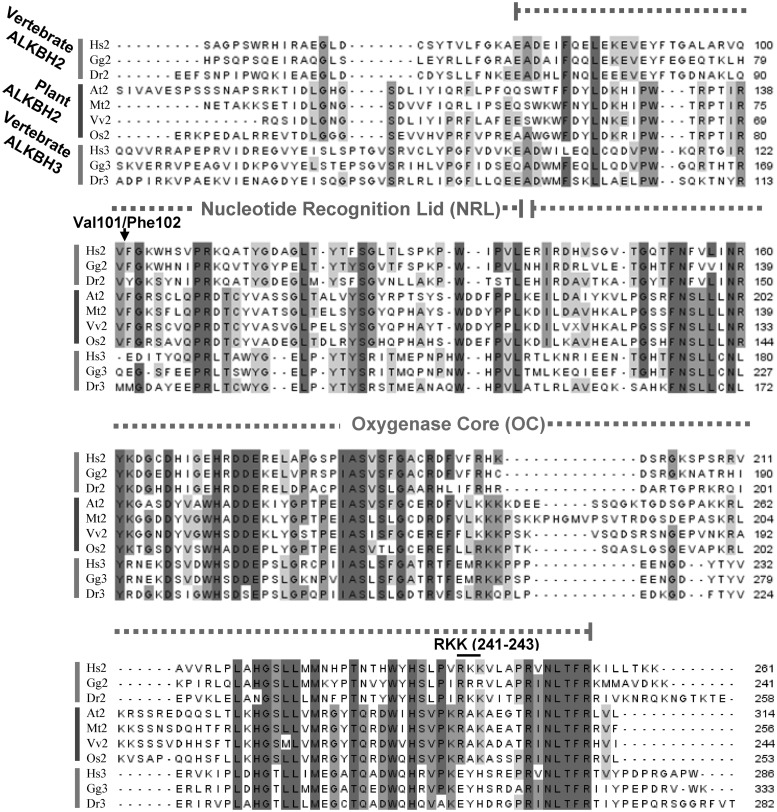
To identify possible nucleic acid repair proteins of the AlkB family in A. thaliana, we performed BLAST searches using human ALKBH2 and ALKBH3 proteins as queries. These two human proteins retrieved the same Arabidopsis protein of 314 amino acids, encoded by the AT2G22260 gene, as the closest hit, and this protein displayed a comparable degree of sequence similarity to human ALKBH2 and ALKBH3 (39% and 37% identity, 50% and 48% similarity, respectively). Further BLAST searches identified putative orthologues of AT2G22260 in other plants, including rice and grapevine, indicating that this function is present in both monocots and dicots. A protein sequence alignment of ALKBH2/ALKBH3 proteins from various plants and vertebrates are shown in Figure 1. The ALKBH family of proteins are defined by a common ‘Oxygenase Core’ (OC), and, in addition, ALKBH proteins involved in DNA/RNA repair, such as ALKBH2, ALKBH3, and EcAlkB, share sequence homology in the N-terminal region shown to interact with the damaged nucleotide, referred to as the ‘Nucleotide Recognition Lid’ (NRL) (53). Interestingly, the AT2G22260 protein and its putative plant orthologues share sequence homology with ALKBH2 and ALKBH3 both in the OC and NRL regions, indicating that these plant proteins are indeed nucleic acid repair proteins. The 3D structure of human ALKBH2 in complex with a dsDNA substrate has revealed that Phe102 is responsible for flipping the methylated base out of the dsDNA helix, while a cluster of positively charged residues, Arg241, Lys242 and Lys243 (RKK241–243) close to the C-terminus is important for interactions with the negatively charged backbone of the complementary strand (53). Moreover, a mutational analysis of human ALKBH2 indicated that Val101, in addition to Phe102, plays an important role in base flipping (54). The AT2G22260 protein and its plant orthologues all have a Val-Phe motif at the position corresponding to Val101–Phe102 in human ALKBH2, and they also have a positively charged four-amino-acid patch (KRAK) at the position corresponding to residues 240–243 (Figure 1). This suggests that these plant proteins have a function similar to that of mammalian ALKBH2, and the AT2G22260 protein will in the following be referred to as ALKBH2.
Figure 1.
Multiple sequence alignment of putative ALKBH2 and ALKBH3 proteins from plants and vertebrates. The alignment was generated with JalView (http://www.jalview.org/) using the embedded MUSCLE algorithm (55,56). The ‘Nucleotide Recognition Lid’ and the ‘Oxygenase Core’, described by Yu et al. (57), are indicated, as well as residues corresponding to Val101–Phe102 (involved in base flipping) and Arg-Lys–Lys (241–243) (involved in interaction with the opposite DNA strand) of human ALKBH2. The non-conserved N-terminal part of the proteins have been omitted from the alignment. Hs, Homo sapiens; Gg, Gallus gallus; Dr, Danio rerio; At, A. thaliana; Mt, Medicago truncatula (barrel medic); Vv, Vitis vinifera (common grape vine); Os, Oryza sativa (rice). GI numbers of sequences: 47124096 (Hs2), 118098574 (Gg2), 68383159 (Dr2), 18399917 (At2), 87162794 (Mt2), 147777784 (Vv2), 222635406 (Os2), 21040275 (Hs3), 118091513 (Gg3), 51011105 (Dr3).
Complementation of alkB mutant E. coli by ALKBH2
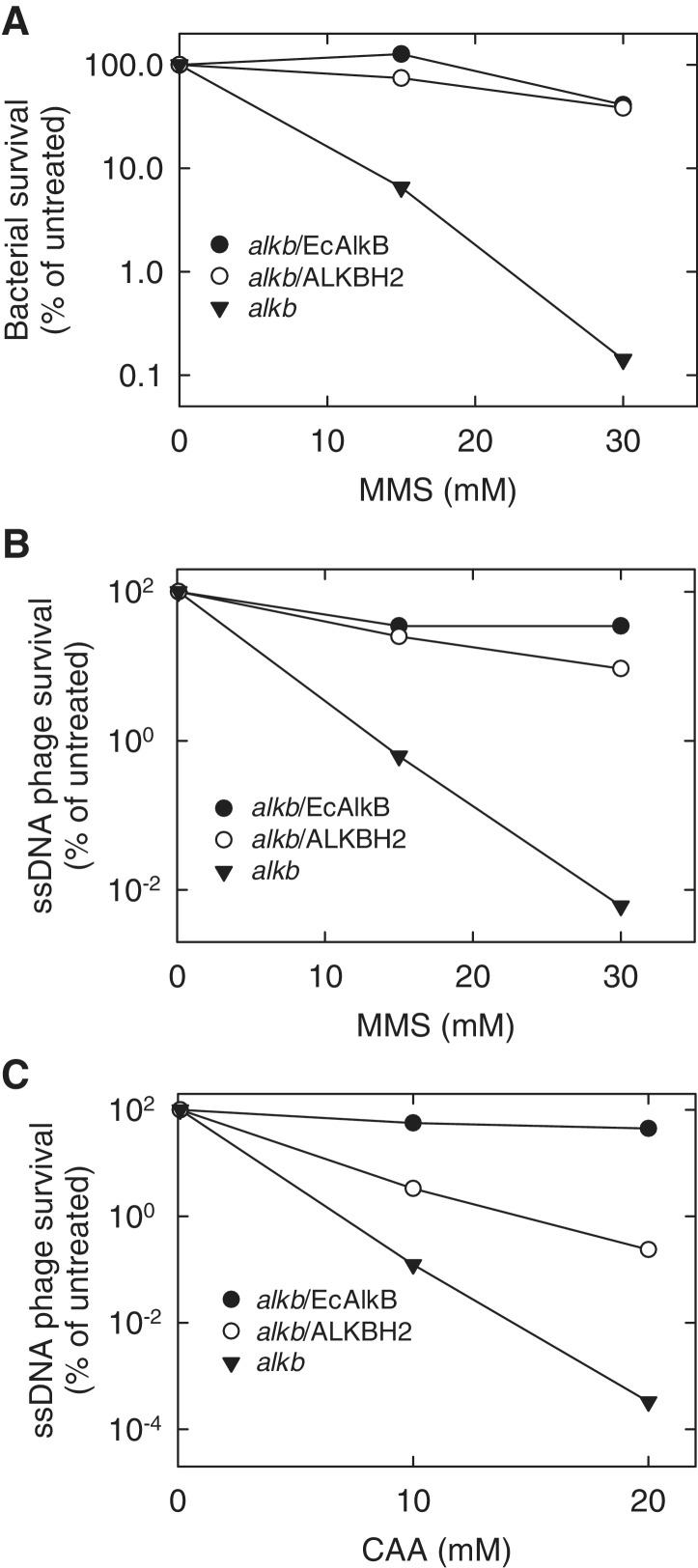
To address whether Arabidopsis ALKBH2 can function as an AlkB homologue in cells, we investigated its ability to complement the MMS sensitive phenotype of the E. coli alkB mutant strain. An expression construct for ALKBH2 was introduced into the alkB strain, and the bacteria were subjected to MMS treatment. In these experiments, ALKBH2 protected the alkB bacteria against MMS induced cell death to the same extent as EcAlkB (Figure 2A), indicating that Arabidopsis ALKBH2 is able to revert MMS-induced cytotoxic lesions that normally are repaired by EcAlkB.
Figure 2.
Arabidopsis ALKBH2-mediated repair of DNA damage in the E. coli alkb mutant. EcAlkB-expressing plasmid and empty expression plasmid were included as positive and negative control, respectively. (A) Complementation of the MMS sensitive phenotype of E. coli alkB mutant bacteria by plasmid-expressed ALKBH2. The bacteria were treated with the indicated concentrations of MMS, plated out on agar plates, and the survival was assessed by colony counting. (B) Ability of ALKBH2 to increase the survival of methylated ssDNA phage. Phage M13 was treated with the indicated concentrations of MMS and mixed with alkb E. coli expressing the indicated proteins. Top agar was then added to the mixture, which was plated out on agar plates. Phage survival was scored by counting resulting plaques. (C) ALKBH2-mediated survival of CAA-treated ssDNA phage. Identical experiment to that in (B), except that CAA was used instead of MMS.
The lesions that are repaired by the AlkB mechanism, such as 1-meA and 3-meC, are introduced much more efficiently by MMS treatment of ssDNA, relative to dsDNA (58). Thus, if a ssDNA phage, such as M13, is treated with MMS and used to infect E. coli, the production of progeny phage (‘phage survival’) is dramatically reduced in an alkB mutant, relative to wild-type cells (59). Accordingly, the expression of a functional AlkB homologue in the alkB mutant has been shown to substantially increase the survival of methylated ssDNA phage, and similar results have been obtained in the case of ssRNA phage (17,28). To investigate the ability of Arabidopsis ALKBH2 to reactivate ssDNA, the effect of ALKBH2 over-expression on the survival of MMS-treated M13 phage in the alkB mutant was investigated. Indeed, expression of ALKBH2 increased the survival of the methylated phage to an extent similar to that observed with EcAlkB (Figure 2B). When M13 phage was treated with chloroacetaldehyde (CAA), a vinyl chloride metabolite that introduces etheno adducts in DNA, ALKBH2 expression increased phage survival, albeit to a lesser extent than EcAlkB. These results indicated that Arabidopsis ALKBH2, like EcAlkB and mammalian ALKBH2, is capable of repairing etheno adducts (Figure 2C). The protein was unable to increase the survival of methylated ssRNA phage (data not shown). In summary, the results above indicate that Arabidopsis ALKBH2 is a functional EcAlkB homologue, involved in reversing methylation damage in DNA.
In vitro repair activity of Arabidopsis ALKBH2
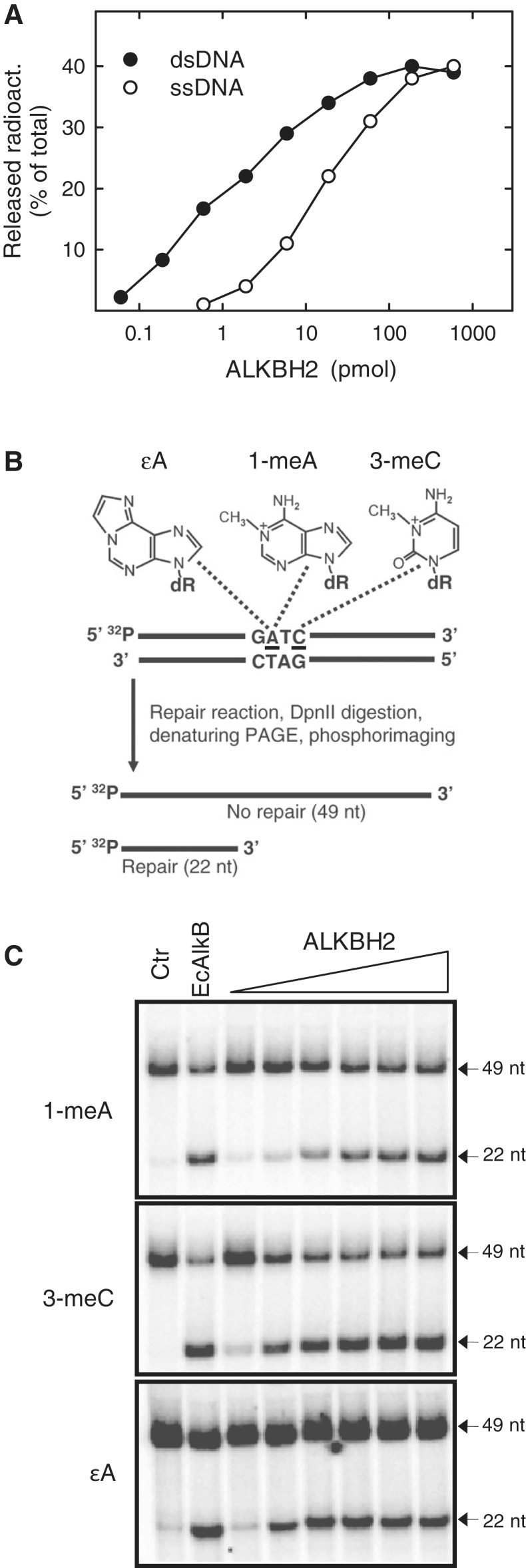
To further characterize the repair activity of Arabidopsis ALKBH2, we investigated the ability of E. coli-expressed recombinant protein to repair various damaged substrates in vitro. Treatment of ssDNA with the radiolabelled methylating agent [3H]methylnitrosourea (MNU) generates 1-meA and 3-meC lesions containing a radiolabelled methyl group. Thus, AlkB-mediated repair can be investigated by measuring the release of ethanol-soluble radioactive formaldehyde from such a substrate, and this type of assay has previously been extensively used to study AlkB-mediated repair (11,12,17,28–30). Indeed, when a [3H]MNU treated oligonucleotide was incubated with 6×His-tagged, recombinant ALKBH2, ∼40% of the total radioactivity was released from the substrate at the highest enzyme concentrations (Figure 3A), similarly to what has previously been observed with bacterial and mammalian AlkB homologues (29). The in vitro activity of Arabidopsis ALKBH2 was also investigated on a dsDNA substrate generated by annealing the [3H]methylated ssDNA to its complementary strand, and the protein displayed a preference for dsDNA over ssDNA.
Figure 3.
Arabidopsis ALKBH2-mediated repair of methylated DNA in vitro. (A) Repair of ssDNA and dsDNA by ALKBH2. A [3H]methylated oligonucleotide, generated by treatment with [3H]MNU, was incubated with varying amounts of recombinant ALKBH2, and then precipitated with ethanol. The released ethanol soluble radioactivity (formaldehyde) was measured by scintillation counting. Open circles represents the [3H]methylated ssDNA oligonucleotide, whereas closed circles represent the corresponding dsDNA, generated by hybridization to the unlabelled complementary strand. (B) Schematic representation of the assay for repair of site-specific lesions in oligonucleotides. The substrate for repair is a 49-nt dsDNA oligonucleotide consisting of one 32P-end-labelled strand containing 1-meA, 3-meC or εA at a specific position, annealed to an unlabelled, lesion-free complementary strand. After incubation with recombinant repair enzyme, the substrate is digested with Dpn II, which is blocked by the presence of lesions in the recognition sequence. The digestion products are then analysed by gel electrophoresis and phosphorimaging, where repaired or unrepaired substrates will give rise to radiolabelled bands of 22 and 49 nt, respectively. (C) Repair activity of ALKBH2 on oligonucleotides containing site-specific lesions. The following amounts of ALKBH2 were used: 2 pmol (lane 3), 5 pmol (lane 4), 10 pmol (lane 5), 30 pmol (lane 6), 50 pmol (lane 7) and 100 pmol (lane 8). As a negative control, the oligonucleotide substrates were incubated with Dpn II without any previous repair reaction. Purified EcAlkB (100 pmol) was included as positive control. Note that it is commonly observed that a proportion of the DNA molecules are refractory to repair in these assays (here, this is particularly prominent for the εA containing substrate).
AlkB substrates generated by treatment of ssDNA with methylating agents such as MMS and MNU will contain both 1-meA and 3-meC. To investigate the activity of Arabidopsis ALKBH2 on individual lesions, we used [32P]-5′-end-labelled oligonucleotides where a single 1-meA, 3-meC or εA lesion had been placed in the recognition sequence for the methylation sensitive restriction enzyme DpnII (Figure 3B). In this assay, the substrate is incubated with repair enzyme, then with DpnII, and the reaction mixture is analysed on an acrylamide gel, which is subsequently subjected to phosphorimaging. If the lesion is repaired, the DpnII site will become susceptible to cleavage, and two fragments are observed. In contrast, an unrepaired substrate will give rise to only a single fragment. In these experiments, ALKBH2 showed activity on 1-meA, 3-meC and εA, which all were repaired to a similar extent as that observed with the EcAlkB protein (Figure 3C). Together, the complementation assays and in vitro experiments demonstrate that Arabidopsis ALKBH2, similarly to its mammalian counterpart, repairs both methyl and etheno lesions in DNA.
Expression pattern of ALKBH2 in Arabidopsis
The expression pattern of the ALKBH2 gene was examined in detail, utilizing an Arabidopsis line carrying a single copy of a transgene (pAtALKBH2::GUS) where the ALKBH2 promoter was coupled to a GUS reporter gene. pAtALKBH2::GUS was shown to be active in all tissues examined (Figure 4), including seedlings, leaves and flowers. In seedlings, the expression was most notable in the shoot meristematic region, as well as in the vasculature of the hypocotyl and root (Figure 4A and B). GUS expression was observed in both primary and lateral root vasculature (Figure 4B). In flowers, GUS is expressed in sepals and stem (Figure 4C and D), as well as in carpel tissue and anther filaments (Figure 4E). GUS expression is also evident in true leaves (Figure 4F). In silico expression analysis using GENEVESTIGATOR (60), indicates that ALKBH2 is expressed in all tissues during development (Figure 4G), in agreement with results from both the GUS line and RT–PCR analysis (Figure 4H). These results indicate that ALKBH2-mediated repair of alkylation damage is important in all parts of the plant.
Figure 4.
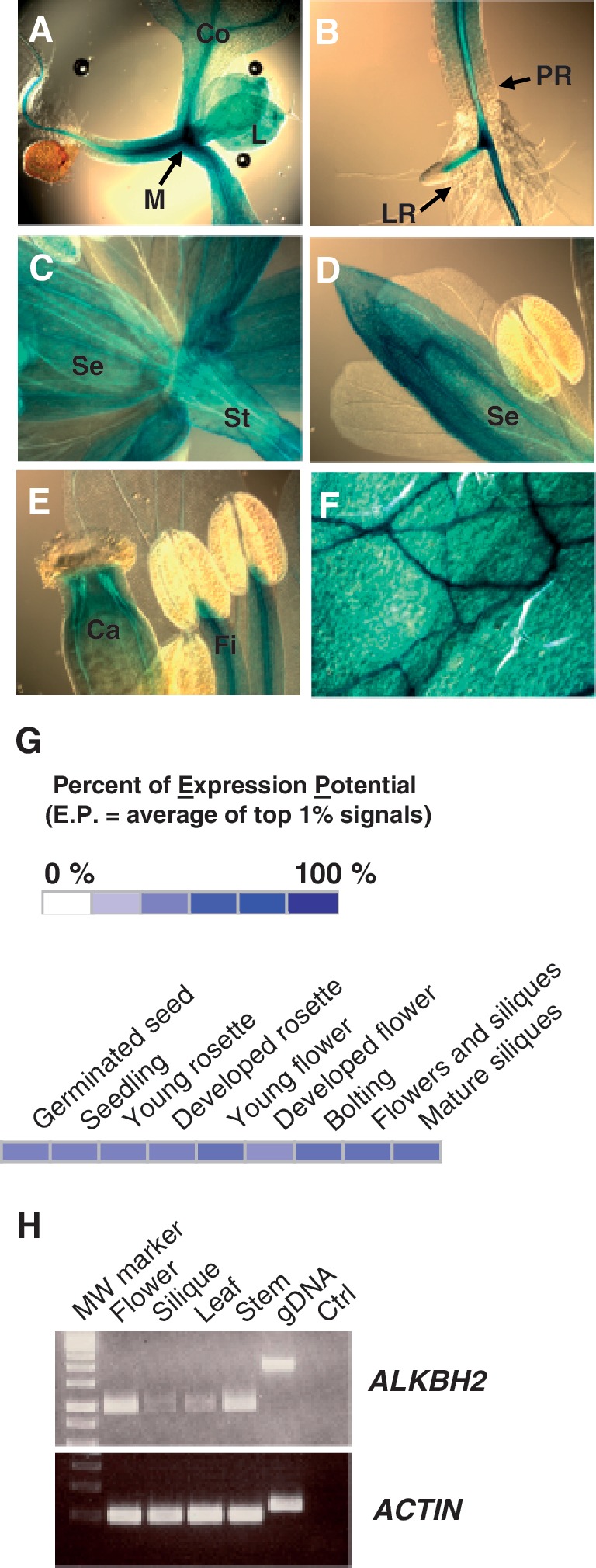
Expression pattern of pALKBH2::GUS. The expression pattern of ALKBH2 was determined using a pALKBH2::GUS transgenic line. The blue GUS staining shows expression in (A) cotyledons (Co), meristematic region (M) and true leaves (L) in seedlings, (B) veins of primary (PR) and lateral roots (LR) in seedlings, (C) stem (St) and sepals (Se), (D) sepal, (E) carpels (Ca) and anther filaments (Fi). (F) Rosette leaf. (G) Expression pattern obtained from in silico analysis using GENEVESTIGATOR (www.genevestigator.com). (H) Expression pattern of ALKBH2 using RT–PCR on cDNA from different tissues, using primers amplifying a fragment of 968 bp. Genomic DNA was used as positive control (fragment in the gDNA lane is larger due to intron sequences). The ACTIN2-7 gene, giving a fragment of 255 bp with primers spanning intron 2, amplified at comparable levels from all tissues. Ctrl, no cDNA.
MMS hypersensitivity of alkbh2-1 plants
Cells with defective alkylation repair enzymes, such as 3-methyladenine DNA glycosylases and AlkB proteins, commonly display an increased sensitivity towards methylating agents, such as MMS. This is particularly prominent in bacteria, where some alkylation repair mutants display a dramatic hypersensitivity, but there are also examples that mammalian knock-out cells lacking specific repair functions display increased sensitivity towards methylation damage. To our knowledge no studies have addressed the role of alkylation repair enzymes in protecting plants against the genotoxic effects of alkylating agents. To study the ALKBH2 function in planta, we took advantage of a mutant line (GT_5_41144, denoted alkbh2-1) annotated to have a Ds-transposon integrated in intron 3 (Figure 5A) and which appeared to represent a true knock-out line for the ALKBH2 function, since no mRNA for ALKBH2 could be detected (Figure 5B). Characterization of the alkbh2-1 line showed integration at position 257 in the ALKBH2 gene, and that a 1-bp duplication had formed at the integration site (Figure 5C). The homozygous mutant plants were viable and did not display any visible phenotype when grown under normal conditions (data not shown). However, when the mutant seeds were germinated on MMS-containing media, the resulting plants displayed aberrant growth, while the wild-type plants were unaffected by the MMS treatment (Figure 5D and E). Since Arabidopsis ALKBH2 was able to reactivate phage DNA treated with CAA, which introduces etheno adducts, and since the protein showed in vitro activity on εA, we also investigated the effects of CAA treatment on alkbh2-1 versus wild-type plants. We observed a negative effect of CAA on plant growth, but could not observe any differences between alkbh2-1 and wild-type plants (data not shown). However, Arabidopsis has several putative DNA alkylbase glycosylases, enzymes shown capable of repairing etheno adducts (61), which may provide functional redundancy for such repair.
Figure 5.
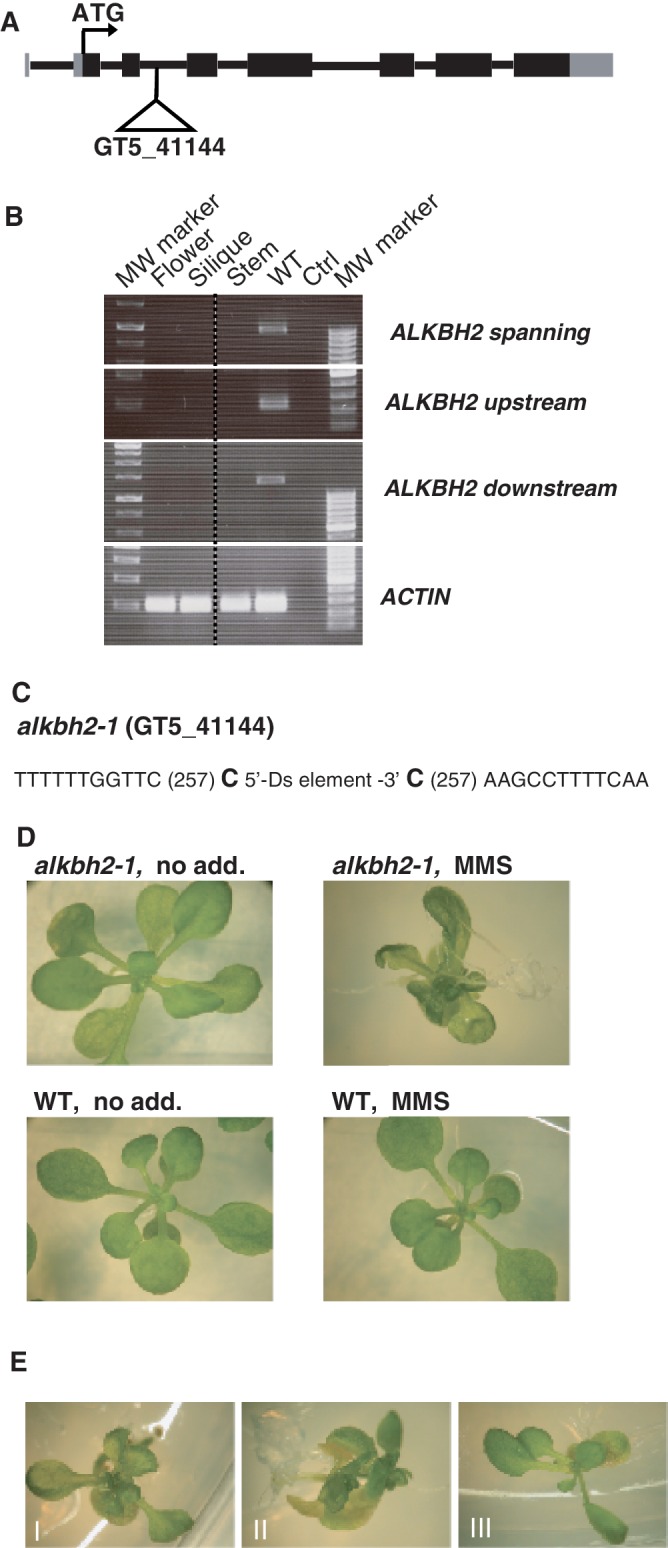
Validation and functional characterization of the alkbh2-1 line. (A) Schematic representation of the Ds insertion in the alkbh2-1 GT_5_41144 line. Exons are represented by boxes, while the intervening lines indicate introns. The untranslated part of exons is indicated in grey. (B) RT–PCR analysis of ALKBH2 expression in the alkbh2-1 line. Primer pairs upstream of, downstream of, or spanning the integration site were used. For each primer set, expression analysis was conducted on cDNA from different tissues from the alkbh2-1 lines as indicated above, as well as on WT flowers as positive control. All samples were also tested with intron spanning ACTIN primers, showing presence of cDNA. Ctrl, no cDNA. (C) Determination of the DNA sequence at the site of Ds element insertion in the alkbh2-1 (GT_5_41144) mutant. The intron sequence flanking the Ds element inserted at position 257 in the ALKBH2 gene is indicated, and the insertion of two extra nucleotides at the integration site is shown in bold. (D) MMS sensitive phenotype of alkbh2-1 plants. Wild-type or alkbh2-1 seeds were plated on medium with or without MMS, and seedlings resulting from 14 days of growth are shown. (E) Representative MMS-induced developmental defects of the alkbh2-1 plants. Seedling with abnormal shape (I); seedling with rumpled leaves (II); seedling with necrosis (III).
Accumulation of methylation damage in the genome of MMS-treated Arabidopsis
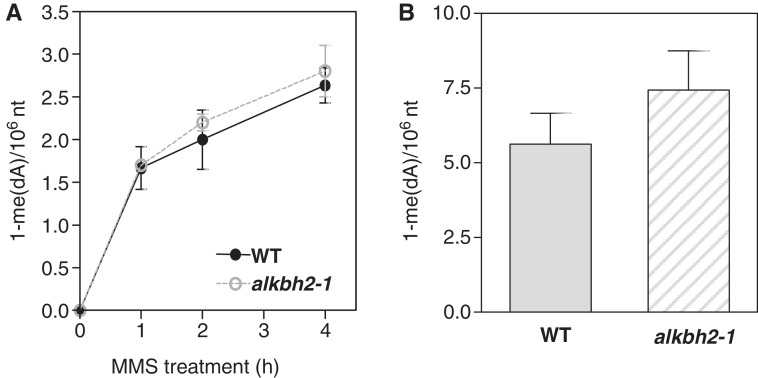
The most obvious explanation for the MMS sensitive phenotype of the alkbh2-1 plants is that unrepaired, MMS-induced lesions block DNA replication and, thereby, cell growth. Thus, we investigated the levels of genomic 1-meA in wild-type and alkbh2-1 plants after exposure to MMS. Seeds were germinated on MMS-free plates, and the resulting plants were transferred to MMS-containing liquid medium 6 days after germination, where they were incubated for up to 4 h before they were harvested, and levels of 1-meA were measured by LC-MS/MS. Clearly, growth of the plants in the presence of MMS caused a time-dependent introduction of 1-meA lesions, but the 1-meA levels were only marginally higher in the alkbh2-1 plants than in the wild-type plants (Figure 6A). Also, when the plants were left in MMS-containing medium for a prolonged time period, the 1-meA levels were only slightly higher than in the wild-type plants (Figure 6B). These results indicate that ALKBH2-mediated repair of MMS-induced 1-meA lesions is inefficient in planta, possibly because the repair capacity is exceeded.
Figure 6.
MMS-mediated induction of 1-meA lesions in genomic DNA of wild-type and alkbh2-1 Arabidopsis. (A) Time course of induction of 1-meA lesions by MMS treatment. Seedlings were treated with 90 ppm MMS at 18°C for the indicated time periods before genomic DNA was isolated and 1-meA content assessed by LC–MS/MS. (B) Induction of 1-meA lesions upon prolonged MMS treatment. Similar experiment as in (A), but the seedlings were grown in liquid media supplemented with MMS for 1 week at 18°C before 1-meA levels were assessed.
DISCUSSION
In this report, we describe the function of the Arabidopsis ALKBH2 protein, encoded by the AT2G22260 gene, thereby establishing the existence of the AlkB mechanism for DNA repair also in plants. The AT2G22260 protein sequence displays similarity to both mammalian ALKBH2 and ALKBH3, but our initial bioinformatics analysis revealed sequence features characteristic of ALKBH2. Moreover, the functional characterization revealed that Arabidopsis ALKBH2 and mammalian ALKBH2 (but not ALKBH3) share several important properties, i.e. exclusive activity on DNA (and not on RNA), preference for dsDNA over ssDNA, and activity on etheno lesions, strongly indicating that these proteins are true orthologues. Furthemore, the observed MMS-sensitive phenotype of ALKBH2 defective Arabidopsis plants indicates that ALKBH2 plays an important role in removing deleterious methyl lesions from the plant genome.
Based on protein sequence searches, the AlkB mechanism for DNA repair shows a rather patchy distribution across the tree of life. However, the genomes of multicellular eukaryotes generally encode several different AlkB homologues, most of which have unknown function, and are likely to have roles other than DNA repair. The mammalian ALKBH proteins represent a classic example of how function is not readily inferred from protein sequence similarity. ALKBH1, which displays the highest degree of sequence similarity to EcAlkB, has not been established as a DNA repair protein, whereas ALKBH2 and ALKBH3, which are less similar to EcAlkB, appear to be genuine repair proteins with EcAlkB-like activities. A similar situation appear to exist in Arabidopsis where the proteins AT3G14160 (NP_566479.5), and AT1G11780 (NP_172643.1), which display sequence homology to EcAlkB and human ALKBH1, failed to show any repair activity in phage reactivation assays (P.Ø. Falnes and A. Bekkelund, unpublished observations) (27). Taken together with previous findings, our results indicate that nucleic acid repair by oxidative demethylation is performed by ALKBH2/ALKBH3-like proteins in eukaryotes, whereas ALKBH1-like proteins have a function other than DNA repair. Notably, many multicellular eukaryotes, including the model organisms Drosophila melanogaster and Caenorhabditis elegans, appear to lack ALKBH2/ALKBH3-like proteins (21). Possibly, these organisms use other enzymes, such as DNA glycosylases, for removing 1-meA lesions from the genome. Indeed, the DNA alkylbase glycosylase AlkA from the archeon Archaeoglobus fulgidus shows activity towards 1-meA and 3-meC lesions, as well as towards the canonical 3-meA substrate (62).
Mutant plants that lack the ALKBH2 function developed normally and were fertile. However, these plants were hypersensitive to MMS, resulting in various developmental defects. Somewhat in contrast, we were not able to detect a significant increase in the levels of the ALKBH2-substrate 1-meA in plants that had been treated with MMS. Probably, the MMS-induced developmental defects displayed by alkbh2-1 plants reflect DNA damage occurring in certain progenitor cells during development, whereas the 1-meA levels were measured as an average across the entire plant. Conceivably, the repair capacity of the expressed ALKBH2 protein may in many tissues be too low to substantially reduce the level of MMS-induced 1-meA lesions, giving rise to a considerable background of unrepaired lesions. Alternatively, the toxic effects caused by MMS in the alkbh2-1 mutant plants are not caused by 1-meA, but by a lesion which is more efficiently repaired. One candidate in this respect is 3-meC, which was a better ALKBH2 substrate than 1-meA in vitro (Figure 3C), but which is unfortunately not detectable by our LC–MS/MS protocol (due to its identical molecular mass and similar LC elution time to the abundant DNA modification 5-methylcytosine). Similar to alkbh2-1 plants, cells from mice with a targeted ablation of the ALKBH2 function displayed hypersensitivity towards MMS, and they also showed a deficiency in repairing MMS-induced 1-meA lesions (32), supporting the notion that plant and mammalian ALKBH2 have similar roles.
Naturally occurring methyl halides, such as methyl chloride, methyl bromide and methyl iodide probably represent the most abundant methylating agents in the environment (6). Such compounds are emitted from plants, and they are generated during degradation of organic matter (3,4). Moreover, methyl bromide has been widely used as a fumigant to eliminate nematodes from crops (63). Methyl halides can induce various lesions in nucleic acids, including the AlkB substrates 1-meA, 3-meC and 1-meG (12,33,58). In Arabidopsis, the methyltransferase AtHOL1 has been shown to catalyse the formation of methyl halides through methylation of halide ions (64). Therefore, as part of the present study, we attempted to investigate whether ALKBH2 has a role in protecting Arabidopsis against the potentially deleterious effects of endogenously generated methyl halides. To this end, alkbh2-1 and wild-type seeds were germinated in the presence of iodide ions, which expectedly would cause the generation of substantial amounts of methyl iodide, a more potent methylating agent than methyl chloride or methyl bromide. The seedlings were analysed after two weeks, but we did not observe any iodide-induced 1-meA lesions in DNA, nor did we observe any difference in sensitivity towards iodide ions between wild-type and ALKBH2-deficient plants (data not shown). This suggested that endogenously generated methyl halides may not represent a major source of methylation damage in the Arabidopsis genome, at least not under the experimental conditions used here.
Mammalian ALKBH2 is a nuclear protein associated with PCNA, which acts as a circular clamp on DNA during replication, coupling its repair function to DNA replication (28). ALKBH2-mediated DNA repair has also been shown to occur when DNA replication is inhibited, indicating that such repair is not strictly dependent on DNA replication, and may be important also in non-dividing tissues (32). Previous studies of the expression of DNA repair genes in plants indicate that some repair proteins are only found in proliferating tissues, whereas others, such as photoreactivation and mismatch repair enzymes, are found in non-proliferating cells as well (2). Our results show that Arabidopsis ALKBH2 is expressed in both proliferating tissues and in leaves, which undergo little proliferation. A recent study, which investigated various AlkB homologues from Arabidopsis, but without detecting any repair activities, showed that Arabidopsis ALKBH2, like its mammalian counterpart, is a nuclear protein (27). Thus, it is reasonable to assume that the ALKBH2 protein is involved in nuclear DNA repair in both proliferating and non-proliferating tissues in Arabidopsis.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1.
FUNDING
FRIBIOMOL and FUGE programs of the Research Council of Norway. Funding for open access charge: Research Council of Norway.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Marius Ulleland for his contribution to the initial part of this work, and to Mari Kjos, Roy Falleth, Charles Albin-Amiot, Solveig Hauge Engebretsen and Anders Bekkelund for providing technical assistance.
REFERENCES
- 1.Tuteja N, Ahmad P, Panda BB, Tuteja R. Genotoxic stress in plants: shedding light on DNA damage, repair and DNA repair helicases. Mutat. Res. 2009;681:134–149. doi: 10.1016/j.mrrev.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 2.Kimura S, Sakaguchi K. DNA repair in plants. Chem. Rev. 2006;106:753–766. doi: 10.1021/cr040482n. [DOI] [PubMed] [Google Scholar]
- 3.Keppler F, Eiden R, Niedan V, Pracht J, Scholer HF. Halocarbons produced by natural oxidation processes during degradation of organic matter. Nature. 2000;403:298–301. doi: 10.1038/35002055. [DOI] [PubMed] [Google Scholar]
- 4.Redeker KR, Wang N, Low JC, McMillan A, Tyler SC, Cicerone RJ. Emissions of methyl halides and methane from rice paddies. Science. 2000;290:966–969. doi: 10.1126/science.290.5493.966. [DOI] [PubMed] [Google Scholar]
- 5.Rydberg B, Lindahl T. Nonenzymatic methylation of DNA by the intracellular methyl group donor S-adenosyl-L-methionine is a potentially mutagenic reaction. EMBO J. 1982;1:211–216. doi: 10.1002/j.1460-2075.1982.tb01149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sedgwick B. Repairing DNA-methylation damage. Nat. Rev. Mol. Cell Biol. 2004;5:148–157. doi: 10.1038/nrm1312. [DOI] [PubMed] [Google Scholar]
- 7.Britt A. Repair of damaged bases. The Arabidopsis Book. 2002 doi: 10.1199/tab.0005. 1:e0005. doi:10.1199/tab.0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Santerre A, Britt AB. Cloning of a 3-methyladenine-DNA glycosylase from Arabidopsis thaliana. Proc. Natl Acad. Sci. USA. 1994;91:2240–2244. doi: 10.1073/pnas.91.6.2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cordoba-Canero D, Morales-Ruiz T, Roldan-Arjona T, Ariza RR. Single-nucleotide and long-patch base excision repair of DNA damage in plants. Plant J. 2009;60:716–728. doi: 10.1111/j.1365-313X.2009.03994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aravind L, Koonin EV. The DNA-repair protein AlkB, EGL-9, and leprecan define new families of 2-oxoglutarate- and iron-dependent dioxygenases. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-3-research0007. RESEARCH0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Falnes PO, Johansen RF, Seeberg E. AlkB-mediated oxidative demethylation reverses DNA damage in Escherichia coli. Nature. 2002;419:178–182. doi: 10.1038/nature01048. [DOI] [PubMed] [Google Scholar]
- 12.Trewick SC, Henshaw TF, Hausinger RP, Lindahl T, Sedgwick B. Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature. 2002;419:174–178. doi: 10.1038/nature00908. [DOI] [PubMed] [Google Scholar]
- 13.Koivisto P, Robins P, Lindahl T, Sedgwick B. Demethylation of 3-methylthymine in DNA by bacterial and human DNA dioxygenases. J. Biol. Chem. 2004;279:40470–40474. doi: 10.1074/jbc.M407960200. [DOI] [PubMed] [Google Scholar]
- 14.Delaney JC, Essigmann JM. Mutagenesis, genotoxicity, and repair of 1-methyladenine, 3-alkylcytosines, 1-methylguanine, and 3-methylthymine in alkB Escherichia coli. Proc. Natl Acad. Sci. USA. 2004;101:14051–14056. doi: 10.1073/pnas.0403489101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Falnes PO. Repair of 3-methylthymine and 1-methylguanine lesions by bacterial and human AlkB proteins. Nucleic Acids Res. 2004;32:6260–6267. doi: 10.1093/nar/gkh964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delaney JC, Smeester L, Wong C, Frick LE, Taghizadeh K, Wishnok JS, Drennan CL, Samson LD, Essigmann JM. AlkB reverses etheno DNA lesions caused by lipid oxidation in vitro and in vivo. Nat. Struct. Mol. Biol. 2005;12:855–860. doi: 10.1038/nsmb996. [DOI] [PubMed] [Google Scholar]
- 17.Duncan T, Trewick SC, Koivisto P, Bates PA, Lindahl T, Sedgwick B. Reversal of DNA alkylation damage by two human dioxygenases. Proc. Natl Acad. Sci. USA. 2002;99:16660–16665. doi: 10.1073/pnas.262589799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frick LE, Delaney JC, Wong C, Drennan CL, Essigmann JM. Alleviation of 1,N6-ethanoadenine genotoxicity by the Escherichia coli adaptive response protein AlkB. Proc. Natl Acad. Sci. USA. 2007;104:755–760. doi: 10.1073/pnas.0607377104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koivisto P, Duncan T, Lindahl T, Sedgwick B. Minimal methylated substrate and extended substrate range of Escherichia coli AlkB protein, a 1-methyladenine-DNA dioxygenase. J. Biol. Chem. 2003;278:44348–44354. doi: 10.1074/jbc.M307361200. [DOI] [PubMed] [Google Scholar]
- 20.Bratlie MS, Drablos F. Bioinformatic mapping of AlkB homology domains in viruses. BMC.Genomics. 2005;6:1. doi: 10.1186/1471-2164-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drablos F, Feyzi E, Aas PA, Vaagbo CB, Kavli B, Bratlie MS, Pena-Diaz J, Otterlei M, Slupphaug G, Krokan HE. Alkylation damage in DNA and RNA–repair mechanisms and medical significance. DNA Repair. 2004;3:1389–1407. doi: 10.1016/j.dnarep.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 22.Falnes PO, Rognes T. DNA repair by bacterial AlkB proteins. Res. Microbiol. 2003;154:531–538. doi: 10.1016/S0923-2508(03)00150-5. [DOI] [PubMed] [Google Scholar]
- 23.van den Born E, Omelchenko MV, Bekkelund A, Leihne V, Koonin EV, Dolja VV, Falnes PO. Viral AlkB proteins repair RNA damage by oxidative demethylation. Nucleic Acids Res. 2008;36:5451–5461. doi: 10.1093/nar/gkn519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van den Born E, Bekkelund A, Moen MN, Omelchenko MV, Klungland A, Falnes PO. Bioinformatics and functional analysis define four distinct groups of AlkB DNA-dioxygenases in bacteria. Nucleic Acids Res. 2009;21:7124–7136. doi: 10.1093/nar/gkp774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gerken T, Girard CA, Tung YC, Webby CJ, Saudek V, Hewitson KS, Yeo GS, McDonough MA, Cunliffe S, McNeill LA, et al. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science. 2007;318:1469–1472. doi: 10.1126/science.1151710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kurowski MA, Bhagwat AS, Papaj G, Bujnicki JM. Phylogenomic identification of five new human homologs of the DNA repair enzyme AlkB. BMC Genomics. 2003;4:48. doi: 10.1186/1471-2164-4-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mielecki D, Zugaj DL, Muszewska A, Piwowarski J, Chojnacka A, Mielecki M, Nieminuszczy J, Grynberg M, Grzesiuk E. Novel AlkB dioxygenases-alternative models for in silico and in vivo studies. PLoS ONE. 2012;7:e30588. doi: 10.1371/journal.pone.0030588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aas PA, Otterlei M, Falnes PO, Vagbo CB, Skorpen F, Akbari M, Sundheim O, Bjoras M, Slupphaug G, Seeberg E, et al. Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature. 2003;421:859–863. doi: 10.1038/nature01363. [DOI] [PubMed] [Google Scholar]
- 29.Falnes PO, Bjoras M, Aas PA, Sundheim O, Seeberg E. Substrate specificities of bacterial and human AlkB proteins. Nucleic Acids Res. 2004;32:3456–3461. doi: 10.1093/nar/gkh655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee DH, Jin SG, Cai S, Chen Y, Pfeifer GP, O'Connor TR. Repair of methylation damage in DNA and RNA by mammalian AlkB homologues. J. Biol. Chem. 2005;280:39448–39459. doi: 10.1074/jbc.M509881200. [DOI] [PubMed] [Google Scholar]
- 31.Ringvoll J, Moen MN, Nordstrand LM, Meira LB, Pang B, Bekkelund A, Dedon PC, Bjelland S, Samson LD, Falnes PO, et al. AlkB homologue 2-mediated repair of ethenoadenine lesions in mammalian DNA. Cancer Res. 2008;68:4142–4149. doi: 10.1158/0008-5472.CAN-08-0796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ringvoll J, Nordstrand LM, Vagbo CB, Talstad V, Reite K, Aas PA, Lauritzen KH, Liabakk NB, Bjork A, Doughty RW, et al. Repair deficient mice reveal mABH2 as the primary oxidative demethylase for repairing 1meA and 3meC lesions in DNA. EMBO J. 2006;25:2189–2198. doi: 10.1038/sj.emboj.7601109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ougland R, Zhang CM, Liiv A, Johansen RF, Seeberg E, Hou YM, Remme J, Falnes PO. AlkB restores the biological function of mRNA and tRNA inactivated by chemical methylation. Mol. Cell. 2004;16:107–116. doi: 10.1016/j.molcel.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 34.Dango S, Mosammaparast N, Sowa ME, Xiong LJ, Wu F, Park K, Rubin M, Gygi S, Harper JW, Shi Y. DNA unwinding by ASCC3 helicase is coupled to ALKBH3-dependent DNA alkylation repair and cancer cell proliferation. Mol. Cell. 2011;44:373–384. doi: 10.1016/j.molcel.2011.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jia G, Yang CG, Yang S, Jian X, Yi C, Zhou Z, He C. Oxidative demethylation of 3-methylthymine and 3-methyluracil in single-stranded DNA and RNA by mouse and human FTO. FEBS Lett. 2008;582:3313–3319. doi: 10.1016/j.febslet.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Westbye MP, Feyzi E, Aas PA, Vagbo CB, Talstad VA, Kavli B, Hagen L, Sundheim O, Akbari M, Liabakk NB, et al. Human AlkB homolog 1 is a mitochondrial protein that demethylates 3-methylcytosine in DNA and RNA. J. Biol. Chem. 2008;283:25046–25056. doi: 10.1074/jbc.M803776200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang YG, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011;7:885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nordstrand LM, Svard J, Larsen E, Nilsen A, Ougland R, Furu K, Lien GF, Rognes T, Namekawa SH, Lee JT, et al. Mice lacking Alkbh1 display sex-ratio distortion and unilateral eye defects. PLoS ONE. 2010;5:e13827. doi: 10.1371/journal.pone.0013827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pan Z, Sikandar S, Witherspoon M, Dizon D, Nguyen T, Benirschke K, Wiley C, Vrana P, Lipkin SM. Impaired placental trophoblast lineage differentiation in Alkbh1(-/-) mice. Dev. Dyn. 2008;237:316–327. doi: 10.1002/dvdy.21418. [DOI] [PubMed] [Google Scholar]
- 40.Fu D, Brophy JA, Chan CT, Atmore KA, Begley U, Paules RS, Dedon PC, Begley TJ, Samson LD. Human AlkB homolog ABH8 Is a tRNA methyltransferase required for wobble uridine modification and DNA damage survival. Mol. Cell Biol. 2010;30:2449–2459. doi: 10.1128/MCB.01604-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fu Y, Dai Q, Zhang W, Ren J, Pan T, He C. The AlkB domain of mammalian ABH8 catalyzes hydroxylation of 5-methoxycarbonylmethyluridine at the wobble position of tRNA. Angew. Chem. Int. Ed Engl. 2010;49:8885–8888. doi: 10.1002/anie.201001242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leihne V, Kirpekar F, Vagbo CB, van den Born E, Krokan HE, Grini PE, Meza TJ, Falnes PO. Roles of Trm9- and ALKBH8-like proteins in the formation of modified wobble uridines in Arabidopsis tRNA. Nucleic Acids Res. 2011;39:7688–7701. doi: 10.1093/nar/gkr406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Songe-Moller L, van den Born E, Leihne V, Vagbo CB, Kristoffersen T, Krokan HE, Kirpekar F, Falnes PO, Klungland A. Mammalian ALKBH8 possesses tRNA methyltransferase activity required for the biogenesis of multiple wobble uridine modifications implicated in translational decoding. Mol.Cell Biol. 2010;30:1814–1827. doi: 10.1128/MCB.01602-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van den Born E, Vagbo CB, Songe-Moller L, Leihne V, Lien GF, Leszczynska G, Malkiewicz A, Krokan HE, Kirpekar F, Klungland A, et al. ALKBH8-mediated formation of a novel diastereomeric pair of wobble nucleosides in mammalian tRNA. Nat. Commun. 2011;2:172. doi: 10.1038/ncomms1173. [DOI] [PubMed] [Google Scholar]
- 45.Blatny JM, Brautaset T, Winther-Larsen HC, Haugan K, Valla S. Construction and use of a versatile set of broad-host-range cloning and expression vectors based on the RK2 replicon. Appl. Environ. Microbiol. 1997;63:370–379. doi: 10.1128/aem.63.2.370-379.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murashige T, Skoog F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiologia Plantarum. 1962;15:473–497. [Google Scholar]
- 47.Grini PE, Jurgens G, Hulskamp M. Embryo and endosperm development is disrupted in the female gametophytic capulet mutants of Arabidopsis. Genetics. 2002;162:1911–1925. doi: 10.1093/genetics/162.4.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bjerkan KN, Jung-Romeo S, Jurgens G, Genschik P, Grini PE. Arabidopsis WD repeat domain55 interacts with DNA damaged binding protein1 and is required for apical patterning in the embryo. Plant Cell. 2012;24:1013–1033. doi: 10.1105/tpc.111.089425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Curtis MD, Grossniklaus U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 2003;133:462–469. doi: 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 51.Mena-Benitez GL, Gandia-Herrero F, Graham S, Larson TR, McQueen-Mason SJ, French CE, Rylott EL, Bruce NC. Engineering a catabolic pathway in plants for the degradation of 1,2-dichloroethane. Plant Physiol. 2008;147:1192–1198. doi: 10.1104/pp.108.119008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Crain PF. Preparation and enzymatic hydrolysis of DNA and RNA for mass spectrometry. Methods Enzymol. 1990;193:782–790. doi: 10.1016/0076-6879(90)93450-y. [DOI] [PubMed] [Google Scholar]
- 53.Yang CG, Yi C, Duguid EM, Sullivan CT, Jian X, Rice PA, He C. Crystal structures of DNA/RNA repair enzymes AlkB and ABH2 bound to dsDNA. Nature. 2008;452:961–965. doi: 10.1038/nature06889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Monsen VT, Sundheim O, Aas PA, Westbye MP, Sousa MM, Slupphaug G, Krokan HE. Divergent ss-hairpins determine double-strand versus single-strand substrate recognition of human AlkB-homologues 2 and 3. Nucleic Acids Res. 2010;38:6447–6455. doi: 10.1093/nar/gkq518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu B, Edstrom WC, Benach J, Hamuro Y, Weber PC, Gibney BR, Hunt JF. Crystal structures of catalytic complexes of the oxidative DNA/RNA repair enzyme AlkB. Nature. 2006;439:879–884. doi: 10.1038/nature04561. [DOI] [PubMed] [Google Scholar]
- 58.Singer B, Grunberger D. Molecular Biology of Mutagens and Carcinogens. New York: Plenum Press; 1983. [Google Scholar]
- 59.Dinglay S, Trewick SC, Lindahl T, Sedgwick B. Defective processing of methylated single-stranded DNA by E. coli AlkB mutants. Genes Dev. 2000;14:2097–2105. [PMC free article] [PubMed] [Google Scholar]
- 60.Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gros L, Ishchenko AA, Saparbaev M. Enzymology of repair of etheno-adducts. Mutat. Res. 2003;531:219–229. doi: 10.1016/j.mrfmmm.2003.07.008. [DOI] [PubMed] [Google Scholar]
- 62.Leiros I, Nabong MP, Grosvik K, Ringvoll J, Haugland GT, Uldal L, Reite K, Olsbu IK, Knaevelsrud I, Moe E, et al. Structural basis for enzymatic excision of N1-methyladenine and N3-methylcytosine from DNA. EMBO J. 2007;26:2206–2217. doi: 10.1038/sj.emboj.7601662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zasada IA, Halbrendt JM, Kokalis-Burelle N, LaMondia J, McKenry MV, Noling JW. Managing nematodes without methyl bromide. Annu. Rev. Phytopathol. 2010;48:311–328. doi: 10.1146/annurev-phyto-073009-114425. [DOI] [PubMed] [Google Scholar]
- 64.Rhew RC, Ostergaard L, Saltzman ES, Yanofsky MF. Genetic control of methyl halide production in Arabidopsis. Curr. Biol. 2003;13:1809–1813. doi: 10.1016/j.cub.2003.09.055. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.