Abstract
RNA-binding proteins regulate multiple steps of RNA metabolism through both dynamic and combined binding. In addition to its crucial roles in cell adhesion and Wnt-activated transcription in cancer cells, β-catenin regulates RNA alternative splicing and stability possibly by binding to target RNA in cells. An RNA aptamer was selected for specific binding to β-catenin to address RNA recognition by β-catenin more specifically. Here, we characterized the structural properties of the RNA aptamer as a model and identified a β-catenin RNA motif. Similar RNA motif was found in cellular RNA, Cyclooxygenase-2 (COX-2) mRNA 3′-untranslated region (3′-UTR). More significantly, the C-terminal domain of β-catenin interacted with HuR and the Armadillo repeat domain associated with RNA to form the RNA–β-catenin–HuR complex in vitro and in cells. Furthermore, the tertiary RNA–protein complex was predominantly found in the cytoplasm of colon cancer cells; thus, it might be related to COX-2 protein level and cancer progression. Taken together, the β-catenin RNA aptamer was valuable for deducing the cellular RNA aptamer and identifying novel and oncogenic RNA–protein networks in colon cancer cells.
INTRODUCTION
Posttranscriptional regulation of RNA is mediated by RNA–protein interactions between RNA-binding proteins (RBPs) and regulatory sequences in RNA (1–4). RBPs are also associated with many other proteins as a ribonucleoprotein (RNP) complex through protein–protein interactions and/or RNA-mediated interactions. Thus, combined binding and dynamic remodeling of RNPs is crucial for regulating various steps in RNA metabolism. There may be many thousands of RBPs in vertebrates, which are usually associated with RNA in a sequence- or structure-dependent manner. The number of RBPs is expected to increase if other types of RNA-binding domains are added to the list (5).
Hu proteins are a family of RBPs with homology to the Drosophila embryonic lethal abnormal vision (ELAV) protein, which include the HuR (HuA), HuB (Hel-N1), HuC and HuD proteins. HuR is ubiquitously expressed, unlike the other members of the ELAV family (HuB, HuC and HuD), which are exclusively found in neuronal tissue (6). HuR is mostly located in the nucleus, but certain events trigger its translocation to the cytoplasm (7,8) where it stabilizes various transcripts following stimulation (9). These transcripts contain AU-rich elements (AREs), and HuR functions as an adaptor protein for the nuclear export of many ARE-containing mRNAs. Regardless of the mechanism, the role of HuR in colon carcinogenesis is crucial (10). Among HuR-regulated oncogenic transcripts, cyclooxygenase-2 (COX-2) expression is critical for colon cancer tumorigenesis (11,12). Many AREs are present in the COX-2 3′-untranslated region (3′-UTR); thus, the identification and mapping of RBPs and their recognition sites on RNA are necessary (13–17).
β-Catenin is a multifunctional protein involved in cell adhesion and transcription downstream of Wnt signaling (18–20). The scaffolding proteins adenomatous polyposis coli (APC) and axin interact with β-catenin at cell adherent junctions, and glycogen synthase kinase-3β phosphorylates and inhibits β-catenin proteolysis. However, mutations are frequently found in the β-catenin gene in colon cancer cells, so its protein level rises and accumulates in the nucleus where it activates the transcription of various oncogenic target genes such as cyclin D1 and c-myc (21–24). Many proteins interact with β-catenin via the central Armadillo (Arm) repeat domain and through the N- or C-terminal domains (25). It was reported recently that β-catenin regulates RNA alternative splicing of estrogen receptor-β and RNA stability of unstable transcripts such as COX-2 mRNA (26–30). More significantly, β-catenin directly interacts with these RNAs in vitro. However, the mechanism behind these unexpected findings has not yet been systematically studied.
To examine the nature of the β-catenin–RNA interaction, we utilized here the RNA aptamer as a model RNA to map the β-catenin-binding RNA motif. We then showed that β-catenin interacts with an RNA motif within the COX-2 3′-UTR, which might be a cellular RNA aptamer. Furthermore, β-catenin interacted with HuR via the C-terminal domain in addition to its association with COX-2 mRNA thru the Arm repeat domain. More interestingly, formation of the tertiary RNP differed depending on tumor progression in colon cancer cells. These findings reveal a novel function of β-catenin, which might explain the altered RNA stabilization of COX-2 mRNA in colon cancer cells.
MATERIALS AND METHODS
Preparation of recombinant proteins and GST pull-down assay
The Arm 1–12 (Arm) bacterial expression vector and full length (FL) Arm have been described previously (30). β-Catenin fragments for C-terminal (C-term, amino acids 685–781) was obtained by polymerase chain reaction (PCR) amplification of FL-β-catenin. The primers used in this study are shown in Supplementary Table S1. PCR fragments were cloned into the pGEX-5X-1 vector. Glutathione-S-transferase (GST)-fusion proteins were purified using glutathione-Sepharose 4B beads (GE Healthcare). Recombinant HuR protein was expressed from pGEX-HuR and treated with thrombin. A GST pull-down assay was performed between GST-tagged FL, Arm, C-term β-catenin and GST and recombinant HuR (1:1 ratio).
Preparation of in vitro transcribed RNA and biotinylation
The pUC19-Aptamer was described previously and was cleaved with BamHI for the in vitro transcription (27). pZEO/Luc-COX-2 3′-UTR was used as a template for PCR amplification of the different fragments of COX-2 mRNA (22). All 5′-primers contained the T7 promoter sequence (T7). To prepare templates for 3′-UTR fragments, U-1, U-2, U-3 and U-4, primer sets in Supplementary Table S1 were used. For in vitro transcription of RNA transcripts, template DNA (1 µg) was incubated with T7 RNA polymerase (Ambion) for 3 h at 37°C. Biotinylation composition buffer (0.2 mM each of ATP, GTP, UTP and 0.12 mM CTP and 0.08 mM Bio-11-CTP) was used to generate biotinylated RNA.
RNase footprinting
RNA that was labeled in vitro at the 5′-end with [γ-32P] ATP (Amersham) was denatured and renatured in binding buffer. Various concentrations (0, 5, 25, 50 and 100 nM) of β-catenin protein were added and incubated at 37°C for 15 min. The RNA–protein complexes were incubated with RNase T1, RNase S1 or with RNase V1 at room temperature for 15 min. The gels were dried and analyzed using a PhosphoImager (FUJIX Bio Image Analyzer System).
Surface plasmon resonance
Surface plasmon resonance (SPR) experiments were carried out using a BIACORE 3000 (GE Healthcare) as recommended by the manufacturer. CM5 sensorchips was used and biotinylated RNA was immobilized on the streptavidin-coated flow cells in HBSEP buffer. For RNA–protein binding analyses, various concentrations (0, 5, 10, 20, 40, 80, 160, 320 and 640 nM) of β-catenin or HuR protein was injected as an analyte in HBSEP buffer. BIA Evaluation 3.1 program was used for evaluation of KD. To test for RNA-mediated protein interaction, 10 nM of protein A was injected into the flow with biotinylated RNA to create RNA–protein complexes on the flow chip, and the background was reset before adding protein B as an analyte.
Biotin RNA pull-down assay
After incubating the biotinylated RNAs (40 nmol) and the whole-cell extracts in TENT buffer [10 mM Tris–HCl (pH 8.0), 1 mM EDTA, 250 mM NaCl and 0.5% Triton X-100] for 30 min at room temperature, the samples were subjected to streptavidin-magnetic bead adsorption and incubated for an additional 30 min. After the incubation, bound beads were washed thrice with PBS.
Cell culture and cell fractionation
Human embryonic kidney 293T (HEK293T) and human colorectal adenocarcinoma HT-29 cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS). HEK293 cells were cultured in MEM with 10% fetal calf serum. The human colorectal cancer cell line LoVo were cultured in RPMI 1640 with 10% FBS. For cell fractionation, cells were rinsed three times in PBS and harvested by scraping into RSB buffer [10 mM Tris–HCl (pH 7.4), 10 mM NaCl, 2.5 mM MgCl2, protease inhibitors and ribonuclease inhibitor] containing digitonin (40 μg/ml). The cells were incubated on ice for 5 min and centrifuged at 2000g for 8 min to obtain the cytosolic supernatant. The remaining pellet was resuspended in RIPA buffer, incubated on ice for 5 min, centrifuged at 14 000g for 8 min, and the supernatant was collected as the nuclear extract.
Reporters and site-directed mutagenesis
The luciferase reporters for the U-4 COX-2 3′-UTR fragment were cloned into the XbaI site for the pZEO/Luc vector and the SacII site for the pDHFR vector. Introduction of a point mutation into the β-catenin element (ACTTT to CCCCC) in the full-length 3′-UTR and U-4 was performed using the sense primer (139–170 of COX-2 3′-UTR). A point mutation was introduced into the β-catenin element (ACTTT to GCGCG) of the pU6-aptamer using the sense primer (33–72 of the β-catenin aptamer). Mutants were generated using the QuickChange™ Site-Directed Mutagenesis kit (Stratagene).
RNP immunoprecipitation and co-immunoprecipitation
The basic RNP immunoprecipitation (RNP-IP) procedure was described previously (30). Briefly, cells were reversibly cross-linked with 0.5% formaldehyde, and immunoprecipitated and bound RNA was analyzed by reverse transcription–polymerase chain reaction (RT–PCR). For co-immunoprecipitation (co-IP), cell lysates were pre-cleared either with Protein G in RIPA buffer for 2 h at 4°C with agitation. After pre-clearing, anti-β-catenin (#610154, BD Biosciences) or anti-HuR (3A2, Santa Cruz Biotechnology) were incubated overnight at 4°C with agitation. Protein G beads were added and tumbled for 3 h. The beads were then washed three times with RIPA buffer. Bound proteins were separated by 10% SDS–PAGE and blotted with monoclonal antibodies that recognize the specific proteins as indicated.
Luciferase reporter assay and siRNA transfection
HEK293 cells were seeded on 12-well plates and transfected in triplicate using Lipofectamine (Invitrogen) containing the luciferase reporter plasmid and the pRL-TK plasmid (as internal control) in the presence of expression clones. After 24 h, luciferase activities were measured using the GloMax® 20/20 luminometer (Promega). HEK293 or LoVo cells were transfected with 40 nM β-catenin or HuR siRNA using Lipofectamine 2000 according to the manufacturer's protocol. Control GFP siRNA was used as the control.
RESULTS
Arm repeat domain of β-catenin binds RNA aptamer via specific RNA element
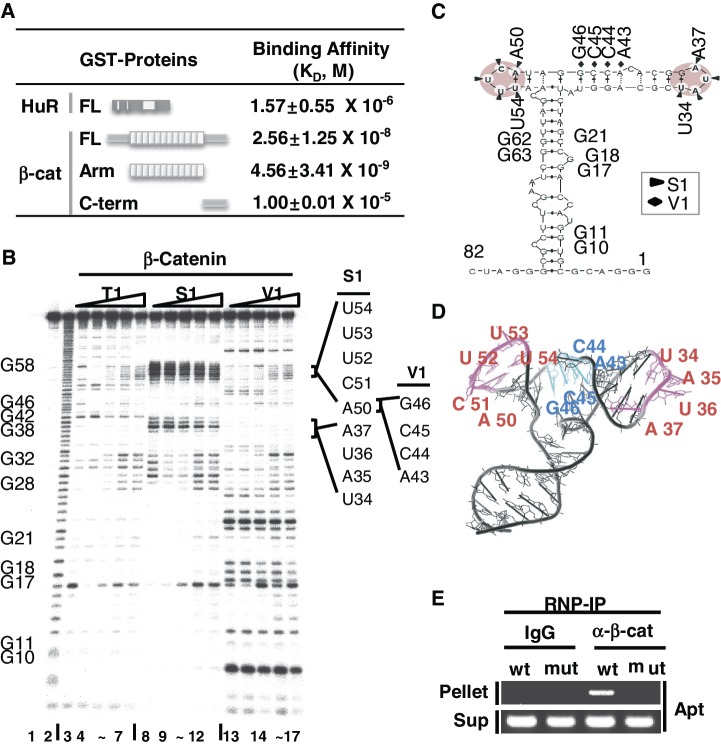
We previously used an RNA aptamer and roughly estimated the binding affinity using the RNA electrophoretic mobility shift assay (R-EMSA) (27). In this study, we further assessed binding affinity and mapped the RNA-binding domain within the β-catenin protein using SPR analysis (Figure 1A). The Arm domain of β-catenin bound to the RNA aptamer with high affinity (KD = 4.56 × 10−9 M), even higher than that of FL β-catenin (KD = 2.56 × 10−8 M). However, the C-terminal (C-term) domain of β-catenin (96 amino acids) did not show any detectable binding to RNA (KD = 1.0 × 10−5 M). This suggests that the RNA-binding potential of β-catenin is largely due to the Arm repeat domain of the protein. SPR analysis also revealed that the RNA aptamer had low-binding affinity to HuR (KD = 1.57 × 10−6 M), confirming specific binding of the RNA aptamer to β-catenin but not to HuR.
Figure 1.
Structural probe for β-catenin-binding site mapping of the RNA aptamer. (A) Determination of in vitro equilibrium dissociation constants, KD, for the RNA aptamer by surface plasmon resonance (SPR) analysis. Schematic diagrams of HuR and β-catenin, FL, Arm and C-term of β-catenin, are shown. KD of FL, Arm and C-term of β-catenin to RNA aptamer were compared to that of HuR binding to the RNA aptamer. RNA was immobilized on a SPR chip and analyte proteins were added. Three independent SPR experiments were performed and the average KD values are shown with the standard deviation. (B) RNase mapping and footprinting of the β-catenin-binding RNA aptamer. The RNA aptamer was end-labeled and incubated with various concentrations of the β-catenin protein, followed by digestion with RNase T1, RNase S1 or RNase V1. Lane 1, no treatment; lane 2, alkaline hydrolysis; lanes 3, 8, 13, RNase treated, no β-catenin protein added; lanes 4–7, 9–12, 14–17, RNase treated with RNA–protein at ratios of 1:1, 1:2, 1:5, 1:10. G nucleotide positions are indicated on the left side of the gel. RNase-protected nucleotides (UAUA and ACUUU) are marked by solid lines on the right side of the gel. (C) MC-fold predicted secondary structure of the RNA aptamer. RNase protected sequences are indicated by triangles (S1) or diamonds (V1) and putative β-catenin-binding stem–loops are shaded. (D) MC-fold predicted tertiary structure of the aptamer was drawn using Pymol. S1 protected single-stranded ARE sequences (magenta, UAUA and ACUUU) and V1 protected sequences (cyan) are shown. (E) Ribonucleoprotein immunoprecipitation (RNP-IP) of the RNA aptamer with an anti-β-catenin antibody. HEK293T cells were transfected with the U6-aptamer containing the wild-type (wt, ACUUU) or mutant (mut, GCGCG) sequence at nucleotides 50–54. Normal IgG was used as a control. Supernatant RNA is shown as a control for RNA aptamer expression.
When we predicted the RNA secondary structure of the RNA aptamer using the Mfold RNA-folding program (31), two structures emerged with similar thermodynamic stabilities (Supplementary Figure S1). To biochemically confirm the predicted RNA structure, RNase mapping analysis was performed (Figure 1B). RNase S1 sensitivity clearly showed two prominent single-stranded regions at 34–37 (UAUA) and around 50–54 (ACUUU) (lanes 3, 8 and 13 in Figure 1B). Interestingly, when the MC-fold was used to predict the RNA secondary structure, an additional double-stranded conformation was predicted, which more accurately fit our biochemical data (Figures 1C) (32). The β-catenin-binding sites were mapped to RNase-protected sequences upon binding to β-catenin. Quantitative analysis of each band showed that nucleotides 34–37 and 50–54 were protected by RNase S1 and nucleotide 43–46 by RNase V1. Prominent binding sites for the β-catenin protein were incorporated into the inferred ternary RNA structure determined by MC-fold [kindly provided by Francois Major (32)], which correlated nicely with the two protruding bulges in the tri-bridged structure (magenta in Figure 1D).
To more clearly demonstrate the critical role of the single-stranded loop with the ACUUU RNA element, site-directed mutagenesis was performed, and the ACUUU sequence on the U6-aptamer was changed to GCGCG (27,28). We expressed wild-type and mutant U6-aptamer in HEK293T cells and performed the RNP-IP. In Figure 1E, the wild-type RNA aptamer, but not the mutant, specifically bound to β-catenin. Since a mutation in ACUUU completely disrupted β-catenin binding to the aptamer, this specific RNA motif must be important to binding.
C-terminal domain of β-catenin interacts with HuR and facilitates formation of the RNA bound protein complex in vitro
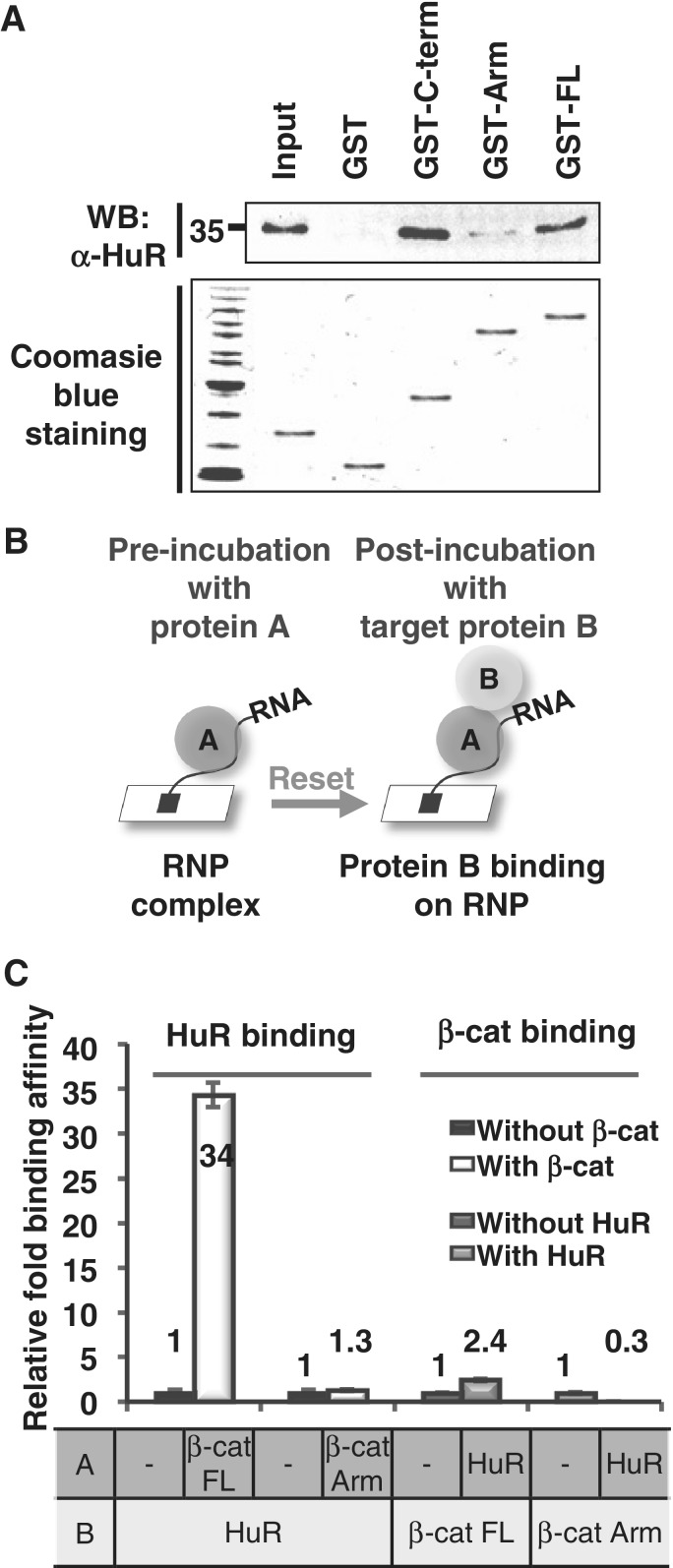
We next investigated whether RNA bound β-catenin interacted with other proteins. As the Arm repeat is the RNA-binding domain, we then asked which β-catenin domain is involved in the protein–protein interaction with HuR, as we have previously reported a protein–protein interaction between β-catenin and HuR in cells (30). GST pull-down analysis with various forms of β-catenin revealed that the C-term domain of the protein directly interacted with HuR (Figure 2A).
Figure 2.
RNA–protein complex between β-catenin and HuR. (A) GST pull-down analysis. Purified GST or various deletions of GST-tagged β-catenin proteins were incubated with HuR recombinant protein. Upper panel, GST pulled down the HuR protein as shown by Western blot analysis with the anti-HuR antibody. Lower panel, HuR and GST-β-catenin proteins were detected by Coomassie blue staining. (B) Schematic diagram of the experiment used to measure tertiary RNA–protein complex formation by SPR. The RNA aptamer was immobilized on the sensor chip, incubated with protein A and followed by the addition of analyte protein B. After resetting the background response to protein A (pre-incubated RNA–protein A complex), the response to target protein B was measured and the binding affinity of protein B was evaluated. (C) Relative binding of HuR (protein B) to the RNA aptamer with or without pre-incubating FL or Arm of β-catenin (protein A). Relative binding of FL or Arm (protein B) to the RNA aptamer with or without pre-incubating HuR (protein A) was also shown. Three independent SPR experiments were performed and the KD values of each experiment were evaluated and shown as relative binding.
Since there were two separate domains for RNA and HuR bindings, it was predicted that RNA bound FL β-catenin could also interact with HuR, possibly through the exposed C-term domain. To test this hypothesis, we designed experiments based on SPR analysis by sequentially adding two recombinant proteins in the presence or absence of one of the other proteins. Protein A was added to the RNA-coated chip for complex formation, and the background was reset. Then, various concentrations of protein B were added as the analytes (Figure 2B). The binding constants were evaluated for protein B binding to the protein A–RNA complex.
The RNA aptamer was coated to the chip, pre-incubated with HuR or β-catenin as protein A, then β-catenin or HuR protein was added as protein B. Strikingly, when FL β-catenin was pre-added as protein A followed by adding HuR as protein B, HuR binding to the RNA–β-catenin complex increased dramatically by more than 34-fold (Figure 2C). In sharp contrast, HuR did not interact with the RNA–Arm complex (Figure 2C). This clearly demonstrated that RNA-bound FL β-catenin could facilitate the formation of a tertiary complex with HuR through the C-term domain. As an opposite control, HuR was pre-incubated as protein A, then FL β-catenin or the Arm domain were added as protein B. No significant changes in binding affinity were observed, because HuR could not bind to the RNA aptamer as protein A (Figure 2C). Taken together, the results indicated that β-catenin and HuR directly comprise the tertiary RNP complex with RNA in vitro, through two separate β-catenin domains, the Arm for RNA binding and the C-term for HuR binding.
β-Catenin and HuR bind non-overlapping RNA sequences in COX-2 3′-UTR
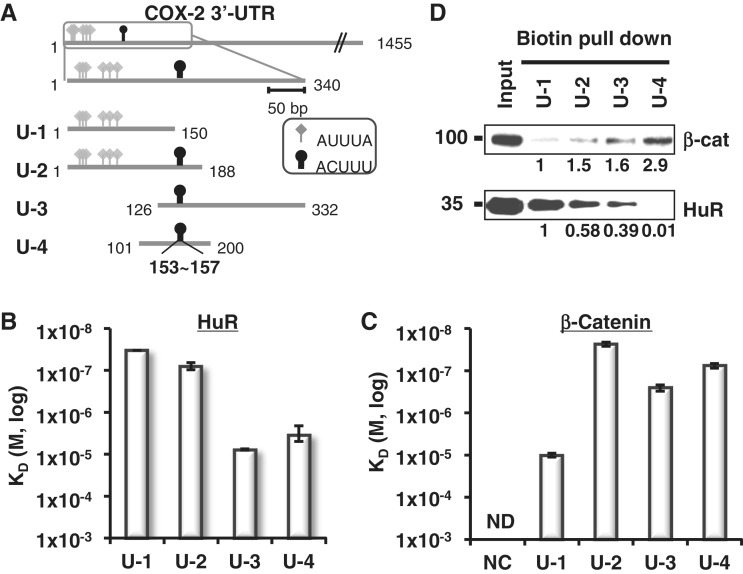
Since we obtained valuable information on β-catenin-binding RNA sequences (Figure 1), we tried to identify the β-catenin and HuR-binding RNA elements in COX-2 3′-UTR. Sequence analysis of the 3′-UTR revealed many prototypic HuR-binding AU-rich elements (AUUUA, diamond arrowhead in Figure 3A) as well as putative β-catenin-binding RNA elements (ACUUU, oval arrowhead in Figure 3A). Many AREs were clustered on the proximal region (shown as a Box), so we generated four different fragments derived from the ARE-rich region (Supplementary Figure 2A). In Figure 3A, six AREs were found in the proximal region of 3′-UTR: Fragments U-1 and U-2 have AUUUA class I/II ARE (shown as a diamond), whereas Fragments U-3 and U-4 have class III ARE (UUUU) only (data not shown). A putative β-catenin-binding sequence ACUUU (shown as a circle) was located in U-2, U-3 and U-4. The predicted locations of these elements were mostly in the loop of the stem-loop structure (Supplementary Figure S2B and C).
Figure 3.
Mapping of β-catenin and HuR binding on COX-2 3′-UTR. (A) Diagram of COX-2 3′-UTR showing ARE clusters (340 nt, box). Four fragments (U-1 to U-4) were generated spanning ARE clusters. HuR binding elements included six class I/II AREs and are shown with diamond arrowheads (AUUUA). Putative β-catenin binding elements are also shown with oval arrowheads (153–157, ACUUU). Starting and terminating nucleotides are shown. (B) HuR-binding affinity (KD) from SPR analysis with immobilized COX-2 3′-UTR fragments. Three independent SPR experiments were performed and the average KD values of each experiment were evaluated and shown with standard deviations. (C) β-Catenin-binding affinity (KD) of the COX-2 3′-UTR fragments and NC RNA. Three independent SPR experiments were performed and are shown as in C, ND, not detectable. (D) Biotin pull-down analysis of U-1 to U-4 using HT-29 colon cancer cell extracts. β-Catenin and HuR proteins in the RNA-bound pellet fractions were detected by Western blot analysis. Relative fold binding of proteins on U-2, U-3 and U-4 RNA was compared with those of U-1 RNA bound proteins.
SPR analysis with U-1 to U-4 RNA fragments and the recombinant HuR protein was performed to measure the binding affinities of HuR to these four UTR fragments. In Figure 3B and Supplementary Table S1, HuR bound U-1 and U-2 with high affinity (KD = 3.34 × 10−8 M and KD = 8.13 × 10−8 M, respectively), whereas its bindings to U-3 and U-4 were moderate (KD = 7.70 × 10−6 M and KD = 3.51 × 10−6 M, respectively). This suggests that HuR prefers the AUUUA sequence over the UUUU sequence in the case of COX-2 3′-UTR. More significantly, recombinant β-catenin bound U-2, U-3 and U-4 with high affinity (KD = 1.58 × 10−7 M, KD = 2.60 × 10−6 M and KD = 1.31 × 10−7 M, respectively) and U-1 with moderate affinity (KD = 1.00 × 10−5 M; Figure 3C). This might explain a previous finding, where the F1 fragment (as same as U-1) of COX-2 3′-UTR was shown to bind β-catenin by the supershift assay (30). In contrast, binding affinity to negative control (NC) RNA (Supplementary Figure 2A) could not be evaluated by SPR.
A biotin RNA pull-down assay was performed to understand the cellular protein binding patterns on the COX-2 3′-UTR (Figure 3D). COX-2 3′-UTR fragments (U-1 to U-4) were biotin labeled and their bindings to cellular β-catenin and HuR proteins from HT-29 colorectal adenocarcinoma were analyzed by western blotting. The cellular HuR protein associated with U-1, U-2 and U-3 but not with U-4, whereas cellular β-catenin bound most of the RNA fragments with different binding affinities (Figure 3D). U-4 was the RNA fragment that was specifically bound by β-catenin but not by HuR. The cellular β-catenin protein also associated with U-1 but not with the coding region (CR) of COX-2 mRNA or the GAPDH mRNA 3′-UTR (Supplementary Figure S2A D). Taken together, we conclude that U-4 is the β-catenin-specific RNA fragment from the COX-2 3′-UTR.
The ACUUU motif is required for β-catenin binding to the cellular RNA aptamer
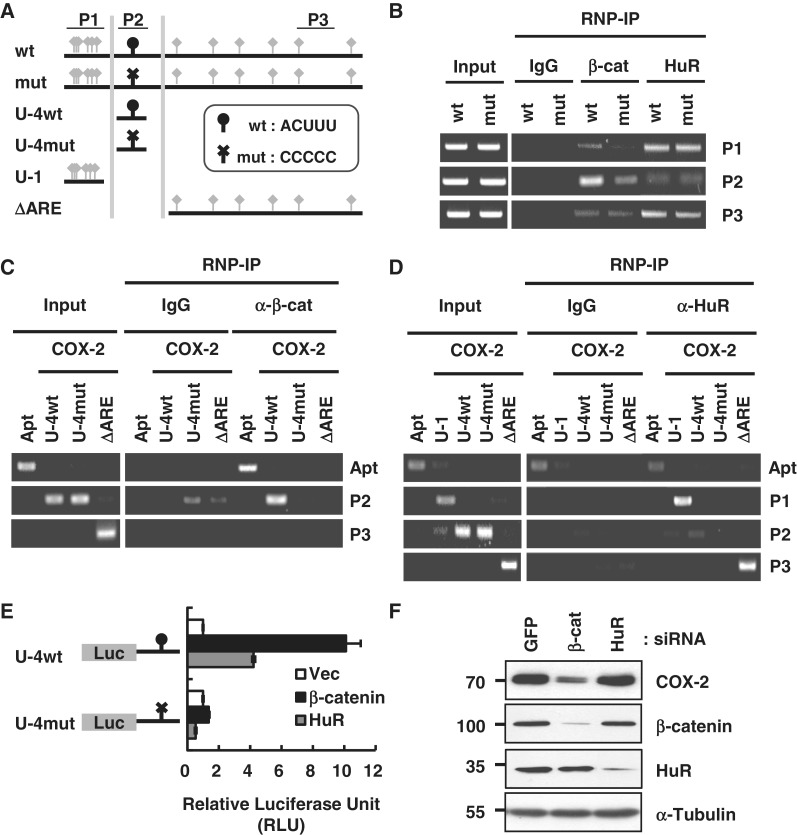
To more clearly demonstrate the critical role of the ACUUU RNA element, luciferase reporters with wild-type (ACUUU) or mutant (CCCCC) sequences were generated in the FL 3′-UTR (wt and mut) and in U-4 (U-4wt and U-4mut). Reporters with U-1 and Delta ARE were also utilized (Figure 4A).
Figure 4.
Specificity of β-catenin-binding RNA motifs in the COX-2 3′-UTR. (A) Diagram of COX-2 3′-UTR fragments with wild-type β-catenin-binding element (wt, ACUUU, oval arrowhead) and HuR-binding element (AUUUA, diamond arrowhead). Mutant β-catenin-binding element (mut, CCCCC) is shown with a cross arrowhead. ΔARE denotes COX-2 3′-UTR without 1–150 nt. U-4 fragments with wild-type (U-4wt) or mutant (U-4mut) β-catenin-binding element were also generated. PCR primer sites used for the analysis are shown as a P1, P2 and P3. (B) RNP-IP with wild-type or mutant FL COX-2 3′-UTR in HEK293T cells. Whole-cell extracts were used for immunoprecipitation with anti-β-catenin or anti-HuR antibody. PCR primers for the analysis of precipitated RNA are indicated on the right side of the gel. (C) RNP-IP with the anti-β-catenin antibody. U-4 fragments with wild-type (U-4wt) or mutant (U-4mut) element, ΔARE COX-2 3′-UTR were expressed in HEK293T cells. The RNA aptamer (Apt) was also expressed as a positive control for β-catenin binding. PCR primers for the analysis of precipitated RNA are indicated on the right side of the gel. (D) RNP-IP with the anti-HuR antibody. U-4 fragments with wild-type (U-4wt) or mutant (U-4mut) site, U-1 and ΔARE COX-2 3′-UTR were transfected in HEK293T cells. RNA aptamer (Apt) was also expressed as a positive control for β-catenin binding. PCR primers for the analysis of precipitated RNA are indicated on the right side of the gel. (E) Luciferase assay of U-4 reporters with wild-type (U-4wt) or mutant (U-4mut) β-catenin-binding element. HEK293 cells were co-transfected with U-4wt or U-4mut luciferase reporters in addition to the vector or the β-catenin or HuR expression clones. Firefly luciferase activity was normalized to Renilla luciferase activity. The results are mean ± SEM of three independent experiments. (F) Knock-down with siRNAs for β-catenin or HuR to test for the expression of COX-2 protein in LoVo colorectal adenocarcinoma cells. siRNA for GFP was used as a control. Western blot analysis was performed using the indicated antibodies and the protein size markers are shown on the left side of the gel. α-Tubulin was used as a loading control.
To test the role of the ACUUU sequence in cellular β-catenin binding, wt or mut FL 3′-UTR were transfected into HEK293 cells and the RNP-IP was performed (Figure 4B). The wt 3′-UTR bound to β-catenin as shown by the P2 primer, which amplified sequences around ACUUU (Figure 4B). More convincingly, when β-catenin was overexpressed, β-catenin binding increased only in response to the wt 3′-UTR (Supplementary Figure 3A). As expected, binding of HuR to its binding sites in the proximal and distal AUUUA sequences in the 3′-UTR was detected with the P1 and P3 primers, respectively (Figure 4B and Supplementary Figure S3B). The PCR primers for the luciferase gene were used as a control for the expression of the reporters.
Since U-4 is a β-catenin-specific minimal fragment, RNA specificity was tested by the RNP-IP with U-4wt and U-4mut reporters and analyzed with the P2 primer (Figure 4C). The RNA aptamer was used as a positive control for β-catenin binding in the cells. The specificity of HuR binding on the typical ARE in the proximal and distal region of the 3′-UTR was again confirmed with the P1 and P3 primers (Figure 4D). We also used the luciferase reporter with the c-myc 3′-UTR as a NC RNA for the RNP-IP (Supplementary Figure S3C), because we have previously reported that β-catenin does not bind c-myc mRNA and is unable to regulate c-myc mRNA stability (28).
The luciferase assay was performed with the U-4wt and U-4mut reporters to test if β-catenin could affect COX-2 protein expression via U-4 in the 3′-UTR (Figure 4E). β-Catenin dramatically increased the luciferase activity by up to 10-fold when the ACUUU element was present in U-4. However, a mutation in U-4 made it completely unresponsive to β-catenin overexpression, which strongly suggests that the ACUUU sequence is a specific functional element for β-catenin. Interestingly, HuR also increased wild-type reporter activity but not mutant reporter activity, albeit to a lesser extent.
To directly test the role of β-catenin and HuR on COX-2 protein level, a knock-down analysis was performed in LoVo colon adenocarcinoma cells. β-Catenin siRNA completely inhibited COX-2 protein expression, whereas HuR siRNA did not alter COX-2 protein levels in LoVo cells (Figure 4F). Taken together, cooperative binding of β-catenin and HuR on COX-2 mRNA seemed to be crucial for COX-2 protein expression in the cells.
β-Catenin–HuR–RNA complex is located in the colon cancer cell cytoplasm
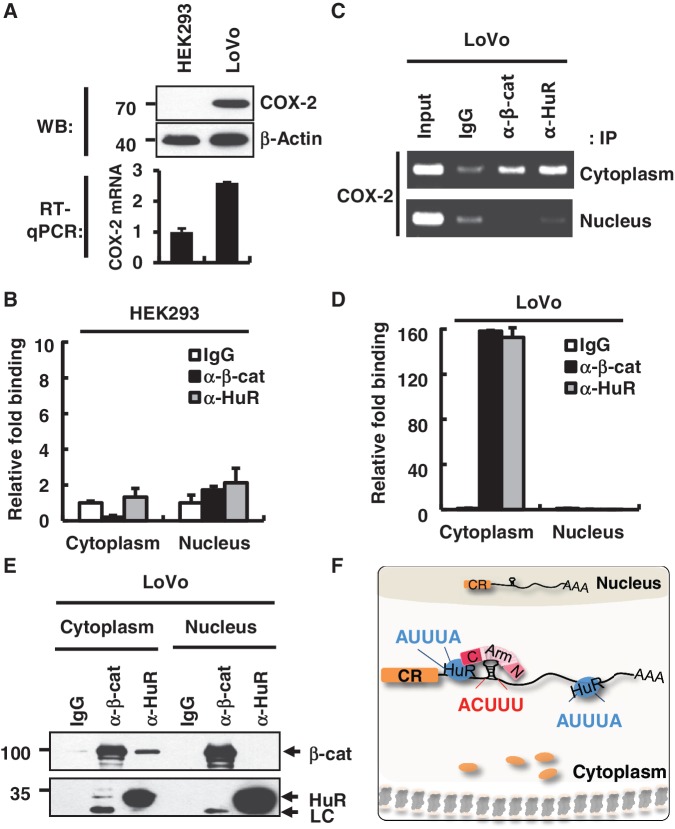
Activation and subcellular re-distribution of β-catenin and HuR were strongly related to cancer progression, and overexpression of the COX-2 protein is one of the characteristics of colon cancer. Therefore, it would be interesting to test whether the tertiary RNP complex formation with COX-2 mRNA is associated with oncogenic localization of these proteins and COX-2 overexpression during tumorigenesis. Here, we used HEK293 cells as a model of normal cells and LoVo as colorectal cancer cells, and found that COX-2 protein and mRNA expression was vastly different between the two cell types (Figure 5A).
Figure 5.
Differential binding patterns of HuR and β-catenin on the COX-2 3′-UTR in normal and tumor cells. (A) Upper panel, western blot analysis of the COX-2 protein in HEK293 and LoVo cells. β-Actin was used as a loading control. Molecular weights are indicated on the left side of the gel. Lower panel, RT–qPCR analysis was performed to analyze the endogenous COX-2 mRNA level in HEK293 and LoVo cells. (B) RNP-IP assay with fractionated HEK293 cells. RNP-IP was performed with anti-β-catenin or anti-HuR antibodies. RT–qPCR analysis was performed to analyze the bound pellet RNA and presented as relative fold binding to the IgG control. Three independent samples were analyzed, and the average and standard deviation are shown. (C) RNP-IP assay with LoVo cells as in B. RT–PCR analysis was performed for the bound pellet RNA and the input RNA. (D) Real-time PCR analysis of RNP-IP pellet RNA with LoVo cells as in C. (E) Co-IP of β-catenin and HuR from nuclear and cytoplasmic fractions of LoVo cells. Protein size markers are shown on the left side of the gel. (F) A model for the β-catenin–HuR–COX-2 mRNA tertiary RNP formation in cancer cells. β-Catenin and HuR bind to the 3′-UTR of COX-2 mRNA via distinct RNA elements, ACUUU and AUUUA, respectively. β-Catenin and HuR may interact with each other through the C-term domain of the β-catenin protein. The tertiary RNP complex was exclusively found in the cytoplasm of colorectal adenocarcinoma cells and promoted COX-2 protein expression in cancer cells. CR, coding region of COX-2 mRNA; black line: 3′-UTR of COX-2 mRNA; blue circle, HuR protein; N, N-terminal domain of β-catenin; Arm, Armadillo domain of β-catenin; C, C-terminal domain of β-catenin; AAA, poly A tail; orange circle, Cyclooxygenase-2 protein.
Subcellular fractionation was combined with RNP-IP to test whether β-catenin and HuR RNP formed differently in normal and cancer cells. In Figure 5B, a relatively low level of β-catenin or HuR binding to COX-2 mRNA was detected both in the nucleus and in the cytoplasm of HEK293 cells (Figure 5B). In contrast, β-catenin and HuR interacted with COX-2 mRNA exclusively in the cytoplasm of LoVo cells (Figure 5C and D). These results suggest that cytoplasmic interaction of β-catenin and HuR on RNA might be important for COX-2 mRNA overexpression in cancer cells in Figure 5A. Moreover, the co-IP analysis with fractionated cells was consistent with the above RNP data as well as our previous reports on β-catenin and HuR interaction in colon cancer cells (30). Protein–protein interactions between β-catenin and HuR occurred exclusively in the cytoplasm of LoVo cells (Figure 5E), as was observed in HT-29 colon adenoma carcinoma cells (30). The distribution of the β-catenin and HuR proteins and proper cellular fractionation was confirmed by western blot analysis (Supplementary Figure S3D). Taken together, these results suggest that the formation and transport of the tertiary RNP complex was different between normal and cancer cells. They also strongly suggest that cytoplasmic RNP might be important for the elevated level of COX-2 protein in cancer cells.
DISCUSSION
How β-catenin binds to RNA inside of cells and how it modulates multiple steps of posttranscription need to be better understood. This study provided biochemical and cell biological evidences that β-catenin could binds the RNA through the ACUUU motif in cellular RNA. In addition, combinatorial binding with HuR could result in the formation of an RNP complex with COX-2 mRNA through distinct and non-overlapping binding sites in 3′-UTR. β-Catenin could also bind to RNA via the Arm domain and HuR could bind to RNA via the C-terminal domain, which facilitated the tertiary RNP complex formation in vitro and in cells. More importantly, this RNP was predominantly found in the cytoplasm of colon cancer cells as shown in the model in Figure 5F. Since β-catenin and HuR localization are greatly changed in cancer cells when compared to normal cells, the identification of β-catenin as a RBP and its binding RNA element could explain the significant impact of oncogenic β-catenin on cancer cell development.
Here, we showed that Arm repeats of the β-catenin protein could be a novel RNA-binding domain with sequence and/or structure-based recognition. As β-catenin might be a novel ARE-binding RBP, the list of potential RBPs may be larger than previously envisioned. Arm repeat domains are common sites for protein binding in cells. Since we have shown here that β-catenin interacts with RNA, RNA-mediated protein–protein interactions might be one form of such diverse intracellular interactions involving the Arm domains. When considering the many interaction partners of β-catenin, the RNA molecule could provide a platform for the complex formation in cells. The structures of the Arm domain and its binding proteins, such as TCF4, TCF3, E-cadherin and APC, have been extensively studied (33–37). Twelve Arm repeats are organized in a right-handed superhelical twist with a shallow, highly positively charged groove that has been proposed to provide a binding pocket for interacting proteins. The positively charged groove of the β-catenin superhelix makes extensive contacts with many negatively charged residues in the extended N-terminus of TCF proteins (38). The striking structural resemblance of Arm repeats to Pumilio and FBF homology protein (PUF) repeats could provide novel insights on the structure of the Arm–RNA interaction based on the RNA recognition patterns of PUF repeats (39,40).
COX-2 gene expression is generally regulated at the posttranscriptional level by multimeric proteins such as ARE-binding proteins (ARE-BPs) (22,23,30). ARE-BPs regulate RNA stability either by recruiting or by excluding exosomes on target transcripts so they are critical regulators of inflammation and cancer (41). Since most ARE-BPs share similar RNA sequences, it would be important to determine whether they compete or cooperate for the same binding sites. Complicated binding patterns of ARE-BPs on the same transcripts could greatly affect RNA stability depending on RNA–protein as well as protein–protein interactions between them (42). For example, this was observed for AUF1 and HuR, which bound to both distinct, non-overlapping sites, and on common sites in a competitive fashion. In the case of AUF1 and HuR ARE-BPs, they interact with the p16 3′-UTR in a cooperative manner and compete for the p21 3′-UTR (43,44). Interestingly, HuR knockdown reduced COX-2 mRNA level but did not lead to a dramatic reduction in protein level (Figure 4F), probably because many distinct RBPs are associated with the COX-2 3′-UTR depending on the cell line, and translational regulation is much more complicated (24,45–47). Thus, our data may reflect the complexity of RNA stability and translation of COX-2 protein expression in cancer cells.
Our data suggest that β-catenin and HuR collaboratively associate with COX-2 3′-UTR by binding to distinct RNA elements to form a tertiary RNP complex. However, it is still possible that they could compete for other transcripts depending on the locations of their targets as well as on the overall structure of RNA. Therefore, more extensive studies on the RNA-mediated interaction between β-catenin and HuR on various target transcripts are needed. Interestingly, several reports have suggested that there are inter-relationships between β-catenin and HuR at the posttranscriptional level (48). Therefore, oncogenic roles of β-catenin and HuR in cancer cells might be related to cooperative or competitive binding of the two proteins depending on mRNA during the course of tumor progression.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2 and Supplementary Figures 1–3.
FUNDING
National Research Foundation of Korea (NRF) grant funded by Ministry Of Education, Science And Technology (MEST) [2011-0018634, 2011-0006428 and 2011-0002169 to S.J.]. Funding for open access charge: NRF [2011-006428].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful Dr. Francois Major (University of Montreal) for predicting 3D structure of RNA aptamer.
REFERENCES
- 1.Lunde BM, Moore C, Varani G. RNA-binding proteins: modular design for efficient function. Nat. Rev. Mol. Cell Biol. 2007;8:479–490. doi: 10.1038/nrm2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Glisovic T, Bachorik JL, Yong J, Dreyfuss G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008;582:1977–1986. doi: 10.1016/j.febslet.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim MY, Hur J, Jeong S. Emerging roles of RNA and RNA-binding protein network in cancer cells. BMB Rep. 2009;42:125–130. doi: 10.5483/bmbrep.2009.42.3.125. [DOI] [PubMed] [Google Scholar]
- 4.Lukong KE, Chang KW, Khandjian EW, Richard S. RNA-binding proteins in human genetic disease. Trends Genet. 2008;24:416–425. doi: 10.1016/j.tig.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 5.Hogan DJ, Riordan DP, Gerber AP, Herschlag D, Brown PO. Diverse RNA-binding proteins interact with functionally related Sets of RNAs, suggesting an extensive regulatory system. PLoS Biol. 2008;6:e255. doi: 10.1371/journal.pbio.0060255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hinman MN, Lou H. Diverse molecular functions of Hu proteins. Cell. Mol. Life. Sci. 2008;65:3168–3181. doi: 10.1007/s00018-008-8252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keene JD. Why is Hu where? Shuttling of early-response-gene messenger RNA subsets. Proc. Natl Acad. Sci. USA. 1999;96:5–7. doi: 10.1073/pnas.96.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doller A, Pfeilschifter J, Eberhardt W. Signaling pathways regulating nucleo-cytoplasmic shuttling of the mRNA-binding protein HuR. Cell Signal. 2008;20:2155–2173. doi: 10.1016/j.cellsig.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 9.López de Silanes I, Zhan M, Lal A, Yang X, Gorospe M. Identification of a target RNA motif for RNA-binding protein HuR. Proc. Natl Acad. Sci. USA. 2004;101:2987–2992. doi: 10.1073/pnas.0306453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.López de Silanes I, Fan J, Yang X, Zonderman AB, Potapova O, Pizer ES, Gorospe M. Role of the RNA-binding protein HuR in colon carcinogenesis. Oncogene. 2003;22:7146–7154. doi: 10.1038/sj.onc.1206862. [DOI] [PubMed] [Google Scholar]
- 11.Araki Y, Okamura S, Hussain P, Nagashima M, He P, Shiseki M, Miura K, Harris CC. Regulation of cyclooxygenase-2 expression by the Wnt and ras pathways. Cancer Res. 2003;63:728–732. [PubMed] [Google Scholar]
- 12.Shao J, Sheng H, Inoue H, Morrow JD, DuBois RN. Regulation of constitutive cyclooxygenase-2 expression in colon carcinoma cells. J. Biol. Chem. 2000;275:33951–33956. doi: 10.1074/jbc.M002324200. [DOI] [PubMed] [Google Scholar]
- 13.Young LE, Dixon DA. Posttranscriptional regulation of cyclooxygenase 2 expression in colorectal cancer. Curr. Colorectal Cancer Rep. 2010;6:60–67. doi: 10.1007/s11888-010-0044-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dixon DA, Kaplan CD, McIntyre TM, Zimmerman GA, Prescott SM. Post-transcriptional control of cyclooxygenase-2 gene expression. The role of the 3′-untranslated region. J. Biol. Chem. 2000;275:11750–11757. doi: 10.1074/jbc.275.16.11750. [DOI] [PubMed] [Google Scholar]
- 15.Dixon DA, Tolley ND, King PH, Nabors LB, McIntyre TM, Zimmerman GA, Prescott SM. Altered expression of the mRNA stability factor HuR promotes cyclooxygenase-2 expression in colon cancer cells. J. Clin. Invest. 2001;108:1657–1665. doi: 10.1172/JCI12973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cok SJ, Acton SJ, Morrison AR. The proximal region of the 3′-untranslated region of cyclooxygenase-2 is recognized by a multimeric protein complex containing HuR, TIA-1, TIAR, and the heterogeneous nuclear ribonucleoprotein U. J. Biol. Chem. 2003;278:36157–36162. doi: 10.1074/jbc.M302547200. [DOI] [PubMed] [Google Scholar]
- 17.Hall-Pogar T, Zhang H, Tian B, Lutz CS. Alternative polyadenylation of cyclooxygenase-2. Nucleic Acids Res. 2005;33:2565–2579. doi: 10.1093/nar/gki544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Willert K, Jones KA. Wnt signaling: is the party in the nucleus? Genes Dev. 2006;20:1394–1404. doi: 10.1101/gad.1424006. [DOI] [PubMed] [Google Scholar]
- 19.MacDonald BT, Tamai K, He X. Wnt/β-catenin signaling: components, mechanisms, and disease. Dev. Cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shitashige M, Hirohashi S, Yamada T. Wnt signaling inside the nucleus. Cancer Sci. 2008;99:631–637. doi: 10.1111/j.1349-7006.2007.00716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tetsu O, McCormick F. β-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999;398:422–426. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- 22.He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, Morin PJ, Vogelstein B, Kinzler KW. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 23.Jho EH, Zhang T, Domon C, Joo CK, Freund JN, Costantini F. Wnt/beta-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol. Cell Biol. 2002;22:1172–1183. doi: 10.1128/MCB.22.4.1172-1183.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Polakis P. Wnt signaling and cancer. Genes Dev. 2000;14:1837–1851. [PubMed] [Google Scholar]
- 25.Shapiro L. β-catenin and its multiple partners: promiscuity explained. Nat. Struct. Biol. 2001;8:484–487. doi: 10.1038/88532. [DOI] [PubMed] [Google Scholar]
- 26.Sato S, Idogawa M, Honda K, Fujii G, Kawashima H, Takekuma K, Hoshika A, Hirohashi S, Yamada T. β-catenin interacts with the FUS proto-oncogene product and regulates pre-mRNA splicing. Gastroenterology. 2005;129:1225–1236. doi: 10.1053/j.gastro.2005.07.025. [DOI] [PubMed] [Google Scholar]
- 27.Lee HK, Choi YS, Park YA, Jeong S. Modulation of oncogenic transcription and alternative splicing by β-catenin and an RNA aptamer in colon cancer cells. Cancer Res. 2006;66:10560–10566. doi: 10.1158/0008-5472.CAN-06-2526. [DOI] [PubMed] [Google Scholar]
- 28.Lee HK, Kwak HY, Hur J, Kim IA, Yang JS, Park MW, Yu J, Jeong S. β-Catenin regulates multiple steps of RNA metabolism as revealed by RNA aptamer in colon cancer cells. Cancer Res. 2007;67:9315–9321. doi: 10.1158/0008-5472.CAN-07-1128. [DOI] [PubMed] [Google Scholar]
- 29.Briata P, Ilengo C, Corte G, Moroni C, Rosenfeld MG, Chen CY, Gherzi R. The Wnt/beta-catenin→Pitx2 pathway controls the turnover of Pitx2 and other unstable mRNAs. Mol. Cell. 2003;12:1201–1211. doi: 10.1016/s1097-2765(03)00407-6. [DOI] [PubMed] [Google Scholar]
- 30.Lee HK, Jeong S. β-Catenin stabilizes cyclooxygenase-2 mRNA by interacting with AU-rich elements of 3′UTR. Nucleic Acids Res. 2006;34:5705–5714. doi: 10.1093/nar/gkl698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parisien M, Major F. The MC-Fold and MC-Sym pipeline infers RNA structure from sequence data. Nature. 2008;452:51–55. doi: 10.1038/nature06684. [DOI] [PubMed] [Google Scholar]
- 33.Graham TA, Weaver C, Mao F, Kimelman D, Xu W. Crystal structure of a β-catenin/Tcf complex. Cell. 2000;103:885–896. doi: 10.1016/s0092-8674(00)00192-6. [DOI] [PubMed] [Google Scholar]
- 34.Poy F, Lepourcelet M, Shivdasani RA, Eck MJ. Structure of a human Tcf4-β-catenin complex. Nat. Struct. Biol. 2001;8:1053–1057. doi: 10.1038/nsb720. [DOI] [PubMed] [Google Scholar]
- 35.Graham TA, Ferkey DM, Mao F, Kimelman D, Xu W. Tcf4 can specifically recognize β-catenin using alternative conformations. Nat. Struct. Biol. 2001;8:1048–1052. doi: 10.1038/nsb718. [DOI] [PubMed] [Google Scholar]
- 36.Huber AH, Weis WI. The structure of the β-catenin/E-cadherin complex and the molecular basis of diverse ligand recognition by β-catenin. Cell. 2001;105:391–402. doi: 10.1016/s0092-8674(01)00330-0. [DOI] [PubMed] [Google Scholar]
- 37.Ha NC, Tonozuka T, Stamos JL, Choi HJ, Weis WI. Mechanism of phosphorylation-dependent binding of APC to β-catenin and its role in β-catenin degradation. Mol. Cell. 2004;15:511–521. doi: 10.1016/j.molcel.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 38.Choi HJ, Huber AH, Weis WI. Thermodynamics of β-catenin-ligand interactions: the roles of the N- and C-terminal tails in modulating binding affinity. J. Biol. Chem. 2006;281:1027–1038. doi: 10.1074/jbc.M511338200. [DOI] [PubMed] [Google Scholar]
- 39.Edwards TA, Pyle SE, Wharton RP, Aggarwal AK. Structure of Pumilio reveals similarity between RNA and peptide binding motifs. Cell. 2001;105:281–289. doi: 10.1016/s0092-8674(01)00318-x. [DOI] [PubMed] [Google Scholar]
- 40.Filipovska A, Razif MF, Nygård KK, Rackham O. A universal code for RNA recognition by PUF proteins. Nat. Chem. Biol. 2011;15:425–427. doi: 10.1038/nchembio.577. [DOI] [PubMed] [Google Scholar]
- 41.Khabar KS. Post-transcriptional control during chronic inflammation and cancer: a focus on AU-rich elements. Cell. Mol. Life Sci. 2010;67:2937–2955. doi: 10.1007/s00018-010-0383-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.David PS, Tanveer R, Port JD. FRET-detectable interactions between the ARE binding proteins, HuR and p37AUF1. RNA. 2007;13:1453–1468. doi: 10.1261/rna.501707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lal A, Mazan-Mamczarz K, Kawai T, Yang X, Martindale JL, Gorospe M. Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. EMBO J. 2004;23:3092–3102. doi: 10.1038/sj.emboj.7600305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chang N, Yi J, Guo G, Liu X, Shang Y, Tong T, Cui Q, Zhan M, Gorospe M, Wang W. HuR uses AUF1 as a cofactor to promote p16INK4 mRNA decay. Mol. Cell Biol. 2010;30:3875–3886. doi: 10.1128/MCB.00169-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anant S, Houchen CW. HuR and TTP: two RNA binding proteins that deliver message from the 3′ end. Gastroenterology. 2009;136:1495–1498. doi: 10.1053/j.gastro.2009.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mukhopadhyay D, Houchen CW, Kennedy S, Dieckgraefe BK, Anant S. Coupled mRNA stabilization and translational silencing of cyclooxygenase-2 by a novel RNA binding protein, CUGBP2. Mol. Cell. 2003;11:113–126. doi: 10.1016/s1097-2765(03)00012-1. [DOI] [PubMed] [Google Scholar]
- 47.Sureban SM, Murmu N, Rodriguez P, May R, Maheshwari R, Dieckgraefe BK, Houchen CW, Anant S. Functional antagonism between RNA binding proteins HuR and CUGBP2 determines the fate of COX-2 mRNA translation. Gastroenterology. 2007;132:1055–1065. doi: 10.1053/j.gastro.2006.12.031. [DOI] [PubMed] [Google Scholar]
- 48.Ale-Agha N, Galban S, Sobieroy C, Abdelmohsen K, Gorospe M, Sies H, Klotz LO. HuR regulates gap junctional intercellular communication by controlling beta-catenin levels and adherens junction integrity. Hepatology. 2009;50:1567–1576. doi: 10.1002/hep.23146. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.