Abstract
Understanding the numerous functions that RNAs play in living cells depends critically on knowledge of their three-dimensional structure. Due to the difficulties in experimentally assessing structures of large RNAs, there is currently great demand for new high-resolution structure prediction methods. We present the novel method for the fully automated prediction of RNA 3D structures from a user-defined secondary structure. The concept is founded on the machine translation system. The translation engine operates on the RNA FRABASE database tailored to the dictionary relating the RNA secondary structure and tertiary structure elements. The translation algorithm is very fast. Initial 3D structure is composed in a range of seconds on a single processor. The method assures the prediction of large RNA 3D structures of high quality. Our approach needs neither structural templates nor RNA sequence alignment, required for comparative methods. This enables the building of unresolved yet native and artificial RNA structures. The method is implemented in a publicly available, user-friendly server RNAComposer. It works in an interactive mode and a batch mode. The batch mode is designed for large-scale modelling and accepts atomic distance restraints. Presently, the server is set to build RNA structures of up to 500 residues.
INTRODUCTION
Recent advances in recognizing the ways RNAs control eukaryotic cell health and disease states make it certain that RNA structures will become increasingly important targets for therapeutic intervention (1,2). However, the 3D structures of most biologically important RNAs are currently unknown. In contrast to the protein field, a much smaller number of RNA tertiary structures has been assessed by X-ray crystallography, NMR spectroscopy and cryo-EM and deposited in structural data banks (3). This situation has led to a great demand in structural biology to envisage the RNA secondary and tertiary structures using prediction methods (4).
Based on different algorithms, in silico RNA secondary structure prediction methods (5,6) have recently been strengthened by incorporating constraints from chemical probing (7), primarily SHAPE (8). This advancement has been reflected in a growing number of reports on the secondary structures of large RNAs with increased accuracy (9–11). However, the ultimate goal of tertiary structure prediction of large RNAs, applying secondary structure information, still remains a significant challenge (12).
Only a few programs and web-accessible tools have been proposed for semi-automated (13,14) and automated prediction of the RNA tertiary structure. Automated methods make use of the coarse-grained and atomic-level molecular dynamics (15–17), internal coordinate space dynamics (18,19), fragment assembly (20,21) and comparative modelling using templates (22). They operate on different input data: RNA sequence, secondary structure, conformational data or structural templates. The reported methods vary considerably in terms of prediction quality across different RNA strand lengths and topologies, processing time and automation levels (4). Full-atomic structure predictions based on dynamics (15–19) and fragment assembly (20,21) are powerful tools for modelling of small RNA molecules but larger structures remain a challenge due to the computational costs. Coarse-grained molecular dynamics can access larger RNAs but addition of atomic details to coarse-grain models is demanding and not fully resolved (17). Knowledge-based, comparative modelling (22) requires access to 3D structural templates and unequivocal sequence alignment. Until now, none of the reported methods has reached the stage of truly full automation, efficient access to large RNA structures and short computing time.
Here, we present a novel approach for the automated RNA 3D structure prediction from a user-defined secondary structure. Its concept is founded on the machine translation system being parallel to that employed in the field of computational linguistics (23). The method takes advantage of our RNA FRABASE database (24,25) tailored to the dictionary relating the RNA secondary structure and tertiary structure elements.
We demonstrate that our approach is characterized by very short computation time, efficiency and easy access to high-resolution models of large RNA structures.
MATERIALS AND METHODS
RNA secondary to tertiary structure machine translation workflow
The workflow of the method is depicted in the Supplementary Figure S1. User-defined RNA secondary structure is fragmented to the elements. The fragmentation algorithm provides the secondary structure elements (25): stems, loops (apical, bulge, internal and n-way junctions) and single strands, all closed by canonical base pair(s). This constitutes the input patterns for an automatic search of related tertiary structure elements in the RNA FRABASE dictionary (see below).
In the preparation step, the 3D structure elements are selected according to the criteria, with the following priority order: secondary structure topology, sequence similarity, pyrimidines/purines compatibility, source structure resolution and the energy (Supplementary Figure S1). If RNA sequence is not matched for the given element, the respective bases are replaced. The base replacement procedure is activated when the RNA FRABASE dictionary contains the element of the correct topology but of dissimilar sequence. Occasionally, it may happen that the dictionary is lacking a particular 3D structure element required to compose a model for a given secondary structure topology. To preclude building incomplete 3D structures in such situations, the machine translation system runs exception procedures that generate stems, single strands and loops of given secondary structure topology and sequence.
The next step is the initial RNA structure building and involves the 3D structure elements produced in the preparation step. The building process is governed by the RNA tree graph representing secondary structure (Supplementary Table S1). The tertiary structure elements are superimposed with reference to common canonical base pairs, and merged to give an initial RNA 3D structure. This structure was subjected to the refinement using energy minimization in torsion angle space and Cartesian coordinates to generate the final, high-quality RNA 3D model. The translation engine can generate a family of closely related 3D models. The first model is always built using all the selection criteria described above. Other models are generated from randomly selected 3D structure elements for which the criteria of structure resolution and energy are ignored.
The RNA FRABASE dictionary
The RNA FRABASE dictionary was developed using the relational database system PostgreSQL 8.0.
The dictionary contains secondary and tertiary structural elements automatically imported from the parent RNA FRABASE 2.0 database (25) and, subsequently, adjusted to fulfil the requirements of the machine translation system. All the 3D structural elements are described by a complete set of atoms, energy, good stereochemistry and structural properties. Presently, the dictionary includes of 14 464 secondary structure elements and as much as 190 928 related tertiary structure elements (Supplementary Table S2).
Upon dictionary design, all the tertiary elements derived from X-ray structures were equipped with missing hydrogen atoms and subjected to energy minimization that optimized their positions. The CHARMM force field implemented in the XPLOR-NIH program (26) was used to conduct 100 steps of the conjugate gradient minimization (the fixed heavy atom positions, use of DNA–RNA-allatom.param and DNA–RNA-allatom.top files). Subsequently, all the 3D structural elements were validated in the terms of their stereochemical quality and energy. Three dimensional elements with too high energy were energy minimized (conditions as above, with all atoms free) and checked whether their structures depart from the original tertiary elements. The engine makes use of only those 3D elements whose heavy atom r.m.s.d. is lower than 1.0 Å when referred to the parent structure. The dictionary does not include elements with modified residues or missing heavy atom coordinates.
Generating 3D structure elements missing in dictionary
The RNA stems and single-strand stretches are build with the fd_helix routine of the NAB 5.0 software (27) and use of the A-RNA structure parameters.
The RNA loop elements being the part of hairpins, bulges, internal loops and n-way junctions are generated in the torsion angle space using the CYANA structure calculation program (28). These structural elements (25) form an uninterrupted cyclic system and are composed of one or more strands linked through canonical base pairs hydrogen bonds. To generate such loops, the standard CYANA residue library (cyana.lib) was expanded (29) to include data for the nucleotide residues within defined canonical base pairs. This ensures exact base pairing when closing the loops in the generated structure and eliminates the use of ‘invisible’ linkers CYANA exploits upon formation of the multi strand structures. To generate the structure, initial values of A-RNA torsion angles extracted from NDB (30) and distance restraints for the closure of sugar rings are applied. Additional distance restraints are followed for hydrogen bonds of the first and last residues closing the loop. The 3D structure elements calculation under standard CYANA conjugate gradient minimization protocol was continued until acquiring the lowest target function value.
All the above procedures take very short processing time and provide the RNA tertiary structure elements with a regular shape. The generated 3D elements show full secondary structure topology conservation and good stereochemical properties. Still, their structure accuracy is much lower than when 3D elements are generated using the RNA FRABASE dictionary.
Base replacement procedure
The base replacement is governed by three mathematical operations: one rigid body translation and two rotations. The coordinates of target bases and the anomeric C1′ atoms are copied from the NAB residues library (27). The respective atoms are rigid body translated into the model of the 3D structure element to overlap anomeric carbon atoms positions. Subsequently, the target base atoms are rotated around the vector perpendicular to the glycosidic bonds in order to impose their overlap. Finally, to preserve the original value of the torsion angle χ an appropriate rotation around the glycosidic bond takes place. After these transformations, the atomic coordinates of the original base are removed. This type of procedure preserves the coordinates of all ribose rings and phosphates atoms, constituting the phosphosugar backbone, as well as the torsion angles of the glycosidic bonds.
Initial RNA structure building
The building starts from the 3D structure element which bears both 5′- and 3′-terminal residues of the target RNA structure. The Kabsch algorithm for least-squares superposition (31) is implemented to overlay 3D structure elements, based on common canonical base pair residues. Subsequently, all atom coordinates within the newly added fragment are removed to avoid the coordinate duplication. After the addition of the last structure element, due to the graph representation, all residues are renumbered according to their sequence.
Final RNA tertiary structure refinement
Two energy minimization steps are conducted to refine the initial RNA structure. In the first step, the atom coordinates are strictly converted to the CYANA program format and energy is minimized in the torsion angle space using the standard protocol (conjugate gradient, 2000 iteration steps), as well as the distance restrains for hydrogen bonds. In the second step, the resultant structure is energy minimized in the Cartesian atom coordinate space using CHARMM force field implemented in the XPLOR-NIH program (conjugate gradient, 1000 iterations) and restraints for hydrogen bonds and base pairs planarity.
RNA 3D structure quality assessment
The stereochemical quality of the predicted 3D structures was assessed using PDB validation tool http://deposit.pdb.org/adit/, X-PLOR program (26) and MolProbity tool (32). The accuracy of the predicted full atomic structures relative to the respective RNA crystal structures is described using two measures. The global and local RNA structure all atoms r.m.s.d. values were computed using XPLOR-NIH program. The interaction network fidelity (INFall) measure (33) was used to check all the canonical and non-canonical base pairing and stacking. Base pair interactions and the base stacking network for the tertiary structures were obtained using RNAView (34) and MC-Annotate (35) programs, respectively. In a similar fashion, we have calculated the parameter INFcbp to inspect the conservation of canonical base pairs.
The architecture of the RNAComposer web server
The architecture of the RNAComposer system comprises two components: a computational server and a web application server. The computational server represents the back-end layer of the RNAComposer system and hosts the translation machine engine (encoded in Java 1.6.0_15, applying the Apache Ant 1.7.0, GSL-Java 1.3-0.6 and Torque 3.0 libraries) and the RNA FRABASE dictionary (implemented in PHP 5, using PostgreSQL 8.0 database and Apache2 web server). The engine implementation integrates selected functionalities from the publicly available software: XPLOR-NIH 2.21, NAB 6.0 (AmberTools 1.3) and the licensed version of CYANA 2.13. The computational server is a 64-bit Intel Xeon (2.33 GHz) processor-based server platform with scalable 8GB memory, operated by openSUSE 11.0 server environment.
The web application server is an Intel Pentium 4 (3.2 GHz) processor-based server platform. It represents the front-end layer of the RNAComposer system and provides a simple, effective and user-friendly web interface (implemented in C# on a ASP). The net platform, using a PostgreSQL 8.4 database and IIS 7.0 web server, is operated by Windows Server 2008 Enterprise environment. The whole system is closed in the Virtual Private Network (OpenVPN 2.0) supporting effective and safe communication between both servers through integrated message brokers (Apache Active MQ 5.3).
The RNAComposer server is publicly available online under http://rnacomposer.cs.put.poznan.pl and http://rnacomposer.ibch.poznan.pl.
The assignment of the 5S rRNA Haloarcula Marismortui secondary structure
The secondary structure of 5S rRNA Haloarcula Marismortui was predicted using the RNAstructure software (36) allowing us to introduce the data from chemical structure probing using the SHAPE protocol (37). 5S rRNA containing flanking 5′ and 3′ sequences to facilitate the analysis of the entire RNA by primer extension (37), was synthesized by in vitro transcription with the Ambion T7-MEGAshortscript. The DNA template was produced based on the PCR (Ambion SuperTaqTM Plus polymerase kit) utilizing single-stranded overlapping oligonucleotides.
The transcripts were purified by denaturing gel electrophoresis (8 M urea), followed by elution and ethanol precipitation. The purified RNAs were dissolved in sterile water and stored at −20°C. The NMIA treatment of RNA was conducted as follows: 20 pmol of RNA were heated at 95°C for 3 min in 20 μl of renaturation buffer [10 mM Tris–HCl (pH 7.5), 100 mM KCl and 0.1 mM EDTA] and slowly cooled to 4°C. Subsequently, 97 μl of water and 29 μl of 5× folding buffer [200 mM Tris–HCl (pH 7.5), 650 mM KCl, 2.5 mM EDTA, 25 mM MgCl2 and 40U RNase inhibitor] were added and the RNA was incubated at 37°C for 10 min. The mixture was divided into equal parts, treated with 7.3 μl of 180 mM NMIA in anhydrous dimethyl sulfoxide (DMSO) (+) or DMSO alone (−) and the reaction was allowed to proceed at 37°C for 50 min. The RNA was precipitated and resuspended in 10 μl of water. Modified sites were detected by primer extension as described earlier (37) except that DTT and betaine were added to final concentrations of 5 mM and 1 M, respectively. Dideoxy sequencing markers were generated using unmodified RNA. Two 5′-end-32P labelled DNA primers complementary to the 3′ RNA-flanking sequence and region +62 to 80 of 5S rRNA were used to analyse the entire RNA. Primer extension reactions were resolved on sequencing gels (8% PAGE with 8 M urea). The results from PAGE were visualized using FLA-5100 with MultiGaugeV 3.0 software (FujiFilm). cDNA band intensities for the (+) and (−) reactions were integrated using SAFA (38) and corrected for stochastic drop-off. The control reaction (−) was subtracted from NMIA reaction (+) and SHAPE reactivities were processed as described (9).
The following 5S rRNA H. Marismortui secondary structure was obtained using SHAPE data and RNA structure software (36):
UUAGGCGGCCACAGCGGUGGGGUUGCCUCCCGUACCCAUCCCGAACACGGAAGAUAAGCCCACCAGCGUUCCGGGGAGUACUGGAGUGCGCGAGCCUCUGGGAAACCCGGUUCGCCGCCACC
...((((((....((((((((.......(((((...(.....)...))))..).....)))))).))...(((((.....((((((.((....))))))))....)))))...))))))...
RESULTS AND DISCUSSION
The RNA 3D structure composition
The presented method for RNA 3D structure prediction from the secondary structure is founded on the machine translation concept, which is parallel to that employed in computational linguistics (23). It needs neither structural templates nor RNA sequence alignment, required for comparative methods. The system operates on the RNA FRABASE database (24,25) tailored to the dictionary relating the RNA secondary structure and tertiary structure elements.
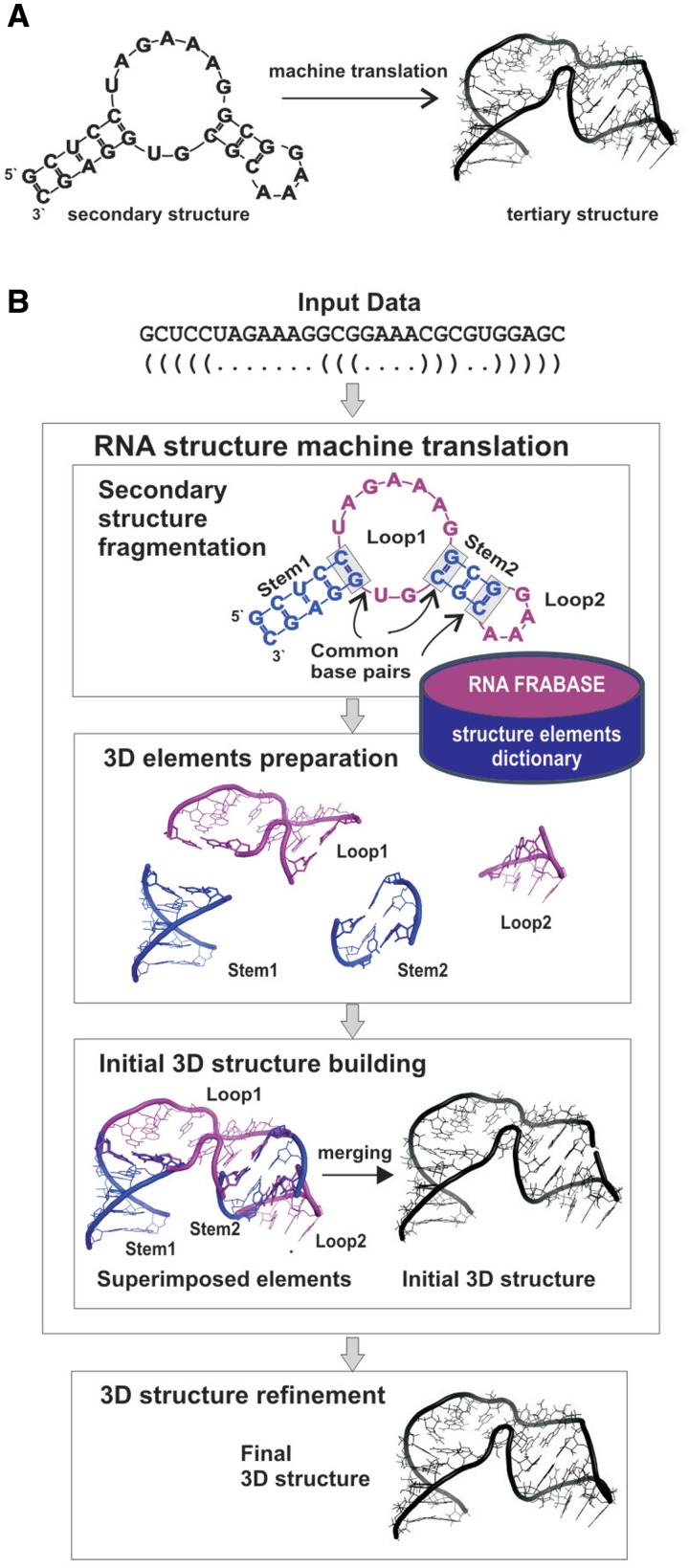
RNA 3D models are built in fully automated way comprised of four major steps depicted on the Figure 1B and exemplified on the structure of 29-mer RNA hairpin with an internal loop. The secondary structure of this hairpin, described by a sequence and a secondary structure topology in the dot-bracket notation, is an input to our system. In the first step, this structure is fragmented into four secondary structure elements, in accordance with its tree graph representation (39): two stems (Figure 1B; Stems 1 and 2: 5 and 3 bp long, respectively), one internal loop (Loop 1: 13 nt size, including two closing base pairs) and one apical tetra-loop (Loop 2: 6 nt size, including closing base pair). These fragments constitute input patterns for the search procedure in the dictionary. The search returns a series of 3D elements for every input pattern: 11 results matching 5 bp stem, 350 results for 3 bp stem, 0 elements matching the pattern of the internal loop and 610 results for apical loop. Since the dictionary does not contain corresponding tertiary structure element for the internal loop, the system performs another search for 3D structures matching secondary structure topology only. The latter run returns 74 elements of a given topology but with various sequences.
Figure 1.
RNA secondary structure to tertiary structure machine translation. (A) The general principle. (B) Schematic of the basic steps of RNA structure machine translation.
The search results (1045 tertiary structure elements) are next processed in the second, preparation step which encompasses multi-criteria selection of the 3D structure elements (Supplementary Figure S1) and base replacement procedure (in this case, for the internal loop). Thus, for this particular hairpin example, this step returns: 3KNO-derived 5 bp stem, 2PWT-derived 3 bp stem, the apical loop extracted from 2R8S and the internal loop from 1VQO. Since the internal loop from 1VQO shows only 54% of sequence similarity with the input secondary structure element, 6 bases are replaced.
In general, the number of 3D structure elements that are prepared for a given RNA depends on fragmentation step. Supplementary Table S1 presents an example for more complex structure of 5S rRNA (122 nt) which fragmentation returns 19 secondary structure elements.
The third step is the initial RNA structure building based on the collection of 3D structure elements produced in the preparation step. The tertiary structure elements are superimposed and merged to give an initial, already well-shaped RNA 3D structure. Its refinement using energy minimization (26,28) leads in the fourth step to the final, high-quality RNA 3D model(s). The computation of the example hairpin 3D structure has taken 7 s in total.
Detailed workflow of the method is described in the ‘Materials and Methods’ section and depicted on the Supplementary Figure S1.
Method evaluation
We conducted two evaluation tests to estimate the scope of the method and the quality of the predicted 3D structure models in terms of the secondary structure topology conservation, their stereochemical properties, energy, precision and accuracy.
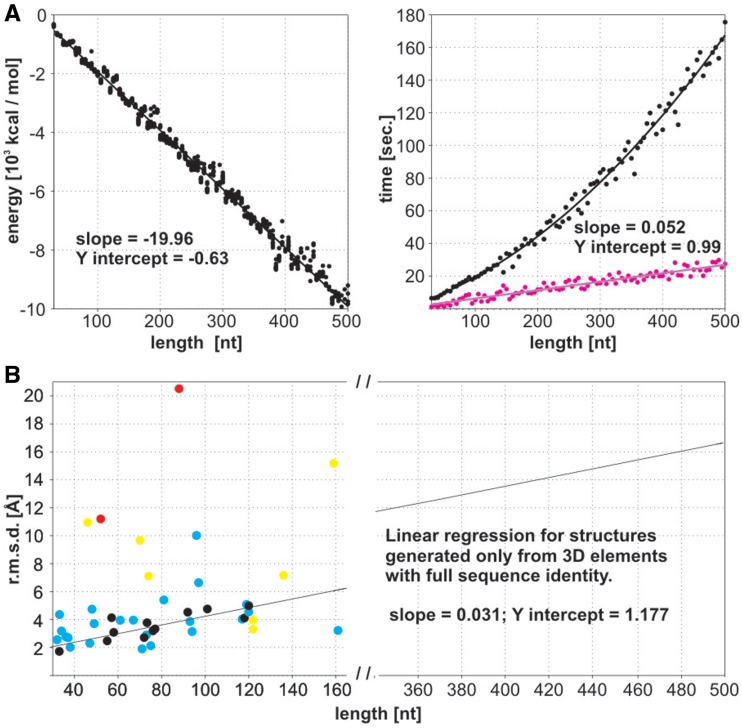
The first test encompasses a set of 95 RNAs of randomized sequences and size up to 500 nt. This set is characterized by a large diversity of secondary structures predicted using RNAfold (40) one of the commonly used programs. Entire input data are presented in the Supplementary Data Set S1. Ten 3D models were generated for each RNA. In all cases, they return accurately the input RNA secondary structure with the Matthews correlation coefficient (MCC) (41) ranging between 0.96 and 1.00 (Supplementary Table S3). Most of the RNA 3D structures show high stereochemical quality including centres of chirality, good energy values, which change linearly with the RNA strand length (Figure 2A) and precision (Figure 2B and Supplementary Table S3). It should be underlined that even for most complex large-branched RNA structures, there was no case observed that all 10 models did not pass X-PLOR energy refinement.
Figure 2.
(A) Energy and prediction time estimation for final 95 RNA 3D structures (length 30–500 nt; marked in black). Computing time for the initial RNA structures is marked in purple. (B) Method accuracy estimation (heavy atom r.m.s.d.) for a benchmark set of 40 RNAs (30–161 nt) indicated in Table 1. The final structures represent four groups: (black) composed of 3D elements with full sequence identity, (blue) with over 50% sequence identity, (yellow) including elements with low sequence identity and (red) including generated elements due to their absence in the dictionary. It should be underlined that, upon this test, all the 3D structure elements comprised by their respective crystal PDB structure were excluded from the dictionary.
For the largest structures in this set, presented evaluation test is inaccessible to other methods because of the computing cost (de novo methods) or the lack of 3D templates (comparative methods). Prediction of RNA structures characteristic of randomized sequence clearly shows the potential of our method to build artificial RNA 3D structures (Supplementary Figure S2).
The machine translation algorithm makes RNA structure predictions in very short processing time. Initial 3D structure is composed in a range of seconds (on a single Intel Xeon 2.33 GHz processor). As shown experimentally (Figure 2A), the building time of the initial structures increases linearly with RNA strand length, opening the door to the prediction of considerably larger structures. Due to the computational time needed for energy minimization, the average total processing time elapsed for the largest single RNA 3D model (500 nt; Supplementary Figure S2 and Table S4) is about 3 min. To the best of our knowledge, this is the most time-efficient RNA 3D structure prediction method reported.
In the second evaluation test, the accuracy of predicted 3D structures was assessed using representative benchmark set of RNAs with the secondary structures derived from the highest resolution X-ray structures. This set encompasses 40 RNAs of different structural complexity like hairpins, first-order pseudoknots, branched RNAs, hammerheads, riboswitches, tRNAs and 5S rRNAs, and of strand length ranged from 30 to 161 nt (Table 1). Only structures with the complete heavy atom coordinates were included. It should be underlined that upon this evaluation test, all the 3D structure elements comprised by the respective crystal PDB structures were excluded from the dictionary. This resulted in observable r.m.s.d. dispersion of the generated RNA models (Figure 2B). The ones assembled from structural elements with high-sequence identity show high accuracy, as reflected by both the r.m.s.d. and interaction network fidelity parameters (33) (INFall and INFcbp), between the predicted and crystal structures (Table 1). Structures containing fragments that were not represented in the dictionary or must have undergone extensive base replacements are characterized by lower accuracy. The average global r.m.s.d. of 5.1 Å is observed for the entire set of RNA 3D structures. The INFcbp and INFall parameter values (33,35) indicate that all canonical and non-canonical base pairing and stacking are well recovered in the predicted structures.
Table 1.
Quality of predicted RNA 3D modelsa
| RNA PDB code and chain | Strand length (nt) | Accuracyb |
Precisionc | ||
|---|---|---|---|---|---|
| r.m.s.d. (Å) | INFall | INFcbp | r.m.s.d. (Å) | ||
| Hairpin | |||||
| 2DR8 B | 33 | 1.7 | 0.88 | 1.00 | 1.4 |
| 3OVA C | 34 | 3.2 | 0.79 | 1.00 | 2.0 |
| Hairpin, internal loop | |||||
| 1JBR D | 31 | 4.4 | 0.74 | 0.98 | 2.7 |
| 2HW8 B | 36 | 2.8 | 0.86 | 1.00 | 1.4 |
| 1ZHO B | 38 | 2.0 | 0.83 | 0.99 | 1.1 |
| 3IAB R | 46 | 11.0 | 0.72 | 0.88 | 1.2 |
| Hairpin, internal loops | |||||
| 1I6U C | 37 | 2.7 | 0.79 | 0.99 | 2.0 |
| 2PXL B | 47 | 2.3 | 0.84 | 1.00 | 2.1 |
| 2VPL B | 48 | 4.7 | 0.81 | 0.97 | 1.7 |
| 2PXB B | 49 | 3.7 | 0.84 | 1.00 | 2.1 |
| 1MZP B | 55 | 2.5 | 0.78 | 0.99 | 1.7 |
| 1KXK A | 70 | 9.7 | 0.75 | 0.97 | 5.0 |
| Three-way junction | |||||
| 1DK1 B | 57 | 4.1 | 0.82 | 0.98 | 2.4 |
| 1MMS C | 58 | 3.1 | 0.73 | 0.97 | 2.3 |
| 1UN6 E | 61 | 4.0 | 0.80 | 0.88 | 2.2 |
| Three-way junction (hammerhead) | |||||
| 2QUS A | 69 | 3.8 | 0.81 | 0.96 | 1.6 |
| Three-way junction (riboswitch) | |||||
| 3LA5 A | 71 | 1.9 | 0.87 | 1.00 | 1.0 |
| 3D2V A | 77 | 3.3 | 0.82 | 0.99 | 2.0 |
| Three-way junction (GMP riboswitch) | |||||
| 3IWN A | 93 | 3.9 | 0.75 | 0.97 | 2.3 |
| Three-way junction (SRP) | |||||
| 2V3C M | 96 | 10.0 | 0.76 | 0.94 | 3.0 |
| 1LNG B | 97 | 6.6 | 0.77 | 0.99 | 2.6 |
| 1Z43 A | 101 | 4.8 | 0.76 | 0.98 | 2.7 |
| 3NDB M | 136 | 7.2 | 0.80 | 0.96 | 4.2 |
| Three-way junction (5S rRNA) | |||||
| 3OFQ B | 117 | 4.0 | 0.75 | 0.90 | 2.3 |
| 3OFR B | 118 | 4.1 | 0.83 | 1.00 | 3.2 |
| 3KIR B | 119 | 5.1 | 0.77 | 0.94 | 4.4 |
| 3I9E B | 120 | 5.0 | 0.81 | 0.96 | 3.3 |
| 1VQO 9 | 122 | 3.3 | 0.84 | 1.00 | 2.6 |
| Four-way junction (tRNA) | |||||
| 1EXD B | 73 | 2.9 | 0.77 | 1.00 | 2.3 |
| 1U0B A | 74 | 7.1 | 0.69 | 0.99 | 2.5 |
| 1FFY T | 75 | 2.1 | 0.82 | 0.96 | 1.4 |
| 2J00 W | 76 | 3.2 | 0.73 | 0.95 | 2.2 |
| Four-way junction (riboswitch) | |||||
| 3IQP A | 94 | 3.1 | 0.81 | 0.99 | 2.2 |
| Five-way junction (tRNA) | |||||
| 3AM1 B | 81 | 5.4 | 0.78 | 0.99 | 3.9 |
| 1WZ2 C | 88 | 20.5 | 0.66 | 1.00 | 5.1 |
| 3ADB C | 92 | 4.7 | 0.67 | 1.00 | 2.8 |
| Pseudoknot | |||||
| 2QWY A | 52 | 11.2 | 0.54 | 0.98 | 2.2 |
| Pseudoknot (HDV ribozyme) | |||||
| 1CX0 B | 72 | 2.7 | 0.83 | 0.97 | 1.6 |
| P4–P6 ribozyme domain | |||||
| 2R8S R | 159 | 15.2 | 0.76 | 0.99 | 7.9 |
| M-box riboswitch | |||||
| 3PDR A | 161 | 3.2 | 0.81 | 1.00 | 1.3 |
aUpon validation, all the 3D structure elements comprised by the respective crystal PDB structure were excluded from the dictionary.
bDescribed as the average heavy-atom r.m.s.d. (in Å) between 10 individual 3D models and the crystal structure, and the average interaction network fidelity (INF) measures (33). INF scores range from 0.00 (worst) to 1.00 (best).
cDescribed as the average heavy-atom r.m.s.d. (in Å) between 10 individual 3D models predicted and their mean coordinate values.
An analysis applying MolProbity tool (32) shows good quality of all but one RNA 3D structures described in Table 1. The bond and angle outliers indicate that our predicted structures in most cases show better stereochemical quality than reference high-resolution X-ray structures (Supplementary Table S5).
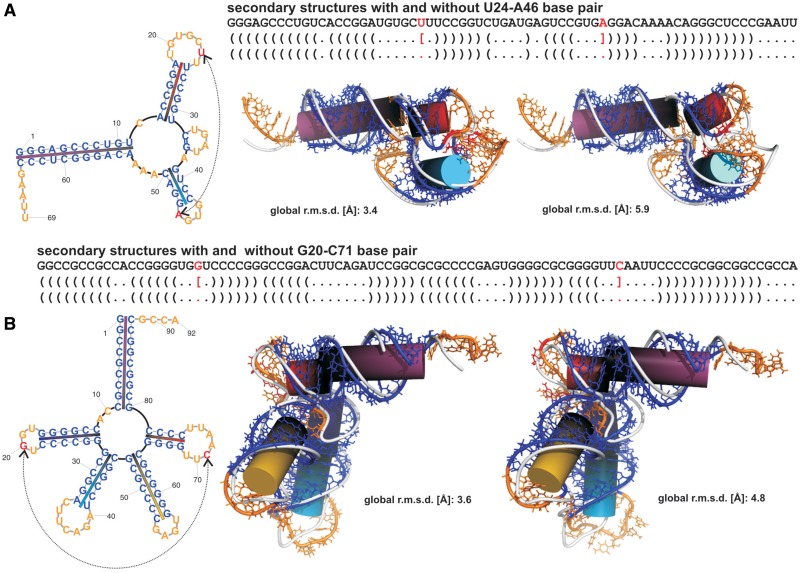
The most complex structures like branched RNAs show proper n-way junction conformation and orientation of the helices. The influence of input RNA secondary structure on the prediction of loop–loop tertiary interactions and the helices orientation in branched RNAs is exemplified on two RNA junctions (Figure 3). In the case of three-way junction of the hammerhead structure (PDB code 2QUS chain A; Figure 3A and Supplementary Table S6), the first input secondary structure was obtained by the back-conversion. In the second input secondary structure, a square bracket annotating the U24–A46 interaction was intentionally removed. As expected, due to the machine translation principle, the 3D model for the first secondary structure contains the U24–A46 base pair. For the RNA 3D structure generated from the later secondary structure, a proper orientation of the helices and the close proximity of loops were observed despite the removal of this base pair. A similar observation was made for the prediction of five-way junction of the tRNA with a long extra arm (PDB code 3ADB chain C; Figure 3B and Supplementary Table S6) characteristic of the G20–C71 base pair involved in a loop–loop interaction.
Figure 3.
The influence of input RNA secondary structure on the prediction of loop–loop tertiary interactions. (A) Three-way junction. The first secondary structure was obtained by the back-conversion of the hammerhead X-ray structure (PDB code 2QUS chain A). In the second, a square bracket annotating the U24–A46 interaction (red) was intentionally removed. In both the secondary structure graphics and the best predicted 3D structure, base paired residues are shown in blue, looped residues in orange. Each base paired segment is a specific colour on the secondary structure graphics and is represented on 3D structure as a correspondingly coloured cylinder. The reference PDB structures are shown in white. The 3D model for the first secondary structure contains the U24–A46 base pair (red). Despite the removal of the U24–A46 base pair from the input secondary structure, the second RNA 3D structure shows a proper orientation of the helices and the close proximity of loops. (B) Five-way junction. A similar observation was made for the prediction of the tRNA (PDB code 3ADB chain C).
Examples of method application
The quality of the predicted RNA 3D structure strongly depends upon the user-defined input secondary structure. In recent years, attempts have been made to incorporate the structure probing data to improve the accuracy of the in silico RNA secondary structure predictions (5,6,42) especially in case of large RNA structures. Here, as with the application examples, we show the predictions of two RNA 3D structures. For the first prediction, the H. Marismortui 122-mer 5S rRNA was chosen. The 3D structure of this RNA molecule is known (PDB 1FFK), but the secondary structure is difficult to access solely in silico. The second prediction example is presented for the 425-mer RNA transport element of the Murine musD retrotransposon. Its RNA 3D structure is unknown and the recently documented secondary structure possesses an exceptionally high level of complexity (43).
3D structure of the 5S rRNA H. Marismortui
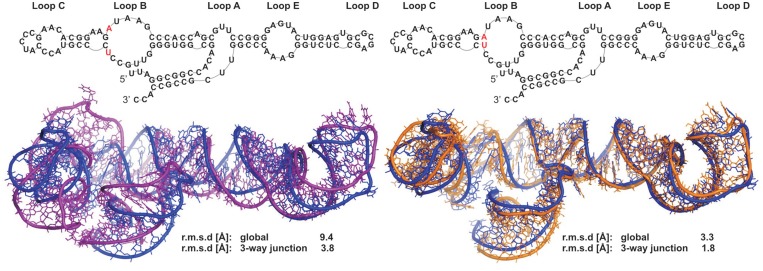
5S rRNA secondary structure was assigned using RNAstructure software (36) and chemical structure probing data (see ‘Materials and Methods’ section and Supplementary Figure S3) from SHAPE (37). The secondary structure assessed (MCC = 0.99), differs at only one point from the RNA FRABASE secondary structure. The size of the internal loop B is larger due to the absence of a U28–A54 base pair. The best out of 10 models predicted (Figure 4 and Supplementary Table S7) has r.m.s.d. of 9.4 Å with respect to the crystallographic coordinates. Most importantly, the closest overall agreement is found in the three-way junction with r.m.s.d. value of 3.8 Å. A correct orientation of the stems and coaxial stacking within the tertiary structure is observed (Figure 4). All base pairing including characteristic non-canonical interactions in the loop E are well predicted. As a reference, a family of structures generated from the ideal RNA FRABASE secondary structure has global structure average r.m.s.d. of 4.0 Å, illustrating the predictive power of our method.
Figure 4.
Prediction of 5S rRNA H. Marismortui tertiary structure as an application example. On the left: superposition of the best 3D model (purple) generated from the secondary structure determined in this work, using the RNAstructure software (36) and the SHAPE chemical structure probing (37) data, over the X-ray structure (PDB 1FFK; blue). On the right: superposition of the best 3D model (orange) generated from the RNA FRABASE secondary structure over the crystal structure (blue).
3D structure of the RNA transport element of the Murine musD retrotransposon
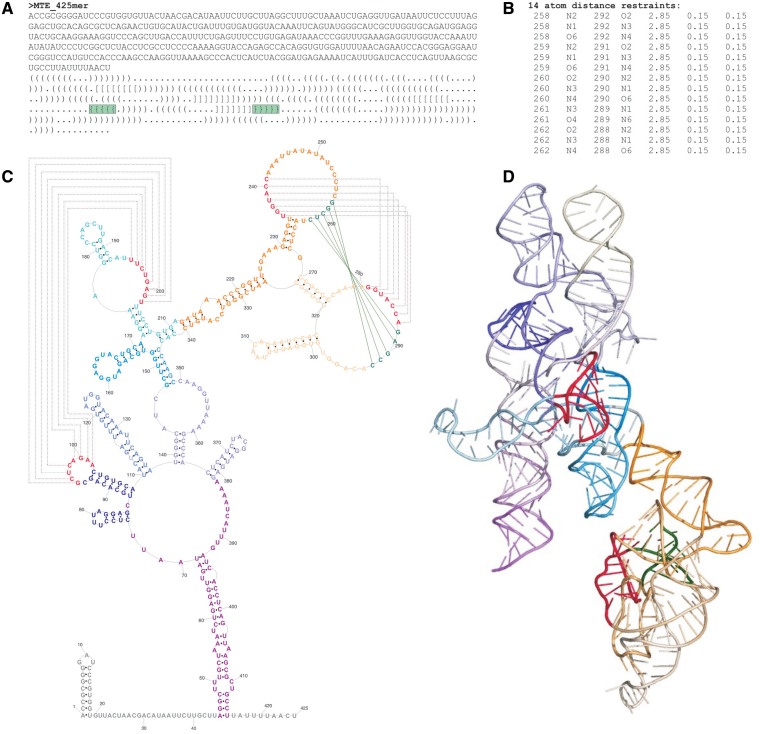
The secondary structure of this RNA transport element was recently presented and encompasses two long range interactions essential for its function (43). The first one corresponds to relatively simple intramolecular kissing loops (H-type pseudoknot) while the second one represents an intricate second-order pseudoknot (Figure 5). The reported secondary structure (43) was manually converted to the dot-bracket notation. To accomplish 3D structure prediction of the second-order pseudoknot, an additional functionality of RNAComposer was utilized, optional introduction of distance restraints. For the respective 5 bp, hydrogen bonding restraints were introduced to enforce the suggested tertiary interaction. Twenty 3D RNA models were generated and the best model of the lowest energy is presented for this RNA structure (Figure 5D). It is interesting to note that the kissing-loops (Figure 5C and D, annotated in red) and the second-order pseudoknot (Figure 5C and D, annotated in red and green) are located on opposite surfaces of this RNA molecule. This may support the previous suggestion (43) that these tertiary motifs function as independent regulatory elements. Presented model allows for rational design of further functional experiments and might be validated using hydroxyl radicals probing to identify solvent inaccessible elements.
Figure 5.
Prediction of the tertiary structure of the MusD RNA transport element. (A) The reported secondary structure (43) in the dot-bracket notation. First-order interactions are represented by square brackets, while the second-order interactions are represented by curl brackets (highlighted in green). In the RNAComoposer input data, the curl brackets were substituted by dots and indicated (B) distance restraints for hydrogen bonds were introduced. (C) Secondary structure model (43) visualized using PseudoViewer 3.0 (44), with the tertiary interactions annotated with lines. Nucleoside residues are renumbered relative to those reported (43). (D) The best of twenty 3D models predicted by RNAComposer. Colour annotation on the depicted secondary structure corresponds to the respective coloured segments (45) on the 3D structure. For clarity, 5′ and 3′ stretches (in secondary structure marked in grey) are not visualized.
RNAComposer web server and its comparison to other automated servers
The described method is implemented in the publicly available web server RNAComposer. It is designed to work with all most commonly used web browsers, such as Microsoft Internet Explorer (8.0 and later), Mozilla Firefox (3.6 and later), Opera (10.53 and later) and Google Chrome (5.0 and later).
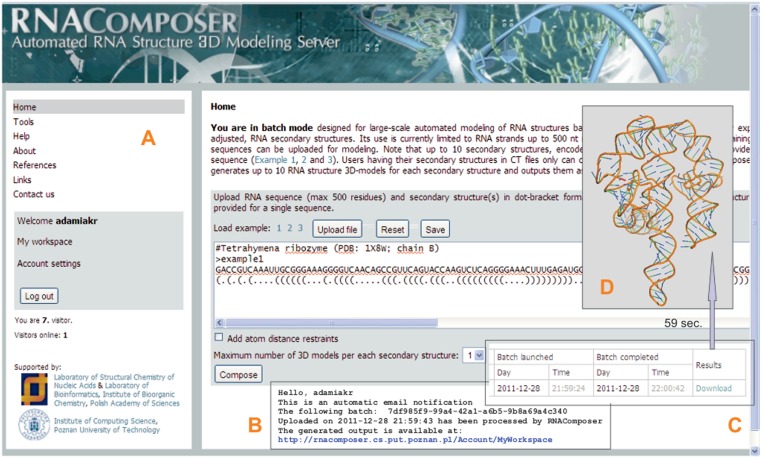
RNAComposer works in two fully automated modes: an interactive mode and a batch mode. The interactive mode enables the prediction of one single 3D model of a short RNA molecule at a time; the PDB file is immediately available and the RNA structure visualization is provided. Moreover, this mode helps to generate RNA secondary structures using integrated prediction tools. The batch mode is designed for large-scale modelling of RNA structures based on a user-defined RNA secondary structure(s) (Figure 6). RNAComposer can generate up to 1000 RNA 3D models from a set of 100 different secondary structures. Presently, the RNAComposer web server is set to build RNA of up to 500 residues. The batch mode also allows a user to enter experimentally derived atom distance restraints, e.g. from NMR data. As the output, a log file describing details of the model generation process is provided along with the pdb files. The log file includes: (i) a list of structure elements resulting from secondary structure fragmentation, (ii) a list of tertiary elements selected for structure assembly with their origin (PDB code) and sequence similarity and (iii) energy of the final structures.
Figure 6.
Selected snapshots of the RNAComposer interface. (Panel A) the batch mode page, clicking on the ‘Compose’ button activates building of a single RNA 3D model (terahymena ribozyme); the addition of the distance restraints window is not checked. (Panel B) automated e-mail notification of the prediction results. (Panel C) the workspace information stating the job status and prediction time. RNA structure coordinates saved in PDB-format files are ready for direct download and visualization at the user site (panel D). Note that in the case of the interactive mode, automated visualization of a single model is provided.
To the best of our knowledge, there are three automated, publicly available web-interfaced methods for the RNA tertiary structure prediction: MC-Fold/MC-Sym Pipeline (21), iFoldRNA (15) and ModeRNA (22).
At the revision stage, we have conducted computational tests and compared our method to the above-mentioned ones, except ModeRNA which as the comparative method requires full-length structure template. Results of comparison concerning eight RNA structures are presented in the Supplementary Tables S8 and 9. Due to intrinsic limitations of MC-Fold/MC-Sym and iFoldRNA servers to deal with large RNA structures, chosen target structures are within the size range from 36 to 81 nt. To level the starting point for the comparison, we have used the ‘ideal’ RNA secondary structures (RNA FRABASE-derived) as the input to RNAComposer and MC-Sym module of MC-Sym/MC-Fold Pipeline. Since iFoldRNA does not accept secondary structure as a user defined input it has been operated from the sequence.
In each case, RNAComposer shows superior performance with respect to the criteria of stereochemical quality and structure accuracy. The average global r.m.s.d. of RNA models generated by our server is about 3.7 Å, while for the same data set MC-Sym reaches 10.2 Å and iFoldRNA reaches 12.0 Å. Interaction network fidelity (INFall) parameter is the average of 0.80 for RNAComposer, 0.71 for structures obtained from MC-Sym and 0.52 for iFoldRNA. As for the conservation of canonical base pairs, the mean values of INFcbp are 1.0 for RNAComposer and MC-Sym, 0.64 for the third server. The quality of predicted models has been evaluated using MolProbity tool (32). The average clash score computed over all atoms was less than 15 for RNAComposer generated models and exceeded 100 for those predicted by the other methods. Moreover, no residues with potentially incorrect bonds and angles were identified in structures composed by our server. Models predicted by MC-Fold/MC-Sym server contained over 66% of residues with potentially incorrect bonds and about 93% of those with incorrect angles in average. The same ratings for iFoldRNA server reached 22 and 74%, respectively. As final remark, we would like to underline that in practice our server makes prediction much faster (in a range of seconds) then other ones.
CONCLUSIONS
We have developed and demonstrated an efficient method for the fully automated prediction of RNA 3D structures from secondary structures. This method is superior to existing ones for users having in hand an experimentally adjusted secondary structure of large RNAs. The accuracy of the method will increase considerably with the growth of the RNA FRABASE dictionary due to the surge of new experimental RNA coordinates.
The results demonstrated in this work allow one to foresee further applications of our method to elucidation of RNA structures using NMR spectroscopy, analysis of RNA/protein complexes based on cryo-EM maps and the prediction of artificial RNA 3D structures. We envisage a further development of the machine translation system to the nucleic acids structure modelling using different dictionaries (databases).
In the future, the RNAComposer server will be equipped with further functionalities permitting prediction of higher order pseudoknots, insertion of additional torsion angle constraints, optimized prediction of long single-stranded stretches and the introduction of user-defined 3D structural elements generated by other methods.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–9, Supplementary Figures 1–3 and Supplementary Data set 1.
FUNDING
Foundation for Polish Science (Mistrz Programme to R.W.A.); Ministry of Science and Higher Education [PBZ-MniSW-07/1/2007/01, NN519314635]; European Regional Development Fund within Innovative Economy Programme [POIG.02.03.00-00-018/08 POWIEW]. Funding for open access charge: European Regional Development Fund within Innovative Economy Programme [POIG.02.03.00-00-018/08 POWIEW].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank D. H. Mathews for the Linux version of RNAstructure and E. Burke and W. Filipowicz for their comments. We wish to thank Katarzyna Pachulska-Wieczorek for her assistance with the RNA structure probing.
REFERENCES
- 1.Sashital DG, Doudna JA. Structural insights into RNA interference. Curr. Opin. Struct. Biol. 2010;20:90–97. doi: 10.1016/j.sbi.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beezhold KJ, Castranova V, Chen F. Microprocessor of microRNAs: regulation and potential for therapeutic intervention. Mol. Cancer. 2010;9:e134. doi: 10.1186/1476-4598-9-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rose PW, Beran B, Bi C, Bluhm WF, Dimitropoulos D, Goodsell DS, Prlic A, Quesada M, Quinn GB, Westbrook JD, et al. The RCSB Protein Data Bank: redesigned web site and web services. Nucleic Acids Res. 2011;39:D392–D401. doi: 10.1093/nar/gkq1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leontis N, Westhof E, editors. Series Nucleic Acids and Molecular Biology. Berlin and Heidelberg: Springer; 2012. RNA 3D structure analysis and prediction. [Google Scholar]
- 5.Mathews DH, Turner DH. Prediction of RNA secondary structure by free energy minimization. Curr. Opin. Struct. Biol. 2006;16:270–278. doi: 10.1016/j.sbi.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 6.Xu Z, Almudevar A, Mathews DH. Statistical evaluation of improvement in RNA secondary structure prediction. Nucleic Acids Res. 2012;40:e26. doi: 10.1093/nar/gkr1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mathews DH, Disney MD, Childs JL, Schroeder SJ, Zuker M, Turner DH. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc. Natl Acad. Sci. USA. 2004;101:7287–7292. doi: 10.1073/pnas.0401799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. RNA structure analysis at single nucleotide resolution by selective 2’-hydroxyl acylation and primer extension (SHAPE) J. Am. Chem. Soc. 2005;127:4223–4231. doi: 10.1021/ja043822v. [DOI] [PubMed] [Google Scholar]
- 9.Wilkinson KA, Gorelick RJ, Vasa SM, Guex N, Rein A, Mathews DH, Giddings MC, Weeks KM. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biol. 2008;6:e96. doi: 10.1371/journal.pbio.0060096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Purzycka KJ, Pachulska-Wieczorek K, Adamiak RW. The in vitro loose dimer structure and rearrangements of the HIV-2 leader RNA. Nucleic Acids Res. 2011;39:7234–7248. doi: 10.1093/nar/gkr385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pang PS, Elazar M, Pham EA, Glenn JS. Simplified RNA secondary structure mapping by automation of SHAPE data analysis. Nucleic Acids Res. 2011;39:e151. doi: 10.1093/nar/gkr773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seetin MG, Mathews DH. Automated RNA tertiary structure prediction from secondary structure and low-resolution restraints. J. Comput. Chem. 2011 doi: 10.1002/jcc.21806. April 21 (doi:10.1002/jcc.21806; epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martinez HM, Maizel JV, Jr, Shapiro BA. RNA2D3D: a program for generating, viewing, and comparing 3-dimensional models of RNA. J. Biomol. Struct. Dyn. 2008;25:669–683. doi: 10.1080/07391102.2008.10531240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jossinet F, Ludwig TE, Westhof E. Assemble: an interactive graphical tool to analyze and build RNA architectures at the 2D and 3D levels. Bioinformatics. 2010;26:2057–2059. doi: 10.1093/bioinformatics/btq321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sharma S, Ding F, Dokholyan NV. iFoldRNA: three-dimensional RNA structure prediction and folding. Bioinformatics. 2008;24:1951–1952. doi: 10.1093/bioinformatics/btn328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jonikas MA, Radmer RJ, Laederach A, Das R, Pearlman S, Herschlag D, Altman RB. Coarse-grained modeling of large RNA molecules with knowledge-based potentials and structural filters. RNA. 2009;15:189–199. doi: 10.1261/rna.1270809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jonikas MA, Radmer RJ, Altman RB. Knowledge-based instantiation of full atomic detail into coarse-grain RNA 3D structural models. Bioinformatics. 2009;25:3259–3266. doi: 10.1093/bioinformatics/btp576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flores SC, Altman RB. Turning limited experimental information into 3D models of RNA. RNA. 2010;16:1769–1778. doi: 10.1261/rna.2112110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flores SC, Sherman MA, Bruns CM, Eastman P, Altman RB. Fast flexible modeling of RNA structure using internal coordinates. IEEE/ACM Trans. Comput. Biol. Bioinform. 2011;8:1247–1257. doi: 10.1109/TCBB.2010.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Das R, Karanicolas J, Baker D. Atomic accuracy in predicting and designing noncanonical RNA structure. Nat. Methods. 2010;7:291–294. doi: 10.1038/nmeth.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parisien M, Major F. The MC-Fold and MC-Sym pipeline infers RNA structure from sequence data. Nature. 2008;452:51–55. doi: 10.1038/nature06684. [DOI] [PubMed] [Google Scholar]
- 22.Rother M, Rother K, Puton T, Bujnicki JM. ModeRNA: a tool for comparative modeling of RNA 3D structure. Nucleic Acids Res. 2011;39:4007–4022. doi: 10.1093/nar/gkq1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilks Y. Machine Translation: Its Scope And Limits. New York: Springer; 2010. [Google Scholar]
- 24.Popenda M, Blazewicz M, Szachniuk M, Adamiak RW. RNA FRABASE version 1.0: an engine with a database to search for the three-dimensional fragments within RNA structures. Nucleic Acids Res. 2008;36:D386–D391. doi: 10.1093/nar/gkm786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Popenda M, Szachniuk M, Blazewicz M, Wasik S, Burke EK, Blazewicz J, Adamiak RW. RNA FRABASE 2.0: an advanced web-accessible database with the capacity to search the three-dimensional fragments within RNA structures. BMC Bioinformatics. 2010;11:e231. doi: 10.1186/1471-2105-11-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 27.Macke T, Case DA. Modeling Unusual Nucleic Acid Structures. In: Leontes NB, Santa Lucia J Jr, editors. Molecular Modeling of Nucleic Acids. American Chemical Society. Washington, DC: 1998. pp. 379–393. [Google Scholar]
- 28.Guntert P, Mumenthaler C, Wuthrich K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 29.Popenda M, Bielecki L, Adamiak RW. High-throughput method for the prediction of low-resolution three-dimensional RNA structures. Nucleic Acids Symp. Ser. (Oxf) 2006;50:67–68. doi: 10.1093/nass/nrl033. [DOI] [PubMed] [Google Scholar]
- 30.Berman HM, Olson WK, Beveridge DL, Westbrook J, Gelbin A, Demeny T, Hsieh SH, Srinivasan AR, Schneider B. The nucleic acid database: a comprehensive relational database of three-dimensional structures of nucleic acids. Biophys. J. 1992;63:751–759. doi: 10.1016/S0006-3495(92)81649-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kabsch W. Solution for best rotation to relate 2 sets of vectors. Acta Crystallogr. A. 1976;32:922–923. [Google Scholar]
- 32.Davis IM, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, III, Snoerink J, Richardson JS. MolProbity: all-atom contacts and structure validation for protein and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parisien M, Cruz JA, Westhof E, Major F. New metrics for comparing and assessing discrepancies between RNA 3D structures and models. RNA. 2009;15:1875–1885. doi: 10.1261/rna.1700409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang H, Jossinet F, Leontis N, Chen L, Westbrook J, Berman H, Westhof E. Tools for the automatic identification and classification of RNA base pairs. Nucleic Acids Res. 2003;31:3450–3460. doi: 10.1093/nar/gkg529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gendron P, Lemieux S, Major F. Quantitative analysis of nucleic acid three-dimensional structures. J. Mol. Biol. 2001;308:919–936. doi: 10.1006/jmbi.2001.4626. [DOI] [PubMed] [Google Scholar]
- 36.Reuter JS, Mathews DH. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics. 2010;11:e129. doi: 10.1186/1471-2105-11-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilkinson KA, Merino EJ, Weeks KM. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat. Protoc. 2006;1:1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- 38.Das R, Laederach A, Pearlman SM, Herschlag D, Altman RB. SAFA: semi-automated footprinting analysis software for high-throughput quantification of nucleic acid footprinting experiments. RNA. 2005;11:344–354. doi: 10.1261/rna.7214405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gan HH, Pasquali S, Schlick T. Exploring the repertoire of RNA secondary motifs using graph theory; implications for RNA design. Nucleic Acids Res. 2003;31:2926–2943. doi: 10.1093/nar/gkg365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hofacker IL, Fontana W, Stadler PF, Bonhoeffer S, Tacker M, Schuster P. Fast folding and comparison of RNA secondary structures. Monastch.. Chem. 1994;125:167–188. [Google Scholar]
- 41.Matthews BW. Comparison of the predicted and observed secondary structure of T4 phage lysozyme. Biochim. Biophys. Acta. 1975;405:442–451. doi: 10.1016/0005-2795(75)90109-9. [DOI] [PubMed] [Google Scholar]
- 42.Deigan KE, Li TW, Mathews DH, Weeks KM. Accurate SHAPE-directed RNA structure determination. Proc. Natl Acad. Sci. USA. 2009;106:97–102. doi: 10.1073/pnas.0806929106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Legiewicz M, Zolotukhin AS, Pilkington GR, Purzycka KJ, Mitchell M, Uranishi H, Bear J, Pavlakis GN, Le Grice SF, Felber BK. The RNA transport element of the murine musD retrotransposon requires long-range intramolecular interactions for function. J. Biol. Chem. 2010;285:42097–42104. doi: 10.1074/jbc.M110.182840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Byun Y, Han K. PseudoViewer3: generating planar drawings of large-scale RNA structures with pseudoknots. Bioinformatics. 2009;25:1435–1437. doi: 10.1093/bioinformatics/btp252. [DOI] [PubMed] [Google Scholar]
- 45.DeLano WL. The PyMOL Molecular Graphics System. San Carlos: DeLano Scientific; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.