Abstract
P27Kip1 (p27) is a member of the Cip/Kip family of cyclin-dependent kinase inhibitors. Recently, a new function of p27 as transcriptional regulator has been reported. It has been shown that p27 regulates the expression of target genes mostly involved in splicing, cell cycle, respiration and translation. We report here that p27 directly binds to the transcriptional coactivator PCAF by a region including amino acids 91–120. PCAF associates with p27 through its catalytic domain and acetylates p27 at lysine 100. Our data showed that overexpression of PCAF induces the degradation of p27 whereas in contrast, the knockdown of PCAF stabilizes the protein. A p27 mutant in which K100 was substituted by arginine (p27-K100R) cannot be acetylated by PCAF and has a half-life much higher than that of p27WT. Moreover, p27-K100R remains stable along cell-cycle progression. Ubiquitylation assays and the use of proteasome inhibitors indicate that PCAF induces p27 degradation via proteasome. We also observed that knockdown of skp2 did not affect the PCAF induced degradation of p27. In conclusion, our data suggest that the p27 acetylation by PCAF regulates its stability.
INTRODUCTION
Cell-cycle progression is regulated by the sequential activation of members of the cyclin-dependent kinase (cdk) family (1). Quiescent cells contain significant levels of cdk4 and cdk2 but only very small amounts of cyclins. Since cyclins associate with and activate cdks, quiescent cells only display very low cdk activity. After mitogenic stimulation cells synthesize cyclin D that associates with cdk4 that as a consequence becomes activated (2). The main function of active cyclin D–cdk4 complexes at early G1 phase of the cell cycle is to phosphorylate transcriptional repressor complexes containing E2F4/p130 or E2F1/pRb that are additionally associated with different transcriptional corepressors (3,4). In quiescent cells, these complexes repress the expression of a number of genes necessary for the onset of DNA replication. Cdk4 phosphorylation of p130, pRb and other proteins of these complexes at early G1, disorganize some of them allowing transcription of a number of genes, including cyclin E that accumulates in the cells at mid-late G1. Then, cyclin E binds to and activates cdk2 that additionally phosphorylate p130 and pRb to finally disrupt the repressor complexes. This leads to the expression of the S phase genes needed for the onset of DNA synthesis.
In addition to the association with cyclins, cdk activity is also regulated by phosphorylation, acetylation and by association with specific cdk inhibitors (CKIs) (5–7). There are two families of CKIs: The Cip/Kip family that includes p21, p27 and p57 and that associate with and inhibit most cyclin–cdk complexes. In contrast, the members of the other family (ink4) including p15, p16, p18 and p19 specifically bind to and inhibit cdk4 and cdk6 (5,8).
Among these CKIs, p27 is specifically relevant because its deficiency is associated with tumorigenesis. Reduced p27 levels are frequently observed in human cancers in association with tumor aggressiveness and poor clinical outcome (9–11). In most of the cases, low levels of p27 in human tumors are due to post-transcriptional regulation that leads to an increased proteasome-dependent degradation (12–14). The role of p27 in tumorigenesis is supported by the evidence that p27−/−—mice spontaneously develop pituitary tumors and are much more susceptible to tumorigenesis induced by chemical carcinogens than p27wt—mice (15–17). Moreover, knock in mice harboring a p27 mutant (p27CK), including four punctual mutations that unables it to interact with and to inhibit cyclin–cdk complexes, develop hyperplasias and tumors in many different organs (18). These data clearly indicate that p27 performs other cellular functions, involved in tumorigenesis, that are independent of its role as a CKI.
We have recently reported that p27 plays a role as a transcriptional regulator (19). Specifically, by ChIP on chip, we have observed that p27 associates with 427 promoters of genes mainly involved in cell-cycle regulation, respiration, translation and RNA processing and splicing. Interestingly, the overexpression of these p27-target genes (p27-TGs) in human tumors correlates with poor clinical outcome. These results suggest that the transcriptional regulatory function of p27 plays an important role in the development of tumors.
On a significant number of these p27-TG promoters p27 is associated with E2F4/p130 repressor complexes. Interestingly, we found that in fact p27 directly interacts with E2F4 and p130 by its carboxyl moiety. Thus, p27 is a structural component of these repressor complexes that operate in quiescent cells to repress the S phase genes. According to that, expression microarrays analysis performed on embryo fibroblasts (MEFs) from p27WT, p27−/− and p27CK− revealed that p27 behaves as a transcriptional repressor of these p27-TGs (19). The specific role of p27 on these repressor complexes still remains to be clarified but a possibility is that in addition to its repressive role it may also participate in the recruitment on cyclin–cdk complexes needed for p130 phosphorylation at early-mid G1 phase. The evidence showing that the interaction of p27 with E2F4/p130 complexes is through its carboxyl moiety indicates that its amino moiety (that contains the cyclin and cdk interacting domains) will be free to associate with cyclin–cdk complexes when they will appear at early G1. However, this possibility still remains to be clarified.
On searching for nuclear proteins that interact with p27 in quiescent cells we observed that the acetyl transferase and transcriptional coactivator PCAF (6) associates with p27 in these cells. We report here that PCAF directly interacts with and acetylates p27 in the K100 residue. We have identified the PCAF interacting domain of p27 and demonstrated that acetylation of p27 by PCAF induces its degradation via proteasome. These results are compatible with a role of PCAF as a coactivator of p27-TGs by inducing the degradation of p27, a transcriptional repressor of these genes.
MATERIALS AND METHODS
Materials
ALLNL and MG132 were purchased from Calbiochem. [14C]acetylCoA from Perkin Elmer. Histones and cycloheximide were from Sigma-Aldrich. Rabbit reticulocyte extracts (L4151) were from Promega. The following antibodies were used in this work: Acetyl-Lys (600-401-939), from Rockland Immunochemicals; p27Kip1 (sc-528 and sc-527) from Santa Cruz, Biotechnology and (610242) from BD transduction labs; cyclin D1 (Sc-20044) and Skp2 (sc-7164) from Santa Cruz Biotechnology; PCAF (P7493), ubiquitin (U5379) and Flag (F7425) from Sigma-Aldrich; actin (69100) from MP Biomedicals; p21Cip1 (OP64) from Calbiochem and GFP (ab290) from Abcam.
Plasmids
The p21 full length, the full-length p27 and the following mutants of p27 were purchased from Genscript: p27WT, p27-K13R in which all lysines of p27 were substituted for arginines, p27–K5R (K47R,K59R,K68R,K73R,K81R). p27-K3R (K25R,K96R,K100R), p27–K2R (K96R,K100R), p27–K100R, p27(Δ91–120), p27(Δ91–100), p27(Δ96–100) and p27(Δ91–95). All these sequences were subsequently subcloned in a pECFP–C1 vector (Clontech). pCDNA3–Flag–PCAF (FlagPCAF), pCDNA3–GFP–Flag–PCAF (GFPCtWT) and pCDNA3–GFP–Flag–PCAFΔHAT (352–832, lacking amino acids 527–547) (GFP–CtΔHAT) have been described elsewhere (20). The shRNA corresponding to PCAF (pLKO-1, puro-PCAF) and the shRNA random (pLKO-1, puro) used as a control were from Sigma-Aldrich. The siRNA corresponding to Skp2 and to GFP were from Dharmacon.
Recombinant proteins
pGEX vectors (Amersham) containing: the full-length p27, the different mutants and fragments of this protein, p21, GST or CBP were transformed into Escherichia coli BL21 strain. The expressed proteins were subsequently purified as previously described (21). The plasmids pGEX–PCAF (GSTPCAF), pGEX–PCAF(352–658) (GSTHAT) and pGEXPCAF(352–832, lacking amino acids 527–547) (GSTCtΔHAT) have been previously described (22,23).
Cell culture and transfection
HEK 293T and HeLa cells were cultured in Dulbecco's modified Eagle's medium (DMEM) (Biological Industries) containing 10% fetal calf serum (FCS). HCT116 cells were cultured in DEMEM:HAM (1:1) complemented with 2 mM Glutamine, 1% non-essential amino acids, 1 mM piruvic acid and 10% FCS. In some experiments the proteasome inhibitors ALLNL (100 µM) or MG132 (10 µM) were added to the culture medium. Transfection of HCT116 and HET293T cells was performed with Lypofectamine 2000 or Lipofect, respectively as directed by the manufacturer (Qiagen). Cells were harvested 24 h after transfection. To synchronize the different cellular types, they were cultured for 48–72 h in 0.5% FCS media. Then, cells were replated in media with 10% FCS and subsequently collected at different times after replating.
Affinity chromatography
To generate the different chromatography columns the purified proteins or peptides were covalently bound to CNBr-activated sepharose 4B, according to the manufacturers (Amersham). The peptides used in these experiments were purchased from GenScript. Cell were lysed with buffer A (50 mM HEPES, pH 7.6, 50 mM KCl, 1 mM EDTA and 1 mM MgCl2). Then, cell extracts (10 mg) were loaded onto the columns and after washing in 50 vol. of buffer A containing 150 mM KCl, the bound proteins were eluted with the same buffer but containing 1.5 M KCl (21). Then, they were subsequently eluted with 0.1 M Na–acetate and 0.5 M NaCl at pH 4.5. To analyze the direct binding of purified proteins to the columns the same protocol was used but loading 5–10 µg of the purified proteins onto the columns.
Immunoprecipitation and western blotting
Cells were lysed in RIPA buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% NP40, 0.5% deoxycolate, 0.1% SDS and 3.3 µM TSA) containing 50 mM NaF, 1 mM phenylmehylsulfonyl fluoride, 0.1 mM Na3VO4, 0.5 µg/µl aprotinin and 10 µg/µl Leupeptin) for 30 min on ice. For the immunoprecipitation (IP) experiments, lysates were incubated with 4 µg of specific antibodies overnight at 4°C. Then, protein A/G agarose beads (Pearce) were added and samples were incubated for 45 min at 4°C. After three washes with RIPA buffer, immunoprecipitates were subjected to western blotting (WB) as described (24).
In vitro acetylation assay
Protein acetyltransferase assays were performed in 30 µl of HAT lysis buffer (50 mM Tris, pH 8.0, 50 mM KCl, 5% glycerol and 1 mM EDTA), 1 µl (0.02 μCi) of [14C]-acetyl-CoA (Perkin Elmer), 6 µM of substrate proteins or histones and 300 ng of recombinant HAT domains of PCAF or CBP. Reactions were performed for 30 min at 30°C. Then, proteins were separated by SDS–polyacrylamide gel electrophoresis (PAGE), transferred to nitrocellulose membranes and exposed to film for 2–4 days.
In vitro ubiquitylation assay
Recombinant GSTp27 and GSTp27K100R were acetylated with recombinant GSTHAT domain of PCAF as described above. After acetylation, 60 µl of reticulocyte cell lysate (Promega) were added to the reaction as a source of ubiquitin and ubiquitylation enzymes. The mixture was incubated for 1 h at 37°C and then, the recombinant GST-fusion substrates were re-purified using glutathione–agarose beads (Amersham) as described by the manufacturer. The GST-fusion substrates were separated by SDS–PAGE and the presence of ubiquitylated p27 forms was analyzed by WB using anti-ubiquitin antibodies. Similar experiments were performed and the proteins transferred to nitrocelulose membranes. Then, the radioactivity incorporated into the proteins was detected by autoradiography for 2–4 days.
Spot mapping analysis
Nine peptides, each one containing one of the lysines present in the N-terminus of p27 were generated (Genosys). The selected sequences were: (i) KSTGGKAPRKQ (histone H3 peptide as a positive control); (ii) RQAEHPKPSACR; (iii) LTRDLEKHCRDM; (iv) EEASQRKWNFDF; (v) NFDFQNHKPLEG; (vi) PLEGKYEWQEVE; (vii) WQEVEKGSLPEF; (viii) YYRPPRPPKGAC; (ix) GACKVPAQESQD. Peptides (5 nmol) were spotted on a nitrocellulose membrane and then the membrane was subjected to an in vitro acetylation assay as described above.
Protein stability assay
Cells were transfected for 20 h with FlagPCAF expression vector or with a pCDNA empty vector as a control. Then, 10 µg/ml of cycloheximide (Sigma) were added to the cell cultures and cells were harvested at different times after cycloheximide addition. Finally, samples were subjected to WB using anti-p27, anti-PCAF or anti-actin that was used as a loading control. Similar experiments were performed to study the stability of the ectopic p27WT or the non-acetylatable mutant p27K100R. In these experiments, HCT116 cells were transfected with pCDNAp27WT or pCDNAK100R expression vectors or with a pCDNA empty vector as a control.
Immunofluorescence
To detect CFP–p27 and YFP–PCAF by immunofluorescence, HCT116 cells with transfected with vectors containing these genes and grown in coverslips, fixed in 4% paraformaldehyde/PBS for 15 min at room temperature and washed in PBS. After fixation, cells were stained with DAPI for the nuclear visualization. Then, coverslips were mounted on glass slides with Mowiol (Calbiochem) and analyzed by fluorescence microscopy.
RESULTS
The protein p27 interacts with the acetyltransferase PCAF
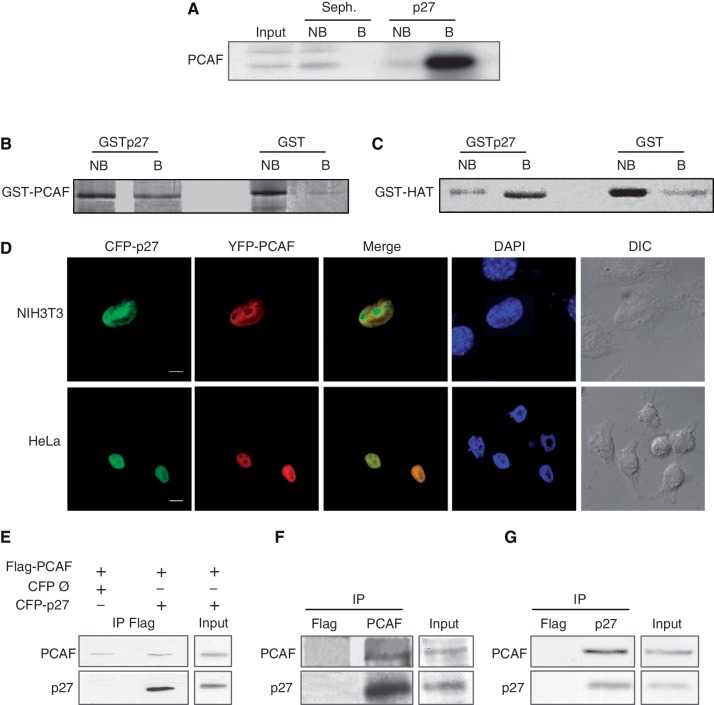
To identify new p27-binding proteins, extracts from HCT116 cells were loaded on a p27–Sepharose 4B column and the proteins associated with p27 were subsequently eluted and analyzed by mass spectrometry. A protein of 89 kDa from the eluates was identified as PCAF by mass spectrometry (data not shown). The binding of PCAF to the p27 column was confirmed by WB (Figure 1A). Direct in vitro interaction between PCAF and p27 was examined by affinity chromatography. Purified recombinant GST–PCAF or the catalytic domain of PCAF (GST–HAT), were separately loaded on a p27 column and the binding analyzed. Results showed that both, full-length PCAF (Figure 1B) and the HAT domain (Figure 1C) associated with the p27 column, indicating that PCAF directly interacts with p27 by its catalytic domain. Colocalization experiments were subsequently performed in HCT116 cells cotransfected with CFP–p27 and YFP–PCAF. As shown in Figure 1D both proteins colocalize in the nucleus. To determine whether p27 and PCAF interact in the cells, we used HEK 293T cells that were cotransfected with YFP–Flag–PCAF and CFP–p27. Then, IP with anti-Flag were performed. Results showed the co-IP of both proteins (Figure 1E). We also observed the co-IP of the endogenous PCAF and p27 by using anti-PCAF (Figure 1F) or anti-p27 antibodies (Figure 1G).
Figure 1.
Interaction of p27 with PCAF. (A) HCT116 cell extracts were loaded on a p27 or a Sepharose 4B column (Seph.). Proteins not bound (NB) or bound (B) to the columns were analyzed by WB using an anti-PCAF antibody. (B) Purified GST–PCAF was loaded on a GST–p27 or on a GST column. Fractions not bound (NB) or bound (B) to the columns were visualized by gel electrophoresis and Coomassie blue staining. (C) Purified GST–HAT was loaded on a GST–p27 or a GST column. Fractions not bound (NB) or bound (B) to the columns were visualized by gel electrophoresis and coomassie blue staining. (D) NIH3T3 and HeLa cells were cotransfected with CFP–p27 and YFP–PCAF, stained with DAPI and subsequently visualized by confocal microscopy. Bar: 5 μm (E) HEK293T cells were transfected with Flag–PCAF or/and CFP–p27. At 24 h post-transfection, cell extracts were immunoprecipitated with anti-Flag. Cells transfected with an empty CFP vector (Ø) and Flag–PCAF were used as a control. The immunoprecipitates were then analyzed by WB using antibodies against PCAF or p27. (F) HEK293T cell extracts were subjected to IP with anti-PCAF or anti-Flag (as a control), then, the immunoprecipitates were analyzed by WB using antibodies against PCAF or p27. (G) HEK293T cell extracts were subjected to IP with anti-p27 or anti-Flag (as a control), then, the immunoprecipitates were analyzed by WB using antibodies against PCAF or p27.
The PCAF-interacting domain of p27 includes amino acids 91–120 and contains a proline-rich domain
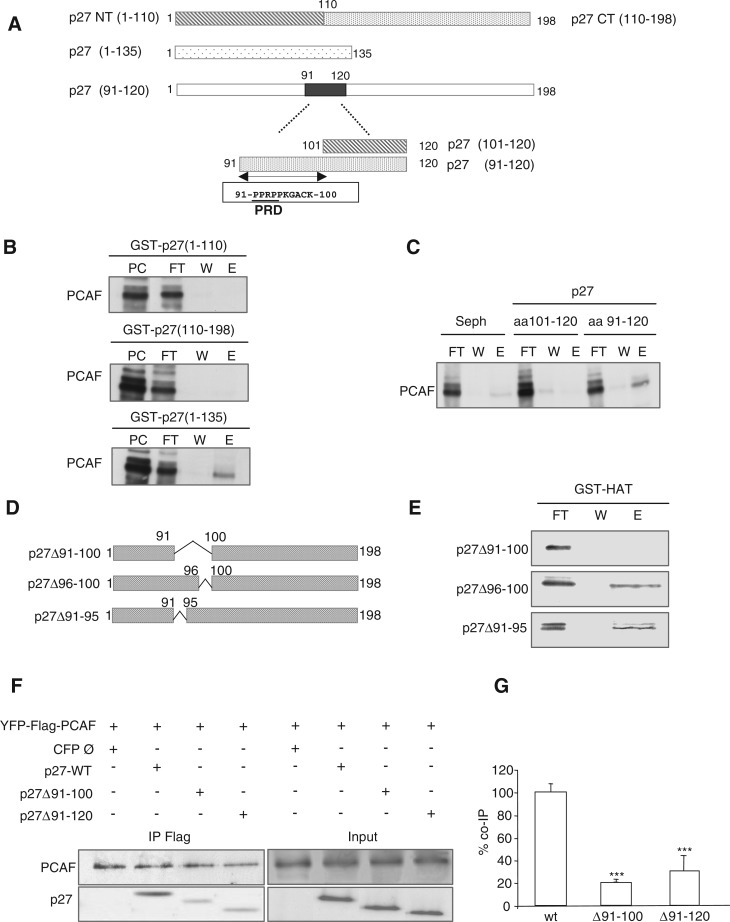
To identify the PCAF-interacting domain (PID) of p27, affinity chromatography experiments using different p27 fragments were performed. We first generated two affinity columns using the fragments p27–NT (amino acids 1–110) and p27–CT (amino acids 110–198) (Figure 2A). Purified PCAF was loaded onto both columns and its association analyzed by WB. As shown in Figure 2B (upper and middle panels), PCAF did not associate with any of both fragments of p27 indicating that the PID was lost with fragmentation. In contrast, we observed that PCAF associates with p27(amino acids 1–135) (Figure 2B, bottom panel). We subsequently studied the association of PCAF with the peptides p27(amino acids 91–120) and p27(amino acids 101–120). Results revealed that p27(91–120) associated with PCAF whereas p27(amino acids 101–120) did not (Figure 2C). These data indicated that the PID is included in the amino acids 91–120 region of p27. To further confirm that this region corresponds to the PID of p27 we generated three deletion mutants, specifically: p27(Δ91–100), p27(Δ96–100) and p27(Δ91–95) (Figure 2D). Purified mutant proteins were loaded onto a GST–HAT column and the interaction analyzed. As shown in Figure 2E when a large part of the PID domain was deleted as in the p27(Δ91–100) mutant, the association with PCAF was totally lost whereas shorter deletions as in p27(Δ96–100) and p27(Δ91–95) still retained the ability to interact with PCAF. Interestingly, this region includes a proline-rich domain (PRD) (amino acids 91–96) (Figure 2A) although as shown in Figure 2E its role in the association with PCAF is not essential. To validate the PID of p27 in the cells, we used HEK293T cells that were cotransfected with the deletion mutants p27(Δ91–100) or p27(Δ91–120) together with Flag–PCAF and subsequently subjected to IP with anti-Flag. Results clearly indicated that the interaction of PCAF with both mutants was strongly reduced (Figure 2F and G).
Figure 2.
The PID of p27 includes amino acids 91–120. (A) Graphic showing different fragments and domains of p27 used in the experiments. (B) Purified PCAF was loaded on GST–p27(1–110), GST–p27(110–198) and GST–p27(1–135) affinity chromatography columns. Then, the input (PC), the flow trough (FT), the last wash (W) and the eluate (E) fractions were subjected to electrophoresis and visualized by WB. (C) Purified PCAF was loaded on p27(101–120) and p27(91–120) affinity chromatography columns. As a control a Sepharose 4B column (Seph.) was used. Then, the flow trough (FT), the last wash (W) and the eluate (E) fractions were subjected to electrophoresis and visualized by WB. (D) Graphic showing the three p27 deletion mutants used in the experiments. (E) Purified p27(Δ91–100), p27(Δ96–100) and p27(Δ91–95) proteins were loaded on a GST–HAT column. Then, the flow trough (FT), the last wash (W) and the eluate (E) fractions were subjected to electrophoresis and visualized by WB. (F) HEK293T cells were transfected with YFP–Flag–PCAF plus p27WT or plus the deletion mutants p27(Δ91-100) or p27(Δ91–120) or woth an empty CFP vector (Ø). Then, cell extracts were subjected to IP with anti-Flag and the immunoprecipitates analyzed by WB with anti-p27 and anti-PCAF. (G) Experiment in (F) was repeated three times and quantified. Results were expressed as the mean value ± SD. Statistical analyses were performed using Student's two-tailed paired t-test. Values of P < 0.05 were considered statistically significant. ***P < 0.001.
PCAF acetylates p27 at Lys 100
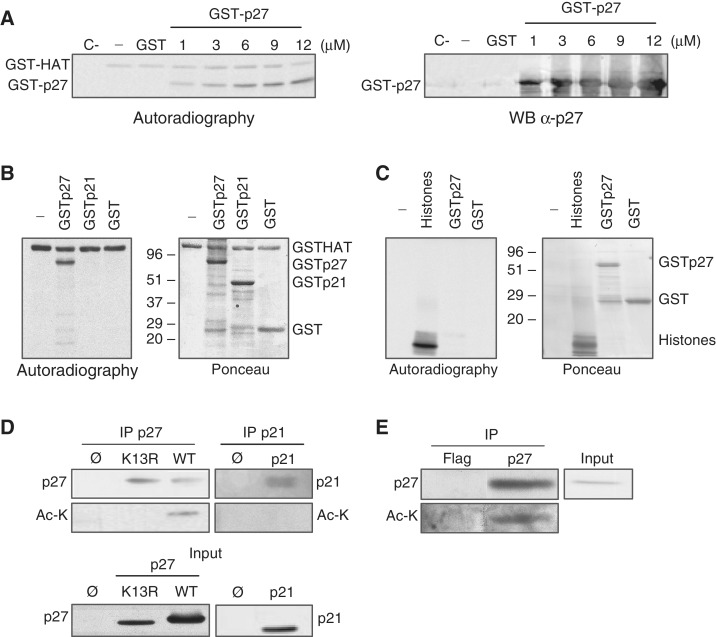
To study whether p27 is acetylated by PCAF, in vitro acetyltransferase assays were performed. Results indicated that p27 was acetylated in vitro by PCAF (Figure 3A). Similar experiments showed that PCAF was not able to acetylate p21Cip1, another member of the Cip/Kip family of inhibitors (Figure 3B). We also observed that p27 could not be acetylated by CBP, a protein belonging to another family of acetyltransferases (Figure 3C).
Figure 3.
PCAF acetylates p27. (A) In vitro acetylation assays were performed by incubating purified GST–HAT with the indicated concentrations of purified GST–p27 in the presence of radioactive acetyl-CoA. Control assays were performed with GST–p27 in the absence of GST–HAT (C−), with GST–HAT in the absence of any substrate (−) or in the presence of GST. Acetylation was examined by autoradiography (left panel). A WB using anti-p27 is also showed (right panel). (B) Purified GST–HAT was incubated with GST–p27, GST–p21 or GST in the presence of radioactive acetyl-CoA. A similar assay was performed in the absence of any substrate (−). Acetylation was examined by autoradiography (left panel). Red Ponceau stain of the membrane is showed in the right panel. (C) Purified GST–CBP was incubated with GST–p27, GST or histones in the presence of radioactive acetyl-CoA. A similar assay was performed in the absence of any substrate (−). Acetylation was examined by autoradiography (left panel). Red Ponceau stain of the membrane is shown in the right panel. (D) HEK293T cells were cotransfected with PCAF plus p27WT or p27K13R, or transfected with an empty vector (ø). Then, cell extracts were subjected to IP with anti-p27. The levels of total or acetylated p27 were examined by WB with anti-p27 or anti-acetyl-lysine antibodies (Ac-K), respectively (upper left panel). A similar experiment was performed using p21 instead of p27 (upper right panel). Expression levels of p27 and p21 were examined by WB (bottom panels). (E) Cell extracts were subjected to IP with anti-p27 or anti-Flag (used as a control) and then the immunoprecipitates were analyzed by anti-p27 and anti-acetyl-Lysine antibodies (Ac-K).
Next, we studied the acetylation of p27 in the cells by IP with anti-p27 followed by WB with anti-acetyl-Lys. To this aim, HEK 293T cells were transfected with CFP–p27 and PCAF. As a negative control, cells were transfected with a p27 mutant in which all lysines were substituted by arginines (p27–K13R). Results indicated that p27WT was acetylated whereas the mutant p27(K13R) did not (Figure 3D, left panel). Similar experiments revealed that p21 was not acetylated in the cells (Figure 3D, right panel). Finally, we also observed the acetylation of endogenous p27 by IP with anti-p27 followed by WB with anti-acetyl-Lys (Figure 3E).
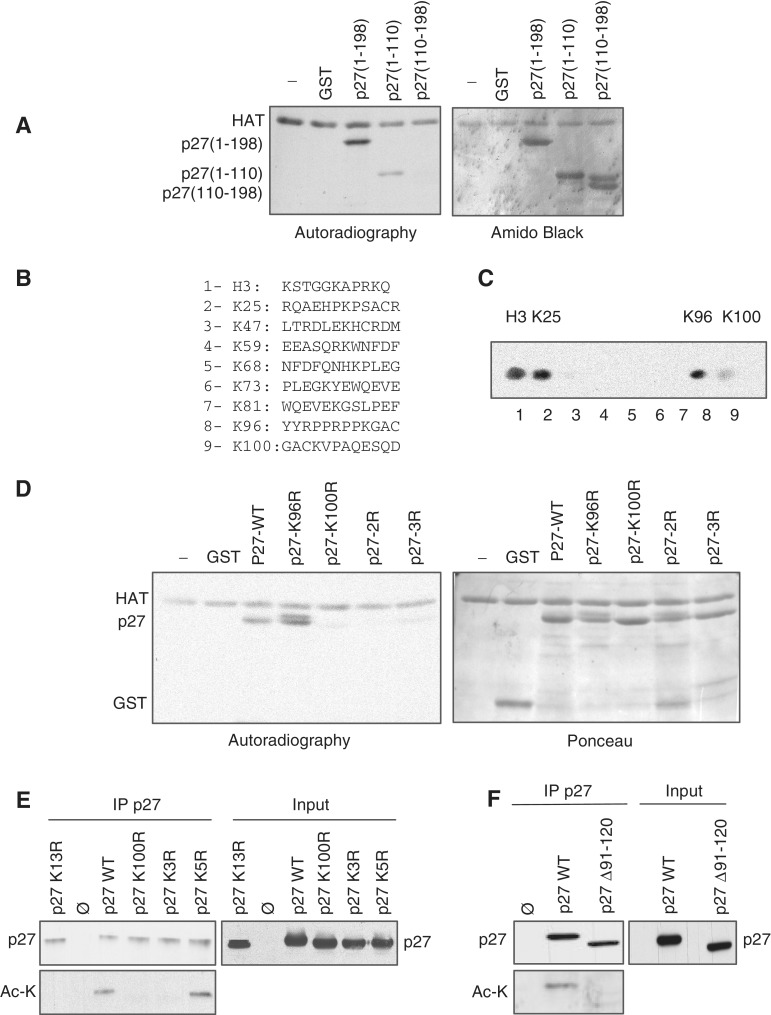
To identify the acetylation site/s of p27 we first performed in vitro acetylation assays using two p27 fragments, p27(1–110) and p27(110–198). Results indicated that p27(1–110) but not p27(110–198) was acetylated in vitro by PCAF (Figure 4A). The p27(1–110) fragment contains eight lysine residues at positions K25, K47, K59, K68, K73, K81, K96 and K100. To identify the residues that might be acetylated by PCAF, in vitro ‘spot mapping’ experiments were performed. Thus, eight peptides, each one containing one of the lysines present in this fragment, were synthesized and spotted on a nitrocellulose membrane. Additionally, a peptide from histone H3 was used as a positive control (Figure 4B). Then, this membrane was subjected to an in vitro acetylation assay using PCAF as acetylase. Results indicated that the control peptide and peptides containing K25, K96 and K100 were acetylated by PCAF (Figure 4C). To analyze whether these lysines were the acetylation sites in the full-length p27 protein, mutational analysis were performed. GST–p27 proteins harboring different K for R substitutions were used as substrates for in vitro acetylation assays. Results revealed that the triple mutant p27–K3R (K25R, K96R and K100R), the double mutant p27–2R (K96R and K100R) and the single mutant p27–K100R were not acetylated by PCAF (Figure 4D). These results indicated that K100 in p27 is the specific acetylation site for PCAF. Finally, acetylation assays were performed in HEK293T cells. Cells were transfected with different forms of p27 and with an empty vector as a control. Then, after an IP with anti-p27 the acetylation status of the protein was determined by WB with an anti-acetyl-lysine. Results revealed that p27WT was acetylated whereas the mutants p27–K3R or p27K100R did not (Figure 4E), indicating that K100 is the specific acetylation site of p27 also in the cells. As an additional positive control we also determined the acetylation status of the mutant p27–K5R (K47R, K59R, K68R, K73R and K81R) that was acetylated as it can be observed in Figure 4E. Finally, as a negative control we tested the acetylation of the mutant p27K13R and of the mutant p27(Δ91–120) that lacks the PID and K100. As shown in Figure 4E and F these mutants were not acetylated.
Figure 4.
PCAF acetylates Lys100 of p27. (A) Purified GST–HAT was incubated with GST, full-length GST–p27, GST–p27(1–110) or GST–p27(110–198), in the presence of radioactive acetyl-CoA. A similar assay was performed in the absence of any substrate (−). Acetylation was examined by autoradiography (left panel). Amido black stain of the membrane is showed in the right panel. (B) Amino acid sequences of peptides used for the identification of the in vitro acetylation sites of p27 by spot mapping analysis. Eight peptides, each one containing one of the lysines present in the N-moiety of p27, were spotted on a nitrocellulose membrane (peptides 2–9). A peptide from histone H3 was also spotted and used as a positive control (peptide 1). (C) The membrane containing the spotted peptides was subjected to an in vitro acetylation assay using GST–HAT as acetylase. Acetylation was examined by autoradiography. (D) Purified GST–HAT was incubated with GST, GST–p27WT, GST–p27–K96R, GST–p27–K100R, GSTp27–K96R,K100R (GST–p27-2R) or GST–p27–K25R,K96R,K100R (GST–p27–3R) in the presence of radioactive acetyl-CoA. A similar assay was performed in the absence of any substrate (−). Acetylation was examined by autoradiography (left panel). Red Pounceau staining of the membrane is showed in the right panel. (E) HEK293T cells were transfected with an empty vector (ø) or cotransfected with p27WT, p27-K13R, p27-K100R, p27-K3R or p27-K5R plus PCAF. Cell extracts were then subjected to IP with anti-p27 and the total levels of p27 or acetylated p27 were analyzed by WB with anti-p27 or anti acetyl-Lysine (Ac-K) antibodies (left panel). Expression levels of the different forms of p27 were determined by WB with anti-p27 (right panel). (F) Cells were transfected with an empty vector or cotransfected with p27WT or p27(Δ91–120) plus PCAF. Cell extracts were then subjected to IP with anti-p27 and the levels of p27 or acetylated p27 were analyzed by WB with anti-p27 or anti-acetyl-Lysine (Ac-K) antibodies (left panel). Expression levels of the different forms of p27 were determined by WB with anti-p27 (right panel).
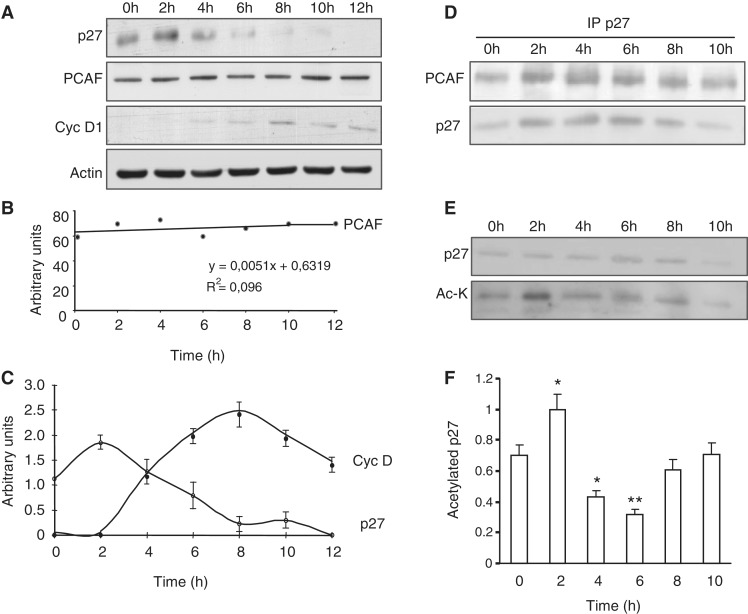
PCAF associates with and acetylates p27 along the cell cycle
We subsequently aimed to analyze the functional relevance of p27 acetylation. Thus, we first studied by WB the levels of PCAF along the cell cycle. HeLa cells were arrested in G0 by serum starvation (0 h) and later on they were induced to enter the cell cycle by serum addition. Then, at different times samples were collected and analyzed by WB with antibodies against PCAF. The levels of cyclin D1 and p27 were also analyzed to monitor cell-cycle entry whereas anti-actin was used as a loading control (Figure 5A). Results from three different WB with anti-PCAF were quantified and represented as a regression line (Figure 5B). Similarly, three different WB with anti-cyclin D1 and anti-p27 were quantified and represented in Figure 5C. These data revealed that the amount of PCAF remained constant at least during the first 12 h after cell-cycle entry. In contrast, levels of p27 were high in arrested cells (0 h) and progressively decreased whereas those of cyclin D1 increased along cell-cycle progression (Figure 5C). We subsequently determined by IP the association of p27 with PCAF at different times after serum addition. As shown in Figure 5D the association between both proteins was similar in quiescent cells and at different times after proliferative activation. Finally, by IP with anti-p27 followed by WB with anti-acetyl-lysine we determined the acetylation of p27 during cell cycle. Results revealed that p27 was slightly acetylated in quiescent cells (0 h) and then acetylation fluctuates: at 2 h it was significantly increased whereas later on at 4–6 h decreased to subsequently recuperate the initial levels of acetylation (Figure 5E). Data from three different experiments were quantified and represented in Figure 5F.
Figure 5.
The p27 protein is acetylated at early G1. (A) HeLa cells were made quiescent by serum starvation for 72 h and then re-plated in the presence of 10% FCS. Total cell extracts were prepared at the indicated times after cell-cycle resumption. The levels of PCAF and p27 were examined by WB with anti-PCAF and anti-p27. WB with anti-cyclin D1 was performed to monitor cell-cycle progression and WB with anti-actin was performed as a loading control. (B) PCAF levels from three different experiments described in (A) were quantified and expressed as a regression line. (C) cyclin D1 and p27 levels from three different experiments described in (A) were quantified and expressed as the mean value ± SD. (D) HeLa cell extracts from the indicated times were subjected to IP with anti-p27 and the levels of PCAF and p27 in the immunoprecipitates were determined by WB. (E) Extracts from HeLa cells at different times after cell-cycle resumption were subjected to IP with anti-p27. Then, the levels of p27 and those of acetylated p27 in the immunoprecipitates were assessed by WB with anti- p27 or with anti-acetyl-lysine (Ac-K), respectively. (F) The acetylation levels of p27 from three different experiments described in (E) were quantified and expressed as the mean value ± SD. Statistical analyses were performed using Student's two-tailed paired t-test. *P < 0.05, **P < 0.01.
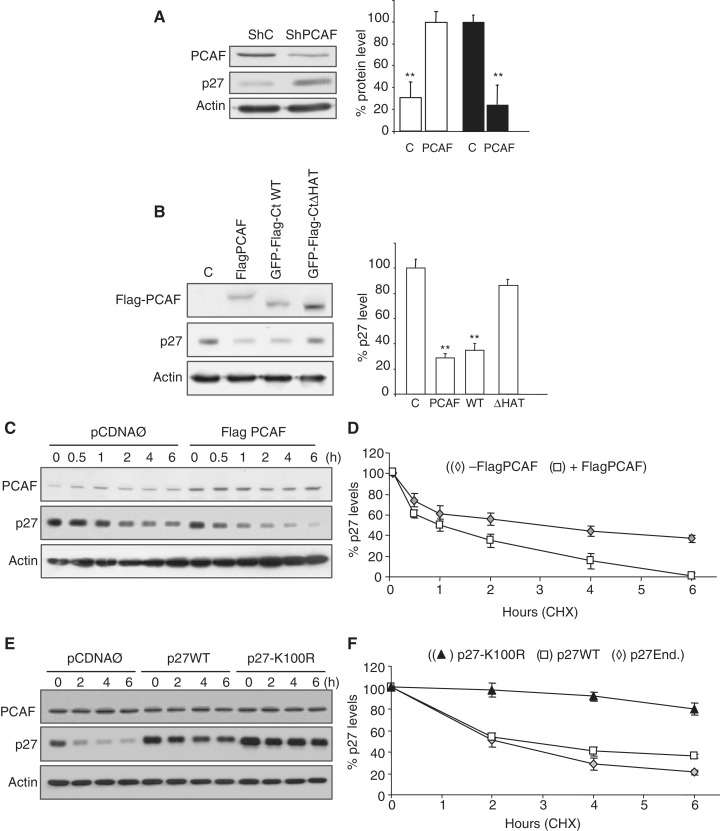
Acetylation of Lysine K100 regulates the Stability of p27
On comparing the kinetics of p27 levels (Figure 5C) and p27 acetylation (Figure 5F), we observed a correlation between both parameters during the first 6 h after serum addition to the cell cultures. Thus, we aimed to study whether acetylation of p27 could play a role on its stability. As shown in Figure 6A, knocking down of PCAF, in HCT116 cells, by a specific shRNA induced a significant increase of p27. Conversely, overexpression of Flag–PCAF induced a significant decrease of p27. Similar results were observed when cells overexpressed an active fragment of PCAF (GFP-CtWT). This decrease of p27 depended on the activity of PCAF because overexpression of an inactive form (GFP-CtΔHAT) did not significantly affect the levels of p27 (Figure 6B). These results suggested that PCAF could regulate the stability of p27. To further analyze this possibility, we determined the half-life of p27 in HCT116 cells overexpressing PCAF. Results revealed that the half-life of p27 in these cells was significantly decreased (1.1 ± 0.2 h) as compared to control cells (3.3 ± 0.4 h), thereby confirming the effect of PCAF on the downregulation of p27 (Figure 6C and D). We also observed that the half-life of the ectopically expressed non-acetylatable mutant p27-K100R was much higher (12.4 ± 0.5 h) than that of the endogenous (1.8 ± 0.2 h) or ectopic p27WT (1.9 ± 0.3 h) (Figure 6E and F), indicating that Lys100 was necessary for the acetylation and stability of p27
Figure 6.
PCAF induces p27 degradation. (A) HCT116 cells were knocked down for PCAF by using a specific shRNA whereas an unrelated shRNA was used as a control (shC). Then, the levels of PCAF and p27 were determined by WB with anti-PCAF and anti-p27. WB with anti-actin was used as a loading control (left panel). Results from three different experiments were quantified and expressed as the mean value ± SD. Statistical analyses were performed using Student's two-tailed paired t-test. **P < 0.01 (right panel). (B) HCT116 cells were transfected with expression vectors for GFP (C), Flag–PCAF, GFP-Flag-CtWT or for its inactive form GFP-Flag-CtΔHAT. At 24 h post-transfection, the levels of p27 and the ectopic Flag–PCAF were examined by WB with anti-p27 or anti-Flag, respectively. WB with anti-actin was performed as a loading control (left panel). Results from three different experiments were quantified and expressed as the mean value ± SD. Statistical analyses were performed using Student's two-tailed paired t-test. **P < 0.01 (right panel). (C) HCT116 cells were transfected with Flag–PCAF or with an empty vector (Ø) as a control. At 24 h post-transfection, cycloheximide (CHX) was added to the cell cultures and then, at the indicated times the levels of PCAF and p27 were examined by WB with an anti-PCAF or anti-p27, respectively. WB with anti-actin was performed as a loading control. (D) The intensities of p27 signals from three independent experiments were quantified, normalized for actin and plotted. Results are expressed as percentage relative to levels observed at Time 0. Statistical analyses were performed using Student's two-tailed paired t-test. In all points P < 0.05. (E) HCT116 cells were transfected with p27WT or p27-K100R. Transfection with the empty vector (Ø) was used as a control. At 24 h post-transfection, CHX was added to the cell cultures and then, at the indicated times the levels of endogenous PCAF and ectopic p27WT or p27-K100R were examined by WB with anti-PCAF or anti-p27, respectively. WB with anti-actin was performed as a loading control. (F) The intensities of p27 signals from three independent experiments were quantified, normalized for actin and plotted. Results are expressed as percentage relative to levels observed at Time 0. Statistical analyses were performed using Student's two-tailed paired t-test. In all points, when comparing ectopic p27WT or endogenous p27 versus p27K100R, P < 0.01.
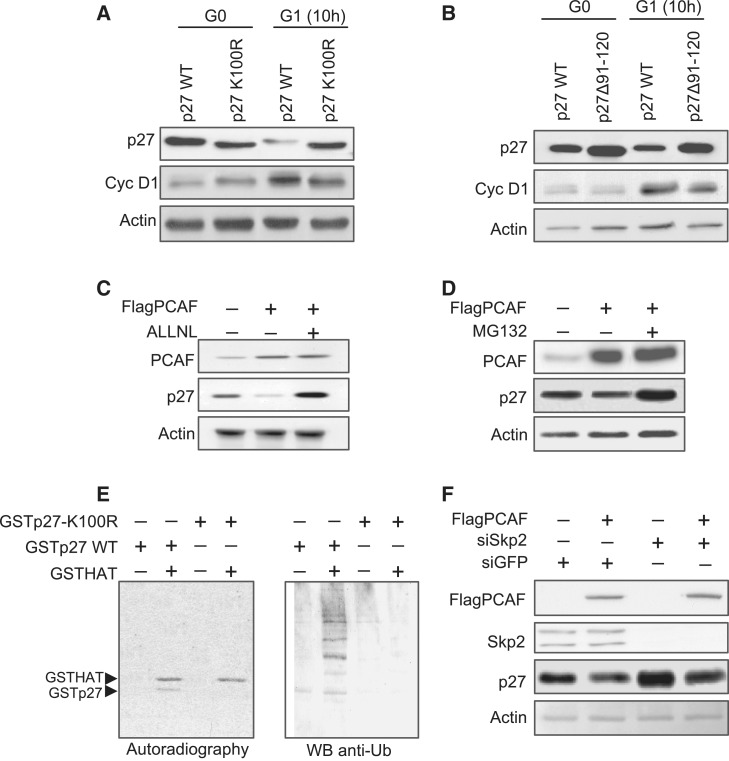
We subsequently analyzed the behavior of ectopic p27WT and p27K100R during cell cycle. HCT116 cells transfected with each one of these forms of p27 were made quiescent by culturing them with 0.5% FCS-containing media during 48 h. Then, they were re-plated and cultured with 10% FCS to induce cell-cycle entry. Cells were then collected at 10 h after serum addition. In these experiments cyclin D1 levels were determined to monitor cell-cycle progression and those of actin as a loading control. As shown in Figure 7A, at 10 h after proliferative activation, the levels of p27WT were decreased whereas those of p27K100R remained similar to the control. These results corroborate that the non-acetylatable mutant of p27 was more stable than p27WT at G1. Similar experiments were performed using the mutant p27(Δ91–120) unable to interact with PCAF and also non-acetylatable. In this case, we also observed that at G1, this mutant was more stable than p27WT.
Figure 7.
Acetylation of K100 in p27 induces its degradation via proteasome. (A) HCT116 cells were transfected with GFPp27WT or GFPp27K100R and then made quiescent by serum starvation for 48 h. Then, they were released from quiescence by serum addition. Cell samples were collected at Time 0 and at 10 h after re-entry into cell cycle. The levels of these two forms of p27 were detected by WB using anti-GFP. Cyclin D1 was detected by WB to monitor cell-cycle progression and WB with anti-actin was performed as a loading control. (B) HCT116 cells were transfected with GFPp27WT or GFPp27Δ91–120 and then made quiescent by serum starvation for 48 h. Then, they were released from quiescence by serum addition. Cell samples were collected at Time 0 and at 10 h after re-entry into cell cycle. The levels of these two forms of p27 were detected by WB using anti-GFP. Cyclin D1 was detected by WB to monitor cell-cycle progression and WB with anti-actin was performed as a loading control. (C) HCT116 cells were transfected with Flag–PCAF or with the empty vector as a control. At 8 h post-transfection, cells were additionally incubated for 16 h in the presence or in the absence of the proteasome inhibitor ALLN. Then, the levels of PCAF and p27 in the total cell extracts were examined by WB. WB with anti-actin was performed as a loading control. (D) HCT116 cells were transfected with Flag–PCAF or with the empty vector as a control. At 8 h post-transfection, cells were additionally incubated for 16 h in the presence or in the absence of the proteasome inhibitor MG132. Then, the levels of PCAF and p27 in the total cell extracts were examined by WB. WB with anti-actin was performed as a loading control. (E) PCAF stimulates the in vitro ubiquitylation of p27. Purified recombinant GSTp27WT or GSTp27-K100R proteins were incubated with radioactive acetyl-CoA and GSTHAT protein for in vitro acetylation reaction in the presence of reticulocyte lysates. After incubation, the proteins from the reticulocyte lysates were eliminated by purifying the GST-containing proteins with a glutathione column. The 50% of the material was analyzed for ubiquitylation by WB with anti-ubiquitin antibody (right panel). The other 50% of the material was used for the analysis of the acetylation of p27 by autoradiography (left panel). (F) The levels of endogenous Skp2 were down-regulated by incubating HCT116 cells with siRNA for Skp2 (siSkp2) during 48 h. Incubation with siRNA for GFP (siGFP) was used as a control. Then, cells were transfected with FlagPCAF expression vector. At 20 h post-transfection, the levels of Flag PCAF, Skp2, p27 and actin were determined in the cell extracts by WB using specific antibodies.
To analyze whether the effect of PCAF on the stability of p27 was depending on proteasome activity, we added ALLNL or MG132, specific inhibitors of the 26 S proteasome, to HCT116 cell cultures previously transfected with Flag–PCAF and then, we checked the levels of p27 protein. Results revealed that in the presence of any of both proteasome inhibitors, the overexpression of PCAF did not decrease the levels of p27 (Figure 7C and D).
We subsequently studied the effect of PCAF on p27 ubiquitylation. Thus, we performed an in vitro ubiquitylation assay using reticulocyte extracts and purified p27Kip1 protein in the presence or in the absence of active PCAF. Results revealed that acetylation by PCAF induced the ubiquitylation of p27 (Figure 7E). This ubiquitylation depends on Lys100 residue in p27 because the mutant p27-K100R was not ubiquitylated in the same experimental conditions (Figure 7E). Finally, we studied whether degradation of p27 induced by PCAF was mediated by Skp2, an F-box protein involved in the recognition of p27 facilitating its ubiquitylation by the SCFSkp2 complex (25). Thus, HCT116 cells were treated with siRNA for Skp2 and then transfected with FlagPCAF. Results indicated that the treatment with siRNA reduced Skp2 and increased the levels of p27 (Figure 7F, lanes 1 and 3). We also observed that the overexpression of PCAF decreased the levels of p27Kip1 in cells with normal amounts of Skp2, but also in cells with low levels of this protein, indicating that degradation of p27 induced by PCAF is independent of Skp2 (Figure 7F, lanes 2 and 4).
DISCUSSION
Here we investigated the role of acetylation on p27 stability. We describe that p27 directly interacts with PCAF forming complexes in the cells, mostly located in the nucleus. This is consistent with previous reports showing that PCAF essentially displays a nuclear localization (26). We provide evidence that p27 associates with the catalytic domain (HAT) of PCAF through a region including amino acids 91–120 that we named PID. Despite the deletion p27 mutant, lacking a region including amino acids 91–100 loses the ability to interact with PCAF (Figure 2E), the fact that PCAF did no interact with the p27NT(1–110) fragment indicates that the domain amino acids 110–120 is also necessary for the interaction and thus the full PID of p27 has to considered to include amino acids 91–120.
Interestingly, this region contains a PRD (amino acids 91–95) known to interact with proteins containing a Src homology 3 (SH3) domain. The PRD of p27 has been reported to interact with other proteins that are relevant for cell-cycle control as for instance Grb2 (27). These data are in agreement with previous observations showing that the HAT domain of GNC5, an acetyltransferase presenting high homology with PCAF, has a structure similar to an SH3 domain (28,29). Due to this high homology, it can be assumed that PCAF also has this structure that would facilitate the association with the PRD of p27. However, as shown in Figure 2E the interaction of PCAF with this domain of p27 is not critical because when it was deleted the association of PCAF with p27 was only slightly affected.
We further confirmed that the PID domain of p27 includes amino acids 91–120 by transfecting the deletion mutants p27(Δ91–100) or p27(Δ91–120) into the cells and analyzing their interaction with PCAF. As shown in Figure 2F and G, the interaction of PCAF with these mutants was strongly reduced. The residual interaction of PCAF with these mutants can be attributed to the indirect interaction through cyclin A–cdk2 complexes. We previously reported that PCAF directly interacts with both cyclin A and cdk2 (6,30). Since these p27 mutants (p27Δ91–100 and p27Δ91–120) still retain the ability to associate with cyclins and cdks, the IP with anti-Flag immunoprecipitate the cyclin A and cdk2 associated with PCAF and additionally the p27 associated with cyclin A and cdk2.
We identified K100 as the specific residue of p27 acetylated by PCAF in vitro and in the cells. K100 is very close to the PRD (amino acids 91–95) and it is included in the PID of p27. Thus, this is consistent with a scenario in which PCAF through its HAT domain associates with the PID of p27 and acetylates K100 included in this region.
Degradation of p27 is produced by ubiquitin-dependent proteolysis in a two-step process (31). The first step takes place during G0/G1 transition and early-mid G1, is independent of phosphorylation at T187 of p27 and it is mediated by the ubiquitin ligase KPC (32,33). However, the molecular signal triggering this degradation still remains unclear.
In contrast, the second proteolytic step is produced during G1/S transition and S and G2 phases and it is dependent on phosphorylation at T187 by cyclin E–Cdk2 (34,35). This second step is preceded by the phosphorylation of p27 at three specific tyrosine residues: Y74, Y88 and Y89. This allows to the p27-associated cyclin E–Cdk2 complexes to be partially activated. Then, these complexes can phosphorylate the T187 of p27 inducing its ubiquitin-dependent degradation (36,37). Interestingly, these tyrosine residues are very close to the PRD of p27. Moreover, the tyrosine kinases, members of the Src family (i.e. c-Src, Bcr-Abl, Lyn) that phosphorylate these p27 tyrosine residues have an SH3 domain that allows the interaction with the PRD of p27 (38–41). Collectively, these results indicate a key participation of the PID (a region including amino acids 91–120) of p27 in the regulation of its stability: by associating with PCAF and acetylating K100 and by interacting with tyrosine kinases that phosphorylate Y74, Y88 and/or Y89 thus allowing the further phosphorylation of T187.
We propose that degradation of p27 mediated by acetylation could operate during G1. We observed that the knockdown of PCAF increases the levels of p27 and conversely overexpression of PCAF decreases its amount. Moreover, we also observed that the half-life of p27 is decreased when PCAF is overexpressed and than that of the non-acetylatable mutant p27K100R is higher than that of p27WT. Moreover, as shown in Figure 7, we observed that differently to p27WT, the non-acetylatable mutants p27K100R and p27Δ91–120 were not degraded during G1.
We also report here several evidences indicating that the acetylation induced p27 degradation could be performed via proteasome. Specifically, we describe that proteasome inhibitors as ALLN and MG132 block the p27 degradation induced by PCAF overexpression and that p27 acetylation by PCAF induced its K100 dependent ubiquitylation, because the non-acetylatable p27K100R mutant was not ubiquitylated under the same experimental conditions (Figure 7E). The observation that the levels of acetylated p27 correlates with the levels of p27 during the first 6 h after mitogenic stimulation of the cells (Figure 5C and F) also supports the relationship between acetylation and degradation during G1. These results indicate that there is a basal acetylation of p27 in quiescent cells. A possibility is that this basal acetylation could be related to a basal turn over of the protein in the nucleus. At 2 h an increase of p27 acetylation was observed by WB. Interestingly, at this point the levels of p27 are still high (Figure 5C). This indicates that at this point degradation still has not been initiated and suggests that p27 is already targeted for its subsequent degradation that will immediately start. This is supported by the fact that at times 4–6 h the total levels of p27 are progressive decreasing concomitantly with the decrease of acetylated p27. This indicates that specifically is the acetylated form of p27 the one that is degraded at that time. Finally, at times 8–10 h the subsequent degradation of p27 is probably performed via phosphorylation of T187 via Skp2 that is produced later on at the late G1 and G1/S transition. At this point acetylation of p27 returns to its basal levels.
Interestingly, we also report here that the acetylation dependent degradation of p27 is independent of Skp2 (Figure 7F). These data also support the role of acetylation induced degradation of p27 during G1. Finally, it has been reported that a domain of p27 including amino acids 42–102 is necessary for the ubiquitylation and degradation of p27 mediated by the E-ubiquitin ligase KPC at G0/G1 (33). Our results indicating that K100 is included in this domain are compatible with the possibility that acetylation could signal p27 for recognition by KPC at G0/G1. Another mechanism involved in the degradation of p27 during G1 related to the activation of the Wnt–Cul4A pathway has been reported (42). However, the possibility that acetylation of p27 could participate in this mechanism remains to be explored.
The functional relevance of p27 acetylation on its role on transcription still remains to be explored. However, because p27 is a transcriptional repressor of its target genes, our data showing that acetylation at K100 by PCAF induces its degradation allows us to speculate that it would facilitate the expression of the p27-TGs. This speculation is compatible with the well-known role of PCAF as a transcriptional coactivator. Experiments to clarify this point are currently underway in our laboratory.
We also speculate that this new mechanism regulating p27 stability could be also relevant during oncogenesis. It is known that p27 is decreased in many human tumors correlating with a bad prognosis (43,44). In a number of cases, this decrease has been correlated with increased levels of Skp2 that mediates p27 degradation at S/G2. However, in other cases, this correlation could not be established. Results reported here allow us to speculate that the decrease of p27 levels in tumor cells could be also produced by alterations in its acetylation status.
In conclusion, we describe here a new mechanism regulating p27 stability that involves acetylation of K100 of p27. We postulate that this new mechanism could be relevant for the transcriptional regulatory role of p27.
FUNDING
The Ministerio de Ciencia e Innovación of Spain [SAF 2006-05212 and SAF 2009-07769 to O.B.] and [BFU2006-01493, CSD2006-00049 and BFU2009-11527 to M.A.M.B.]; the Instituto de Salud Carlos III: RETICS [RD06/0020/0010 to O.B.]; Generalitat de Catalunya [SGR 09-1382 to O.B.]. Funding for open access charge: Ministerio de Ciencia e Innovación of Spain [SAF 2009-07769].
Conflict of interest statement. None declared.
REFERENCES
- 1.Morgan DO. Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu. Rev. Cell Dev. Biol. 1997;13:261–291. doi: 10.1146/annurev.cellbio.13.1.261. [DOI] [PubMed] [Google Scholar]
- 2.Malumbres M, Barbacid M. Mammalian cyclin-dependent kinases. Trends Biochem. Sci. 2005;30:630–641. doi: 10.1016/j.tibs.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 3.Paternot S, Bockstaele L, Bisteau X, Kooken H, Coulonval K, Roger PP. Rb inactivation in cell cycle and cancer: the puzzle of highly regulated activating phosphorylation of CDK4 versus constitutively active CDK-activating kinase. Cell Cycle. 2010;9:689–699. doi: 10.4161/cc.9.4.10611. [DOI] [PubMed] [Google Scholar]
- 4.Blais A, Dynlacht BD. E2F-associated chromatin modifiers and cell cycle control. Curr. Opin. Cell Biol. 2007;19:658–662. doi: 10.1016/j.ceb.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Besson A, Dowdy SF, Roberts JM. CDK inhibitors: cell cycle regulators and beyond. Dev. Cell. 2008;14:159–169. doi: 10.1016/j.devcel.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 6.Mateo F, Vidal-Laliena M, Canela N, Zecchin A, Martinez-Balbas M, Agell N, Giacca M, Pujol MJ, Bachs O. The transcriptional co-activator PCAF regulates cdk2 activity. Nucleic Acids Res. 2009;37:7072–7084. doi: 10.1093/nar/gkp777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Echalier A, Endicott JA, Noble ME. Recent developments in cyclin-dependent kinase biochemical and structural studies. Biochim. Biophys. Acta. 2010;1804:511–519. doi: 10.1016/j.bbapap.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 8.Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 9.Porter PL, Malone KE, Heagerty PJ, Alexander GM, Gatti LA, Firpo EJ, Daling JR, Roberts JM. Expression of cell-cycle regulators p27Kip1 and cyclin E, alone and in combination, correlate with survival in young breast cancer patients. Nat. Med. 1997;3:222–225. doi: 10.1038/nm0297-222. [DOI] [PubMed] [Google Scholar]
- 10.Chu IM, Hengst L, Slingerland JM. The Cdk inhibitor p27 in human cancer: prognostic potential and relevance to anticancer therapy. Nat. Rev. Cancer. 2008;8:253–267. doi: 10.1038/nrc2347. [DOI] [PubMed] [Google Scholar]
- 11.Slingerland J, Pagano M. Regulation of the cdk inhibitor p27 and its deregulation in cancer. J. Cell Physiol. 2000;183:10–17. doi: 10.1002/(SICI)1097-4652(200004)183:1<10::AID-JCP2>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 12.Pagano M, Tam SW, Theodoras AM, Beer-Romero P, Del Sal G, Chau V, Yew PR, Draetta GF, Rolfe M. Role of the ubiquitin-proteasome pathway in regulating abundance of the cyclin-dependent kinase inhibitor p27. Science. 1995;269:682–685. doi: 10.1126/science.7624798. [DOI] [PubMed] [Google Scholar]
- 13.Catzavelos C, Bhattacharya N, Ung YC, Wilson JA, Roncari L, Sandhu C, Shaw P, Yeger H, Morava-Protzner I, Kapusta L, et al. Decreased levels of the cell-cycle inhibitor p27Kip1 protein: prognostic implications in primary breast cancer. Nat. Med. 1997;3:227–230. doi: 10.1038/nm0297-227. [DOI] [PubMed] [Google Scholar]
- 14.Belletti B, Nicoloso MS, Schiappacassi M, Chimienti E, Berton S, Lovat F, Colombatti A, Baldassarre G. p27(kip1) functional regulation in human cancer: a potential target for therapeutic designs. Curr. Med. Chem. 2005;12:1589–1605. doi: 10.2174/0929867054367149. [DOI] [PubMed] [Google Scholar]
- 15.Fero ML, Rivkin M, Tasch M, Porter P, Carow CE, Firpo E, Polyak K, Tsai LH, Broudy V, Perlmutter RM, et al. A syndrome of multiorgan hyperplasia with features of gigantism, tumorigenesis, and female sterility in p27(Kip1)-deficient mice. Cell. 1996;85:733–744. doi: 10.1016/s0092-8674(00)81239-8. [DOI] [PubMed] [Google Scholar]
- 16.Kiyokawa H, Kineman RD, Manova-Todorova KO, Soares VC, Hoffman ES, Ono M, Khanam D, Hayday AC, Frohman LA, Koff A. Enhanced growth of mice lacking the cyclin-dependent kinase inhibitor function of p27(Kip1) Cell. 1996;85:721–732. doi: 10.1016/s0092-8674(00)81238-6. [DOI] [PubMed] [Google Scholar]
- 17.Fero ML, Randel E, Gurley KE, Roberts JM, Kemp CJ. The murine gene p27Kip1 is haplo-insufficient for tumour suppression. Nature. 1998;396:177–180. doi: 10.1038/24179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Besson A, Hwang HC, Cicero S, Donovan SL, Gurian-West M, Johnson D, Clurman BE, Dyer MA, Roberts JM. Discovery of an oncogenic activity in p27Kip1 that causes stem cell expansion and a multiple tumor phenotype. Genes Dev. 2007;21:1731–1746. doi: 10.1101/gad.1556607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pippa R, Espinosa L, Gundem G, Garcia-Escudero R, Dominguez A, Orlando S, Gallastegui E, Saiz C, Besson A, Pujol MJ, et al. p27(Kip1) represses transcription by direct interaction with p130/E2F4 at the promoters of target genes. Oncogene. 2011 doi: 10.1038/onc.2011.582. December 19 (doi:10.1038/onc.2011.582; epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 20.Santos-Rosa H, Valls E, Kouzarides T, Martinez-Balbas M. Mechanisms of P/CAF auto-acetylation. Nucleic Acids Res. 2003;31:4285–4292. doi: 10.1093/nar/gkg655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Estanyol JM, Jaumot M, Casanovas O, Rodriguez-Vilarrupla A, Agell N, Bachs O. The protein SET regulates the inhibitory effect of p21(Cip1) on cyclin E-cyclin-dependent kinase 2 activity. J. Biol. Chem. 1999;274:33161–33165. doi: 10.1074/jbc.274.46.33161. [DOI] [PubMed] [Google Scholar]
- 22.Martinez-Balbas MA, Bannister AJ, Martin K, Haus-Seuffert P, Meisterernst M, Kouzarides T. The acetyltransferase activity of CBP stimulates transcription. EMBO J. 1998;17:2886–2893. doi: 10.1093/emboj/17.10.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bannister AJ, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 24.Canela N, Rodriguez-Vilarrupla A, Estanyol JM, Diaz C, Pujol MJ, Agell N, Bachs O. The SET protein regulates G2/M transition by modulating cyclin B-cyclin-dependent kinase 1 activity. J. Biol. Chem. 2003;278:1158–1164. doi: 10.1074/jbc.M207497200. [DOI] [PubMed] [Google Scholar]
- 25.Kossatz U, Dietrich N, Zender L, Buer J, Manns MP, Malek NP. Skp2-dependent degradation of p27kip1 is essential for cell cycle progression. Genes Dev. 2004;18:2602–2607. doi: 10.1101/gad.321004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blanco-Garcia N, Asensio-Juan E, de la Cruz X, Martinez-Balbas MA. Autoacetylation regulates P/CAF nuclear localization. J. Biol. Chem. 2009;284:1343–1352. doi: 10.1074/jbc.M806075200. [DOI] [PubMed] [Google Scholar]
- 27.Sugiyama Y, Tomoda K, Tanaka T, Arata Y, Yoneda-Kato N, Kato J. Direct binding of the signal-transducing adaptor Grb2 facilitates down-regulation of the cyclin-dependent kinase inhibitor p27Kip1. J. Biol. Chem. 2001;276:12084–12090. doi: 10.1074/jbc.M010811200. [DOI] [PubMed] [Google Scholar]
- 28.Poux AN, Cebrat M, Kim CM, Cole PA, Marmorstein R. Structure of the GCN5 histone acetyltransferase bound to a bisubstrate inhibitor. Proc. Natl Acad. Sci. USA. 2002;99:14065–14070. doi: 10.1073/pnas.222373899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schuetz A, Bernstein G, Dong A, Antoshenko T, Wu H, Loppnau P, Bochkarev A, Plotnikov AN. Crystal structure of a binary complex between human GCN5 histone acetyltransferase domain and acetyl coenzyme A. Proteins. 2007;68:403–407. doi: 10.1002/prot.21407. [DOI] [PubMed] [Google Scholar]
- 30.Mateo F, Vidal-Laliena M, Canela N, Busino L, Martinez-Balbas MA, Pagano M, Agell N, Bachs O. Degradation of cyclin A is regulated by acetylation. Oncogene. 2009;28:2654–2666. doi: 10.1038/onc.2009.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakayama KI, Nakayama K. Regulation of the cell cycle by SCF-type ubiquitin ligases. Semin. Cell Dev. Biol. 2005;16:323–333. doi: 10.1016/j.semcdb.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 32.Kamura T, Hara T, Matsumoto M, Ishida N, Okumura F, Hatakeyama S, Yoshida M, Nakayama K, Nakayama KI. Cytoplasmic ubiquitin ligase KPC regulates proteolysis of p27(Kip1) at G1 phase. Nat. Cell Biol. 2004;6:1229–1235. doi: 10.1038/ncb1194. [DOI] [PubMed] [Google Scholar]
- 33.Kotoshiba S, Kamura T, Hara T, Ishida N, Nakayama KI. Molecular dissection of the interaction between p27 and Kip1 ubiquitylation-promoting complex, the ubiquitin ligase that regulates proteolysis of p27 in G1 phase. J. Biol. Chem. 2005;280:17694–17700. doi: 10.1074/jbc.M500866200. [DOI] [PubMed] [Google Scholar]
- 34.Malek NP, Sundberg H, McGrew S, Nakayama K, Kyriakides TR, Roberts JM, Kyriakidis TR. A mouse knock-in model exposes sequential proteolytic pathways that regulate p27Kip1 in G1 and S phase. Nature. 2001;413:323–327. doi: 10.1038/35095083. [DOI] [PubMed] [Google Scholar]
- 35.Hara T, Kamura T, Nakayama K, Oshikawa K, Hatakeyama S, Nakayama K. Degradation of p27(Kip1) at the G(0)-G(1) transition mediated by a Skp2-independent ubiquitination pathway. J. Biol. Chem. 2001;276:48937–48943. doi: 10.1074/jbc.M107274200. [DOI] [PubMed] [Google Scholar]
- 36.Grimmler M, Wang Y, Mund T, Cilensek Z, Keidel EM, Waddell MB, Jakel H, Kullmann M, Kriwacki RW, Hengst L. Cdk-inhibitory activity and stability of p27Kip1 are directly regulated by oncogenic tyrosine kinases. Cell. 2007;128:269–280. doi: 10.1016/j.cell.2006.11.047. [DOI] [PubMed] [Google Scholar]
- 37.Chu I, Sun J, Arnaout A, Kahn H, Hanna W, Narod S, Sun P, Tan CK, Hengst L, Slingerland J. p27 phosphorylation by Src regulates inhibition of cyclin E-Cdk2. Cell. 2007;128:281–294. doi: 10.1016/j.cell.2006.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mayer BJ. SH3 domains: complexity in moderation. J. Cell Sci. 2001;114:1253–1263. doi: 10.1242/jcs.114.7.1253. [DOI] [PubMed] [Google Scholar]
- 39.Borriello A, Cucciolla V, Oliva A, Zappia V, Della RF. p27Kip1 metabolism: a fascinating labyrinth. Cell Cycle. 2007;6:1053–1061. doi: 10.4161/cc.6.9.4142. [DOI] [PubMed] [Google Scholar]
- 40.Kinoshita T, Miyano N, Nakai R, Yokota K, Ishiguro H, Tada T. Protein purification and preliminary crystallographic analysis of human Lyn tyrosine kinase. Protein Expr. Purif. 2008;58:318–324. doi: 10.1016/j.pep.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 41.Vervoorts J, Luscher B. Post-translational regulation of the tumor suppressor p27(KIP1) Cell Mol. Life Sci. 2008;65:3255–3264. doi: 10.1007/s00018-008-8296-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miranda-Carboni GA, Krum SA, Yee K, Nava M, Deng QE, Pervin S, Collado-Hidalgo A, Galic Z, Zack JA, Nakayama K, et al. A functional link between Wnt signaling and SKP2-independent p27 turnover in mammary tumors. Genes Dev. 2008;22:3121–3134. doi: 10.1101/gad.1692808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsihlias J, Kapusta L, Slingerland J. The prognostic significance of altered cyclin-dependent kinase inhibitors in human cancer. Annu. Rev. Med. 1999;50:401–423. doi: 10.1146/annurev.med.50.1.401. [DOI] [PubMed] [Google Scholar]
- 44.Blain SW, Scher HI, Cordon-Cardo C, Koff A. p27 as a target for cancer therapeutics. Cancer Cell. 2003;3:111–115. doi: 10.1016/s1535-6108(03)00026-6. [DOI] [PubMed] [Google Scholar]