Abstract
Upf1 is a highly conserved RNA helicase essential for nonsense-mediated mRNA decay (NMD), an mRNA quality-control mechanism that degrades aberrant mRNAs harboring premature termination codons (PTCs). For the activation of NMD, UPF1 interacts first with a translation–terminating ribosome and then with a downstream exon–junction complex (EJC), which is deposited at exon–exon junctions during splicing. Although the helicase activity of Upf1 is indispensable for NMD, its roles and substrates have yet to be fully elucidated. Here we show that stable RNA secondary structures between a PTC and a downstream exon–exon junction increase the levels of potential NMD substrates. We also demonstrate that a stable secondary structure within the 3′-untranslated region (UTR) induces the binding of Upf1 to mRNA in a translation-dependent manner and that the Upf1-related molecules are accumulated at the 5′-side of such a structure. Furthermore, we present evidence that the helicase activity of Upf1 is used to bridge the spatial gap between a translation–termination codon and a downstream exon–exon junction for the activation of NMD. Based on these findings, we propose a model that the Upf1-related molecular motor scans the 3′-UTR in the 5′-to-3′ direction for the mRNA-binding factors including EJCs to ensure mRNA integrity.
INTRODUCTION
RNA helicases comprise a large family of enzymes that are thought to unwind double-stranded RNA molecules through the hydrolysis of NTP and participate in a variety of essential physiological processes. Several members of the family have been demonstrated in vitro to unwind RNA duplexes and translocate unidirectionally as processive molecular motors along the RNA track (1). Despite tremendous biochemical studies that revealed much about these enzymatic properties, the mechanisms through which RNA helicases exert influences on a variety of physiological processes are poorly understood.
Upf1 is a highly conserved RNA helicase that is essential for nonsense-mediated mRNA decay (NMD), an mRNA quality-control mechanism that protects eukaryotic cells from the potentially deleterious effects of truncated proteins (2,3). NMD has evolved to recognize and specifically degrade aberrant mRNAs whose open reading frames (ORFs) are truncated by the presence of premature termination codons (PTCs) (3,4). NMD is important not only for the suppression of mutated gene expression, but also for the regulation of physiological gene function during development and homeostasis [reviewed in (5)]. The NMD pathway is activated through a translation-dependent mechanism, in which trans-acting factors including Upf1 are recruited to a translation–terminating ribosome (6,7). Previous studies demonstrated that the purified Upf1 protein exhibits the RNA-dependent ATPase activity that leads to the unwinding of RNA duplexes in vitro (8–10), and mutations affecting its ATPase/helicase activity impair NMD (8,11). Although a recent report indicates that the helicase activity of Upf1 promotes disassembly of mRNP undergoing the final mRNA-degradation steps of NMD (12), the roles and substrates of the Upf1 helicase activity during the induction phase of NMD still remain unkonwn.
In vertebrates, a translation–termination codon located >50–55 nt upstream of an exon–exon junction is generally recognized as premature (13,14), based on the crucial function of the exon–junction complex (EJC) (15,16). The EJC is deposited during splicing at 20–24 nt upstream of each exon–exon junction as the signature of intron removal (15,16) and displaced from an mRNA by a translating ribosome in the cytoplasm (17,18). While all EJCs are removed from normal mRNAs during the first round of translation, the PTC-containing mRNAs, in which the ribosome does not pass all of the exon–exon junctions, remain associated with EJCs during the translation–termination process (17,18). According to the current model of the EJC-dependent NMD, Upf1 is first recruited to a translation–terminating ribosome as a component of the SMG1-Upf1-eukaryotic release factor 3 (eRF3)–eRF1 (SURF) complex, and the subsequent interaction between the Upf1-containing SURF complex and the downstream EJCs, with which two of the other essential NMD factors Upf2 and Upf3 are associated, activates the mRNA-degradation pathway (7,10,19). Several lines of evidence indicate that the interaction between the SURF complex and the EJC is a highly efficient and accurate process. Even if the distance between a PTC and a downstream exon–exon junction is extremely long (e.g. 1–5 kb), the mRNA undergoes NMD, indicating the presence of a mechanism to bridge the spatial gap between a translation–termination site and a downstream exon–exon junction on an mRNA molecule (20,21).
In this article, we show that stable RNA secondary structures inserted between a PTC and a downstream exon–exon junction operate as cis-acting elements to increase the levels of potential NMD targets. We also demonstrate that the general translation–termination processes induce the binding of Upf1 to mRNAs containing a stable secondary structure within the 3′-untranslated regions (UTRs) in a splicing-independent manner and reveal the accumulation of the Upf1-related molecule(s) at the 5′-side of the secondary structure. We further show that the helicase activity of Upf1 is required for the activation of NMD only when a PTC is located significantly apart from a downstream exon–exon junction. Based on these findings, we propose a model, in which Upf1 acts as a helicase-driven molecular motor that translocates along the 3′-UTR of an mRNA molecule in the 5′-to-3′ direction, scanning for the mRNA-binding factors including EJCs to ensure mRNA integrity.
MATERIALS AND METHODS
Plasmids
Plasmids for expression of the enhanced green fluorescent protein (EGFP) reporter genes were generated by inserting a splicing unit derived from the mouse Hprt1 gene (exon 8–intron 8–exon 9) into the multi-cloning site downstream of EGFP–ORF in pEGFP–C1 (Clontech). The internal ribosome entry site (IRES) and its downstream sequence are derived from the UPATrap vector (22) and the hp7 stem loop is generated by using synthetic oligonucleotides. For secreted alkaline phosphatase (SEAP) reporter plasmids, the SEAP ORF was inserted into the 3′-exon of the EGFP reporter constructs, and the intron was removed to avoid the effect of NMD. Spacer fragments of various lengths between the EGFP–ORF and the exon–exon junction are derived from mouse Hprt1 cDNA. Nucleotide sequences of the 3′-UTR of model mRNAs are shown in Supplementary Data. The free energy of each RNA secondary structure within the model mRNAs was calculated using the CentroidFold algorithm (http://www.ncrna.org/centroidfold).
Quantification of mRNAs and siRNA transfection
For analysis of reporter mRNAs containing various elements downstream of the PTC, we prepared polyclonal transfectants of HEK293 cells stably expressing the reporter mRNAs to avoid the construct-specific negative effects that are sometimes observed in our transient transfection experiments, probably due to the stable secondary structures in the constructs. The HEK293 cells were cotransfected with the linearized pGK-hygro vector and one of the plasmid DNAs for reporter mRNAs. The transfected cells were selected in medium containing hygromycin B, and the resulting 150–300 colonies were mixed and subjected to several rounds of passages before subsequent analysis in order to exclude the positional effect of the various vector integration sites. RNA was extracted with the RNeasy Plus kit (Qiagen) or Sepasol RNA 1 (Nacalai), and cDNA was synthesized using the Moloney murine leukemia virus (MMLV) reverse transcriptase (RT) (Invitrogen). For NMD inhibition experiment, we treated 80% confluent cells with 100 μg/ml of emetine dihydrochloride hydrate (Sigma) at 37°C for 8 h before preparation of total RNA. To quantify the levels of spliced transcripts, real-time RT–PCR was carried out by using Power SYBR Green PCR Master Mix (Applied Biosystems) or LightCycler 480 SYBR Green I Master (Roche) on ABI7700 (Applied Biosystems) or on LC480 (Roche). For analysis of the stable transfectants, values in real-time RT–PCR were normalized by the amount of insert DNA contained in the genomic DNA samples by using the primers for EGFP. To quantify reporter mRNAs with various lengths of spacer sequences in transiently transfected HeLa cells, the levels of reporter mRNAs were normalized by real-time RT–PCR using the primers designed for the unique 3′-UTR sequence of the NEO cassette in the plasmids. Nucleotide sequences of the primers are available in Supplementary Table S1. Poly(A) RNA was isolated by Oligotex-dT30 super (Takara), and northern blot analysis was performed with digoxigenin (DIG) probes for EGFP and SDF1 according to the manufacturer’s protocol (Roche Applied Science). Transient transfections of siRNAs were carried out at a final concentration of 100 nM, using the Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s instructions. Twenty-four hours later, cells were transfected again with 100 nM siRNAs. Forty-eight hours later (72 h after the first transfection), cells were harvested for analysis. The siRNA oligonucleotides used for transfections [Luciferase (CGUACGCGGAAUACUUCGAdTdT) and hUPF1 (GAUGCAGUUCCGCUCCAUUdTdT)] were purchased as annealed, ready-to-use duplexes from Dharmacon.
Flow cytometric and chemiluminescent analyses
The flow cytometric analysis of EGFP-expressing cells was performed using FACS Calibur (Becton Dickinson). Viable cells were pre-gated in FSC/SSC channels, and the EGFP fluorescence was analyzed on the fluorescence channel-1 (FL-1). For the reporter analysis, we detected SEAP in the culture supernatant of the transfected cells by using Great EscAPe SEAP Chemiluminescence Kit 2.0 (TAKARA) and Lumat LB 9501(Berthold, TN).
RNA immunoprecipitation
Cells were lysed in a lysis buffer [50 mM Tris–HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1 U/μl RNasin Plus (Promega) and 1% Triton X-100] and then incubated with anti-FLAG M2 beads (Sigma) for 2 h at 4°C. The bead–RNP complexes were washed two times in the 1 ml lysis buffer and one time in the 1 ml TBS buffer (50 mM Tris–HCl, pH 7.6 and 150 mM NaCl). RNA was extracted with the RNeasy Plus kit and subjected to real-time RT–PCR. The immunoprecipitation (IP) efficiency of GAPDH mRNA was used as an internal control to confirm that all of the samples were equally treated. Nucleotide sequences of the primers are available in Supplementary Table S1.
Oligonucleotide-targeted RNase H protection assay
HEK293T cells were lysed in an assay buffer [0.4% Triton X-100, 2.5 mM MgCl2, 130 mM KCl, 25 mM Tris–HCl, pH 8.0, 1 mM DTT, protease inhibitor cocktail (Roche) and 1.5 U/μl RNasin Plus] at 4°C and then centrifuged. An amount of 25 μl of the supernatant was mixed with 25 μl of the reaction buffer [40 U RNase H (Takara), 1 μg oligonucleotide DNA, 30 mM Tris–HCl, pH 8.0, 3 mM MgCl2, 150 mM KCl and 1 mM DTT] and was incubated for 1 h at 37°C. RNA was extracted using the RNeasy Plus kit and was reverse-transcribed using MMLV-RT. For this assay, the indicator mRNAs were designed to minimize secondary structures within the target sites for oligonucleotides annealing because such structures hamper the accessibility of the oligonucleotides to the RNA molecules. The sequence data for the target RNA, DNA oligonucleotides and primers is available in Supplementary Data.
In vitro translation, pull-down of protein-biotinylated RNA complexes and immunoblotting
Capped and poly(A)-tailed biotin-RNA was synthesized using the mMESSAGE mMACHINE T7 Ultra kit (Ambion) in the presence of biotin-11-UTP. In vitro translation was carried out in 72% (v/v) of the nuclease-treated Flexi rabbit reticulocyte lysate (RRL) (Promega) in the presence of 2.0 mM DTT, 520 μM MgOAc, 72 mM KCl, the amino-acid mixure and RNasin Plus at 30°C for 40 min. The RRL was pretreated with either nuclease free water or cycloheximide, and pre-cleaned with magnetic streptavidin-conjugated Dynabeads M-280 (Dynal) before addition of RNA. The translation was stopped by cooling the reaction on ice and adding a buffer (50 mM Tris–HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA and 1% Triton X-100). Complexes were isolated by incubating them with Dynal M-280 for 1 h at 4°C. After washing the isolated bead-RNP, proteins were eluted by boiling with an SDS loading buffer and analyzed by SDS–PAGE. Proteins transferred on PVDF membrane were probed with primary and secondary HRP-conjugated antibodies (GE Healthcare), and developed using the ECL Advance (GE Healthcare). Protein band intensities were quantified using Multi Gauge Version 3.2 (Fujifilm). The antibodies used are as follows: anti-RENT1 (Upf1) (Bethyl Laboratories), anti-eRF1 (Sigma), anti-eRF3 (Abcam), anti-RENT2 (Upf2) and anti-PABP (Santa Cruz Biotechnology) and anti-Y14 (ImmuQuest).
Preparation of proteins
HEK293T cells were transfected with the vector expressing FLAG-tagged human Upf1 by using Polyfect (Qiagen), and cell lysates prepared were incubated with 800 μl of FLAG M2 beads at 4°C for 2 h. The beads were washed with TBS, and Upf1 proteins were eluted from the resin by incubation with the FLAG peptide (Sigma) for 30 min at 4°C in TBS. The volume of the eluate was reduced to 100 μl by using Centriprep and Microcon YM3 (Millipore) sequentially.
RESULTS
Stable RNA secondary structures between a PTC and an exon–exon junction increase the levels of PTC-containing mRNAs
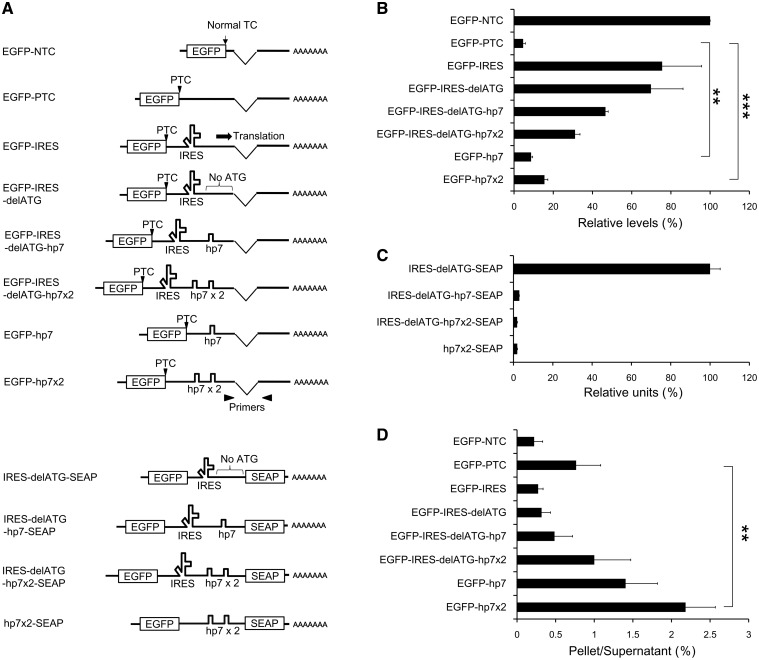
We reported previously that the IRES sequence of encephalomyocarditis virus (EMCV) inserted between a PTC and downstream exon–exon junctions protects the mRNA from NMD (22). To study the underlying mechanism, we prepared a set of reporter constructs containing various elements inserted between the ORF of EGFP and a downstream exon–exon junction (Figure 1A). The mRNA level of the EGFP–PTC construct with a translation–termination codon located >200 nt upstream of the exon–exon junction was significantly lower than that of the EGFP–NTC construct carrying a termination codon at only 45 nt upstream of the exon–exon junction, suggesting that the steady-state levels reflect the mRNA degradation due to NMD (Figure 1B). As we observed previously (22), EGFP–IRES, containing EMCV–IRES between the EGFP–ORF and the downstream exon–exon junction, showed marked recovery in its mRNA level compared to that of EGFP–PTC (Figure 1B and Supplementary Figure S1A). Interestingly, EGFP–IRES–delATG, in which all ATG triplets between IRES and the downstream exon–exon junction were mutated (Supplementary Figure S2), also showed the increased level of the PTC-containing mRNA (Figure 1B), suggesting that a mechanism independent of the IRES-mediated internal translation is responsible for the upregulation of the PTC-containing mRNA. Since a previous study indicated that the internal translation from EMCV–IRES is initiated with the scanning activity of the ribosomal small subunit (23), we investigated the relationship between the level of the PTC-containing mRNA and the translocation of the small subunit by introducing a stable stem-loop structure (hp7), which was reported to completely block the movement of the small subunit (24,25). To confirm the effect of hp7, we monitored the expression of the SEAP reporter in culture supernatant of the cells transfected with one of the plasmids, each of which contains the SEAP ORF downstream of the ATG-less IRES (Figure 1A). Although the insertion of one or two copies of hp7 between the ATG-less IRES and the SEAP ORF completely eliminate the SEAP activity (Figure 1C), it does not abolish the IRES-mediated upregulation of the PTC-containing mRNAs (Figure 1B and Supplementary Figure S1A), indicating that the EMCV–IRES increases the levels of the PTC-containing mRNAs by a mechanism independent of the internal translation or the translocation of the ribosomal small subunit.
Figure 1.
Stable RNA secondary structures between a PTC and a downstream exon–exon junction operate as cis-acting elements that increase the levels of the potential NMD substrates. (A) Diagram of the reporter mRNAs with different 3′-UTRs. The arrowheads represent the primer pairs used in real-time RT–PCR. (B) The steady-state levels of reporter mRNAs expressed in stably transfected polyclonal HEK293 cells as measured by the real-time RT–PCR. The data, expressed as mean ± SD (n = 3), represent percentages relative to EGFP–NTC. (C) The result of the SEAP-reporter analysis using culture supernatants of transfected 293T cells. The data, expressed as mean ± SD (n = 2), represent relative luminescence units. (D) The result of RNA immunoprecipitation from the lysate of HEK293 cells transiently expressing FLAG-Y14. The data, expressed as mean ± SD (n = 3), represent the ratios (IP pellet/post-IP supernatant) of the mRNA levels. Statistical analyses were done with unpaired two-tailed Student’s t-test (**P < 0.01; ***P < 0.001).
We next examined the possibility that the RNA secondary structure between the PTC and the exon–exon junction might be important for the upregulation of the PTC-containing mRNAs since the EMCV–IRES sequence could generate a highly complex form at the RNA level (ΔG = −199 kcal/mol). To investigate the effect of a secondary-structure element other than IRES on the levels of mRNAs, we tested constructs containing hp7 (but lacking IRES) downstream of the PTC because hp7 itself forms a stable secondary structure (40 nt, ΔG = −45 kcal/mol). The mRNA containing a single copy of hp7 (EGFP–hp7) showed a modest but significant recovery, and the mRNA with two copies of hp7 (EGFP–hp7x2) attained a more marked increase in its level (Figure 1B and Supplementary Figure S1A), supporting the hypothesis that RNA secondary structures between the PTC and the exon–exon junction upregulate the PTC-containing mRNA. Furthermore, the flow cytometric analysis of the EGFP fluorescence in the cells expressing one of the reporter constructs (Figure 1A) confirmed that the stable RNA secondary structures increase the levels of translation-competent mRNAs (Figure 1B and Supplementary Figure S3).
Stable RNA secondary structures in the model RNAs operate as cis-acting elements to suppress NMD
To investigate whether the upregulation of the PTC-containing mRNAs by the cis-acting elements is due to the suppression of NMD, we analyzed the levels of these mRNAs under condition in which NMD is inhibited by a translational blocker or a siRNA against Upf1. As previously shown (26), treatment of the cells with a translational blocker, emetine, increased the levels of the PTC-containing mRNAs more effectively than siRNA-mediated knockdown of Upf1, and such procedures equalized the levels of model mRNAs (Supplementary Figure S4). These results demonstrate that NMD is actually responsible for the reduced mRNA levels of the PTC-containing model mRNAs and suggest that the stable secondary structures in the model RNAs operate as cis-acting elements to suppress NMD.
Next, we asked whether these cis-acting elements affect the EJC formation/removal or not, since the EJCs retained downstream of a PTC is one of the key features of NMD substrates in mammalian cells (17,18). To investigate the association of the model mRNAs with EJCs, we performed the RNA immunoprecipitation using the cells coexpressing one of the reporter constructs and FLAG-tagged Y14, a core component of the EJC (16,17) (Supplementary Figure S5). While none of the EGFP–IRES nor EGFP–IRES–delATG mRNA showed the significant level of coprecipitation with Y14, inserting one or two copies of hp7 downstream of the ATG-less IRES resulted in a significant increase of Y14 association (Figure 1D). This can be explained by assuming that the EJCs could be removed by the translating ribosome or the translocation of the ribosomal small subunit originated from the IRES, and the insertion of hp7 would substantially block it. Remarkably, mRNAs containing one or two copies of hp7 but not the IRES element associated with Y14 much more efficiently than did the other mRNAs (Figure 1D), suggesting that the insertion of secondary structures between a PTC and an exon–exon junction increases the levels of potential NMD substrates through a mechanism independent of the EJC removal.
A stable RNA secondary structure within the 3′-UTR elicits the binding of Upf1 in a translation-dependent manner
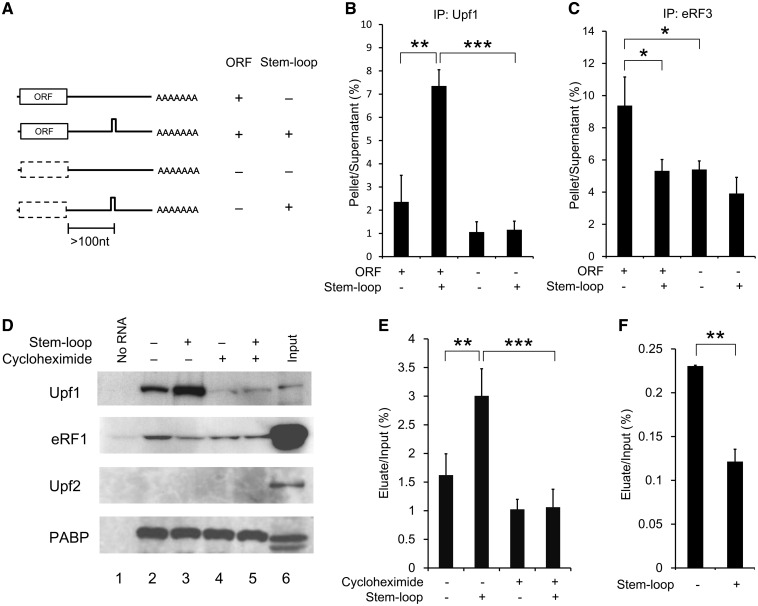
To elucidate the molecular mechanism underlying the NMD suppression, we focused on the influence of RNA secondary structures within the 3′-UTR on the core NMD factors. We first investigated the association of the Upf1 protein with intron-less model mRNAs that contain the ORF of the mouse SDF-1 gene and a 3′-UTR with or without a stable stem-loop structure (66 nt, ΔG = −81 Kcal/mol) (Figure 2A and Supplementary Figures S1B and S2). Since the SDF-1-ORF has only two ATG triplets in any of the three reading frames, the elimination of these ATGs directly allows us to evaluate the significance of translation in the assay system (Supplementary Figure S2). After the RNA immunoprecipitation using the lysate of the HeLa cells expressing a FLAG-tagged human Upf1 protein and one of the model mRNAs (Supplementary Figure S5), the mRNA levels in IP pellets (Ppt) and post-IP supernatants (Sup) were determined by real-time RT–PCR to evaluate the binding efficiency (Ppt/Sup). As a result, we found that the mRNA containing a stable stem-loop structure was associated with Upf1 at a significantly higher rate than the mRNA lacking such a structure (Figure 2B). In addition, the result of the analysis on the mRNA without the protein-coding capability clearly indicated that the preferential association of Upf1 depends on prior mRNA translation (Figure 2B). On the other hand, FLAG-Y14 did not show significant association with any of these intron-less model mRNAs (Ppt/Sup < 0.5%, data not shown), confirming the specificity of the RNA immunoprecipitation.
Figure 2.
A stable RNA secondary structure within the 3′-UTR elicits the binding of Upf1 in a translation-dependent manner. (A) Schematic representation of model mRNAs containing mouse SDF-1-ORF with different 3′-UTRs. The dashed box indicates the SDF-1 coding region harboring mutations in the two ATG codons, which disrupt all ATG codons upstream of the stem-loop structure (Supplementary Figure S2). (B) The result of RNA immunoprecipitation using the HeLa cells transiently expressing FLAG-Upf1 and the model mRNA with (ORF−) or without (ORF+) mutations in the two ATG codons. (C) The result of RNA immunoprecipitation using the HEK293T cells transiently expressing FLAG-eRF3 and the model mRNA. The immunoprecipitation data, expressed as mean ± SD (n = 3), represent the ratios (IP pellet/post-IP supernatant) of the mRNA levels. (D) Immunoblot analyses after the pull-down experiments of protein-biotinylated RNA complexes formed during in vitro translation. Proteins were eluted and analyzed by immunoblot using antibodies against indicated proteins. Lanes 1, control samples eluted from streptavidin-beads incubated in RRL without the RNA. Lanes 2–5, protein samples eluted from the RNP–bead complexes after incubation in RRL containing the biotinylated RNA with (lanes 3 and 5) or without (lanes 2 and 4) the stem-loop structure, in the presence (lanes 4 and 5) or absence (lanes 2 and 3) of cycloheximide. Lanes 6, input (0.12% volume of lysate was loaded). (E and F) The ratios of the Upf1 protein (E) or eRF1 protein (F) recovered in the pull-down eluates to the total inputs were calculated from the intensity of immunoblot bands. Statistical analyses were done with unpaired two-tailed Student’s t-test (*P < 0.05; **P < 0.01; ***P < 0.001).
Next, to examine whether the preferential association of Upf1 with mRNAs containing stable stem-loop structures within the 3′-UTR is universally coupled to eukaryotic translation termination, we utilized an in vitro translation system derived from RRL. We produced the biotinylated model mRNAs in vitro that contained the stem-loop structure (Supplementary Figure S1D), subjected them to the in vitro translation in RRL, and carried out a pull-down analysis of the protein–mRNA complex by using streptavidin-coated magnetic beads. Through the western blotting of poly(A)-binding protein (PABP) eluted from the beads, we confirmed that the samples were equally treated (Figure 2D). The results of the same pull-down experiments followed by the detection using anti-Upf1 antibody clearly showed that endogenous rabbit Upf1 associated at a higher rate with the mRNA containing a stem-loop in the 3′-UTR than with the mRNA lacking such a structure (Figure 2D and E). In the presence of a translational inhibitor cycloheximide, the Upf1 binding was significantly impaired (Figure 2D and E), as in the case of the ORF-less mRNAs (Figure 2A and B). Furthermore, the RNA-immunoprecipitation experiment using RRL that had been supplemented with the purified FLAG-Upf1 protein also showed the same pattern of Upf1 binding (Supplementary Figure S6). On the other hand, we could not detect any mRNA binding for endogenous Upf2, which is known to be recruited to the EJC platform after the nuclear export of mRNPs in mammalian cells (Figure 2D). These results suggest that the association of the Upf1 with mRNAs containing a stable secondary structure within the 3′-UTR occurs independently of splicing or the activation of NMD, and is coupled to general translation–termination processes. Interestingly, the pull-down experiment with RRL revealed that the secondary structure in the 3′-UTR reduced the binding of eRF1 to the mRNA, in accordance with the results of the in vivo immunoprecipitation experiments, in which we observed basically the same binding pattern for eRF3 (Figure 2C, D and F).
Accumulation of the Upf1-related molecule(s) at the 5′-side of the stable stem-loop structure within the 3′-UTR
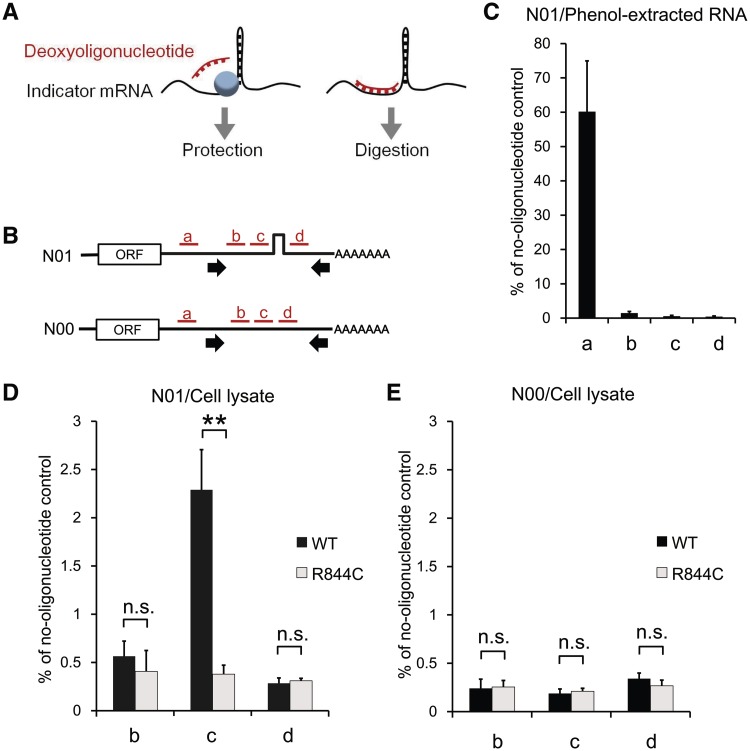
The translation-mediated association of Upf1 to the stem-loop-containing mRNA led us to hypothesize that, after being recruited to a translation–termination site as a component of the SURF complex (7), the Upf1 protein translocates along the mRNA strand from the translation–termination site to the downstream stem-loop structure. To address this possibility, we examined the presence of proteins bound around a stem-loop structure, by performing an oligonucleotide-targeted RNase H protection analysis (Figure 3A). The experiment was carried out using the lysate of HEK293T cells coexpressing an intron-less indicator RNA (Figure 3B and Supplementary Figure S7) and either the wild-type or a helicase-deficient (R844C) Upf1 protein (11,27,28). One of the different deoxyoligonucleotides (15-nt long) complementary to the sequences around the stem-loop (a, b, c and d in Figure 3B) was added to the lysate, and the resulting DNA:RNA hybrid was treated with RNase H. Protein complexes present on the eluted RNP interfere with deoxyoligonucleotides’ annealing, thereby protecting the corresponding region from the cleavage by RNase H (Figure 3A). To detect the RNase H protection with high sensitivity, we developed a real-time RT–PCR method in which protected RNAs are specifically amplified by using a primer pair flanking the target sites. We first carried out control RNase H reactions in which protein-free RNAs purified from cells expressing an indicator RNA was treated to distinguish whether RNase H protection was caused by the binding of proteins or by the RNA conformations. The result of the RT–PCR confirmed that the oligonucleotides b, c, and d directed RNase H cleavage of the protein-free RNA with a comparable efficiency, while the control oligonucleotide a, which binds to a target site upstream of the PCR-amplified region, showed little effect (Figure 3C).
Figure 3.
Accumulation of the Upf1-related molecule(s) at the 5′ side of the stable stem-loop structure within the 3′-UTR. (A) A schematic representation of the oligonucleotide-targeted RNase H protection analysis. (B) The model mRNAs (N00 and N01) used for RNase H protection and the target sites for oligonucleotide binding. The oligonucleotides (15 nt each) are indicated by red bars, and the positions of primers used for real-time RT–PCR are indicated by arrows. (C–E) The levels of protected/undigested mRNAs after RNase H reactions using the phenol-extracted RNA (C) or the RNP (D and E). The RNP/RNA was extracted from the HEK293T cells transiently expressing the wild-type (black bars) or the R844C mutant (gray bars) of Upf1 and the N01 (C and D) or N00 RNA (E). The data, expressed as mean ± SD (n = 3), represent the percentages relative to the no-oligonucleotide control. Statistical analyses were done with unpaired two-tailed t-test (**P < 0.01; n.s., P > 0.1).
When the same experiments were carried out using the RNP lysate from the cells expressing the wild-type Upf1, we observed significant protection of the RNP against the RNase H cleavage only at the position immediately 5′ to the stem-loop (c in Figure 3D), but not at the other regions (b and d in Figure 3D). On the other hand, all three target positions were accessible to the oigonucleotides in the RNP extracted from the cells expressing a helicase mutant (R844C) of Upf1 (Figure 3D), indicating that the protection at the 5′-side of the stable stem-loop structure depends on the helicase activity of Upf1. The RNP lacking the stem-loop structure did not exhibit any protection regardless of the Upf1 status (Figure 3E). Furthermore, we observed the same protection patterns depending on the helicase activity of Upf1 by using another indicator mRNA in which the nucleotide sequences of a stem-loop structure and target sites for deoxyoligonucleotides’ annealing are completely changed from those used in Figure 3, excluding the possibility that the results depend on the specific nucleotide sequence of the indicator mRNAs (Supplementary Figures S7 and S8). Although the proteins accumulated on the indicator mRNAs cannot be identified through the RNase H protection analysis, these results, combined with those from the RNA-immunoprecipitation and in vitro pull-down experiments (Figure 2), provide evidence supporting our hypothesis that the Upf1 protein translocates along an mRNA strand in the 5′-to-3′ direction by using its helicase activity and stalls immediately at the 5′ side of the stem-loop structure.
Close proximity between a PTC and a downstream exon–exon junction bypasses the need of the helicase activity of Upf1
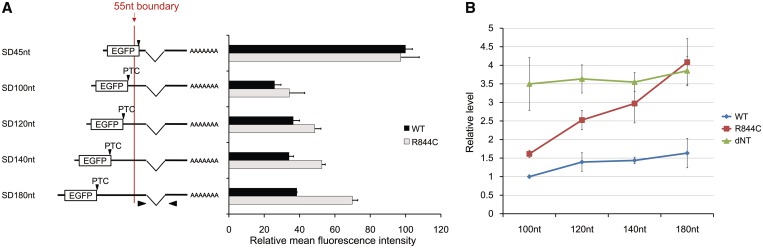
The cis-acting effect of stable stem-loop structures between a PTC and an exon–exon junction (Figure 1 and Supplementary Figure S3) can be reasonably explained by assuming that the 5′-to-3′ translocation of Upf1, which is mediated by its helicase activity, ensures proper interaction between the SURF complex and downstream EJCs, and the stalling of Upf1 at the stem-loop structure causes impaired activation of NMD. To examine whether the helicase activity of Upf1 in fact plays an important role in proper interaction between these two complexes, we investigated the dominant-negative effect of a helicase mutant of Upf1 on NMD activation for reporter mRNAs containing various lengths of spacer sequences between the EGFP–ORF and a downstream exon–exon junction (Figure 4A and Supplementary Figure S1C). Since the EJCs are deposited 20–24 nt upstream of exon–exon junctions, the binding positions of the SURF complex and the EJC are determined exactly by the sites of the translation–termination codon and the exon–exon junction, respectively (15).
Figure 4.
Close proximity between a PTC and a downstream exon–exon junction bypasses the need of the helicase activity of Upf1. (A) The left diagram shows reporter mRNAs with different spacer lengths between the EGFP–ORF and the exon–exon junction. Arrowheads represent the primer pairs used in real-time RT–PCR. The right panel indicates the mean fluorescence intensities of EGFP obtained by flow cytometric analysis of the HEK293T cells expressing one of the reporter mRNAs and the wild-type (black bars) or the R844C mutant (gray bars) of Upf1. The data represent mean ± standard error of the mean (SEM) (n = 3). (B) The levels of reporter mRNAs expressed in HeLa cells. The data, expressed as mean ± SEM (n = 3), represent a fold difference of the mRNA levels compared to that of SD100nt with the wild-type of Upf1.
From the results of the flow cytometric analysis and real-time RT–PCR for cells expressing the wild-type Upf1 and reporter mRNAs, we confirmed that the mRNAs other than SD45nt are degraded to the comparable extent, following the 50 - to 55-nt boundary rule of PTC recognition (Figure 4A, B and Supplementary Figure S9). Interestingly, the efficiency of NMD suppression by the R844C helicase mutant of Upf1 varied significantly depending on the spacer length. When the exon–exon junction was placed at 180-nt downstream of the EGFP termination codon, the helicase mutant was able to suppress NMD as previously reported (11,27,28). However, as the distance between the binding sites for the SURF complex and the EJC became shorter, the weaker levels of NMD suppression were achieved, and the Upf1 mutant no longer showed the apparent dominant-negative effect for the SD100nt mRNA (Figure 4A and B and Supplementary Figure S9). These observations strongly suggest that the close proximity between the SURF complex and the EJC bypasses the need of the helicase activity of Upf1 for the activation of NMD because the overexpressed helicase-defficient mutant of Upf1 still permitted NMD to take place when a translation–termination codon and a downstream exon–exon junction were located close to each other. In contrast, an N-terminal deletion mutant of Upf1 (dNT), which exhibits the reduced ability to associate with SMG-5, a downstream element of the NMD pathway (19), suppressed NMD equally well, regardless of the spacer lengths (Figure 4B), suggesting that the dominant-negative effect dependent upon the distance between the two complexes (i.e. SURF and EJC) would be specific to the helicase-deficient mutant of Upf1. These results indicate that the helicase activity of Upf1 is required for the activation of NMD only when the two complexes are formed at the spatially separated sites on an mRNA molecule and provide evidence that the helicase activity is used for the translocation of the Upf1-related molecular motor along an mRNA to bridge the spatial gap between SURF and EJC.
DISCUSSION
Based on the results of our current study, we propose a model that an Upf1-related molecular motor translocates along the 3′-UTR of eukaryotic mRNA molecules to ensure proper interaction between the SURF complex and the downstream EJC for the activation of NMD.
Recent studies showed that the helicase activity of Upf1 is enhanced by its interaction with Upf2 (10) and promotes disassembly of mRNP undergoing the final mRNA-degradation steps of NMD (12). However, there are substantial biochemical studies demonstrating that the purified Upf1 protein exhibits the RNA unwinding activity by itself (8,9), even without the aid of Upf2. Our present findings on intron-less model mRNAs indicate that the helicase activity of Upf1 plays an important role in the molecular events either upstream or independent of the SURF–EJC interaction and shed light on a novel function of the helicase activity.
Furthermore, our in vitro study using RRL suggests that the association of Upf1 with the stem-loop-containing mRNA is not a phenomenon unique to the pioneer round of translation (29) or to translation termination at PTCs because the in vitro translation in RRL and the pioneer round of translation in mammalian cells were previously shown to be the distinguishable molecular processes, dependent on the distinct translation–initiation factors eIF4E (30) and CBP80/20 (29), respectively. In addition, the ratios of the proteins recovered in the pull-down eluate to those in the total input in the in vitro experiment reveal that mRNAs are associated with a much higher rate with endogenous Upf1 than with endogenous eRF1 (Figure 2D). Taken together, the above findings in our current study suggest that, in spite of the lower intra-cellular abundance of Upf1 compared to that of eRFs (eRF1 and eRF3), the Upf1 helicase participates in the general translation–termination processes regardless of the presence or absence of PTCs in mRNAs.
It was previously demonstrated that Upf1 enhances translation–termination through its interaction with eRFs (6,31), and the interaction between eRF3 and PABPC1 promotes efficient translation probably by facilitating ribosome recycling on circularized mRNA molecules (32,33). Importantly, however, it has been elusive how spatially separated eRF3 and PABPC1 interact with each other on an mRNA molecule to complete normal translation termination. Our findings, together with those of previous studies, lead us to propose a hypothesis that the 5′-to-3′ sliding of Upf1 along the 3′-UTR of an mRNA molecule assists not only the interaction between the SURF complex and the EJC for NMD activation, but also the association between SURF (including eRF3) and PABPC1 to complete normal translation termination. If this is the case, it would be reasonable to assume that the unwinding and translocation of an mRNA strand occur while Upf1 stays associated with a translation–terminating ribosome (Figure 5), and blockade of the ‘reeling-in’ activity of Upf1 reduces the efficient induction of NMD (Figure 1 and Supplementary Figure S4). This model reasonably explains our observations that the secondary structure downstream of a termination codon reduced the association of eRFs with an mRNA, contrary to the enhanced binding of Upf1 to such a molecule (Figure 2). Since it has been demonstrated that eRF3 has a much higher affinity for PABPC1 than for Upf1 (34), the impaired interaction between eRF3 and PABPC1 caused by the blockade of the ‘reeling-in’ activity of Upf1 should result in the reduction of the PABPC1-mediated high-affinity binding of eRFs to mRNAs (Figure 5).
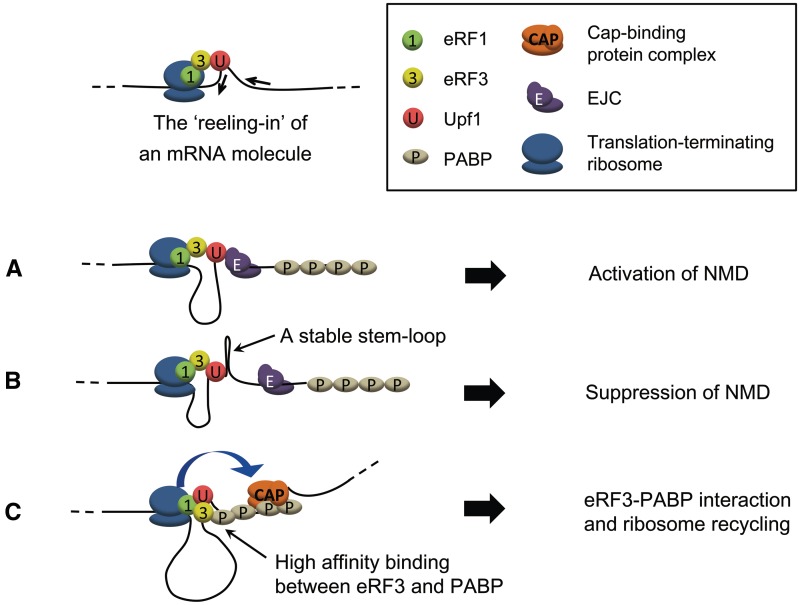
Figure 5.
A model for the roles of the Upf1-related molecular motor. The sliding of the Upf1-related molecular motor occurs while Upf1 stays associated with a translation–terminating ribosome, leading to the ‘reeling-in’ of a line of the 3′-UTR portion of an mRNA molecule. (A) On the NMD substrates, the 5′-to-3′ translocation of the Upf1-related motor allows rapid interaction between the SURF complex and the EJC, avoiding the premature interaction between SURF and PABPC1. (B) If the RNA secondary structure between the PTC and the downstream exon–exon junction is extremely stable, it causes the Upf1 motor to stall and suppresses the activation of NMD. (C) On a normal mRNA without a PTC, the Upf1 motor contributes to the ribosome recycling through facilitating the interaction between SURF and PABPC1.
In our present study, the stable stem-loop structures downstream of an ORF are shown to be predominantly associated with Upf1, but their effect on the levels of PTC-containing mRNAs was moderate (Figure 1). The fact that the levels of the mRNAs containing the ATG-less IRES and hp7 (EGFP–IRES–delATG–hp7 or EGFP–IRES–delATG–hp7x2) are significantly higher than that of the mRNAs containing hp7 but not the IRES (EGFP–hp7x2 or EGFP–hp7x2) raises the possibility that the result simply reflects the stability or the complexity of RNA secondary structures (Figure 1B). Another possibility is that, although blockade of the Upf1 translocation inhibits the SURF–EJC interaction, the concurrent interruption of the eRF3–PABP interaction might activate an alternative pathway of NMD. In yeast whose protein-coding genes often lack introns, it was previously demonstrated that unusually long/contextually wrong (i.e. faux) 3′-UTRs of mRNAs elicit the EJC-independent NMD through interfering with the eRF3–PABP interaction (35). Our study based on the conventional poly(A) trapping in mouse embryonic stem (ES) cells (22,36) suggests that, at least in mammals, the EJCs downstream of termination codons are the more critical determinant for the PTC recognition than is the length of 3′-UTRs (Supplementary Figure S10), a number of observations suggest that the EJC-independent/3′-UTR length-dependent mechanism of NMD is also conserved in mammals (37). The authors of a recent related study made an interesting observation that Upf1 associates with mammalian mRNAs in a 3′-UTR length-dependent manner, raising the hypothesis that Upf1 ‘senses’ the length of 3′-UTRs to be able to recognize PTC (38). This finding can be explained by using our model, since Upf1 should stay associated with the translation–terminating ribosome for a longer period of time on mRNAs with long 3′-UTRs than on mRNAs with short 3′-UTRs (Figure 5). In this regard, it is intriguing to consider the possibility that the strength of the secondary structures within a 3′-UTR, as well as the EJCs remaining on the mRNA and/or the 3′-UTR length, is one of the crucial determinants for the PTC recognition, and the Upf1 molecular motor contributes to the sensing of such features.
Recently, it has been demonstrated that PABPC1 tethered upstream of the EJCs can overcome NMD, proposing that the SURF (with Upf1)–EJC (with Upf2/Upf3) interaction competes with translation termination mediated by the SURF (with eRF3)–PABPC1 interaction (34,39). These findings strongly suggest that the SURF complex interacts exclusively with the EJC prior to its interaction with PABPC1 on the PTC-containing mRNAs in spite of the structural flexibility of the RNP molecules. This sequence of events can be explained by our model because the restricted linear movement of the Upf1-related motor along an mRNA molecule should always result in the SURF complex first associating with the EJC, avoiding the premature interaction between SURF and PABPC1 on the NMD substrates, which would otherwise lead to multiple rounds of translation of aberrantly truncated proteins before the execution of NMD.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1, Supplementary Figures 1–10 and Supplementary References [40,41].
FUNDING
The Japan Society for the Promotion of Science (JSPS) [20710152 to T.S., 19310130 and 21310128 to Y.I.]; Ministry of Education, Culture, Sports, Science and Technology (MEXT) [grants under the Fundamental Technologies Upgrading Program of the National Bio-Resource Project (NBRP) to Y.I.]. Funding for open access charge: A Kaken-hi grant from MEXT, Japan (to Y.I.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank L.E. Maquat for kindly providing us with the UPF1 cDNAs. The authors are grateful to the former graduate students at Nara Institute of Science and Technology (NAIST), especially T. Inoue, Y. Yonekawa, K. Nuta and K. Uesugi, for their contributions. The authors also thank Y. Hirose, J. Naritomi, K. Ashida and Y. Kawakami for their technical assistance.
REFERENCES
- 1.Jankowsky E, Gross CH, Shuman S, Pyle AM. The DExH protein NPH-II is a processive and directional motor for unwinding RNA. Nature. 2000;403:447–451. doi: 10.1038/35000239. [DOI] [PubMed] [Google Scholar]
- 2.Culbertson MR, Underbrink KM, Fink GR. Frameshift suppression Saccharomyces cerevisiae. II. Genetic properties of group II suppressors. Genetics. 1980;95:833–853. doi: 10.1093/genetics/95.4.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maquat LE. When cells stop making sense - effects of nonsense codons on RNA-metabolism in vertebrate cells. RNA. 1995;1:453–465. [PMC free article] [PubMed] [Google Scholar]
- 4.Conti E, Izaurralde E. Nonsense-mediated mRNA decay: molecular insights and mechanistic variations across species. Curr. Opin. Cell. Biol. 2005;17:316–325. doi: 10.1016/j.ceb.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 5.Isken O, Maquat LE. The multiple lives of NMD factors: balancing roles in gene and genome regulation. Nat. Rev. Genet. 2008;9:699–712. doi: 10.1038/nrg2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Czaplinski K, Ruiz-Echevarria MJ, Paushkin SV, Han X, Weng Y, Perlick HA, Dietz HC, Ter-Avanesyan MD, Peltz SW. The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev. 1998;12:1665–1677. doi: 10.1101/gad.12.11.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R, Hoshino S, Ohno M, Dreyfuss G, Ohno S. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev. 2006;20:355–367. doi: 10.1101/gad.1389006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weng YM, Czaplinski K, Peltz SW. Genetic and biochemical characterization of mutations in the ATPase and helicase regions of the Upf1 protein. Mol. Cell. Biol. 1996;16:5477–5490. doi: 10.1128/mcb.16.10.5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhattacharya A, Czaplinski K, Trifillis P, He F, Jacobson A, Peltz SW. Characterization of the biochemical properties of the human Upf1 gene product that is involved in nonsense-mediated mRNA decay. RNA. 2000;6:1226–1235. doi: 10.1017/s1355838200000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chamieh H, Ballut L, Bonneau F, Le Hir H. NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nat. Struct. Mol. Biol. 2008;15:85–93. doi: 10.1038/nsmb1330. [DOI] [PubMed] [Google Scholar]
- 11.Sun XL, Perlick HA, Dietz HC, Maquat LE. A mutated human homologue to yeast Upf1 protein has a dominant-negative effect on the decay of nonsense-containing mRNAs in mammalian cells. Proc. Natl Acad. Sci. USA. 1998;95:10009–10014. doi: 10.1073/pnas.95.17.10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franks TM, Singh G, Lykke-Andersen J. Upf1 ATPase-dependent mRNP disassembly is required for completion of nonsense-mediated mRNA decay. Cell. 2010;143:863–865. doi: 10.1016/j.cell.2010.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nagy E, Maquat LE. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem. Sci. 1998;23:198–199. doi: 10.1016/s0968-0004(98)01208-0. [DOI] [PubMed] [Google Scholar]
- 14.Thermann R, Neu-Yilik G, Deters A, Frede U, Wehr K, Hagemeier C, Hentze MW, Kulozik AE. Binary specification of nonsense codons by splicing and cytoplasmic translation. EMBO J. 1998;17:3484–3494. doi: 10.1093/emboj/17.12.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Le Hir H, Izaurralde E, Maquat LE, Moore MJ. The spliceosome deposits multiple proteins 20-24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 2000;19:6860–6869. doi: 10.1093/emboj/19.24.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gehring NH, Neu-Yilik G, Schell T, Hentze MW, Kulozik AE. Y14 and hUpf3b form an NMD-activating complex. Mol. Cell. 2003;11:939–949. doi: 10.1016/s1097-2765(03)00142-4. [DOI] [PubMed] [Google Scholar]
- 17.Dostie J, Dreyfuss G. Translation is required to remove Y14 from mRNAs in the cytoplasm. Curr. Biol. 2002;12:1060–1067. doi: 10.1016/s0960-9822(02)00902-8. [DOI] [PubMed] [Google Scholar]
- 18.Gehring NH, Lamprinaki S, Kulozik AE, Hentze MW. Disassembly of exon junction complexes by PYM. Cell. 2009;137:536–548. doi: 10.1016/j.cell.2009.02.042. [DOI] [PubMed] [Google Scholar]
- 19.Yamashita A, Izumi N, Kashima I, Ohnishi T, Saari B, Katsuhata Y, Muramatsu R, Morita T, Iwamatsu A, Hachiya T, et al. SMG-8 and SMG-9, two novel subunits of the SMG-1 complex, regulate remodeling of the mRNA surveillance complex during nonsense-mediated mRNA decay. Genes Dev. 2009;23:1091–1105. doi: 10.1101/gad.1767209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chester A, Somasekaram A, Tzimina M, Jarmuz A, Gisbourne J, O'Keefe R, Scott J, Navaratnam N. The apolipoprotein B mRNA editing complex performs a multifunctional cycle and suppresses nonsense-mediated decay. EMBO J. 2003;22:3971–3982. doi: 10.1093/emboj/cdg369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maquat LE. Nonsense-mediated mRNA decay: Splicing, translation and mRNP dynamics. Nat. Rev. Mol. Cell Biol. 2004;5:89–99. doi: 10.1038/nrm1310. [DOI] [PubMed] [Google Scholar]
- 22.Shigeoka T, Kawaichi M, Ishida Y. Suppression of nonsense-mediated mRNA decay permits unbiased gene trapping in mouse embryonic stem cells. Nucleic Acids Res. 2005;33:e20. doi: 10.1093/nar/gni022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lancaster AM, Jan E, Sarnow P. Initiation factor-independent translation mediated by the hepatitis C virus internal ribosome entry site. RNA. 2006;12:894–902. doi: 10.1261/rna.2342306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kozak M. Circumstances and mechanisms of inhibition of translation by secondary structure in eukaryotic messenger-RNAs. Mol. Cell. Biol. 1989;9:5134–5142. doi: 10.1128/mcb.9.11.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Belgrader P, Cheng J, Maquat LE. Evidence to implicate translation by ribosomes in the mechanism by which nonsense codons reduce the nuclear-level of human triosephosphate isomerase messenger-RNA. Proc. Natl Acad. Sci. USA. 1993;90:482–486. doi: 10.1073/pnas.90.2.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ni JZ, Grate L, Donohue JP, Preston C, Nobida N, O'Brien G, Shiue L, Clark TA, Blume JE, Ares M., Jr Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev. 2007;21:708–718. doi: 10.1101/gad.1525507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lykke-Andersen J, Shu MD, Steitz JA. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell. 2000;103:1121–1131. doi: 10.1016/s0092-8674(00)00214-2. [DOI] [PubMed] [Google Scholar]
- 28.Lykke-Andersen J, Shu MD, Steitz JA. Communication of the position of exon-exon junctions to the mRNA surveillance machinery by the protein RNPS1. Science. 2001;293:1836–1839. doi: 10.1126/science.1062786. [DOI] [PubMed] [Google Scholar]
- 29.Ishigaki Y, Li XJ, Serin G, Maquat LE. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell. 2001;106:607–617. doi: 10.1016/s0092-8674(01)00475-5. [DOI] [PubMed] [Google Scholar]
- 30.Mochizuki K, Oguro A, Ohtsu T, Sonenberg N, Nakamura Y. High affinity RNA for mammalian initiation factor 4E interferes with mRNA-cap binding and inhibits translation. RNA. 2005;11:77–89. doi: 10.1261/rna.7108205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weng YM, Czaplinski K, Peltz SW. ATP is a cofactor of the Upf1 protein that modulates its translation termination and RNA binding activities. RNA. 1998;4:205–214. [PMC free article] [PubMed] [Google Scholar]
- 32.Uchida N, Hoshino S, Imataka H, Sonenberg N, Katada T. A novel role of the mammalian GSPT/eRF3 associating with poly(A)-binding protein in cap/poly(A)-dependent translation. J. Biol. Chem. 2002;277:50286–50292. doi: 10.1074/jbc.M203029200. [DOI] [PubMed] [Google Scholar]
- 33.Hoshino S, Imai M, Kobayashi T, Uchida N, Katada T. The eukaryotic polypeptide chain releasing factor (eRF3/GSPT) carrying the translation termination signal to the 3′-poly(A) tail of mRNA - direct association of eRF3/GSPT with polyadenylate-binding protein. J. Biol. Chem. 1999;274:16677–16680. doi: 10.1074/jbc.274.24.16677. [DOI] [PubMed] [Google Scholar]
- 34.Singh G, Rebbapragada I, Lykke-Andersen J. A competition between stimulators and antagonists of Upf complex recruitment governs human nonsense-mediated mRNA decay. PLoS Biol. 2008;6:e111. doi: 10.1371/journal.pbio.0060111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Amrani N, Ganesan R, Kervestin S, Mangus DA, Ghosh S, Jacobson A. A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature. 2004;432:112–118. doi: 10.1038/nature03060. [DOI] [PubMed] [Google Scholar]
- 36.Matsuda E, Shigeoka T, Iida R, Yamanaka S, Kawaichi M, Ishida Y. Expression profiling with arrays of randomly disrupted genes in mouse embryonic stem cells leads to in vivo functional analysis. Proc. Natl Acad. Sci. USA. 2004;101:4170–4174. doi: 10.1073/pnas.0400604101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Silva AL, Ribeiro P, Inacio A, Liebhaber SA, Romao L. Proximity of the poly(A)-binding protein to a premature termination codon inhibits mammalian nonsense-mediated mRNA decay. RNA. 2008;14:563–576. doi: 10.1261/rna.815108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hogg JR, Goff SP. Upf1 senses 3′ UTR length to potentiate mRNA decay. Cell. 2010;143:349–389. doi: 10.1016/j.cell.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ivanov PV, Gehring NH, Kunz JB, Hentze MW, Kulozik AE. Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways. EMBO J. 2008;27:736–747. doi: 10.1038/emboj.2008.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brogna S, Wen J. Nonsense-mediated mRNA decay (NMD) mechanisms. Nat. Struct. Mol. Biol. 2009;16:107–113. doi: 10.1038/nsmb.1550. [DOI] [PubMed] [Google Scholar]
- 41.Ishida Y, Leder P. RET: a poly A-trap retrovirus vector for reversible disruption and expression monitoring of genes in living cells. Nucleic Acids Res. 1999;27:e35. doi: 10.1093/nar/27.24.e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.